Summary
The killer immunoglobulin‐like receptors (KIR) as well as their MHC class I ligands display enormous genetic diversity and polymorphism in macaque species. Signals resulting from interaction between KIR or CD94/NKG2 receptors and their cognate MHC class I proteins essentially regulate the activity of natural killer (NK) cells. Macaque and human KIR share many features, such as clonal expression patterns, gene copy number variations, specificity for particular MHC class I allotypes, or epistasis between KIR and MHC class I genes that influence susceptibility and resistance to immunodeficiency virus infection. In this review article we also annotated publicly available rhesus macaque BAC clone sequences and provide the first description of the CD94–NKG2 genomic region. Besides the presence of genes that are orthologous to human NKG2A and NKG2F, this region contains three NKG2C paralogues. Hence, the genome of rhesus macaques contains moderately expanded and diversified NKG2 genes in addition to highly diversified KIR genes. The presence of two diversified NK cell receptor families in one species has not been described before and is expected to require a complex MHC‐dependent regulation of NK cells.
Keywords: genomics, killer immunoglobulin‐like receptors, MHC, natural killer cells
Introduction
Macaques belong to the Cercopithecidae family (Old World monkeys) of primates with 23 currently known macaque species. The predominant macaque species for biomedical research are the rhesus macaque (Macaca mulatta), the long‐tailed (or cynomolgus) macaque (Macaca fascicularis), the pig‐tailed macaque (Macaca nemestrina), and the Japanese macaque (Macaca fuscata). Macaques are particularly used as non‐human primate models of human infectious diseases.1 For example, experimental infection with the simian immunodeficiency virus (SIV) is an excellent and established non‐human primate model of HIV infection and AIDS.2, 3
Natural killer (NK) cells play an important role in fighting infectious diseases through direct killing of infected cells and regulation of adaptive immune responses.4, 5 The activity of NK cells is regulated by expression of specific receptors that initiate stimulatory or inhibitory signal transduction. A fine‐tuned balance of this signalling ensures tolerance against healthy cells and immune effector reactivity against unhealthy (e.g. infected or malignant) cells. Almost all NK cell receptors fall into two distinct protein families and harbour either immunoglobulin‐like domains or C‐type lectin‐like domains in their extracellular part.6 The activity of NK cells is particularly regulated by those receptors that bind to MHC class I proteins such as the killer immunoglobulin‐like receptors (KIR), members of the leukocyte immunoglobulin‐like receptors (LILR) or the killer cell lectin‐like receptors (KLR). All these receptors come in two functionally distinct types and are either stimulatory or inhibitory. Hence, the presence or absence of MHC class I ligands on target cells is perceived through such receptors and this essentially determines the NK cells’ effector functions.
A further characteristic feature of primate (and other mammalian) NK cell receptors is their variegated expression on NK cell and T cell subsets,7, 8 with most NK cells expressing only a single receptor.9 As the various receptors are specific for their cognate MHC class I ligands, this expression pattern considerably sharpens the ability of NK cells to recognize diseased cells. For example, infection may lead to virus‐induced down‐regulation of host MHC class I proteins and discontinuation of inhibitory receptor engagement and may lead to host cell‐induced expression of certain MHC class I‐like proteins (MIC and ULBP family members) that engage activating receptors and induce activating signalling.10 Hence, the interplay between MHC class I ligands and their cognate NK cell receptors plays a central role in the regulation of NK cell activity. The interaction between inhibitory receptors and their cognate MHC class I ligands is important in two aspects: (i) binding of MHC class I proteins on other cells mediates tolerance of the NK cell, (ii) the absence of cognate MHC class I on the target cell is a strong trigger of NK cell activity. This latter point is known as the ‘missing‐self hypothesis’.11 To avoid unwanted intolerance of those NK cell clones that clonally express a receptor for which the host does not encode an appropriate ligand, the NK cells undergo an educational process that requires the signalling via at least one inhibitory receptor specific for self MHC class I. This process produces so‐called licensed or armed NK cells.12, 13
Macaque NK cells were defined by absence of T cell and B cell markers (CD3, CD20) and specific expression of NKG2A/C or alternatively NKp80.14 However, there are NKG2A/C‐negative NK cells in macaques and a more complete definition was given by Webster and Johnson15 who classified macaque NK cells as CD3− CD8bright CD20−/dim cells.
We here review the current knowledge of MHC class I‐dependent regulation of NK cells in macaques with focus on the KIR and CD94/NKG2 family.
The KIR and MHC class I gene families of macaques
Macaque KIR and MHC class I genes
Analyses of rhesus macaque cDNA and genomic DNA revealed the presence of 22 KIR genes (Table 1), which can all be assigned to established KIR lineages. Phylogenetic analyses suggest that cynomolgus macaques and pig‐tailed macaques have similar sets of KIR genes with few species‐specific differences.16, 17 Similar to human KIR haplotypes, macaque KIR haplotypes also vary in gene content, giving rise to a substantial degree of genomic diversity, in particular of the KIR3D genes.17, 18, 19, 20 The MHC class I genes of macaques have undergone significant expansions during evolution.21 Although an HLA‐C orthologue is not present, multiple copies of HLA‐A and HLA‐B paralogues can be found in macaques. Contrasting a fixed number of class I genes on HLA haplotypes in humans are extreme copy number variations of MHC‐A and MHC‐B genes of macaque MHC haplotypes.22, 23, 24, 25 In addition, the degree of their polymorphism (allelic variation) is usually much smaller compared with their human paralogues, suggesting that variability of the macaque MHC class I system is more focused on copy number variation than on allelic polymorphism.21
Table 1.
Rhesus macaque killer immunoglobulin‐like receptor (KIR) genes
| Receptor | KIR lineage | Putative function |
|---|---|---|
| Mamu‐KIR1D | III | n.d. (no cytoplasmic domain) |
| Mamu‐KIR2DL4 | I | Activation/Inhibition |
| Mamu‐KIR3DL01 | II | Inhibition |
| Mamu‐KIR3DL02 | II | Inhibition |
| Mamu‐KIR3DLW03 | II | Inhibition |
| Mamu‐KIR3DL04 | II | Inhibition |
| Mamu‐KIR3DL05 | II | Inhibition |
| Mamu‐KIR3DL06 | II | Inhibition |
| Mamu‐KIR3DL07 | II | Inhibition |
| Mamu‐KIR3DL08 | II | Inhibition |
| Mamu‐KIR3DL10 | II | Inhibition |
| Mamu‐KIR3DL11 | II | Inhibition |
| Mamu‐KIR3DL20 | V | Inhibition |
| Mamu‐KIR3DS01 | II | Activation |
| Mamu‐KIR3DS02 | II | Activation |
| Mamu‐KIR3DS03 | II | Activation |
| Mamu‐KIR3DS04 | II | Activation |
| Mamu‐KIR3DS05 | II | Activation |
| Mamu‐KIR3DS06 | II | Activation |
| Mamu‐KIR3DS07 | II | Activation |
| Mamu‐KIR3DSW08 | II | Activation |
| Mamu‐KIR3DS09 | II | Activation |
KIR and MHC class I in macaque disease models
In rhesus macaques experimentally infected with SIV, several MHC class I alleles were identified that influence the disease in this important animal model (for review see ref. 26). Among the alleles with a beneficial effect in SIV infection were Mamu‐A1 alleles *001, *002, *011 and Mamu‐B alleles *008, *017, *029, *047, and *069, whereas A1*004 and B*001 were associated with susceptibility.27, 28, 29, 30, 31, 32 Recently, also the presence/absence of KIR genes was included in such SIV disease association studies. Hellmann et al.33 reported an association of high copy numbers of activating KIR3DS genes with lower peak viral load in animals negative for Mamu‐A1*001 and positive for a protective TRIM5 allele. In a further study, this group demonstrated high KIR2DL4 copy numbers to be associated with a better preservation of CD4+ T cells and an increased interferon‐γ production of stimulated CD56− CD16− NK cells in SIV‐infected rhesus macaques.34 Albrecht et al.32 identified individual KIR genes with either a protective or a detrimental effect in SIV‐infected rhesus macaques. KIR3DL02, KIR3DSW08 and most probably also KIR3DS01 are associated with slow disease progression, longer survival times, higher numbers of blood NK cells, and higher lytic capability of NK cells.32 In contrast, KIR3DS02, KIR3DL10 and KIR3DL05 were more frequently found in animals with fast disease progression, shorter survival times, lower numbers of blood NK cells, and decreased lytic capability of NK cells.32 Furthermore, analyses of epistasis identified combinations of certain KIR and MHC class I genes with progression to AIDS: KIR3DL05, KIR3DS05 as well as KIR3DL10 are associated with faster progression to AIDS, but only in combination with presence of Mamu‐B*012 or absence of Mamu‐A1*001.32
KIR‐mediated regulation of NK cell activity
Rhesus macaque KIR proteins are expressed in a clonal manner, but in contrast to humans the frequencies of rhesus macaque KIR‐expressing NK cells do not appear to depend on the presence of their cognate MHC class I ligands.8, 10 Specific interactions between macaque KIR3D proteins and MHC class I ligands have been described for rhesus and pig‐tailed macaques (Table 2). Mamu‐A1 allotypes are ligands of the activating receptor KIR3DS0535 (Table 2), but unlike inhibitory KIR, these interactions are characterized by low avidity. Lower avidity of activating receptors for their cognate ligands is also evident for activating human KIR and activating Ly49 of mice.36
Table 2.
Interactions between macaque KIR3D and MHC class I proteins
| Receptor | Ligands | Ref. |
|---|---|---|
| Mamu‐KIR3DL01 | Mamu‐B*007:01, ‐B*041:01, ‐B*058:02, ‐B*065:01 | 36 |
| Mamu‐KIR3DLW03 | Mamu‐A1*001:01, ‐A1*008:01, ‐A1*011:01 | 35 |
| Mamu‐KIR3DL05 | Mamu‐A1*001:01, ‐A1*002:01, ‐A3*13:11 | 35, 37 |
| Mamu‐KIR3DL11 | Mamu‐A1*008:01 | 35 |
| Mamu‐KIR3DS05 | Mamu‐A1*001:01, ‐A1*011:01 | 35 |
| Mane‐KIR049‐4 | Mane‐A1*082, ‐A1*084 | 17 |
In comparison to human inhibitory KIR (iKIR), the iKIR–MHC class I interactions in macaques are characterized by lower avidity and broader reactivity.17, 35, 37, 38 Although the Bw4/Bw6 motifs play a role in binding macaque KIR3D proteins,17, 35, 37 there are some KIR3D with restricted specificity for Bw4 such as KIR3DL0137 or KIR3DLW03,35 whereas other iKIR bind to both motifs such as KIR3DL0535, 38 or KIR049‐4.17 One reason for this broader reactivity of some iKIR might be the enormous genetic diversity and copy number variation in macaque species that might favour the evolution of KIR with broad interaction specificity.
The KIR‐mediated NK cell activity is also modulated by antigenic peptides presented by MHC class I proteins, e.g. HLA‐B‐ or HLA‐C‐bound peptides were shown to modulate binding of their cognate KIR.39 Support for the role of peptides in modulation of KIR‐mediated NK cell responses came from a study that demonstrated certain HIV‐1 amino acid polymorphisms to be particularly frequent in KIR2DL2‐positive individuals.40 Peptides containing such a polymorphic position mediated particularly strong inhibition of KIR2DL2‐expressing NK cells.40 Hence, mutation of HIV‐1 peptides resulted in increased avidity of HLA‐C (presenting these HIV‐1 peptides) to inhibitory KIR2DL2. This is interpreted as a strategy to avoid immune recognition and underscores the importance of NK cells in the defence of HIV‐1.40 An influence of MHC class I‐bound peptides on binding avidity of KIR3D proteins was also demonstrated in rhesus and pig‐tailed macaques.17, 37, 38, 41 Hence, there is accumulating evidence that the repertoire of presented peptides is an important factor in the KIR‐mediated control of NK cell activity and that viruses might exploit this. On the other hand, primate activating KIR genes originate from inhibitory KIR genes and are rapidly evolving.42 The reason for this is probably an important role of activating KIR in fighting pathogens. Consistent with this assumption are the above‐mentioned epidemiological studies showing a beneficial role of activating KIR3DS genes in infections with immunodeficiency viruses in humans43 and macaques.32 In line with this, the interaction between human KIR3DS1 and its ligand Bw4+ HLA‐B*57 is restricted and is dependent on the presence of B*57‐bound peptides with aromatic residues (tryptophan, phenylalanine or tyrosine, but not histidine) at P8.44 It is thought that theses residues might compensate the interaction disabling arginine residue at position 166 of KIR3DS1 and that the frequency of such peptides increases in diseased (infected, transformed) cells.44 Hence, the activating receptor KIR3DS1 does not interact with B*57 of healthy cells and thereby ensures immune tolerance, but engages B*57 upon disease‐mediated changes of the peptide repertoire (‘altered‐self’ recognition).
The CD94/NKG2 gene family of macaques
The NKG2 genes of macaques revisited
The human inhibitory NKG2A as well as both the activating NKG2C and NKG2E form heterodimers with CD94. The function of NKG2F is less clear. The distantly related NKG2D forms homodimers and binds stress‐inducible ligands of the MHC class I‐related families of MIC and ULBP proteins. In contrast to CD94/NKG2A and CD94/NKG2C, the CD94/NKG2E heterodimer is not expressed at the cell surface but is retained intracellularly due to hydrophobic residues at the C terminus, which do not exist in other human NKG2 molecules.45 All human CD94/NKG2 heterodimers bind the non‐polymorphic HLA‐E ligand.46
Although the rhesus macaque genome is sequenced, regions containing immune gene families are in general often incomplete and incorrectly displayed in genome browsers due to high sequence similarities of gene family members, difficulties in automated gene annotations, or gene copy number variations. Hence, also the rhesus macaque CD94‐NKG2 genomic region is not properly represented in the rhesus macaque reference genome. We identified two overlapping rhesus macaque BAC clones from the GenBank sequence database and annotated the CD94‐NKG2 genomic region (Fig. 1). Besides CD94 and NKG2D, we identified NKG2A, NKG2F, and three genes that we tentatively named NKG2C‐1, NKG2C‐2 and NKG2C‐3 (Fig. 1). A summary of the putative functions of the rhesus macaque NKG2 genes is shown in Table 3. According to phylogenetic analyses these three rhesus macaque NKG2C genes appear to be more closely related to human NKG2C than to NKG2E, both in trees based on the complete amino acid sequences (Fig. 2a) and on the cytoplasmic and transmembrane part (Fig. 2b). The extracellular C‐type lectin‐like domains show the known clustering by species (Fig. 2c) as the exons encoding these domains most likely underlie homogenization.47 The NKG2A and NKG2F genes of human and rhesus macaque both form separate clusters with high bootstrap support when the complete sequences or the cytoplasmic/transmembrane parts are considered (Fig. 2a,b), indicating that these genes are orthologous between human and rhesus macaque.
Figure 1.
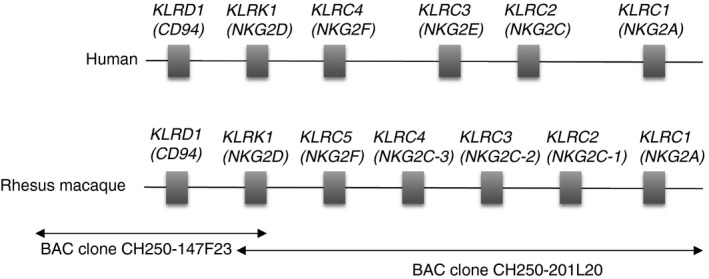
Genomic organization of the human and rhesus macaque CD94 and NKG2 region. The maps are not to scale. Orthologous genes CD94,NKG2D,NKG2F, and NKG2A are ordered vertically.
Table 3.
Summary of rhesus macaque NKG2 receptors
| Receptor | Putative function |
|---|---|
| NKG2A | Inhibition |
| NKG2C‐1 | Activation |
| NKG2C‐2 | Activation |
| NKG2C‐3 | Activation |
| NKG2D | Activation |
| NKG2F | Activation |
Figure 2.
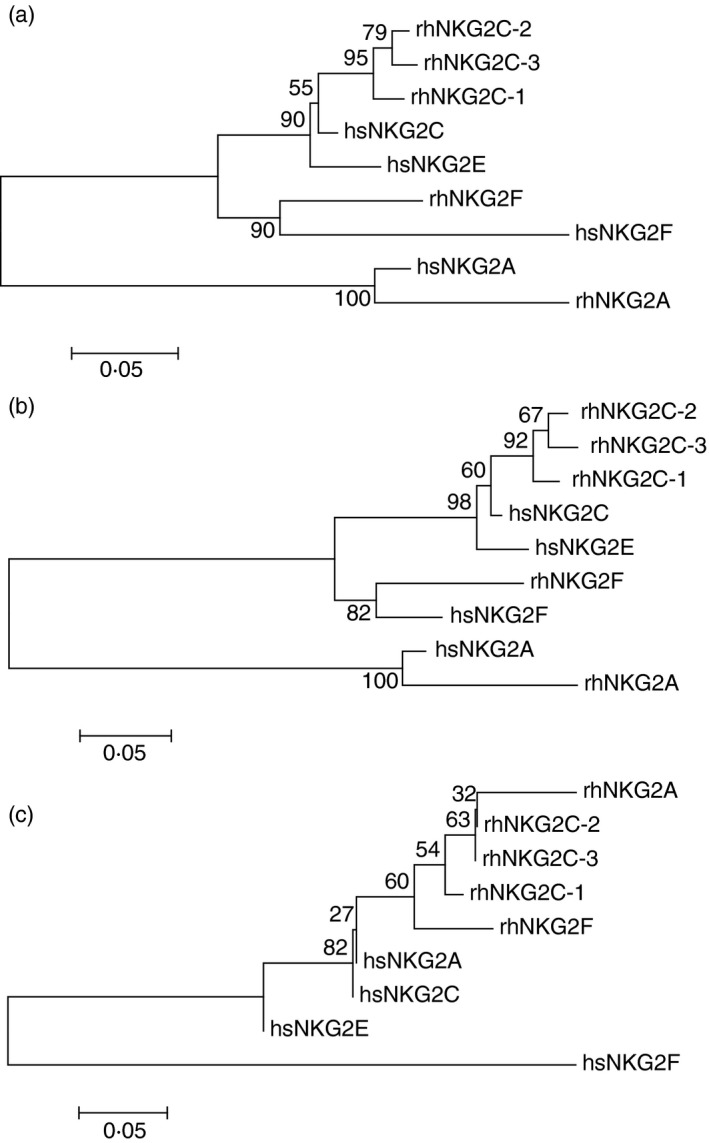
Phylogenetic tree analysis of human (hs) and rhesus macaque (rh) NKG2 amino acid sequences. Sequences were aligned and neighbour‐joining trees using the Jones–Taylor–Thornton model with 1000 bootstraps were constructed. Scale bars denote substitutions per site and bootstrap support in per cent is indicated at nodes. Trees were based on (a) complete amino acid sequences, (b) transmembrane and cytoplasmic regions only, (c) C‐type lectin‐like domains only.
How do previously published rhesus macaque NKG2 sequences relate to these BAC clone‐derived sequences? The rhesus macaque NKG2A and NKG2C alleles identified by Biassoni et al.48 show 96% and 99% (one non‐synonymous substitution) sequence similarity with the BAC clone‐derived NKG2A and NKG2C‐1 alleles. Kravitz et al.49 identified several NKG2 clones from a rhesus macaque decidua cDNA library: the NKG2A allele shows 97% sequence similarity with the NKG2A allele of the BAC clone, and NKG2C clones 65 and 74 display 99% identity with the BAC clone‐derived alleles of NKG2C‐1 and NKG2C‐2, respectively. LaBonte et al.50, 51 described a single NKG2A sequence and a couple of transcripts that were similar to human NKG2C, NKG2E and NKG2F: the NKG2C sequences (clones 10/1#66 and 10/1#2) display sequence similarities of 99% and 97% with BAC‐derived NKG2C1, the NKG2‐C2 sequences (clones 10/1#69 and 10/1#81) are 99% identical with the BAC‐derived NKG2C‐2, the NKG2Ce2 sequence is 99% identical with the BAC‐derived NKG2C‐3, and the NKG2Fe2, NKG2F2 and NKG2F3 sequences are almost identical (97–99%) to NKG2F.
Therefore, we conclude that the rhesus macaque genome contains genes that encode CD94, NKG2A, NKG2C‐1, NKG2C‐2, NKG2C‐3, NKG2D and NKG2‐F. Interestingly, none of the rhesus macaque NKG2 sequences contains the hydrophobic amino acids that are present at the C terminus of human (see Fig. 3), chimpanzee, or orang‐utan NKG2E and we hypothesize that none of the three rhesus macaque NKG2C proteins is retained intracellularly.
Figure 3.

Amino acid sequence alignment of human (hs) and rhesus macaque (rh) NKG2 sequences. Identical residues are indicated by a dot and gaps by a dash. Residues known to be involved in binding of human NKG2A to HLA‐E53, 54, 55 are marked. The ITIM motifs of inhibitory receptors and the lysine residue in the transmembrane region of activating receptors are marked in yellow and red, respectively.
Biassoni et al.48 reported a cynomolgus macaque NKG2C sequence. Sequence comparison indicates that it is most closely related to the rhesus macaque genomic sequence NKG2C‐1 reported here (not shown).
CD94/NKG2‐mediated regulation of macaque NK cell responses
Experiments by Biassoni et al.48 showed that both rhesus and cynomolgus macaque CD94/NKG2A as well as CD94/NKG2C‐1 heterodimers are expressed at the cell surface upon transfection and react with anti‐human NKG2A antibody Z199. Hence, unlike in humans, this monoclonal antibody does not discriminate between NKG2A and NKG2C in macaques.48 These data were confirmed and extended by LaBonte et al.,52 who showed that also CD94/NKG2C‐2 (alias NKG2‐C2) are expressed at the cell surface and react with Z199. Hence, besides the inhibitory CD94/NKG2A heterodimer, at least two stimulatory CD94/NKG2C can be expressed at the cell surface and react with Z199. LaBonte et al. also expressed NKG2Ce, which is an alternatively spliced and C‐terminally shorter form of NKG2Ce2 (= NKG2C‐3), but did not detect binding of Z199.52 A possible reason for the lack of antibody binding might be that the shorter isoform was used.
The three‐dimensional structure of the human CD94/NKG2A heterodimer in contact with its ligand HLA‐E was reported by different groups.53, 54, 55 Comparison of the known interacting amino acid residues of human NKG2A and its ligand HLA‐E with those residues present in rhesus macaque NKG2A, NKG2C‐1, NKG2C‐2 and NKG2C‐3 suggests that Mamu‐E is the ligand of the respective heterodimeric CD94/NKG2 receptors (Fig. 3). In accord with this hypothesis, previous experiments with HLA‐E tetramers folded with an HLA‐A2 leader peptide showed binding to rhesus macaque CD3− CD16+ cells.56 Interestingly, the putative MHC‐E ligand is moderately polymorphic in macaque species, yet the polymorphisms appear to map outside of peptide‐binding regions.57, 58
Published reports on the involvement of CD94/NKG2 heterodimers in the regulation of macaque NK cell responses are rather scarce, which is probably due to the lack of specific antibodies that distinguish between macaque NKG2A and the various NKG2C proteins. However, Reeves et al.59 demonstrated recently that a specific memory NK cell response (killing) of SHIV‐ or SIV‐infected macaques against Gag‐ and Env‐pulsed dendritic cells is dependent on NKG2A/C but not on NKp46. These data indicate an essential role of NKG2A and/or NKG2C molecules in the regulation of NK cell memory responses.
A moderately diversified (NKG2) combined with a highly diversified (KIR) NK cell receptor system is interesting and to the best of our knowledge has not been described in any other mammal. Such a highly diversified NK cell receptor system implicates a complex MHC‐dependent regulation of NK cells and possibly of other lymphocytes expressing these receptors such as CD8+ T cells. Reasons for the development of expanded and diversified NKG2C genes in macaques might be that the complex genetic systems of KIR and MHC class I with numerous gene copy number variations of both systems requires additional activating receptors without extensive copy number variations. Alternatively, the Mamu‐E ligand plays a more prominent role in macaque immunology compared with its human orthologue HLA‐E, requiring the expansion and evolution of NKG2 genes in macaques. Another scenario could be that the system of two diverse NK cell receptors was the original state in the evolution of catarrhine primates. Selection during evolution of Hominidae may have forced the development of restricted immunological functions of the MHC‐E/NKG2 system and of extended immunological functions of the MHC‐I/KIR system, coming along with a reduced set of NKG2 genes, a highly diverse set of KIR genes, and fixed numbers of MHC class I genes in humans and apes.
Disclosures
The authors do not have any competing interests.
Both authors contributed to data analysis and writing of the manuscript.
References
- 1. Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR J 2008; 49:220–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nehete PN, Singh S, Sastry KJ. Lessons on non‐progression of HIV disease from monkeys. Front Immunol 2013; 4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Misra A, Thippeshappa R, Kimata JT. Macaques as model hosts for studies of HIV‐1 infection. Front Microbiol 2013; 4:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol 2008; 8:259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watzl C, Urlaub D, Fasbender F, Claus M. Natural killer cell regulation – beyond the receptors. F1000Prime Rep 2014; 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet 2005; 1:129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manser AR, Weinhold S, Uhrberg M. Human KIR repertoires: shaped by genetic diversity and evolution. Immunol Rev 2015; 267:178–96. [DOI] [PubMed] [Google Scholar]
- 8. Hermes M, Albrecht C, Schrod A, Brameier M, Walter L. Expression patterns of killer cell immunoglobulin‐like receptors (KIR) of NK‐cell and T‐cell subsets in old world monkeys. PLoS ONE 2013; 8:e64936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schönberg K, Sribar M, Enczmann J, Fischer JC, Uhrberg M. Analyses of HLA‐C‐specific KIR repertoires in donors with group A and B haplotypes suggest a ligand‐instructed model of NK cell receptor acquisition. Blood 2011; 117:98–107. [DOI] [PubMed] [Google Scholar]
- 10. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008; 9:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ljunggren HG, Kärre K. In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today 1990; 11:237–44. [DOI] [PubMed] [Google Scholar]
- 12. Yokoyama WM, Kim S. Licensing of natural killer cells by self‐major histocompatibility complex class I. Immunol Rev 2006; 214:143–54. [DOI] [PubMed] [Google Scholar]
- 13. Raulet DH, Vance RE. Self‐tolerance of natural killer cells. Nat Rev Immunol 2006; 6:520–31. [DOI] [PubMed] [Google Scholar]
- 14. Mavilio D, Benjamin J, Kim D, Lombardo G, Daucher M, Kinter A et al Identification of NKG2A and NKp80 as specific natural killer cell markers in rhesus and pigtailed monkeys. Blood 2005; 106:1718–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Webster RL, Johnson RP. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology 2005; 115:206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O'Connor DH. Complete characterization of killer Ig‐like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol 2008; 181:6301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maloveste SM, Chen D, Gostick E, Vivian JP, Plishka RJ, Iyengar R et al Degenerate recognition of MHC class I molecules with Bw4 and Bw6 motifs by a killer cell Ig‐like receptor 3DL expressed by macaque NK cells. J Immunol 2012; 189:4338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moreland AJ, Guethlein LA, Reeves RK, Broman KW, Johnson RP, Parham P et al Characterization of killer immunoglobulin‐like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genom 2011; 12:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blokhuis JH, van der Wiel MK, Doxiadis GGM, Bontrop RE. The extreme plasticity of killer cell Ig‐like receptor (KIR) haplotypes differentiates rhesus macaques from humans. Eur J Immunol 2011; 41:2719–28. [DOI] [PubMed] [Google Scholar]
- 20. Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics 2010; 62:281–93. [DOI] [PubMed] [Google Scholar]
- 21. de Groot NG, Blokhuis JH, Otting N, Doxiadis GGM, Bontrop RE. Co‐evolution of the MHC class I and KIR gene families in rhesus macaques: ancestry and plasticity. Immunol Rev 2015; 267:228–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otting N, Doxiadis GGM, Bontrop RE. Definition of Mafa‐A and ‐B haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 2009; 61:745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Leary CE, Wiseman RW, Karl JA, Bimber BN, Lank SM, Tuscher JJ et al Identification of novel MHC class I sequences in pig‐tailed macaques by amplicon pyrosequencing and full‐length cDNA cloning and sequencing. Immunogenetics 2009; 61:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pratt BF, O'Connor DH, Lafont BAP, Mankowski JL, Fernandez CS, Triastuti R et al MHC class I allele frequencies in pigtail macaques of diverse origin. Immunogenetics 2006; 58:995–1001. [DOI] [PubMed] [Google Scholar]
- 25. Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ et al Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3: Genes ‐ Genomes ‐ Genetics 2013; 3:1195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walter L, Ansari AA. MHC and KIR polymorphisms in rhesus macaque SIV infection. Front Immunol 2015; 6:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR et al ALVAC‐SIV‐gag‐pol‐env‐based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac‐induced immunodeficiency. J Virol 2002; 76:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mühl T, Krawczak M, ten Haaft P, Hunsmann G, Sauermann U. MHC class I alleles influence set‐point viral load and survival time in simian immunodeficiency virus‐infected rhesus monkeys. J Immunol 2002; 169:3438–46. [DOI] [PubMed] [Google Scholar]
- 29. Sauermann U, Siddiqui R, Suh Y‐S, Platzer M, Leuchte N, Meyer H et al MHC class I haplotypes associated with survival time in simian immunodeficiency virus (SIV)‐infected rhesus macaques. Genes Immun 2008; 9:69–80. [DOI] [PubMed] [Google Scholar]
- 30. Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T et al Mamu‐B*08‐positive macaques control simian immunodeficiency virus replication. J Virol 2007; 81:8827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM et al The high‐frequency major histocompatibility complex class I allele Mamu‐B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol 2006; 80:5074–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Albrecht C, Malzahn D, Brameier M, Hermes M, Ansari AA, Walter L. Progression to AIDS in SIV‐infected rhesus macaques is associated with distinct KIR and MHC class I polymorphisms and NK cell dysfunction. Front Immunol 2014; 5:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hellmann I, Lim S‐Y, Gelman RS, Letvin NL. Association of activating KIR copy number variation of NK cells with containment of SIV replication in rhesus monkeys. PLoS Pathog 2011; 7:e1002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hellmann I, Letvin NL, Schmitz JE. KIR2DL4 copy number variation is associated with CD4+ T‐cell depletion and function of cytokine‐producing NK cell subsets in SIV‐infected mamu‐A*01‐negative rhesus macaques. J Virol 2013; 87:5305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosner C, Kruse PH, Hermes M, Otto N, Walter L. Rhesus macaque inhibitory and activating KIR3D interact with Mamu‐A‐encoded ligands. J Immunol 2011; 186:2156–63. [DOI] [PubMed] [Google Scholar]
- 36. Lanier LL. NK cell recognition. Annu Rev Immunol 2005; 23:225–74. [DOI] [PubMed] [Google Scholar]
- 37. Schafer JL, Colantonio AD, Neidermyer WJ, Dudley DM, Connole M, O'Connor DH et al KIR3DL01 recognition of Bw4 ligands in the rhesus macaque: maintenance of Bw4 specificity since the divergence of apes and old world monkeys. J Immunol 2014; 192:1907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colantonio AD, Bimber BN, Neidermyer WJ, Reeves RK, Alter G, Altfeld M et al KIR polymorphisms modulate peptide‐dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog 2011; 7:e1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cassidy SA, Cheent KS, Khakoo SI. Effects of peptide on NK cell‐mediated MHC I recognition. Front Immunol 2014; 5:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM et al HIV‐1 adaptation to NK‐cell‐mediated immune pressure. Nature 2011; 476:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schafer JL, Ries M, Guha N, Connole M, Colantonio AD, Wiertz EJ et al Suppression of a natural killer cell response by simian immunodeficiency virus peptides. PLoS Pathog 2015; 11:e1005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abi‐Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin‐like receptor and Ly49 from inhibitory homologues. J Exp Med 2005; 201:1319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F et al Innate partnership of HLA‐B and KIR3DL1 subtypes against HIV‐1. Nat Genet 2007; 39:733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Connor GM, Vivian JP, Gostick E, Pymm P, Lafont BAP, Price DA et al Peptide‐dependent recognition of HLA‐B*57:01 by KIR3DS1. J Virol 2015; 89:5213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Orbelyan GA, Tang F, Sally B, Solus J, Meresse B, Ciszewski C et al Human NKG2E Is expressed and forms an intracytoplasmic complex with CD94 and DAP12. J Immunol 2014; 193:610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Braud VM, Allan DS, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS et al HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998; 391:795–9. [DOI] [PubMed] [Google Scholar]
- 47. Averdam A, Kuhl H, Sontag M, Becker T, Hughes AL, Reinhardt R et al Genomics and diversity of the common marmoset monkey NK complex. J Immunol 2007; 178:7151–61. [DOI] [PubMed] [Google Scholar]
- 48. Biassoni R, Fogli M, Cantoni C, Costa P, Conte R, Koopman G et al Molecular and functional characterization of NKG2D, NKp80, and NKG2C triggering NK cell receptors in rhesus and cynomolgus macaques: monitoring of NK cell function during simian HIV infection. J Immunol 2005; 174:5695–705. [DOI] [PubMed] [Google Scholar]
- 49. Kravitz RH, Grendell RL, Slukvin II, Golos TG. Selective expression of NKG2‐A and NKG2‐C mRNAs and novel alternative splicing of 5’ exons in rhesus monkey decidua. Immunogenetics 2001; 53:69–73. [DOI] [PubMed] [Google Scholar]
- 50. LaBonte ML, Levy DB, Letvin NL. Characterization of rhesus monkey CD94/NKG2 family members and identification of novel transmembrane‐deleted forms of NKG2‐A, B, C, and D. Immunogenetics 2000; 51:496–9. [DOI] [PubMed] [Google Scholar]
- 51. LaBonte ML, Hershberger KL, Korber B, Letvin NL. The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol Rev 2002; 183:25–40. [DOI] [PubMed] [Google Scholar]
- 52. LaBonte ML, McKay PF, Letvin NL. Evidence of NK cell dysfunction in SIV‐infected rhesus monkeys: impairment of cytokine secretion and NKG2C/C2 expression. Eur J Immunol 2006; 36:2424–33. [DOI] [PubMed] [Google Scholar]
- 53. Sullivan LC, Clements CS, Beddoe T, Johnson D, Hoare HL, Lin J et al The heterodimeric assembly of the CD94‐NKG2 receptor family and implications for human leukocyte antigen‐E recognition. Immunity 2007; 27:900–11. [DOI] [PubMed] [Google Scholar]
- 54. Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA‐E. Proc Natl Acad Sci USA 2008; 105:6696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petrie E, Clements C, Lin J, Sullivan L, Johnson D, Huyton T et al CD94‐NKG2A recognition of human leukocyte antigen (HLA)‐E bound to an HLA class I leader sequence. J Exp Med 2008; 205:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miller JD, Weber DA, Ibegbu C, Pohl J, Altman JD, Jensen PE. Analysis of HLA‐E peptide‐binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J Immunol 2003; 171:1369–75. [DOI] [PubMed] [Google Scholar]
- 57. Boyson JE, McAdam SN, Gallimore A, Golos TG, Liu X, Gotch FM et al The MHC E locus in macaques is polymorphic and is conserved between macaques and humans. Immunogenetics 1995; 41:59–68. [DOI] [PubMed] [Google Scholar]
- 58. Lafont BAP, Buckler‐White A, Plishka R, Buckler C, Martin MA. Pig‐tailed macaques (Macaca nemestrina) possess six MHC‐E families that are conserved among macaque species: implication for their binding to natural killer receptor variants. Immunogenetics 2004; 56:142–54. [DOI] [PubMed] [Google Scholar]
- 59. Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL et al Antigen‐specific NK cell memory in rhesus macaques. Nat Immunol 2015; 16:927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]


