ABSTRACT
Ipilimumab, the first immune-checkpoint inhibitor extending overall survival (OS) in metastatic melanoma patients, has a survival benefit only in a proportion of patients and the development of reliable predictive biomarkers is still an unmet need. To meet this request, we used a multivariate statistical approach to test whether myeloid-derived suppressor cells (MDSC) or other tumor-associated and immunological parameters may serve as predictive or prognostic biomarkers in melanoma patients receiving ipilimumab. By using a standardized approach to determine the circulating levels of four MDSC subsets, we observed a significant expansion of three MDSC subsets at baseline, as compared to controls and, upon treatment, that high levels of CD14+/IL4Rα+ MDSCs were an independent prognostic factor of reduced OS. On the contrary, longer OS was associated to low levels of the proinflammatory proteins IL-6 and CRP and tumor-associated factors S100B and LDH both at baseline and after treatment. Increasing number of total T cells and especially of PD-1+/CD4+ T cells were associated with better prognosis, and upregulation of PD-1+ expression on CD4+ T cells upon treatment was associated with lower toxicity. As several parameters were associated to OS, we included these factors in a multivariate survival model, and we identified IL-6 and ECOG PS as independent biomarkers associated with improved OS, whereas high levels of LDH and CD14+/IL4Rα+ MDSCs were negative independent markers of reduced OS.
KEYWORDS: Biomarkers, CTLA-4, ipilimumab, melanoma, myeloid-derived suppressor cells
Abbreviations
- ADR
adverse drug reaction
- CRP
C-reactive protein
- CTLA-4
cytotoxic T-lymphocyte antigen 4
- DCM
death cell marker
- ECOG PS
Eastern Cooperative Oncology Group Performance Status
- EMA
European Medicine Agency
- FDA
Food and Drug Administration
- FMO
fluorescence minus one
- FSC
forward scatter
- G3–G4
grade 3 and 4 adverse drug reactions
- HD
healthy donors
- HR
Hazard ratio
- ICI
immune checkpoint inhibitor
- IL
interleukin
- IL4Rα
α chain of IL-4 receptor
- ir-ADR
immune-related adverse drug reaction
- L1
CD3+/CD4+
- L2
CD3+/CD8+
- L3
CD3+/CD4+/PD-1+
- L4
CD3+/CD8+/PD-1+
- L5
PD-1+ within CD3+/CD4+
- L6
PD-1+ within CD3+/CD8+
- L7
CD4+/CD8+
- L8
CD3+
- LDH
lactate dehydrogenase
- L/D
live/dead, Lin: lineage
- MDSC
myeloid-derived suppressor cell
- MDSC 1
CD14+/IL4Rα+
- MDSC 2
CD15+/IL4Rα+
- MDSC 3
Lin−/HLA-DR−/CD33+/CD11b+
- MDSC 4
CD14+/HLA-DRlow/−
- N
absolute number
- OS
overall survival
- PD-1
programmed death-1
- PFS
progression-free survival
- PT
patient
- SSC
side-scatter
- TIPs
tumor-associated and immunological parameters
- Treg
regulatory T cell
- ULN
upper limit normal
- VEGF
vascular endothelial growth factor
- W
week
Introduction
The therapeutic management of metastatic melanoma was confined to palliative chemotherapeutic regimens until 2011 when a major breakthrough in this field was achieved by the introduction in the clinical practice of ipilimumab (reviewed in ref.1). In fact, this anti-CTLA-4 monoclonal antibody was the first treatment producing a significant survival benefit in metastatic melanoma and a long-term survival in 18–26% of treated patients.2 Its efficacy has been proven both in untreated and in pre-treated patients, in wild-type melanoma as well as in BRAF and NRAS mutated tumors, and even in patients with brain metastasis when not requiring steroids. Two independent prospective clinical trials proved the efficacy of ipilimumab in controlling the metastatic burden of melanoma and in increasing the lifespan of patients. In these trials, ipilimumab showed an efficacy in extending overall survival (OS) superior than either a vaccination protocol, using the melanoma-associated gp100 peptide,3 or the standard chemotherapy regimen based on administration of dacarbazine.4
Ipilimumab is a fully human monoclonal antibody targeting cytotoxic-T lymphocytes antigen-4 (CTLA-4), an immune checkpoint molecule that mediates the physiological inhibition of activated T cells (reviewed in ref.5).6 Ipilimumab acts by releasing the brake induced by CTLA-4, thus enabling activated and tumor-specific T cells to build an effective immune response toward tumor antigens. Along the same line, several antibodies targeting immune checkpoint molecules are under clinical evaluation (reviewed in ref.7). In particular, the FDA and EMA recently approved pembrolizumab and nivolumab, two antibodies targeting PD-1, for the treatment of metastatic melanoma, lung and renal cell cancer. The introduction of these antibodies expands the range of available treatments for metastatic melanoma and paves the way for combinatorial approaches, especially for patients who have a poor or ineffective antitumor response. Recently, a deeper molecular characterization of ipilimumab mechanism of action was pursued and two independent studies found that the antigenic drivers of the immune rejection are mutant neo-epitopes arising from the genetic instability of the tumor itself.8 However, albeit the generation of neo-antigens depends on the higher tumor-intrinsic mutational load, a hypermutated genetic landscape is not sufficient to predict clinical response to ipilimumab, thus suggesting that also the immunological profile of patients plays a pivotal role in generating a strong response to CTLA-4 blockade. Indeed, a recent study performing gene expression profiling on melanoma biopsies from patients undergoing ipilimumab treatment revealed that clinical response can be predicted by a peculiar immunological pattern rather than by dysregulation of the genetic profile within malignant melanocytes.9
In the last years, a great effort has been dedicated to the identification of immunological biomarkers predictive of the efficacy of ipilimumab treatment. As expected, several studies searched for predictive markers within the T cell compartment, as this is the effector player of antitumor response. Indeed, it has been demonstrated that increased lymphocytes count10-14 and expression of activation markers, like ICOS, are powerful indicators of a good prognosis.15-17 Instead conflicting results emerged on the role of the regulatory T cells (Treg),10,13,18,19 since CTLA-4 is constitutively expressed by Tregs.3 Recently, Speiser and collaborators suggested that Tregs can be depleted by ADCC triggered by alternative monocytes.20 In parallel to these findings, clinical prognostic factors were identified especially serum lactate dehydrogenase (LDH) vascular endothelial growth factor (VEGF), and neutrophil/lymphocyte ratio, besides sex and ECOG performance status.11,12 In this context, there was also an increasing interest in monitoring suppressive populations able to divert T cell functions, and one of the key suppressive players expanded in cancer patients are myeloid-derived suppressor cells (MDSCs).21
Immunophenotyping of MDSCs is characterized by a high degree of complexity given the fact that several myeloid subsets have been described, ranging from immature cells to more differentiated cells such as monocytes and granulocytes, and a specific marker of such cells is still missing (reviewed in ref.22). In this context, some studies associated the circulating levels of MDSCs to prognosis in ipilimumab-treated patients but these studies often considered only one MDSC subset rather than describing the variations of the different MDSC subsets in response to ipilimumab.18,19,23,24
Overall, observational studies indicated a set of pharmacodynamic markers as potential surrogate predictors of survival in response to ipilimumab, but a clear picture of the immunological profile of responding patients is still missing.
In this study, we monitored the changes of a set of tumor-associated and immunological parameters (TIPs) in a cohort of ipilimumab-treated patients and set out to identify the immune profiles modulated by ipilimumab and associated with longer OS or lower toxicity.
Results
Clinical characteristic of the cohort
The median age of the 44 enrolled patients was 63 y (range 33–83 y). All patients were metastatic with a prevalence of M1c patients (n = 29, 66%) and a lower number of M1a (n = 6, 14%) and M1b (n = 9, 20%). Albeit most of the patients had a high metastatic burden, 70% of the patients presented an ECOG PS value of 0–1; the disease affected seriously the daily living ability (ECOG PS = 2) in 13 patients, corresponding to one-third of the cohort. Ipilimumab treatment was well tolerated and the majority of patients (n = 35, 80%) completed the scheduled four cycles of treatment. Disease control rate was 25% at week 12 (PR + SD), while a higher number of patients presented stable disease (SD) at the first evaluation at week 24, in which a complete response (CR) was observed, along with four partial responses (PR) and six SD. One-year survival rate was 45%. All patients' characteristics and response information are in Table 1.
Table 1.
Baseline characteristics of melanoma patients and healthy donors.
| Patients | Healthy donors | |||
|---|---|---|---|---|
| N | 44 | 21 | ||
| Age | 63 (33–83) | 58 (37–87) | ||
| Gender M/F | 29/15 | 14/7 | ||
| N | % | |||
| ECOG PS | ≤ 1 | 31 | 70 | |
| 2 | 13 | 30 | ||
| Stage | M1aM1bM1c | 6929 | ||
| Ipilimumab therapy | completed | 35 | 80 | |
| not completed | 9 | 20 | ||
| Response | Median/Mean TTP | 12/28 weeks | ||
| Response W12 | CR | 0 | 0 | |
| PR | 5 | 11 | ||
| SD | 6 | 14 | ||
| PD | 32 | 73 | ||
| NV | 1 | 2 | ||
| Response W24 | CR | 1 | 2 | |
| PR | 4 | 9 | ||
| SD | 6 | 14 | ||
| PD | 30 | 68 | ||
| NV | 3 | 7 | ||
| Survival | Median/Mean OS | 46/47 weeks | ||
| 1 year OS n = 40 | 18 | 45% | ||
Response was assessed 12 weeks upon enrolment according to immune-related response criteria and classified as follows: complete response (CR), partial response (PR), stable disease (SD), progressive disease (PD). Eastern Cooperative Oncology Performance Status (ECOG PS), week 0 (W0), week 12 (W12).
Immunological status at baseline
The first aim of this study was to investigate the presence of predictive and prognostic factors for OS in ipilimumab-treated patients and thus we evaluated baseline characteristics of the immune system of these patients. To this end, we measured by flow cytometry a set of immunological parameters belonging to both the lymphoid and the myeloid compartment, and we compared these values with a cohort of healthy donors (HD) matched for age and gender (Table 1). At baseline, melanoma patients showed a significant expansion of 3 out of 4 MDSC subsets investigated (Figs. 1A-D). In particular, the percentages of monocytic subsets CD14+/IL4Rα+ (Fig. 1A, %MDSC 1, HD = 3.3% vs. PT = 6.9% p <0.001) and CD14+/HLA-DRlow/− (Fig. 1D, %MDSC 4, HD= 0.7% vs. PT = 2.3%, P = 0.0015) were significantly higher compared to HD, together with the granulocytic subset CD15+/IL4Rα+ (Fig. 1B, %MDSC 2, HD = 0.4% vs. PT = 1.4%, p = 0.008). The immature subset of Lin−/HLA-DR−/CD33+/CD11b+ MDSCs was not significantly altered in melanoma patients (Fig. 1C, %MDSC 3, HD = 1.9 vs. PT = 2.2%, p = 0.15). Of note, we observed a significant correlation between the levels of MDSC 1 and MDSC 2 which are the two MDSC subsets expressing IL4Rα (Table 2, R = 0.344, p = 0.043).
Figure 1.
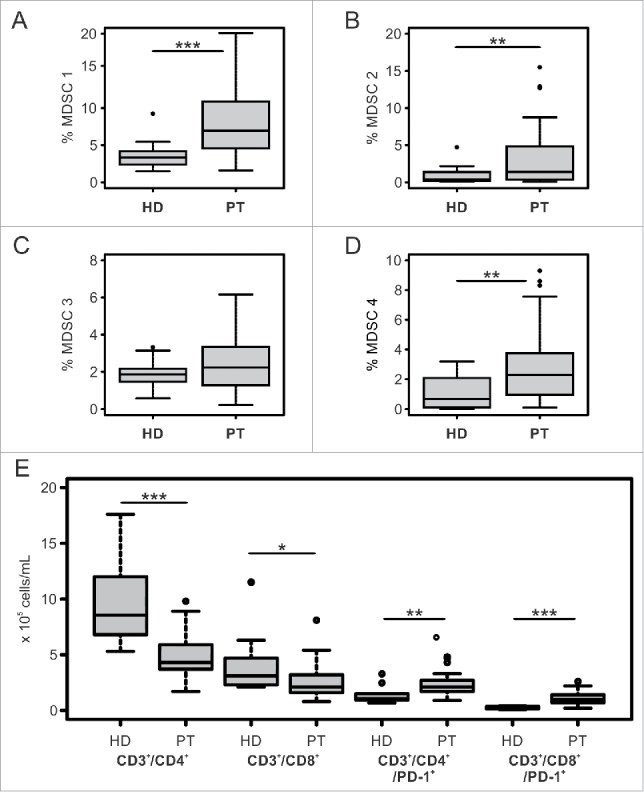
Immunological profile of melanoma patients and healthy donors at baseline. Panels A,B,C,D: Baseline levels of subsets in melanoma patients (PT, n = 40 to 35 depending on the myeloid subset considered) and age- and gender-matched healthy donors (HD n = 21) are shown. Panel E: Baseline absolute count of T cell subsets in melanoma patients (PT, n = 29) and age- and gender-matched healthy donors (HD n = 10). Each box plot shows first and third quartiles and median values of the subset indicated. Outlier are plotted as individual points. Wilcoxon Rank Sum test was used to compare the frequencies of myeloid or T cell subsets between the two groups (*p ≤ 0.05, **p < 0.01, ***p < 0.001). Values were considered statistically significant for p < 0.05.
Table 2.
Correlations between TIPs at baseline.
| CORRELATION BETWEEN TIPS AT WO | ||
|---|---|---|
| Correlation between X and Y | R | p |
| MDSC 1 – MDSC 2 | 0.344 | 0.043 |
| CRP – IL-6 | 0.9 | <0.001 |
| CRP – LDH | 0.8 | <0.001 |
| CRP – MDSC 1 | 0.58 | <0.001 |
| IL-6 – LDH | 0.9 | <0.001 |
| IL-6 – MDSC 1 | 0.46 | 0.006 |
| LDH – MDSC 1 | 0.57 | <0.001 |
Pearson Correlation was used to assess correlation between baseline levels of TIPs. The pair subjected to correlation is indicated in the first column and the coefficient of correlation (R) and the level of significance (p) are shown in the second and third columns, respectively. Values were considered statistically significant for p < 0.05.
Concerning the lymphoid cells, at baseline, melanoma patients had a significant reduction in the total number of CD3+ cells (data not shown) which depended on a decrease of both T cell subsets, i.e., CD3+/CD4+ (HD = 8.6 vs. PT = 4.3 × 105/mL, p <0.001) and CD3+/CD8+ (HD = 3.1 vs. PT = 2.1 × 105/mL, p = 0.029). We also evaluated PD-1 expression on both CD4+ and CD8+ T cell subsets, and, in contrast, we observed that the number of CD3+/CD4+/PD-1+ (Fig. 1E, HD = 1.1 vs. PT = 2.1 × 105/mL, p = 0.006) and CD3+/CD8+/PD-1+ (HD = 0.2. vs. PT = 1.0 × 105/mL, p <0.001) was significantly expanded in patients compared to HD.
We also evaluated haematological indexes, such as C-reactive protein (CRP), interleukin-6 (IL-6), vascular endothelial growth factor (VEGF), S100B protein and LDH, proteins that are known to correlate with the inflammatory status of the patients and tumor burden. When we evaluated at baseline the correlation between TIPs, we found that the levels of CRP, LDH and IL-6 were significantly associated (Table 2). Of note, this inflammatory loop also involved one subset of MDSC, CD14+/IL4Rα+ (MDSC1) which was significantly correlated to CRP, IL-6 and LDH (Table 2, R = 0.58, p < 0.001 for MDSC1-CRP, R = 0.46, p = 0.006 for MDSC1-IL-6, R = 0.57, p < 0.001 for MDSC1-LDH).
Changes in levels of immunological and tumor-associated parameters following ipilimumab treatment
We evaluated the levels of the different TIPs at baseline and following ipilimumab treatment at W12. In addition, to unveil possible correlations between TIPs and OS, we divided the cohort of patients in two groups based on the survival status at 1 year.
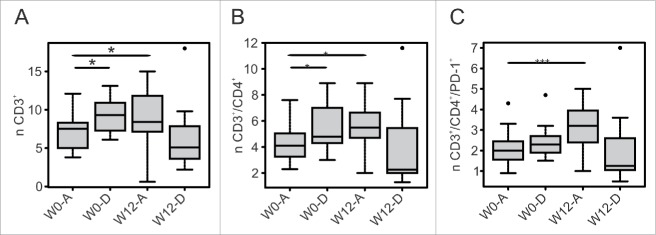
We observed that patients alive 1 year after enrolment had a significant increase in the number of T cells following treatment (W12), (Fig. 2A, 7.5 vs. 8.4 × 105/mL, p = 0.027), which was mainly due to CD3+/CD4+ cells (Fig. 2B). Of note, the number of CD4+ T cells expressing PD-1 increased significantly only in patients alive (Fig. 2C, 2.0 vs. 3.2 × 105/mL, p = 0.009). The CD3+/CD8+ T cell subset did not change significantly after ipilimumab treatment in both groups (data not shown).
Figure 2.
T cell subsets varied differently in response to ipilimumab according to the survival status of patients. W0 and W12 absolute count (n) of T cell subsets are reported according to the survival status of patients at 1 year (A = alive, D = deceased). Box plot report median, first and third quartiles. Outlier are plotted as individual points. Significance of intra-group variations between baseline and post-treatment values were assed using Wilcoxon Signed Rank test, while the Wilcoxon Rank Sum Test was used to compare inter-group variations (*p ≤ 0.05, **p < 0.01, ***p < 0.001). Values were considered statistically significant for p < 0.05.
Although ipilimumab acts directly on T cell, we also monitored the myeloid compartment upon treatment and observed that patients with worst clinical outcome had a significant increase in MDSC4 subset following treatment (Fig. 3D W0 D = 2.45% vs. W12 D = 3.63%, p = 0.039). We also observed that patients with worst clinical outcome had higher levels of MDSC 1 after treatment, as compared to patients still alive after 1 year (Fig. 3A, W12 A = 6.1% vs. W12 D = 10.7%, p = 0.035). On the contrary, patients alive 1 year after treatment had stable levels of MDSC 1 and MDSC 4 (Figs. 3A and D) and decreasing levels of MDSC3 (Fig. 3C, W0 A = 2.68% vs. W12 A = 1.9%, p = 0.008), while they upregulated MDSC2 similarly to patients with worst prognosis (Fig. 3B W0 A = 2.7% vs. W12 A = 5.1% p = 0.03).
Figure 3.
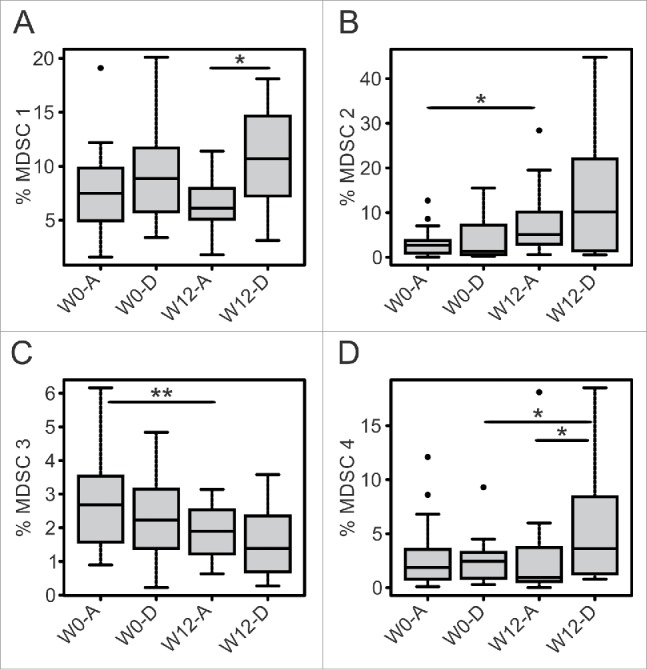
Levels of MDSC subsets before and after ipilimumab treatment in patients with different survival status. W0 and W12 frequencies (%) of MDSC subsets are reported according to the survival status of patients at 1 year (A = alive, D = deceased). Box plot report median, first and third quartiles. Outlier are plotted as individual points. Significance of intra-group variations between baseline and post-treatment values were assed using Wilcoxon Signed Rank test, while the Wilcoxon Rank Sum Test was used to compare inter-group variations (*p ≤ 0.05, **p < 0.01, ***p < 0.001). Values were considered statistically significant for p < 0.05).
When we analyzed the correlation between MDSC subset and hematological parameters after treatment, we confirmed the significant correlation between the inflammatory markers CRP and IL-6 with MDSC1 (Table 3, R = 0.45, p = 0.015 for MDSC 1-CRP and R = 0.45, p = 0.013 for MDSC1 and IL-6) and we observed an additional correlation between CRP and MDSC 4 subsets (Table 3, R = 0.56, p = 0.001).
Table 3.
Correlations between TIPs after ipilimumab treatment.
| CORRELATION BETWEEN TIPS AT W12 | ||
|---|---|---|
| Correlation between X and Y | R | p |
| MDSC 1 – MDSC 4 | 0.43 | 0.018 |
| MDSC 1 – CD3+ | −0.36 | 0.05 |
| MDSC 1 – IL-6 | 0.45 | 0.013 |
| MDSC 1 – CRP | 0.45 | 0.015 |
| MDSC 4 – CD3+ | −0.5 | 0.003 |
| MDSC 4 – CRP | 0.56 | 0.001 |
| CRP – IL-6 | 0.73 | <0.001 |
Pearson Correlation was used to assess correlation between post-treatment (W12) levels of TIPs. The pair subjected to correlation is indicated in the first column and the coefficient of correlation (R) and the level of significance (p) are shown in the second and third columns, respectively. Values were considered statistically significant for p < 0.05.
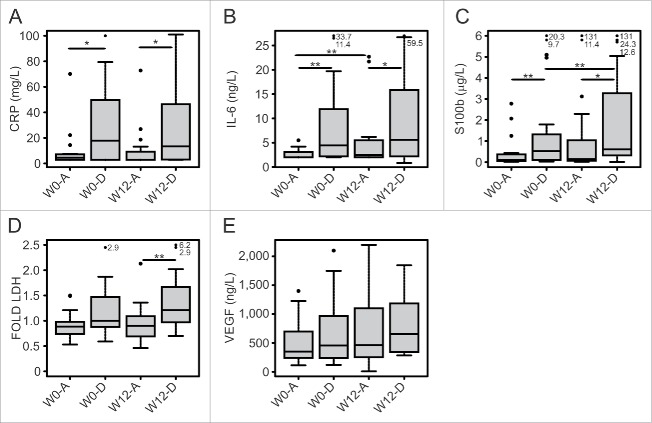
An analysis of CRP and IL-6 concentration showed that these factors were significantly lower in patients with better prognosis both at baseline and after treatment (Fig. 4A, 4.6 vs. 17.9 mg/L, p = 0.04 at baseline and 2.9 vs. 13.4 mg/L, p = 0.03 at W12 for CRP) (Fig. 4B, 2 vs. 4.5 ng/L, p = 0.003 at baseline and 2.5 vs. 5.6 ng/L, p = 0.05 at W12 for IL-6). On the other hand, patients with worse prognosis had significantly higher levels of S100B and of LDH both at baseline and after treatment (Fig. 4C, 0.1 vs. 0.5 μg/L, p = 0.004 at baseline and 0.2 vs. 0.6 μg/L, p = 0.022 at W12 for S100B) (Fig. 4D, 252 vs. 342.5, p = 0.06 at baseline and 223 vs. 323, p = 0.005 at W12 for LDH). Finally, VEGF concentration was not significantly associated with prognosis and was not correlated with other TIPs (Fig. 4E and data not shown).
Figure 4.
Profiles of haematological parameters of patients treated with ipilimumab represented according to their survival status. W0 and W12 values of haematological parameters (CRP, IL-6, S100B, fold LDH, VEGF) are reported, dividing the cohort of patients in two groups on the basis of the survival status at 1 year (A = alive, D = deceased). Box plot report median, first and third quartiles. Outlier are plotted as individual points. Significance of intra-group variations between baseline and post-treatment values were assed using Wilcoxon Signed Rank test, while the Wilcoxon Rank Sum test was used to compare inter-group variations (*p ≤ 0.05, **p < 0.01, ***p < 0.001). Values were considered statistically significant for p < 0.05).
Baseline and post-treatment levels of TIPs are associated with OS in response to ipilimumab
The analysis of the changes of TIP levels following ipilimumab treatment and the network of correlations found among TIPs prompted us to investigate possible associations between TIPs and OS. To this aim, we developed a statistical model to perform the multivariate analysis of association between TIPs and OS and applied Cox proportional hazard univariate and multivariate regression models. We included in the model the variables that were more significantly associated with OS from our preliminary analysis based on the hypothesis-testing step performed that divides the cohort in two groups based on their survival status at 1 year. We tested separately the possible association between OS and TIPs measured either at W0 or at W12. In the first case, for the analysis at baseline, we included in the multivariate analysis a set of hematological parameters that were tightly associated with prognosis in the preliminary step of analysis. The model requires binary variables and therefore we used the upper limit of normal (ULN) or, when ULN was not applicable, the medium value for clusterization. We divided the patients with ECOG PS < 1 from those with an ECOG PS above 1.
We first investigated possible associations between baseline values of TIPs and OS searching for predictive factors. To this end, we considered baseline values of IL-6, LDH, S100B, CRP and included the ECOG PS that is a classical predictive factor for OS in cancer patients, and, as expected, the analysis revealed that an ECOG PS above one is an independent predictive factor for reduced OS (Table 4A). Of note, the levels of IL-6 were similarly predictive for OS. Although the Hazard ratio (HR) for ECOG PS is equal to 3 (HR = 3 (1.2–7.9), p = 0.023), levels of IL-6 above the ULN expose the patients to a risk of premature death 4.76 times higher (HR = 4.76 (1.3–16.8), p = 0.015) than patients with IL-6 values below the ULN. Other factors failed to demonstrate a significant predictive power for OS in our cohort of patients.
(B)
| FACTOR | Pr(>|z|) | HR | lower 95 | upper 95 |
|---|---|---|---|---|
| S100B | 0.53369 | 0.9916 | 0.9656 | 1.018 |
| LDH | 0.00452** | 4.7604 | 1.6214 | 13.977 |
| IL-6 | 0.52059 | 1.0354 | 0.9311 | 1.151 |
| L3 | 0.50013 | 0.9263 | 0.7415 | 1.157 |
| MDSC 1 | 0.02649* | 1.2556 | 1.0269 | 1.535 |
| MDSC 2 | 0.98174 | 0.9994 | 0.9467 | 1.055 |
Tables A and B show the results of the Cox multivariate analysis using baseline or W12 levels of TIPS, respectively. The TIPs included in the statistical model are indicated in the first column. We reported the levels of significance (Pr(>|z|), the Hazard Ratio (HR) and the 95% confidence interval (lower 95 – upper 95). Values were considered statistically significant for p < 0.05.
Table 4.
Multivariate survival analysis.
(A)
| FACTOR | Pr(>|z|) | HR | lower 95 | upper 95 |
|---|---|---|---|---|
| S100B | 0.5560 | 1.338 | 0.5073 | 3.531 |
| LDH | 0.0891 | 2.271 | 0.8821 | 5.849 |
| ECOG-PS | 0.0234* | 3.038 | 1.1624 | 7.939 |
| IL-6 | 0.0152* | 4.755 | 1.3495 | 16.758 |
| CRP | 0.5917 | 0.715 | 0.2098 | 2.436 |
In a similar manner, we built the multivariate survival analysis model for post-treatment values of TIPs in order to identify prognostic factors for OS in response to ipilimumab. We included IL-6, S100B, LDH, CD3+/CD4+/PD-1+ T cells, MDSC 1 and MDSC 2 subsets as factors (Table 4B). Our analysis confirmed that LDH is a potent independent prognostic factor for ipilimumab treatment (HR = 4.76 (1.6–14), p = 0.005). Interestingly, also MDSC 1 levels are an independent prognostic factor for OS in patients treated with ipilimumab (HR = 1.26 (1–1.5), p = 0.026).
We tested the possible predictive and prognostic significance of the other TIPs, and in particular of the other MDSC subsets which seemed associated with prognosis from our preliminary analysis; however, the model did not find any other significant associations between OS and TIPs.
The levels of TIPs are predictive of toxicity
The management of immune-related toxicities of checkpoint inhibitors is now well known, but adverse drug reactions (ADR) of moderate to high intensity must be managed readily in order to avoid severe toxicity. Since most of the ADR induced by ipilimumab are immune-mediated, we investigated whether immune parameters can be predictive of the onset of toxicity. To this end, we divided the cohort of patients in two groups based on the development of G3-4 ADR. We then compared baseline and post-treatment values of TIPs of the two groups.
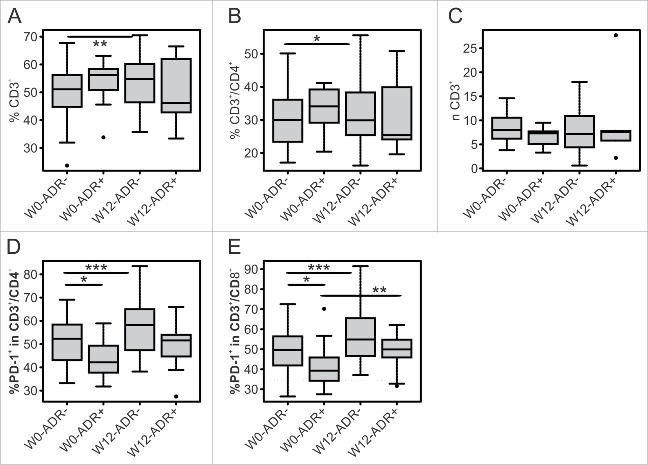
We observed that the lymphoid compartment could be predictive for toxicity. Indeed, patients without severe toxicity presented a boost of the percentage of CD3+ T cells (L8), in particular of the CD4+ subset (L1), albeit the number (n) of total T cells is equal in both groups (Fig. 5A, %L8 51.1% vs. 54.8%, p = 0.008, Fig. 5B %L1 30% vs. 30%, p = 0.03, Fig. 5C nL8, 8 vs. 7.2 × 105/mL, p = NS). Of note, we observed that patients that did not develop ADR presented higher percentage of PD-1 expressing T cells on both CD4+ (L5) and CD8+ (L6) subsets at baseline (Fig. 5D, %L5, 52.3% vs. 42.2%, p = 0.025, Fig. 5E %L6, 49.6% vs. 39.3%, p = 0.022). In addition, both groups upregulated PD-1 expression upon treatment but this upregulation on CD4+ T cells was significant only for patients with lower toxicity (Fig. 5D, %L5, 52.3% vs. 58.2%, p = 0.0007).
Figure 5.
T cell subsets as early predictors of toxicity. Box plots report median, first and third quartiles of T cell subsets (frequency = %, absolute number = n) measured at W0 and W12 in patients without (ADR−) or with (ADR+) adverse drug reactions. Outlier are plotted as individual points. Significance of intra-group variations between W0 and W12 values were assed using Wilcoxon Signed Rank test, while Wilcoxon Rank Sum test was used to compare inter-group variations (*p ≤ 0.05, **p < 0.01, ***p < 0.001). Values were considered statistically significant for p < 0.05).
Finally, we investigated whether the TIPs associated with OS were significantly associated with toxicity, but we did not find any statistically significant association.
Discussion
There is a growing interest in identifying circulating factors as surrogate biomarkers of clinical response to ipilimumab treatment (reviewed in refs.25,26). One potential biomarker is MDSC level because the presence of these cells has been correlated to tumor burden and OS in different types of cancer, and some studies demonstrated its prognostic role for the outcome of different chemotherapeutic regimens (reviewed in ref.21). Several non-overlapping MDSC phenotypes have been described, classified as immature, monocytic and granulocytic subsets, but most of the studies reduced MDSC monitoring to only one phenotype.22 The phenotypic complexity of human MDSCs complicates the validation of the predictive significance of MDSCs in multicenter studies, and creates a challenge in finding a consensus on the minimal requirements for MDSC monitoring.27 We developed a standardized approach to monitor the circulating levels of four MDSC subsets in melanoma patients receiving ipilimumab. To date, only one study monitored six MDSC subsets simultaneously, and found that five out of six subsets were significantly expanded in the blood of renal cell carcinoma patients; moreover, baseline levels of two of these subsets were negatively associated with OS in response to a multipeptide-based vaccination protocol.28 In line with these results, we observed that also melanoma patients significantly expanded more than one MDSC subset at baseline, thus suggesting the presence of immune suppression in these patients. In particular, the cohort under investigation expanded three MDSC subsets (MDSC1-2-4) which were originally discovered in melanoma patients and further described in other types of cancer (reviewed in ref.21).
In view of the immune-mediated effect of ipilimumab, lower levels of suppressive cells could represent not only an estimator of clinical benefit but also a pharmacodynamics biomarker, reflecting the shift from immune escape to immune-response. Indeed, the role of MDSCs as a biomarker for ipilimumab treatment was demonstrated in a number of studies, mostly reporting correlations between monocytic MDSCs and clinical response.18,23,24,29-32 Kitano and colleagues developed a computational algorithm-based method to perform a uniform analysis of the levels of a monocytic subset in melanoma patients treated with ipilimumab at 3 or 10 mg/kg and found a significant association between levels of Lin− CD14+CD11b+ HLA-DRlow/−MDSCs evaluated at baseline and at week 6 and OS, albeit the prognostic significance of MDSCs was restricted only to patients receiving the higher dose of ipilimumab.23 Along the same line, Martens et al. introduced MDSCs defined as CD14+/HLA-DRlow/− cells, together with other peripheral blood biomarkers, in a combination model able to identify a biomarker signature at baseline in advanced melanoma patients experiencing clinical benefit from ipilimumab treatment.24 In another study, granulocytic MDSCs defined as Lin−/HLA-DR−/CD15+/CD11b+ cells were monitored in seven patients that received ipilimumab treatment at 3 mg/kg or 10 mg/kg doses and it was demonstrated a significant reduction of this subset, along with the downregulation of Arginase-1 expression on the whole myeloid compartment.19 In our study, in which ipilimumab was administered at the standard dose of 3 mg/kg, we identified post-treatment levels of the monocytic subset CD14+/IL4Rα+ cells (MDSC 1) as a strong independent prognostic factor for OS in a multivariate model comprehending six TIPs, but we did not find a significant association between baseline levels of MDSCs and OS. Besides, two MDSC subsets presented a significant inverse correlation with CD3+T cells at W12 (Table 3). Such inverse correlation was also observed in two independent studies, conducted in non-treated lung cancer patients33 and in melanoma patients receiving ipilimumab.23
Lin−/HLA-DR−/CD33+/CD11b+ subset (MDSC3), which is the most immature MDSC population, presented a peculiar behavior in this study. We previously demonstrated that this population correlates with OS and progresson-free survival of patients with solid tumor and with levels of circulating tumor cells.34 However, in the present cohort of melanoma patients, MDSC3 subset is not expanded at baseline, but it shows a tendency to increase during treatment; moreover, it does not have a particular correlation with TIPs or survival. An intriguing explanation for this result is that ipilimumab treatment affects, directly or indirectly, the maturation of MDSC, but, to the best of our knowledge, this possibility has never been explored
All together these findings suggest the presence of a balance between suppressive and effector leukocytes characterizing the immunological profile of cancer patients, and in this scenario the boosting T cell responses by ipilimumab could alter this equilibrium, thus leading to override the action of immune suppression exerted by MDSCs. Along this line of reasoning, a suitable strategy to track the onset of immune rejection triggered by ipilimumab is to monitor the activation and functional status of T cells. In fact, absolute lymphocyte count is one of the first parameter identified as strongly associated to clinical effect of ipilimumab and many studies characterized the immune correlates of this drug.10,11,13,14,23,35 In our cohort, patients presented a reduced number of CD3+ T cells at baseline and ipilimumab treatment significantly boosted the level of these cells but only in patients demonstrating a clinical benefit. Of note, the increase in T cell number was due mainly to the CD4+ subset while the CD8+ counterpart did not significantly change upon ipilimumab treatment.
Several studies investigated the phenotype of T cells in patients receiving ipilimumab and found that an activated phenotype, expressing ICOS or HLA-DR,17,36,37 increased antigen-specific responses11,36 and boosted IFNγ production38 are hallmarks of response to the treatment. However, less is known about the expression of other immune checkpoint molecules in response to the blockade of CTLA-4. To this end, we also monitored the levels of PD-1 expression on T cells in response to ipilimumab and found that melanoma patients presented significantly higher numbers of PD-1 in both CD4+ and CD8+ compartments at baseline. Of note, an expansion of PD-1+/CD4+ T lymphocytes' number was observed upon treatment only in patients with longer survival.
Since in our cohort of patients the expansion of CD4+/PD-1+ T cells was associated also with reduced toxicity, we speculated that increasing frequencies of these cells may be due to an expansion of Tregs. This hypothesis is based on the fact that PD-1 and CTLA-4 play an important role in Treg induction and functional activity.39 The presence of Tregs was monitored in ipilimumab-treated patients with conflicting results which makes the predictive power of Treg not extensively reliable.10,13,18,19
In addition to circulating levels of MDSC and T cell subsets, we observed that a number of TIPs are good estimator of OS like ECOG PS, the pro-inflammatory cytokine IL-6 and LDH levels. While the association between OS and ECOG PS is well known, our results are in line with a study that demonstrated the predictive potential of LDH levels in ipilimumab-treated patients.12 Of note, we identified baseline level of IL-6 as a new strong independent estimator of OS in response to ipilimumab; and, in fact, patients with IL-6 levels above the ULN had a 4.7-fold higher risk of mortality compared to patients with lower levels of this cytokine, independently from LDH, ECOG-PS, S100B and CRP levels.
Given the predictive and prognostic power of IL-6, LDH and of CD14+/IL4Rα+ MDSCs underscored by our study, it is interesting to observe that baseline levels of these parameters were significantly interconnected, thus suggesting the existence of an immunological loop associated with OS in response to ipilimumab. In fact, IL-6 levels correlated significantly with the circulating levels of CD14+/IL4Rα+ MDSCs, LDH and CRP.
In addition, CD15+/IL4Rα+ cells (MDSC2) correlate with LDH and with MDSC1, the subset that shares the expression of IL4Rα with MDSC2. On the contrary, MDSC3 and MDSC4 did not correlate with pro-inflammatory and tumor-associated markers and they seem to be excluded from the network in which MDSC1 participates. In a similar context, Meyers et al. investigated the possible correlation between MDSC4 and LDH levels in ipilimumab-treated melanoma patients and they found no correlation between these factors, as well.29
We did not investigate whether high levels of TIPs may play a preferential effect on MDSC1 expansion in our cohort of patients, but we found that only the MDSC1 subset is associated with overall survival, thus suggesting a critical role for this cell subset in ipilimumab-treated patients. Indeed, the presence of a suppressive network, in which inflammatory proteins induce MDSC expansion and regulate their suppressive functions has been already demonstrated in murine models40 (reviewed in ref.41) and in human cells in vitro.42,43 In addition, the downstream mediator of IL-6 signaling, signal transducer and activator of transcription 3 (STAT-3), regulated the suppressive activity of tumor-infiltrating MDSCs from head and neck squamous cell carcinoma patients through modulation of arginase 1 activity.44 Finally, we recently demonstrated that STAT-3 plays a pivotal role in the mechanism of immune-suppression of in vitro derived MDSCs that in turns induced the expression of PD-1 and Lag-3 on activated T cells upon contact.45
The connection observed in our study between IL-6, MDSC levels and OS suggests the hypothesis of a combination therapy of anti-IL-6 mAb and immune-checkpoint inhibitors. From one side, IL-6 blockade could enhance the efficacy of immune checkpoint inhibitors by disrupting the mechanisms of immune escape at the tumor site, and from the other side, it could also reduce the toxicity of this treatment, which is characterized by autoimmune responses. Indeed, blockade of IL-6 pathway is currently approved for the treatment of autoimmune diseases, as rheumatoid arthritis, and combinations of anti-IL-6 and chemotherapy are already under development in phase I studies for cancer treatment (reviewed in ref.46).
In addition, we designed the present study also to identify an immune profile associated with the development of immune-related adverse drug reactions (irADR) due to ipilimumab treatment. Albeit early reports indicated that the onset of irADR was prognostic of response to ipilimumab the potential link between irADR and clinical benefit was not confirmed by large perspective studies.47,48 Our results indicate that the development of toxicity was not predictive for longer OS in response to ipilimumab. However, in our study the immune profile of patients protected by toxicity was characterized by a boost in the percentage of T cells, mainly CD4+ T cells, and a higher expression of PD-1 on CD4+ T cells at baseline. In addition, patients with lower toxicity significantly expanded PD-1+ T lymphocytes compared to those experiencing grade 3 irADR. Our results constitute a first attempt to characterize the immune profile of patients prone to toxicity and are supported by the study of Wang and colleagues,16 in which it was observed that the onset of irADR was associated with lower frequencies of proliferating Ki67+, CD4+ and CD8+ T cells. In particular, patients free from toxicity expanded a subset of CD4+ T cells expressing eomesodermin, a transcription factor that regulates the generation of memory T cells49 and, similarly to our results, was associated to increased PD-1 expression.50
In conclusion, this study explored different aspects of immunomonitoring in metastatic melanoma defining the immune profile of patients that most likely benefit from ipilimumab treatment. Our results require further validation in an independent cohort, but shed light on the complex immunological scenario in which immune checkpoint inhibitors act. A better understanding of the cross-talk between patient's immunity and tumor is a necessary step to tailor the most suitable combination of treatments on the immunological characteristics of each patient, also in view of the future therapies based on the combination of immune checkpoint inhibitors.51,52
Patients and methods
Patients
Forty-four patients with a diagnosis of stage IV melanoma were enrolled in this study. A description of the clinical characteristics of these patients is reported in Table 1. Exclusion criteria for the administration of ipilimumab were the presence of symptomatic brain metastases and a history of autoimmune disease requiring immunosuppressive therapy. Ipilimumab was administered after at least another line of treatment, consisting in systemic chemotherapy, or BRAF inhibitors. Twenty-one age- and gender-matched HD were used as controls. The study was approved by the ethical committee of Istituto Oncologico Veneto and all the patients enrolled provided a written informed consent before entering in this study. Patients received four doses of ipilimumab (3 mg/kg) every 3 weeks, as approved by the Italian Medicine Agency. In case of onset of immune-related adverse reactions to the treatment, requiring corticosteroids treatment, the therapy was discontinued and eventually resumed when patients recovered from toxicity. Disease evaluation was done comparing baseline computered axial tomography (CAT) with a CAT done at week 12, 16, 24, 36, 48, 60 and thereafter every 6 months, or before if clinically indicated. Response to the treatment was assessed using the immune-related Response Criteria.53 Patients presenting SD were computed together with patients obtaining partial or complete remission. Follow-up of patients continued until death or subsequent therapy for disease progression or recurrence. Patients who presented, at any time within the observation period, X′ with severity above grade 3 (G3–G4) were named ADR+ while X severity below G3 were named ADR−. The grading of ADR was assessed according to Common Terminology Criteria for Adverse Events (CTCAE v.4.0).
Study design
Peripheral blood from melanoma patients was collected at baseline (W0) and 12 weeks after the first dose of ipilimumab was administered (W12). Additional blood samples were collected from patients every 12 weeks until disease progression. Blood samples were withdrawn in EDTA-treated vacutainer tubes (BD Bioscience) and processed immediately from HD and from patients. Moreover, a set of parameters associated with the immune system and tumor burden were monitored at W0 and W12. Circulating levels of myeloid and T cell subsets were evaluated by multicolor flow cytometry, whereas hematological parameters were evaluated by the Central Laboratory of the University Hospital of Padova. As the normal value range for LDH varied during our observation time, in this study we considered the normalized value of LDH that is the raw value of LDH/ ULN. All collected parameters are listed in Table S1
Characterization of the immune profile by flow cytometry
The presence of circulating myeloid and T cell subsets was assessed in whole blood samples by multi-color flow-cytometry, in order to identify four subsets of MDSCs and different subsets of T lymphocytes. To identify MDSC subsets, an eight-color staining cocktail was used containing: anti-CD11b Alexa 700 (clone ICRF44, BD PharMingen), anti-CD14 APC-H7 (clone MϕP9, BD Bioscience), anti-CD15 V450 (clone MMA, BD Biosciences), anti-CD33 PECy7 (clone P67.6, BD Biosciences), anti-IL4Rα PE (clone 25463, R&D SYSTEMS), Lineage cocktail (anti-CD3-14-19-56) (clone UCHT1, M5E2, HIB19, NCAM16.2, BD Biosciences and BD PharMingen), anti-HLA-DR APC (clone L243, BD Biosciences). The live/dead (L/D), an amine-reactive dye, was chosen as dead cell marker (DCM) because it resists to cell-fixation with unaltered staining capability. The staining panel for T cell subsets identification is composed of: anti-CD3 ECD (clone UCHT1, Beckman Coulter), anti-CD4 FITC (clone SK3, BD Bioscience) anti-CD8 APC-H7 (clone SK1, BD Bioscience), anti-PD-1 PE (clone PD1.3.1.3, Miltenyi Biotec). For myeloid subsets phenotyping, 150 μL of fresh blood were washed with staining buffer (Hanks' Balanced Salt Solution for Flow Cytometry supplemented of 1% FBS: 137 mM NaCl (Sigma-Aldrich), 5 mM KCl (Sigma-Aldrich), 0.3 mM Na2HPO4 (Sigma-Aldrich), 0.7 mM KH2PO4 (Sigma-Aldrich), 0.4 mM MgSO4 (Sigma-Aldrich), 0.3 mM MgCl2 (Sigma-Aldrich), 5 mM Glucose (Sigma-Aldrich), 4 mM NaHCO3 (Sigma-Aldrich), 1 mM EDTA (Sigma-Aldrich)) and subsequently incubated with Fc-Receptor Blocking reagent (Miltenyi Biotec) at 4°C for 15 min. Afterwards, cells were stained with anti-IL4Rα (anti-CD124) PE antibody and incubated at 4°C for 10 min. Later, the mixture of properly diluted antibodies (plus L/D) was added to the tubes and incubated at 4°C for 20 min. Cells were then washed with staining buffer and centrifuged at 1,300 rpm for 6 min at 4°C. For T lymphocytes phenotyping, the procedure was the same with the exception that 50 μL of whole blood were used per tube and the total incubation time for antibody-staining was reduced from 30 to 20 min. After the washing step, red blood cells were lysed using Cal-Lyse whole blood lysing solution (Life Technologies) according to manufacturer's instructions. Absolute counts of T cell subsets was determined using TruCount tubes (BD Bioscience). Data acquisition was performed using a LSRII flow cytometer (BD Biosciences) equipped with four lasers (405 nm, 488 nm, 561 nm, 640 nm). Data were analyzed using FlowJo software (Three Star Inc.). Autofluorescence and fluorescence minus one (FMO) controls for HLA-DR, IL4Rα and PD-1 were used as negative controls. Exemplary gating strategies for phenotyping of myeloid and T cell subsets are shown in Fig. S1.
Standardization of the immunophenotyping assay
To standardize the staining panels for myeloid and T lymphocytes' subsets, a dilution of antibodies that maximizes the signal to noise ratio was chosen based on single antibodies titration. In addition, a protocol to monitor the performance of antibodies against HLA-DR and IL4Rα was set-up. Briefly, an EBV-B cell line that constitutively expresses these markers at high expression intensity was used as reference. To reduce inter-assay variance, we used a single batch of the B cell-line, which was fixed and permeabilized before cryopreservation. For each staining, the control cell-line was run in parallel to blood staining, labeling it with the same amount of anti-HLA-DR or anti-IL4Rα antibodies used for blood staining. We acquired the control cells before acquiring the blood sample, in order to determine whether the mean fluorescence intensity (MFI) of HLA-DR or IL4Rα were included in the range of tolerance. The range of tolerance was built by repeated staining of the control cells performed before the beginning of the study. To this end, the MFI of HLA-DR and IL4Rα of control cells was calculated in repeated measurements and the borders of tolerance were set within the mean ± 2 × standard deviation of our measures. In order to monitor the performance of the flow-cytometer, the potential variation of the performance of the flow cytometer was assessed, using a protocol after Perfetto et al.54
Statistical analysis
Clinical characteristics of melanoma patients and HD were reported as frequencies and percentages and with median and quartiles, for categorical and continuous variables, respectively. Comparisons of a set of immunological parameters able to describe both the lymphoid and the myeloid compartment, between melanoma patients at baseline and HD were performed using Wilcoxon–Mann–Whitney test. In order to assess a possible association between TIPs and OS, the cohort of patients has been divided in two groups based on the survival status 1 year after enrolment. Comparisons of levels of TIPs at baseline and post-treatment between patients alive and deceased at 1 year were performed using Wilcoxon–Mann–Whitney test, whereas differences of TIPs between the two occasions (baseline and post-treatment) separately for patients alive and deceased at 1 year, have been tested by Wilcoxon Signed Rank Sum test, used when comparing two related samples.55 In order to investigate the association between TIP values and presence of ADR provoked by ipilimumab treatment, the cohort of patients has been divided in two groups on the basis of the development of G3-4 ADR. Wilcoxon–Mann–Whitney and Wilcoxon Signed Rank Sum tests have again been adopted to compare different groups of patients (with and without ADR) and the same patients at baseline and post-treatment, respectively. In order to identify factors associated with mortality, Cox proportional hazard univariate and multivariate regression models were applied and results were reported as hazard ratio (HRs) along with their 95% confidence intervals (95% CIs). Cox proportional hazard model has been adopted to put in relation OS with values of TIPs both at baseline and post-treatment. We included in the multivariate analysis a set of hematological parameters, which seemed to be the most tightly associated with prognosis from the preliminary step of analysis. Variables selected have been preliminary transformed in binary variables based on the ULN or, when ULN was not applicable, the median value. In this way, variables were coded as 0 when values were less than ULN (or median) and as 1 when values were greater than ULN (or median). We also divided the patients with ECOG PS < 1 from those with an ECOG PS above 1. HR in Table 4 have to be interpreted as relative hazard of death being in class 1 (>ULN, or >MED, or >1) with respect to being in class 0 (<ULN, <MED, <1). Correlations between hematological and immunological parameters were assessed at baseline and after ipilimumab treatment using Person Correlation analysis. One-sided alternatives with a significance level α = 0.05 were considered for all the tests.
The analyses were performed using StatXact-5® (Cytel Software), SAS Release 9.2 (SAS Institute, Cary, NC), Sigmaplot version 12 (Systat Software) and R version 3.2.1.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from Italian Association for Cancer Research (AIRC grant #17400 to S.M.). L.P. was a recipient of an AIRC fellowship, and S.S. a recipient of a Pezcoller Fondation fellowship.
References
- 1.Jang S, Atkins MB. Which drug, and when, for patients with BRAF-mutant melanoma? Lancet Oncol 2013; 14:e60-9; PMID:23369684; http://dx.doi.org/ 10.1016/S1470-2045(12)70539-9 [DOI] [PubMed] [Google Scholar]
- 2.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC et al.. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363:711-23; PMID:20525992; http://dx.doi.org/ 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature 2007; 445:771-5; PMID:17220874; http://dx.doi.org/ 10.1038/nature05543 [DOI] [PubMed] [Google Scholar]
- 4.Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ et al.. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 2011; 364:2517-26; PMID:21639810; http://dx.doi.org/ 10.1056/NEJMoa1104621 [DOI] [PubMed] [Google Scholar]
- 5.Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015; 348:56-61; PMID:25838373; http://dx.doi.org/ 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 6.Linsley PS, Greene JL, Tan P, Bradshaw J, Ledbetter JA, Anasetti C, Damle NK. Coexpression and functional cooperation of CTLA-4 and CD28 on activated T lymphocytes. J Exp Med 1992; 176:1595-604; PMID:1334116; http://dx.doi.org/ 10.1084/jem.176.6.1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 2015; 14:561-84; PMID:26228759; http://dx.doi.org/ 10.1038/nrd4591 [DOI] [PubMed] [Google Scholar]
- 8.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ et al.. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014; 515:577-81; PMID:25428507; http://dx.doi.org/ 10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS et al.. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014; 371:2189-99; PMID:25409260; http://dx.doi.org/ 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simeone E, Gentilcore G, Giannarelli D, Grimaldi AM, Caraco C, Curvietto M, Esposito A, Paone M, Palla M, Cavalcanti E et al.. Immunological and biological changes during ipilimumab treatment and their potential correlation with clinical response and survival in patients with advanced melanoma. Cancer Immunol Immunother 2014; 63:675-83; PMID:24695951; http://dx.doi.org/ 10.1007/s00262-014-1545-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan J, Zhou J, Dong Z, Tandon S, Kuk D, Panageas KS, Wong P, Wu X, Naidoo J, Page DB et al.. Pretreatment serum VEGF is associated with clinical response and overall survival in advanced melanoma patients treated with ipilimumab. Cancer Immunol Res 2014; 2:127-32; PMID:24778276; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelderman S, Heemskerk B, van Tinteren H, van den Brom RR, Hospers GA, van den Eertwegh AJ, Kapiteijn EW, de Groot JW, Soetekouw P, Jansen RL et al.. Lactate dehydrogenase as a selection criterion for ipilimumab treatment in metastatic melanoma. Cancer Immunol Immunother 2014; 63:449-58; PMID:24609989; http://dx.doi.org/ 10.1007/s00262-014-1528-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamid O, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, Guida M, Hyams DM, Gomez H, Bastholt L et al.. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med 2011; 9:204; PMID:22123319; http://dx.doi.org/ 10.1186/1479-5876-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens A, Wistuba-Hamprecht K, Yuan J, Postow MA, Wong P, Capone M, Madonna G, Khammari A, Schilling B, Sucker A et al.. Increases in absolute lymphocytes and circulating CD4+ and CD8+ T cells are associated with positive clinical outcome of melanoma patients treated with ipilimumab. Clin Cancer Res 2016; 22(19):4848-58; PMID:27169993; http://dx.doi.org/21708958 10.1158/1078-0432.CCR-16-0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu T, He Q, Sharma P. The ICOS/ICOSL pathway is required for optimal antitumor responses mediated by anti-CTLA-4 therapy. Cancer Res 2011; 71:5445-54; PMID:21708958; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-1138 [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Yu D, Sarnaik AA, Yu B, Hall M, Morelli D, Zhang Y, Zhao X, Weber JS. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J Transl Med 2012; 10:146; PMID:22788688; http://dx.doi.org/ 10.1186/1479-5876-10-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liakou CI, Kamat A, Tang DN, Chen H, Sun J, Troncoso P, Logothetis C, Sharma P. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Natl Acad Sci U S A 2008; 105:14987-92; PMID:18818309; http://dx.doi.org/ 10.1073/pnas.0806075105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarhini AA, Edington H, Butterfield LH, Lin Y, Shuai Y, Tawbi H, Sander C, Yin Y, Holtzman M, Johnson J et al.. Immune monitoring of the circulation and the tumor microenvironment in patients with regionally advanced melanoma receiving neoadjuvant ipilimumab. PLoS One 2014; 9:e87705; PMID:24498358; http://dx.doi.org/ 10.1371/journal.pone.0087705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pico de Coana Y, Poschke I, Gentilcore G, Mao Y, Nystrom M, Hansson J, Masucci GV, Kiessling R. Ipilimumab treatment results in an early decrease in the frequency of circulating granulocytic myeloid-derived suppressor cells as well as their Arginase1 production. Cancer Immunol Res 2013; 1:158-62; PMID:24777678; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0016 [DOI] [PubMed] [Google Scholar]
- 20.Romano E, Kusio-Kobialka M, Foukas PG, Baumgaertner P, Meyer C, Ballabeni P, Michielin O, Weide B, Romero P, Speiser DE. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc Natl Acad Sci U S A 2015; 112:6140-5; PMID:25918390; http://dx.doi.org/ 10.1073/pnas.1417320112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solito S, Marigo I, Pinton L, Damuzzo V, Mandruzzato S, Bronte V. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann N Y Acad Sci 2014; 1319:47-65; PMID:24965257; http://dx.doi.org/ 10.1111/nyas.12469 [DOI] [PubMed] [Google Scholar]
- 22.Damuzzo V, Pinton L, Desantis G, Solito S, Marigo I, Bronte V, Mandruzzato S. Complexity and challenges in defining myeloid-derived suppressor cells. Cytometry B Clin Cytom 2014; 88(2):77-91; PMID:25504825; http://dx.doi.org/24844912 10.1002/cytob.21206 [DOI] [PubMed] [Google Scholar]
- 23.Kitano S, Postow MA, Ziegler CG, Kuk D, Panageas KS, Cortez C, Rasalan T, Adamow M, Yuan J, Wong P et al.. Computational algorithm-driven evaluation of monocytic myeloid-derived suppressor cell frequency for prediction of clinical outcomes. Cancer Immunol Res 2014; 2:812-21; PMID:24844912; http://dx.doi.org/ 10.1158/2326-6066.CIR-14-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martens A, Wistuba-Hamprecht K, Geukes Foppen M, Yuan J, Postow MA, Wong P, Romano E, Khammari A, Dreno B, Capone M et al.. Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin Cancer Res 2016; 22:2908-18; PMID:26787752; http://dx.doi.org/ 10.1158/1078-0432.CCR-15-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ascierto PA, Kalos M, Schaer DA, Callahan MK, Wolchok JD. Biomarkers for immunostimulatory monoclonal antibodies in combination strategies for melanoma and other tumor types. Clin Cancer Res 2013; 19:1009-20; PMID:23460532; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-2982 [DOI] [PubMed] [Google Scholar]
- 26.Umansky V, Utikal J, Gebhardt C. Predictive immune markers in advanced melanoma patients treated with ipilimumab. Oncoimmunology 2016; 5:e1158901; PMID:27471626; http://dx.doi.org/ 10.1080/2162402X.2016.1158901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandruzzato S, Brandau S, Britten CM, Bronte V, Damuzzo V, Gouttefangeas C, Maurer D, Ottensmeier C, van der Burg SH, Welters MJ et al.. Toward harmonized phenotyping of human myeloid-derived suppressor cells by flow cytometry: results from an interim study. Cancer Immunol Immunother 2016; 65:161-9; PMID:26728481; http://dx.doi.org/ 10.1007/s00262-015-1782-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, Staehler M, Brugger W, Dietrich PY, Mendrzyk R et al.. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 2012; 18:1254-61; PMID:22842478; http://dx.doi.org/ 10.1038/nm.2883 [DOI] [PubMed] [Google Scholar]
- 29.Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, Michielin O, Romano E, Speiser DE. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother 2014; 63:247-57; PMID:24357148; http://dx.doi.org/ 10.1007/s00262-013-1508-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postow MA, Callahan MK, Barker CA, Yamada Y, Yuan J, Kitano S, Mu Z, Rasalan T, Adamow M, Ritter E et al.. Immunologic correlates of the abscopal effect in a patient with melanoma. N Engl J Med 2012; 366:925-31; PMID:22397654; http://dx.doi.org/ 10.1056/NEJMoa1112824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjoern J, Juul Nitschke N, Zeeberg Iversen T, Schmidt H, Fode K, Svane IM. Immunological correlates of treatment and response in stage IV malignant melanoma patients treated with Ipilimumab. Oncoimmunology 2016; 5:e1100788; PMID:27141381; http://dx.doi.org/ 10.1080/2162402X.2015.1100788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sade-Feldman M, Kanterman J, Klieger Y, Ish-Shalom E, Mizrahi O, Saragovi A, Shtainberg H, Lotem M, Baniyash M. Clinical significance of circulating CD33+CD11b+HLA-DR- myeloid cells in Stage-IV melanoma patients treated with ipilimumab. Clin Cancer Res 2016; PMID:27178742; http://dx.doi.org/ 10.1158/1078-0432.CCR-15-3104 [DOI] [PubMed] [Google Scholar]
- 33.Liu CY, Wang YM, Wang CL, Feng PH, Ko HW, Liu YH, Wu YC, Chu Y, Chung FT, Kuo CH et al.. Population alterations of L-arginase- and inducible nitric oxide synthase-expressed CD11b+/CD14(-)/CD15+/CD33+ myeloid-derived suppressor cells and CD8+ T lymphocytes in patients with advanced-stage non-small cell lung cancer. J Cancer Res Clin Oncol 2010; 136:35-45; PMID:19572148; http://dx.doi.org/ 10.1007/s00432-009-0634-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solito S, Falisi E, Diaz-Montero CM, Doni A, Pinton L, Rosato A, Francescato S, Basso G, Zanovello P, Onicescu G et al.. A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood 2011; 118:2254-65; PMID:21734236; http://dx.doi.org/ 10.1182/blood-2010-12-325753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ku GY, Yuan J, Page DB, Schroeder SE, Panageas KS, Carvajal RD, Chapman PB, Schwartz GK, Allison JP, Wolchok JD. Single-institution experience with ipilimumab in advanced melanoma patients in the compassionate use setting: lymphocyte count after 2 doses correlates with survival. Cancer 2010; 116:1767-75; PMID:20143434; http://dx.doi.org/ 10.1002/cncr.24951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber JS, Hamid O, Chasalow SD, Wu DY, Parker SM, Galbraith S, Gnjatic S, Berman D. Ipilimumab increases activated T cells and enhances humoral immunity in patients with advanced melanoma. J Immunother 2012; 35:89-97; PMID:22130166; http://dx.doi.org/ 10.1097/CJI.0b013e31823aa41c [DOI] [PubMed] [Google Scholar]
- 37.Chen H, Liakou CI, Kamat A, Pettaway C, Ward JF, Tang DN, Sun J, Jungbluth AA, Troncoso P, Logothetis C et al.. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFN-gamma levels in both nonmalignant and malignant prostate tissues. Proc Natl Acad Sci U S A 2009; 106:2729-34; PMID:19202079; http://dx.doi.org/ 10.1073/pnas.0813175106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO et al.. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother 2012; 61:1019-31; PMID:22146893; http://dx.doi.org/ 10.1007/s00262-011-1172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 2010; 236:219-42; PMID:20636820; http://dx.doi.org/ 10.1111/j.1600-065X.2010.00923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunt SK, Yang L, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Reduced inflammation in the tumor microenvironment delays the accumulation of myeloid-derived suppressor cells and limits tumor progression. Cancer Res 2007; 67:10019-26; PMID:17942936; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 2012; 12:253-68; PMID:22437938; http://dx.doi.org/ 10.1038/nri3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, Ugel S, Sonda N, Bicciato S, Falisi E et al.. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity 2010; 32:790-802; PMID:20605485; http://dx.doi.org/ 10.1016/j.immuni.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 43.Lechner MG, Liebertz DJ, Epstein AL. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol 2010; 185:2273-84; PMID:20644162; http://dx.doi.org/ 10.4049/jimmunol.1000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang H et al.. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest 2013; 123:1580-9; PMID:23454751; http://dx.doi.org/ 10.1172/JCI60083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinton L, Solito S, Damuzzo V, Francescato S, Pozzuoli A, Berizzi A, Mocellin S, Rossi CR, Bronte V, Mandruzzato S. Activated T cells sustain myeloid-derived suppressor cell-mediated immune suppression. Oncotarget 2016; 7:1168-84; PMID:26700461; http://dx.doi.org/ 10.18632/oncotarget.6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao X, Huang J, Zhong H, Shen N, Faggioni R, Fung M, Yao Y. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther 2014; 141:125-39; PMID:24076269; http://dx.doi.org/ 10.1016/j.pharmthera.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 47.Attia P, Phan GQ, Maker AV, Robinson MR, Quezado MM, Yang JC, Sherry RM, Topalian SL, Kammula US, Royal RE et al.. Autoimmunity correlates with tumor regression in patients with metastatic melanoma treated with anti-cytotoxic T-lymphocyte antigen-4. J Clin Oncol 2005; 23:6043-53; PMID:16087944; http://dx.doi.org/ 10.1200/JCO.2005.06.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lutzky JWJ, Hamid O, Lebbe C, Pehamberger H, Linette G, de Pril V, Ibrahim R, Hoos A. Association between immune-related adverse events (irAEs) and disease control or overall survival in patients (pts) with advanced melanoma treated with 10 mg/kg ipilimumab in three phase II clinical trials. Journal of Clinical Oncology 2009; 27:15s; PMID:1991785019917850 [Google Scholar]
- 49.McLane LM, Banerjee PP, Cosma GL, Makedonas G, Wherry EJ, Orange JS, Betts MR. Differential localization of T-bet and Eomes in CD8 T cell memory populations. J Immunol 2013; 190:3207-15; PMID:23455505; http://dx.doi.org/ 10.4049/jimmunol.1201556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buggert M, Tauriainen J, Yamamoto T, Frederiksen J, Ivarsson MA, Michaelsson J, Lund O, Hejdeman B, Jansson M, Sonnerborg A et al.. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog 2014; 10:e1004251; PMID:25032686; http://dx.doi.org/ 10.1371/journal.ppat.1004251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P et al.. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015; 373:23-34; PMID:26027431; http://dx.doi.org/ 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kyi C, Postow MA. Immune checkpoint inhibitor combinations in solid tumors: opportunities and challenges. Immunotherapy 2016; 8:821-37; PMID:27349981; http://dx.doi.org/ 10.2217/imt-2016-0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolchok JD, Hoos A, O'Day S, Weber JS, Hamid O, Lebbe C, Maio M, Binder M, Bohnsack O, Nichol G et al.. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res 2009; 15:7412-20; PMID:19934295; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-1624 [DOI] [PubMed] [Google Scholar]
- 54.Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay P, Roederer M. Quality assurance for polychromatic flow cytometry. Nat Protoc 2006; 1:1522-30; PMID:17406444; http://dx.doi.org/ 10.1038/nprot.2006.250 [DOI] [PubMed] [Google Scholar]
- 55.Myles Hollander DAW, Eric Chicken Nonparametric Statistical Methods, 3rd Edition John Wiley & Sons, Hoboken, New Jersey, USA, 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.