ABSTRACT
Human centromeres contain large amounts of repetitive DNA sequences known as α satellite DNA, which can be difficult to replicate and whose functional role is unclear. Recently, we have characterized protein composition, structural organization and checkpoint response to stalled replication forks of centromeric chromatin reconstituted in Xenopus laevis egg extract. We showed that centromeric DNA has high affinity for SMC2-4 subunits of condensins and for CENP-A, it is enriched for DNA repair factors and suppresses the ATR checkpoint to ensure its efficient replication. We also showed that centromeric chromatin forms condensins enriched and topologically constrained DNA loops, which likely contribute to the overall structure of the centromere. These findings have important implications on how chromosomes are organized and genome stability is maintained in mammalian cells.
KEYWORDS: ATR pathway, DNA damage, genomic-genetic instability, DNA replication, stress response
Chromosome segregation is an essential process that enables transmission of hereditary information during cell division. Key sites required for proper segregation of chromosomes are centromeres, which support kinetochore assembly and hold sister chromatids together during metaphase ensuring their proper alignment on mitotic spindles.1 Centromere integrity is required for optimal function of kinetochores. Correct function of the kinetochore and centromere apparatus prevents chromosome segregation errors that could lead to a wide range of diseases, including cancer and aneuploidy-related disorders such as Down syndrome.2
Centromeres of most eukaryotic chromosomes are characterized by: 1) the presence of centromeric chromatin containing specific proteins among which CENP-A and CENP-B; 2) duplicated tandem DNA elements, also known as α satellite DNA in human cells organized in high order repeats (HORs); 3) a pattern of post-translational histone modifications that contribute to regulate centromere transcription and assembly of centromere proteins.3
Repetitive sequences are generally unstable and prone to recombination.4 Repetitive DNA can also form hairpin-like secondary structures, which could induce replication fork stalling or lead to fully replicated but still intertwined DNA.5 In addition, the compact structure of centromeric chromatin can act as barrier to DNA replication machinery. The problematic progression of replication fork in centromeric regions could therefore induce DNA breakage under replication stress. Indeed, centromeres are known to harbor endogenous sites of replication fork pausing in yeast,6 and hotspots for chromosomal breakage and rearrangements in mammalian cells.7 This intrinsic fragility could contribute to centromere breakage-fusion cycles observed in many solid tumors.8,9 Interestingly, proteins involved in the maintenance of chromosomes stability such as the Werner Helicase (WRN), whose loss leads to premature aging and cellular senescence, bind centromeric and pericentromeric chromatin and promote stability of critical epigenetic marks such as histone H3K9 methylation.10
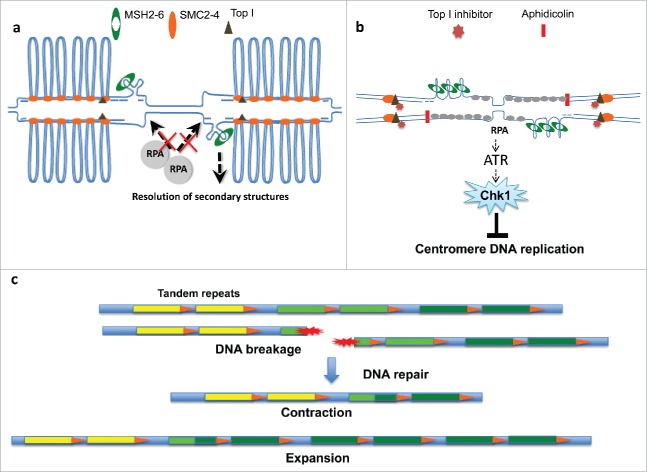
Recently, using the vertebrate model system Xenopus laevis, we revealed striking new features of centromeric DNA.11 We reconstituted the replication of repetitive human centromeric DNA using bacterial artificial chromosomes (BACs) bearing centromeric α satellite DNA sequences of different human chromosomes. Such BACs were able to induce efficient formation of nuclear structures in egg extract, a pre-requisite for semiconservative DNA replication.11 Centromeric DNA was efficiently replicated and replication efficiency was comparable to BACs containing non-repetitive sequences. The high replication efficiency of the centromeric DNA was surprising given the presence of repetitive DNA. Mass spectrometry analysis of the proteome associated with centromeric DNA revealed the enrichment of several DNA repair and DNA structural proteins among which MSH2-6, the MRE11-RAD50 complex, HMGB1-3, XRCC1, XRCC5/DNAPK, PARP1, ERCC6L/PICH helicase and MUS81 endonuclease.11 Mass spectrometry also revealed the enrichment of condensins components SMC2-4, which play a fundamental role in the structural and functional organization of chromosomes. Theses results were consistent with proteomic studies on mammalian centromeres.12 In contrast, other common replication players were under-represented, such as the single-stranded DNA binding (ssDNA) complex RPA and ATR activator TopBP1. Accumulation on DNA of these proteins following induction of stalled replication forks was also diminished. Consistent with these findings the ATR-dependent checkpoint leading to Chk1 phosphorylation, which requires both TopBP1 and RPA to be activated, was not induced by stalled forks arising on repetitive DNA. To understand the cause of this poor checkpoint response we performed structural analysis of replicating DNA using electron microscopy (EM).11 The rationale of this analysis was to detect potential hairpins formed by misaligned repetitive DNA sequences, which could hinder formation of open ssDNA regions upon fork stalling, preventing RPA and TopBP1 binding. We were expecting to find such secondary structures also after observing the enrichment of MSH2-6, which can bind DNA hairpins. However, we were unable to detect hairpins. Instead, we discovered the presence of numerous single stranded DNA bubbles, which were highly enriched on replicating centromeric DNA. Inhibition of DNA replication by geminin prevented their appearance, indicating that replication was required for their formation. We hypothesized that these bubbles were due to inability of psoralen to efficiently cross-link centromeric DNA. Psoralen is usually added to chromatin before isolation to cross-link doubled stranded DNA and freeze replication intermediates present at the moment of the cross-linking.13 This method prevents unwanted melting and branch migration of replication intermediates. Spreading of DNA on EM grids is instead performed in the presence of formamide, a mild denaturing agent required to prevent aggregation and uneven distribution of long DNA molecules.13 Although the psoralen cross-linking efficiency is usually fairly high on naked DNA, it decreases on DNA wrapped around nucleosomes. Decreased crosslinking efficiency can be easily observed after mild heat denaturation of DNA, which gives rise to wide appearance of ssDNA bubbles. However, centromeric DNA formed ssDNA bubbles, although less frequently than heat-denatured DNA, even in the absence of the heat denaturation. When we looked for the causes of this poor cross-linking we found that it was due to the presence of positively supercoiled DNA. Positively supercoiled DNA is known to strongly prevent psoralen cross-linking.14 Psoralen-biotin, which is poorly incorporated in positively supercoiled DNA, is used to map positively supercoiled genomic regions.15 When using psoralen-biotin, we found that psoralen was poorly incorporated in centromeric DNA, confirming that ssDNA bubbles were formed by poorly cross-linked DNA regions, which melted during DNA spreading. Intriguingly, positively supercoiled DNA has been documented on centromeric chromatin assembled in vitro with purified components in other species16 and our findings seem to confirm original observations extending the analysis to more physiological conditions. We then hypothesized that positively supercoiled DNA was responsible for inhibition of checkpoint activation. To test this hypothesis we inhibited topoisomerase activity and we showed that this treatment was able to restore ATR checkpoint activation during centromeric DNA replication halted by aphidicolin. These results confirmed that the topological arrangement of centromeric DNA was involved in the suppression of ATR activation.
To understand whether formation of positively supercoiled DNA was an active phenomenon we looked for proteins enriched on centromeric DNA with a possible structural role. Among these there were SMC2-4 subunits of the condensin complex.17 Condensin I has been shown to form positively supercoiled DNA in closed circular plasmids in the presence of Topoisomerase I and this activity has been linked to the ability of condensins to condense DNA.17 The structural conformation of condensed DNA is unclear at the moment. In order to visualize the structure of centromeric chromatin we sought to process partially digested chromatin for EM. Strikingly, this approach revealed the presence of large double stranded DNA loops embedded in a protein matrix11 (Fig. 1a). Complete protein digestion was able to dissolve such loops. Most interestingly, formation of loops was inhibited by replication inhibitor geminin. The link with the replisome is unclear at the moment. However, it is possible that the assembly of the replisome and DNA replication forks are required to load factors involved in this process. Importantly, inhibition of topoisomerases also prevented formation of the loops, indicating that topoisomerase activity, probably in conjunction with condensins, was required for loop formation (Fig. 1a). Loops were likely formed behind replication forks as DNA replication progressed on centromeric DNA, whereas non-centromeric genomic DNA did not form loops. Organization of centromeric chromatin in loops kept by condensins and cohesins has been predicted based on biophysical studies.18 Loops could be formed by a DNA extrusion mechanism with condensins playing an active role while standing at the base of the extruding loop.19 In this model the presence of DNA wrapped around nucleosomes might require the action of topoisomerases to facilitate DNA extrusion. Loops might confer resistance and flexibility to centromeric chromatin and might contribute to centromeric chromatin condensation, which could start from centromere and spread to other parts of the chromosome. Contextual loading of SMCs and loop formation might also help to disentangle chromosomes after replication and prevent recombination between them.20
Figure 1.
(A) General model showing organization of centromeric DNA in loops promoted and kept by condensins (SMC2-4) in the presence of Topoisomerase activity (Top I). Secondary DNA structures containing misaligned repeats could be repaired by MSH2-6. This overall arrangement prevents RPA loading onto chromatin and ATR activation, resulting in Chk1 activity suppression and more efficient repetitive DNA replication. (B) Disruption of centromeric DNA organization induced by inhibitors of TopI restores ATR activation following replication fork progression slow down induced by aphidicolin, which inhibits DNA polymerase activity. (C) When DNA breakage occurs due to replication or mechanical stress tandem repeats might facilitate repair through SSA and SDSA based recombination pathways (see text) leading to contraction or expansion of centromeric repeats.
In addition to these functions we speculated that the loop arrangement was linked to the suppression of the ATR checkpoint and was involved in the efficient replication of repetitive centromeric DNA. To test this hypothesis we used small amounts of aphidicolin to trigger low levels of ATR activity in conditions where such loops were disrupted by Topoisomerase I inhibition. This was sufficient to restore ATR activation and inhibit centromeric DNA replication, whereas DNA replication on non-repetitive DNA was unaffected (Fig. 1b). These observations might indicate that local suppression of ATR signaling facilitates replication of centromeric repetitive DNA, which would otherwise trigger continuous activation of ATR inhibiting replication origin firing. According to this hypothesis general activation of the ATR checkpoint should selectively compromises centromeric DNA replication in intact cells. This is currently under investigation in the lab. Significantly, similar to centromeric DNA other repetitive DNA regions such as the telomeres suppress checkpoint activation, form DNA loops and are positively supercoiled.21
It is intriguing that the α satellite DNA can trigger a series of complex events in vitro. However, this is not surprising as Xenopus egg extract contains all the factors required to support all the major events related to chromosome formation and metabolism. Furthermore, human centromeric DNA can be recognized in Xenopus egg extract. Although the overall human centromeric DNA sequence is different from Xenopus centromeric DNA, it contains a conserved CENP-B box, a 17 nt element which functions as binding site for CENP-B protein.22 CENP-B protein is required for the stable loading of CENP-A histone H3 variant on α satellite DNA.22
CENP-A is the major determinant of kinetochore formation and it usually co-localizes with centromeric DNA, although it can bind also areas of the genome lacking α satellite DNA, triggering there the formation of functional neocentromeres.22,23 We were able to show that CENP-A protein is selectively loaded onto human centromeric DNA when incubated in egg extract. The loading of CENP-A takes place during DNA replication and continues after replication has been completed. This is different from CENP-A loading onto chromosomes in intact cells, which takes place between the end of mitosis and subsequent G1 phase and requires the presence of CENP-A protein in the same location, which epigenetically marks the sites for loading of new CENP-A on replicated chromosome.22,23 However, our system was not designed to recapitulate in vivo loading of CENP-A as there is no pre-existing CENP-A on naked DNA introduced in egg extract. Nonetheless our experiments clearly showed a preferential affinity of CENP-A for α satellite DNA sequences. These observations do not exclude that CENP-A can be loaded onto other α satellite free sites such as in the case of neocentromeres when primary centromeric DNA is deleted.24
It is possible that CENP-A stable binding requires structural constraints induced by condensins on α satellite DNA. Condensins have indeed been shown to promote CENP-A chromatin loading in Xenopus egg extract.25 Condensins accumulate on centromeric and pericentromeric chromatin in yeast and condensin subunits have been directly linked to tandem repeats.26 The role of CENP-B in CENP-A localization Xenopus is instead unclear at the moment. Proteomic analysis identified a Xenopus laevis protein with limited similarity to human CENP-B. However, further studies are needed to validate this as true CENP-B ortholog.
The ability of satellite DNA to induce selective loading of centromeric proteins was in line with extensive work performed on human artificial chromosomes (HACs), which are built with human satellite DNA and are able to trigger de novo assembly of a functional centromere capable of nucleating a kinetochore when introduced into the cell.27-29 It is worth mentioning that despite differences in checkpoint activation replication origin assembly and inter-origin distance on centromeric DNA was similar to non-centromeric regions in the Xenopus system and that these results were consistent with data obtained from studies on HAC replication in intact cells.30 Therefore, the combination of in vitro centromere assembly in Xenopus together with HACs might provide unique tools to understand the fine mechanisms underlying centromeric chromatin metabolism.
A surprising aspect of the proteomic analysis of centromeric chromatin was the elevated presence of a number of DNA repair factors. Some of these factors might contribute to replication fidelity of these regions in the absence of the checkpoint. Indeed, while ssDNA binding proteins RPA and TopBP1 did not accumulate onto centromeric chromatin following induction of replication fork arrest MSH2-6 heterodimer showed increased chromatin recruitment. MSH6 was also required for efficient replication of centromeric DNA11 although it is unclear whether this applies also to intact cells. It is therefore possible that MSH6 is recruited onto misaligned repetitive sequences containing mismatches to promote their repair. However, how this results in more efficient replication of repetitive DNA is unclear. It is also possible that MSH2-6 heterodimer is recruited to promote other types of recombination based DNA repair between misaligned repeats.
Among centromere enriched DNA repair factors we indeed found DNA-PK, PARP1, MRE11 and MUS81,11 which are also required for double strand break (DSB) processing and repair. The presence of these proteins might indicate the occurrence of ongoing DSB repair. We did not directly test the hypothesis, as this was difficult to assess with available assays due to the repetitive nature of the DNA, the size of the DNA and the lack of a clear DNA damage response activation. Therefore, the function of DSB repair factors on centromeric chromatin remains to be clarified. However, it is tempting to speculate that these factors have increased affinity for DNA regions being under constant mechanical stress due to topological constraints and pulling forces exerted during chromosomes segregation, which might cause centromere DNA breakage. Intriguingly, DSB preferentially accumulate at centromeres in cancer cell lines following replication stress induced by aphidicolin.31 It is also worth mentioning that CENP-A has high affinity also for DSBs.22
Induction of replication stress by replication fork stalling agents has been associated to the formation of ultrafine DNA bridges (UFBs) between segregating chromosomes. These structures are commonly seen at telomeres and fragile sites. Replication fork stalling does not seem to induce UFB at centromeres, which are instead formed when DNA decatenation is prevented with Topoisomerase II inhibitors.32 The absence of UFBs at centromeres when fork stall might be due to the existence of alternative and more efficient process to bypass fork stalling such breakage and rejoining of the broken DNA or looping out and excision of the structures produced by the pulling of the unreplicated DNA regions during chromosome segregation. These repair events might be facilitated by the presence of tandem repeats. Compatibly with this hypothesis many extra-chromosomal circles of centromeric DNA accumulate during cell cycle after replication stress in several species.33,34 Such extra-chromosomal circles might be the results of centromeric DNA loop excision or circularization of linear centromeric DNA fragments formed during DNA repair events.
Centromere DNA breakage has been proposed to be an early event in cellular transformation leading to breakage-fusion cycles generating further chromosome instability.8,9 Centromere breakage-fusion cycles are indeed frequent in tumors harboring whole chromosome arm translocations.8,9 Although it is unclear how such breakages might be generated, it is important to note that tandem repeats could favor repair of such DSBs by single strand annealing (SSA) and synthesis dependent strand annealing (SDSA), types of recombination pathways occurring at repetitive sequences.35,36 Such processes would require most of the repair proteins enriched on centromeric repeats.
Evolutionary analysis of centromeric DNA repeats shows cycles of contraction and expansion associated to independent evolution of new tandem repeats. These events might be compatible with multiple SSA and SDSA events in which DNA molecules can undergo expansion and contraction of the intervening repeats35,36 (Fig 1c). SDSA could also explain how homogenous centromeric repeats are generated and maintained on different chromosomes.37
Tandem repeats could therefore ensure efficient repair, conferring special status to the centromeric region of the chromosomes. Interestingly, formation of neocentromeres triggered by CENP-A binding frequently occurs in genomic areas enriched for duplicated sequences.24 Pre-existing duplications might be advantageous for the formation of neocentromeres due to the ability to promote efficient repair of DNA breaks. It is also interesting to note that α- satellite sequences are subsequently incorporated in evolutionary young and satellite-free neocentromeres.24 Alpha satellite DNA transposition onto neocentromeres could confer an adaptive advantage possibly by increasing the accuracy of chromosome segregation at these sites. As such these sequences would be then selected and fixed in the population thanks to meiotic drive.24 Interestingly, satellite sequences do not gradually integrate throughout the neocentromere, but rather expand from a single location, compatibly with a SDSA mediated DNA repeats expansion.24 On the other hand loss of satellite DNA repeats at centromeres that become inactive in favor of neocentromere formed on the same chromosome24 might be due to contraction events following DSB repair.
There is still a lot to learn about the genomic structure of centromeric DNA. Answering the many unknowns about satellite DNA might help to understand why these DNA sequences are retained despite their problematic replication. Unfortunately, most of genomic regions harboring α satellite DNA are still poorly annotated due to their repetitive nature, which hampers any attempt to align them correctly. We are currently investigating the possibility that cycles of contraction and expansion occur at repetitive centromeric DNA during unperturbed and challenged DNA replication. Given our results we favor the possibility that repetitive satellite DNA might be advantageous for a number of reasons such as its ability to increase the loading of CENP-A at centromeres, to promote chromatin arrangement favorable for sister chromatid cohesion and to increase resistance to microtubule tension ensuring the stability of the chromosome during mitosis and meiosis. Suppression of ATR activity and enrichment of DNA repair factors might contribute to maintain and stabilize these regions during DNA replication. Loss of faithful DNA repair and chronic activation of ATR might predispose to genomic rearrangements in which centromeric breakage-fusion cycles might constitute an early event on the way to cellular transformation. Understanding how these processes occur at molecular level will therefore be essential to clarify the origin of genome instability predisposing to cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The work described here was funded by the Associazione Italiana per la Ricerca sul Cancro (AIRC), a European Research Council (ERC) consolidator grant (614541), a Giovanni Armenise-Harvard foundation award, the EPIGEN Progetto Bandiera (4.7), AICR-Worldwide Cancer Research (13-0026) and a Fondazione Telethon (GGP13-071) grant awarded to V.C. A. B. is funded by AIRC grant IG 14578.
Reference
- [1].Westhorpe FGS, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol 2013; 25:334-40; PMID:23490282; http://dx.doi.org/ 10.1016/j.ceb.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011; 333:1895-8; PMID:21960636; http://dx.doi.org/ 10.1126/science.1210214 [DOI] [PubMed] [Google Scholar]
- [3].Biscotti MA, Olmo E, Heslop-Harrison JS. Repetitive DNA in eukaryotic genomes. Chromosome Res 2015; 23:415-20; PMID:26514350; http://dx.doi.org/ 10.1007/s10577-015-9499-z [DOI] [PubMed] [Google Scholar]
- [4].Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 2010; 11:208-19; PMID:20177396; http://dx.doi.org/ 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- [5].McFarlane RJ, Humphrey TC. A role for recombination in centromere function. Trends Genet 2010; 26:209-13; PMID:20382440; http://dx.doi.org/ 10.1016/j.tig.2010.02.005 [DOI] [PubMed] [Google Scholar]
- [6].Greenfeder SA, Newlon CS. Replication forks pause at yeast centromeres. Mol Cell Biol 1992; 12:4056-66; PMID:1508202; http://dx.doi.org/ 10.1128/MCB.12.9.4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Simi S, Simili M, Bonatti S, Campagna M, Abbondandolo A. Fragile sites at the centromere of Chinese hamster chromosomes: A possible mechanism of chromosome loss. Mutat Res 1998; 397:239-46; PMID:9541649; http://dx.doi.org/ 10.1016/S0027-5107(97)00219-4 [DOI] [PubMed] [Google Scholar]
- [8].Forsburg SL. The CINs of the centromere. Biochem Soc Trans 2013; 41:1706-11; PMID:24256279; http://dx.doi.org/ 10.1042/BST20130146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Martinez AC, van Wely KH. Centromere fission, not telomere erosion, triggers chromosomal instability in human carcinomas. Carcinogenesis 2011; 32:796-803; PMID:21478459; http://dx.doi.org/ 10.1093/carcin/bgr069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, et al.. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 2015; 348:1160-3; PMID:25931448; http://dx.doi.org/ 10.1126/science.aaa1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat Cell Biol 2016; 18:684-91; PMID:27111843; http://dx.doi.org/ 10.1038/ncb3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla ME, et al.. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 2014; 56:580-94; PMID:25457167; http://dx.doi.org/ 10.1016/j.molcel.2014.10.001 [DOI] [PubMed] [Google Scholar]
- [13].Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17:1305-11; PMID:20935632; http://dx.doi.org/ 10.1038/nsmb.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bermudez I, Garcia-Martinez J, Perez-Ortin JE, Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res 2010; 38:e182; PMID:20685815; http://dx.doi.org/ 10.1093/nar/gkq687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol 2013; 20:387-95; PMID:23416946; http://dx.doi.org/ 10.1038/nsmb.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell 2009; 138:104-13; PMID:19596238; http://dx.doi.org/ 10.1016/j.cell.2009.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 2012; 26:1659-78; PMID:22855829; http://dx.doi.org/ 10.1101/gad.194746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bloom KS. Centromeric heterochromatin: the primordial segregation machine. Ann Rev Genet 2014; 48:457-84; PMID:25251850; http://dx.doi.org/ 10.1146/annurev-genet-120213-092033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al.. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A 2015; 112:E6456-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang X, Le TB, Lajoie BR, Dekker J, Laub MT, Rudner DZ. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev 2015; 29:1661-75; PMID:26253537; http://dx.doi.org/ 10.1101/gad.265876.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, Bauwens S, Djerbi N, Latrick CM, Fraisier V, Pei B, Gay A, et al.. TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Mol Cell 2016; 61:274-86; PMID:26774283; http://dx.doi.org/ 10.1016/j.molcel.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev Cell 2015; 33:314-27; PMID:25942623; http://dx.doi.org/ 10.1016/j.devcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 2016; 17:16-29; PMID:26601620; http://dx.doi.org/ 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 2008; 82:261-82; PMID:18252209; http://dx.doi.org/ 10.1016/j.ajhg.2007.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, et al.. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol 2011; 192:569-82; PMID:21321101; http://dx.doi.org/ 10.1083/jcb.201005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].He H, Zhang S, Wang D, Hochwagen A, Li F. Condensin Promotes Position Effects within Tandem DNA Repeats via the RITS Complex. Cell Rep 2016; 14:1018-24; PMID:26832414; http://dx.doi.org/ 10.1016/j.celrep.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kouprina N, Earnshaw WC, Masumoto H, Larionov V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cell Mol Life Sci 2013; 70:1135-48; PMID:22907415; http://dx.doi.org/ 10.1007/s00018-012-1113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 2008; 14:507-22; PMID:18410728; http://dx.doi.org/ 10.1016/j.devcel.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 2011; 30:328-40; PMID:21157429; http://dx.doi.org/ 10.1038/emboj.2010.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Erliandri I, Fu H, Nakano M, Kim JH, Miga KH, Liskovykh M, Earnshaw WC, Masumoto H, Kouprina N, Aladjem MI, et al.. Replication of α-satellite DNA arrays in endogenous human centromeric regions and in human artificial chromosome. Nucleic Acids Res 2014; 42:11502-16; PMID:25228468; http://dx.doi.org/ 10.1093/nar/gku835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al.. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 2013; 10:361-5; PMID:23503052; http://dx.doi.org/ 10.1038/nmeth.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu Y, Nielsen CF, Yao Q, Hickson ID. The origins and processing of ultra fine anaphase DNA bridges. Curr Opin Genet Dev 2014; 26:1-5; PMID:24795279; http://dx.doi.org/ 10.1016/j.gde.2014.03.003 [DOI] [PubMed] [Google Scholar]
- [33].Cohen S, Segal D. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenetic Genome Res 2009; 124:327-38; PMID:19556784; http://dx.doi.org/ 10.1159/000218136 [DOI] [PubMed] [Google Scholar]
- [34].Cohen S, Yacobi K, Segal D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res 2003; 13:1133-45; PMID:12799349; http://dx.doi.org/ 10.1101/gr.907603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Paques F, Leung WY, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol 1998; 18:2045-54; PMID:9528777; http://dx.doi.org/ 10.1128/MCB.18.4.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 1999; 63:349-404; PMID:10357855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Plohl M, Luchetti A, Mestrovic N, Mantovani B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 2008; 409:72-82; PMID:18182173; http://dx.doi.org/ 10.1016/j.gene.2007.11.013 [DOI] [PubMed] [Google Scholar]