ABSTRACT
Optimal expansion protocols for adoptive human T-cell therapy often include interleukin (IL)-15; however, the mechanism by which IL-15 improves the in vivo antitumor effect of T cells remains to be elucidated. Using human T cells generated from HLA-A2+ donors against novel T-cell epitopes derived from the human U266 myeloma cell line Ig light chain V-region (idiotype) as a model, we found that T cells cultured with IL-15 provided superior resistance to tumor growth in vivo, compared with IL-2, after adoptive transfer into immunodeficient hosts. This effect of IL-15 was associated with delayed/reversed senescence in tumor antigen-specific memory CD8+ T cells mediated through downregulation of P21WAF1, P16INK4a, and P53 expression. Compared to IL-2, IL-15 stimulation dramatically activated JAK3-STAT5 signaling and inhibited the expression of DNA damage genes. Thus, our study elucidates a new mechanism for IL-15 in the regulation of STAT signaling pathways and CD8+ T-cell senescence.
Keywords: Idiotype, IL-15, immunotherapy, myeloma, senescence, T cells
Introduction
In 2016, it is predicted that a total of 1,685,210 new cancer cases and 595,690 cancer deaths will occur in the United States.1 One promising strategy to improve the survival of cancer patients is adoptive transfer with tumor antigen-specific T cells. In a variety of clinical trials, with both solid and hematologic cancers, adoptive T-cell transfer has emerged as one of the most effective immunotherapies.2 Early clinical studies have demonstrated a 50–70% clinical response in patients.3-5 However, the optimal protocols for expansion of T cells, especially antigen-specific CD8+ T cells, remain to be determined.
Cytokines have substantial effects on T-cell phenotype and function.6 For example, IL-2 is widely used for T-cell growth because it can actively drive the expansion of T cells and the contraction phase of immune response.7,8 IL-7 and IL-15 are required for the initiation of immune response and the survival of T cells.9-11 IL-21 can promote the development of both Th17 and Tfh T cells that play roles in antitumour and antiviral responses.12 Detailed studies revealed these cytokines have a distinguished effect on different T-cells subsets. For example, IL-2 is required for the in vitro growth of CD4+ T cells, but is not required for normal clonal expansion of antigen-specific CD8+ T cells.13 In vivo studies revealed IL-2 induces the apoptosis of effector memory CD4+ T cells and IL-15 can enhance the in vivo function of CD8+ T cells.14-16 Interestingly, it is known that most cytokines like IL-2, IL-15, IL-21, and IL-7 can activate the JAK-STATs signaling pathway; however, it is not yet clear how these cytokines exert their individual functions through one common signaling pathway.
In this study, we used T cells generated against the Ig light chain V-region epitopes (Idiotype, Id) of the human myeloma U266 cell line as a model to test the effect of cytokines on the generation of T-cells for adoptive therapy. We found that IL-15-expanded, Id-specific T cells mediate long-term antitumor effects in vivo, which were associated with delayed/reversed memory CD8+ T-cell senescence. The effect of IL-15 in memory CD8+ T-cell senescence delay is through the downregulation of P21WAF1, P16INK4a, and P53. Specifically, we found that IL-15 strongly activated the JAK3-STAT5 signaling pathway and inhibited the expression of DNA damage genes. Our study provides a new mechanism for IL-15 regulation in the CD8+ T-cells senescence process.
Results
In vivo antitumor effects of adoptively transferred Id L-chain-specific T cells expanded by IL-2 or IL-15
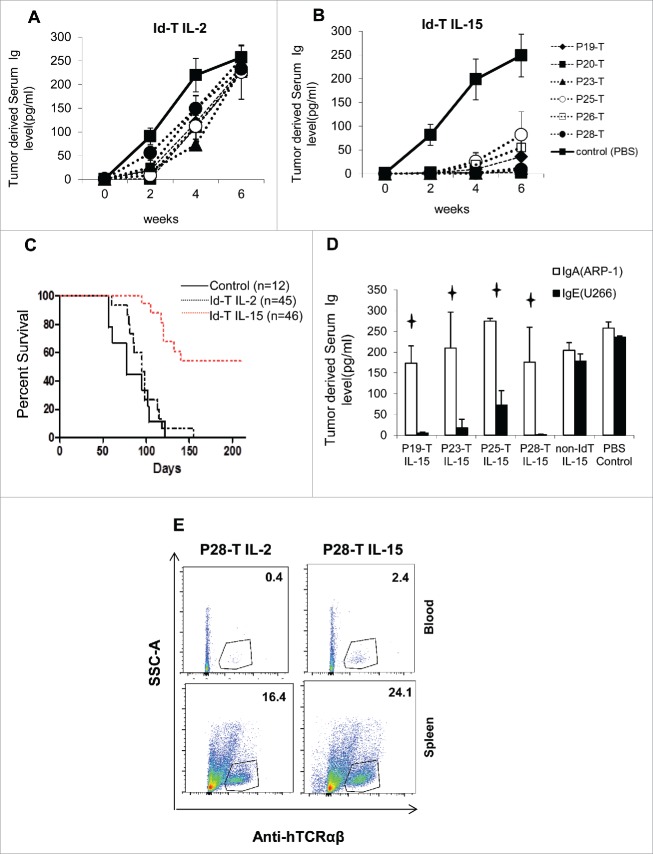
We have previously reported the identification of novel immunogenic CD8+ T-cell epitopes in the V-region of the Ig light chain (L-chain, Idiotype antigen) of the U266 human myeloma cell line and primary human lymphomas.17,18 In order to test the in vivo function of these L-chain-specific T cells, we stimulated HLA A2+ normal donors' T cells as previously reported,19 and purified Id L-chain, peptide-specific CD8+ T cells and expanded them with IL-2 (180 IU/mL) or IL-15 (50 ng/mL) using the rapid expansion protocol (REP).20,21 After 14 d, we subsequently transferred the same number of T cells (1 × 107) into the immune-deficient mice, bearing 3 d U266 xenografts.21 Tumor growth was monitored by U266-specific IgE protein secretion in mouse serum.22,23 While IL-2-expanded L-chain-specific CD8+ T cells can lyse the tumor cells very well in vitro,17 these T cells only temporarily inhibited tumor cell growth in vivo (Fig. 1A). By contrast, mice receiving IL-15-expanded, L-chain-specific CD8+ T cells demonstrated significantly lower IgE serum concentrations, compared with IL-2-expanded T cells (Fig. 1B), and about 53% of mice remained alive at the end of observation (Fig. 1C). The inhibition was tumor-specific, as the Id L-chain-specific T cells expanded by IL-15 did not inhibit IgA-secreting ARP-1 myeloma xenografts and the non-U266-idiotype-specific T-cells expanded by IL-15 did not inhibit U266 tumor growth in vivo (Fig. 1D). To determine whether the antitumor effect of IL-15-expanded T cells is associated with increased proliferation and persistence of Id L-chain-specific CD8+ T cells, we adoptively transferred 1 × 107 L-chain-specific T cells into tumor-free mice and collected the blood and spleens on day 7. We found that significantly more IL-15-expanded, L-chain-specific CD8+ T cells were detectable in both the blood and spleens of mice, compared with IL-2-expanded L-chain-specific CD8+ T cells, suggesting that IL-15-expanded CD8+ T cells have superior proliferation and persistence in vivo (Fig. 1E).
Figure 1.
Specific in vivo tumor inhibition by adoptively transferred Ig L-chain, V-region (Idiotype, Id)-peptide-specific T cells against U266 xenografts. (A) IL-2-expanded, or (B) IL-15-expanded, L-chain peptide-specific (P19, 20, 23, 25, 26, 28) T cells (1 × 107) were transferred to SCID γc chain knockout (NSG) mice bearing day 3 U266 (105) xenografts. U266-derived IgE was monitored as a serum marker of tumor growth by ELISA. (C) Kaplan–Meier survival curves of 103 experimental mice-bearing U266 xenografts treated with either IL-2- or IL-15-expanded, L-chain-specific T cells. (D) Inhibition of tumor growth by IL-15-expanded, L-chain peptide-specific (P19, 23, 25, 28) T cells (1 × 107) against day-3 U266 (IgE secreting) or ARP-1(IgA secreting) (105) xenografts, which were injected simultaneously into the same mice. (E) Flow cytometry detection of Id L-chain-specific CD8+ T cells (P28, hCD3+) in the blood and spleens of non-tumor bearing NSG mice that had received 1 × 107 L-chain peptide-specific (P28) T cells 7 d earlier. Panels A, B, and D shown are indicated as mean ± SD of 5–7 mice per group. p < 0.05.
IL-15-expanded, Id L-chain-specific T cells exhibited delayed cellular senescence
Senescence is a special cell cycle mechanism that living cells become unresponsive to growth stimulation, permanently withdraw from cell cycle and exist with a pattern of specific gene signatures and phenotypes.24,25 To investigate if the IL-15-expanded T cells have delayed senescence process compared to IL-2-expanded T cells, we performed cell cycle analysis of day 14 IL-2 or IL-15-expanded, L-chain-specific T cells after anti-CD3 antibody (OKT3) stimulation for 72 h, before adoptive transfer. We found that IL-15-expanded, CD8+ central memory (CD8+ Tcm: CD62L+, CD45RA−, p < 0.01) and CD8+ effector memory (CD8+ Tem: CD62L−, CD45RA−, p < 0.01) L-chain-specific T cells have a significantly higher percentage of cells in S/G2 phase compared with IL-2-expanded T cells after stimulation (Fig. 2A). We also analyzed the expression of cell cycle inhibitors P21WAF1, P16INK4a, and P53 in the day 14, L-chain-specific T cells, before the adoptive transfer. We found the expression of P21WAF1, P16INK4a, and P53 was significantly lower in IL-15-expanded T cells compared to IL-2-expanded T cells (Fig. 2B). Recent studies found that senescence immune cells can secret a large amount of the senescence-associated proinflammatory cytokines,26 we performed intracellular cytokines assays and observed that IL-15-expanded day 14 L-chain specific CD8+ Tcm and CD8+ Tem cells expressed lower amounts of IL-8, TNFα, IFNγ, and TGF-β1 after PMA and ionomycin stimulation, compared with IL-2-expanded T cells (Fig. 2C). We also performed cell surface staining of day 14 T cells just before adoptive transfer, which showed IL-15-expanded L-chain-specific CD8+ T cells have significantly higher expression of CD27 and CD28 compared with IL-2-expanded T cells (p < 0.05) (Fig. 2D). Finally, we extracted RNA from IL-2 or IL-15-expanded, L-chain specific T cells before adoptive transfer and reverse transcribed the RNA into cDNA. We performed real-time PCR microarrays with senescence signaling pathway gene-specific primers. We found that the expression of 85% (71 out of 84) cellular senescence biomarker genes was significantly decreased in IL-15-expanded T cells (Fig. 2E). These genes include the following: 53BP1 (TP53BP1), ATM, BMI1, CDK6, ETS1, CDKN1A, CDKN1B, CDKN1C, CDKN2A, CDKN2B, E2F1,MDM2, RB1, RBL2, MDC1, and TWIST1, which have been reported to play important roles in the regulation of the initiation and progression of cellular senescence and cell cycle inhibition (Table 1 and Fig. S1).27-29 Taken together, we found IL-15-expanded, L-chain-specific T cells have a higher percentage of S/G2 phase cells after stimulation, lower expression of cell cycle inhibitors, less production of senescence-associated proinflammatory cytokines, higher expression of CD27 and CD28, and downregulation of cellular senescence biomarker genes, suggesting that IL-15-expanded, L-chain-specific T cells exhibit senescence delay.24,26,30,31
Figure 2.
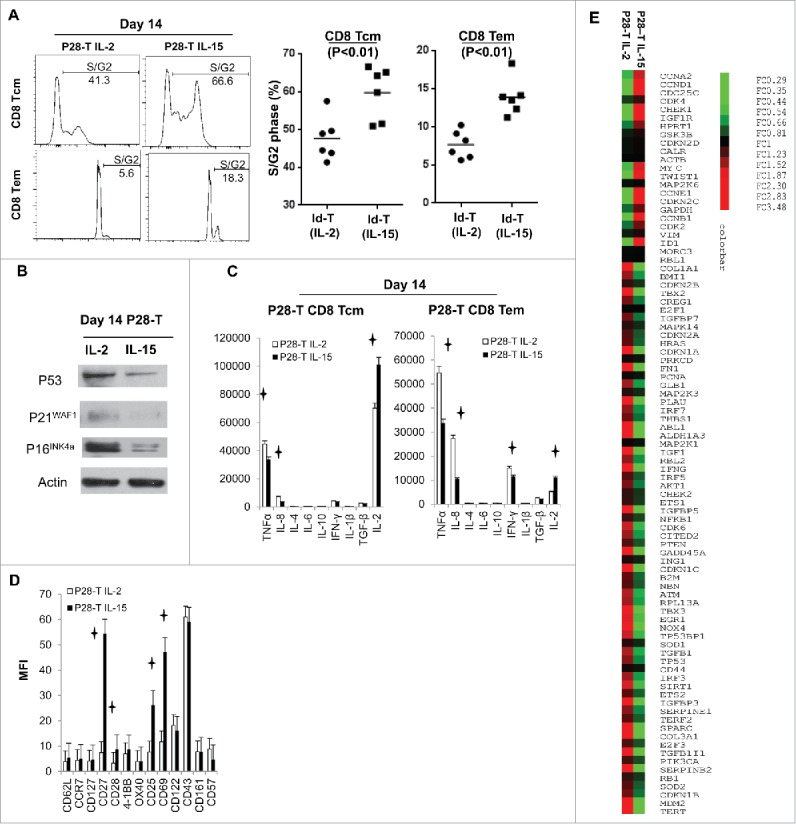
IL-15-expanded, Id L-chain-specific human Tcells exhibit delayed cellular senescence. (A) Cell cycle analysis of IL-2- or IL-15-expanded idiotype-specific memory CD8+ T cells after stimulation with OKT3 (1 µg/mL, plate-bound) for 72 h. CD8+ Tcm (CD8+, CD62L+, CD45RA−); CD8+ Tem (CD8+, CD62L−, CD45RA−). (B) Western blot analysis for P53, P21WAF1, and P16INK4a expression of idiotype-specific memory CD8+ T cells expanded by IL-2 or IL-15, before transfer into mice. (C) Intracellular cytokine staining of IL-2- or IL-15-expanded idiotype-specific (P28) memory CD8+ T cells after 5 h of stimulation with PMA (50 ng/mL) and ionomycin (250 ng/mL) in the presence of 10 ug/mL Brefeldin A. (D) Flow cytometry analysis of cell surface markers of IL-2- or IL-15-expanded, L-chain peptide-specific (P28) day 14 T cells, before transfer into mice. (E) Heat map showing the expression of 84 cellular senescence biomarkers by real-time RT-PCR array assays in IL-15- or IL-2-expanded Id L-chain-specific (P28) T cells, before transfer into mice (List of genes is shown in Table 1). MFI: Mean fluorescence intensity. p < 0.05. Tcm = central memory T cells. Tem = effector memory T cells.
Table 1.
Relative expression levels of cellular senescence biomarker genes in IL-2/IL-15-expanded Id L-chain-specific T cells.
| Gene table |
Relative expression (unit) |
|||||
|---|---|---|---|---|---|---|
| Position | Unigene | GeneBank | Symbol | Description | P28 (IL-2) | P28 (IL-15) |
| A01 | Hs.431048 | NM_005157 | ABL1 | C-abl oncogene 1, non-receptor tyrosine kinase | 0.009894 | 0.001658 |
| A02 | Hs.525622 | NM_005163 | AKT1 | V-akt murine thymoma viral oncogene homolog 1 | 0.054965 | 0.028693 |
| A03 | Hs.459538 | NM_000693 | ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | 0.006871 | 0.000016 |
| A04 | Hs.367437 | NM_000051 | ATM | Ataxia telangiectasia mutated | 0.044658 | 0.017633 |
| A05 | Hs.380403 | NM_005180 | BMI1 | BMI1 polycomb ring finger oncogene | 0.141755 | 0.065053 |
| A06 | Hs.515162 | NM_004343 | CALR | Calreticulin | 0.253652 | 0.275238 |
| A07 | Hs.58974 | NM_001237 | CCNA2 | Cyclin A2 | 0.014048 | 0.042295 |
| A08 | Hs.23960 | NM_031966 | CCNB1 | Cyclin B1 | 0.008914 | 0.03789 |
| A09 | Hs.523852 | NM_053056 | CCND1 | Cyclin D1 | 0.006871 | 0.000206 |
| A10 | Hs.244723 | NM_001238 | CCNE1 | Cyclin E1 | 0.006871 | 0.008538 |
| A11 | Hs.502328 | NM_000610 | CD44 | CD44 molecule (Indian blood group) | 0.159753 | 0.147131 |
| A12 | Hs.656 | NM_001790 | CDC25C | Cell division cycle 25 homolog C (S. pombe) | 0.006871 | 0.001533 |
| B01 | Hs.19192 | NM_001798 | CDK2 | Cyclin-dependent kinase 2 | 0.018076 | 0.034652 |
| B02 | Hs.95577 | NM_000075 | CDK4 | Cyclin-dependent kinase 4 | 0.039731 | 0.056178 |
| B03 | Hs.119882 | NM_001259 | CDK6 | Cyclin-dependent kinase 6 | 0.191699 | 0.066696 |
| B04 | Hs.370771 | NM_000389 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 0.405019 | 0.075319 |
| B05 | Hs.238990 | NM_004064 | CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 0.092236 | 0.050659 |
| B06 | Hs.106070 | NM_000076 | CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 0.006871 | 0.000016 |
| B07 | Hs.512599 | NM_000077 | CDKN2A | Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 0.017 | 0.011094 |
| B08 | Hs.72901 | NM_004936 | CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 0.035951 | 0.028407 |
| B09 | Hs.728783 | NM_078626 | CDKN2C | Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 0.006871 | 0.017095 |
| B10 | Hs.435051 | NM_001800 | CDKN2D | Cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 0.009557 | 0.01036 |
| B11 | Hs.24529 | NM_001274 | CHEK1 | CHK1 checkpoint homolog (S. pombe) | 0.006871 | 0.021533 |
| B12 | Hs.291363 | NM_007194 | CHEK2 | CHK2 checkpoint homolog (S. pombe) | 0.006871 | 0.003549 |
| C01 | Hs.82071 | NM_006079 | CITED2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 0.049903 | 0.02201 |
| C02 | Hs.172928 | NM_000088 | COL1A1 | Collagen, type I, α 1 | 0.006871 | 0.000016 |
| C03 | Hs.443625 | NM_000090 | COL3A1 | Collagen, type III, α 1 | 0.006871 | 0.000016 |
| C04 | Hs.5710 | NM_003851 | CREG1 | Cellular repressor of E1A-stimulated genes 1 | 0.019568 | 0.010543 |
| C05 | Hs.654393 | NM_005225 | E2F1 | E2F transcription factor 1 | 0.006871 | 0.006707 |
| C06 | Hs.269408 | NM_001949 | E2F3 | E2F transcription factor 3 | 0.006871 | 0.002488 |
| C07 | Hs.326035 | NM_001964 | EGR1 | Early growth response 1 | 0.006871 | 0.001447 |
| C08 | Hs.369438 | NM_005238 | ETS1 | V-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 0.406507 | 0.307562 |
| C09 | Hs.644231 | NM_005239 | ETS2 | V-Ets erythroblastosis virus E26 oncogene homolog 2 (avian) | 0.010578 | 0.006555 |
| C10 | Hs.203717 | NM_002026 | FN1 | Fibronectin 1 | 0.007966 | 0.000016 |
| C11 | Hs.80409 | NM_001924 | GADD45A | Growth arrest and DNA-damage-inducible, α | 0.019251 | 0.004137 |
| C12 | Hs.443031 | NM_000404 | GLB1 | Galactosidase, β 1 | 0.039809 | 0.017389 |
| D01 | Hs.445733 | NM_002093 | GSK3B | Glycogen synthase kinase 3 β | 0.020953 | 0.024772 |
| D02 | Hs.37003 | NM_005343 | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | 0.030779 | 0.018645 |
| D03 | Hs.504609 | NM_002165 | ID1 | Inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | 0.006871 | 0.00338 |
| D04 | Hs.856 | NM_000619 | IFNG | Interferon, gamma | 0.069941 | 0.012135 |
| D05 | Hs.160562 | NM_000618 | IGF1 | Insulin-like growth factor 1 (somatomedin C) | 0.007121 | 0.00072 |
| D06 | Hs.643120 | NM_000875 | IGF1R | Insulin-like growth factor 1 receptor | 0.006871 | 0.003122 |
| D07 | Hs.450230 | NM_000598 | IGFBP3 | Insulin-like growth factor binding protein 3 | 0.008874 | 0.002167 |
| D08 | Hs.607212 | NM_000599 | IGFBP5 | Insulin-like growth factor binding protein 5 | 0.049616 | 0.000016 |
| D09 | Hs.479808 | NM_001553 | IGFBP7 | Insulin-like growth factor binding protein 7 | 0.009151 | 0.00502 |
| D10 | Hs.46700 | NM_005537 | ING1 | Inhibitor of growth family, member 1 | 0.008724 | 0.007556 |
| D11 | Hs.75254 | NM_001571 | IRF3 | Interferon regulatory factor 3 | 0.094596 | 0.041639 |
| D12 | Hs.521181 | NM_001098629 | IRF5 | Interferon regulatory factor 5 | 0.006871 | 0.003375 |
| E01 | Hs.166120 | NM_001572 | IRF7 | Interferon regulatory factor 7 | 0.019498 | 0.009162 |
| E02 | Hs.145442 | NM_002755 | MAP2K1 | Mitogen-activated protein kinase kinase 1 | 0.137061 | 0.132296 |
| E03 | Hs.514012 | NM_002756 | MAP2K3 | Mitogen-activated protein kinase kinase 3 | 0.019382 | 0.016122 |
| E04 | Hs.463978 | NM_002758 | MAP2K6 | Mitogen-activated protein kinase kinase 6 | 0.009738 | 0.010533 |
| E05 | Hs.485233 | NM_001315 | MAPK14 | Mitogen-activated protein kinase 14 | 0.039407 | 0.032542 |
| E06 | Hs.484551 | NM_002392 | MDM2 | Mdm2 p53 binding protein homolog (mouse) | 0.56428 | 0.137308 |
| E07 | Hs.421150 | NM_015358 | MORC3 | MORC family CW-type zinc finger 3 | 0.061078 | 0.064593 |
| E08 | Hs.202453 | NM_002467 | MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | 0.006871 | 0.00261 |
| E09 | Hs.492208 | NM_002485 | NBN | Nibrin | 0.016481 | 0.010458 |
| E10 | Hs.654408 | NM_003998 | NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | 0.141479 | 0.101153 |
| E11 | Hs.371036 | NM_016931 | NOX4 | NADPH oxidase 4 | 0.006871 | 0.000016 |
| E12 | Hs.728886 | NM_182649 | PCNA | Proliferating cell nuclear antigen | 0.195372 | 0.184122 |
| F01 | Hs.553498 | NM_006218 | PIK3CA | Phosphoinositide-3-kinase, catalytic, α polypeptide | 0.073685 | 0.050435 |
| F02 | Hs.77274 | NM_002658 | PLAU | Plasminogen activator, urokinase | 0.006871 | 0.000414 |
| F03 | Hs.155342 | NM_006254 | PRKCD | Protein kinase C, delta | 0.015161 | 0.014037 |
| F04 | Hs.500466 | NM_000314 | PTEN | Phosphatase and tensin homolog | 0.16335 | 0.10826 |
| F05 | Hs.408528 | NM_000321 | RB1 | Retinoblastoma 1 | 0.121739 | 0.09276 |
| F06 | Hs.207745 | NM_002895 | RBL1 | Retinoblastoma-like 1 (p107) | 0.036618 | 0.038316 |
| F07 | Hs.513609 | NM_005611 | RBL2 | Retinoblastoma-like 2 (p130) | 0.473698 | 0.222004 |
| F08 | Hs.594481 | NM_002575 | SERPINB2 | Serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 0.006871 | 0.000016 |
| F09 | Hs.414795 | NM_000602 | SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 0.006871 | 0.000167 |
| F10 | Hs.369779 | NM_012238 | SIRT1 | Sirtuin 1 | 0.066963 | 0.021391 |
| F11 | Hs.443914 | NM_000454 | SOD1 | Superoxide dismutase 1, soluble | 0.207349 | 0.174491 |
| F12 | Hs.487046 | NM_000636 | SOD2 | Superoxide dismutase 2, mitochondrial | 0.055267 | 0.034729 |
| G01 | Hs.111779 | NM_003118 | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 0.006871 | 0.000024 |
| G02 | Hs.531085 | NM_005994 | TBX2 | T-box 2 | 0.006871 | 0.000016 |
| G03 | Hs.714737 | NM_016569 | TBX3 | T-box 3 | 0.006871 | 0.000016 |
| G04 | Hs.63335 | NM_005652 | TERF2 | Telomeric repeat binding factor 2 | 0.053443 | 0.033128 |
| G05 | Hs.492203 | NM_198253 | TERT | Telomerase reverse transcriptase | 0.006871 | 0.000016 |
| G06 | Hs.645227 | NM_000660 | TGFB1 | Transforming growth factor, β 1 | 0.312842 | 0.126748 |
| G07 | Hs.513530 | NM_015927 | TGFB1I1 | Transforming growth factor β 1 induced transcript 1 | 0.006871 | 0.000016 |
| G08 | Hs.164226 | NM_003246 | THBS1 | Thrombospondin 1 | 0.006871 | 0.001149 |
| G09 | Hs.654481 | NM_000546 | TP53 | Tumor protein p53 | 0.105234 | 0.051741 |
| G10 | Hs.440968 | NM_005657 | TP53BP1 | Tumor protein p53 binding protein 1 | 0.064137 | 0.022034 |
| G11 | Hs.66744 | NM_000474 | TWIST1 | Twist homolog 1 (Drosophila) | 0.006871 | 0.002333 |
Data is representative of three independent experiments with three Id-specific T-cell lines.
IL-15 regulates senescence delay in antigen-specific T cells through the JAK3-STAT5 signaling pathway
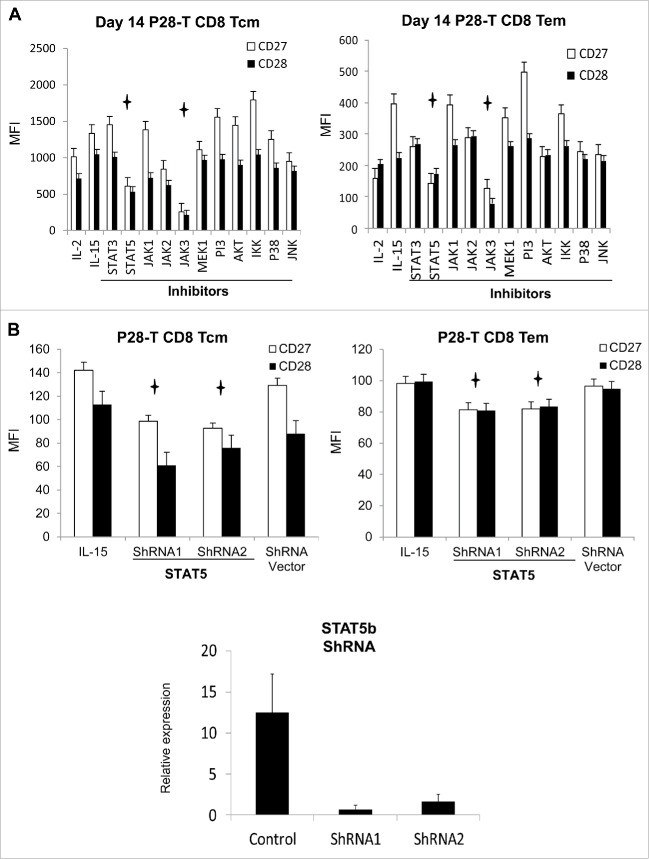
To determine the molecular mechanism underlining IL-15 regulation of senescence delay, we expanded the Id L-chain-specific T cells with IL-15 (50 ng/mL) by REP, and added the candidate signaling pathway inhibitors on day 12. On day 14, we analyzed the expression of CD27 and CD28 in these expanded T cells and observed that JAK3 and STAT5 inhibitors significantly downregulated CD27 and CD28 expression (Fig. 3A). The JAK1 and JAK2 inhibitors also partially downregulated the expression of the CD27 and CD28 of L-chain-specific CD8+ Tcm, but not CD8+ Tem cells. The signaling pathway inhibitors MEK1/2, PI3, AKT, IKK, P38, and JNK did not have a significant effect on CD27 or CD28 expression in L-chain-specific T cells. Next, the effect of STAT5 in the regulation of senescence delay was confirmed by ShRNA knockdown. We found that knockdown of STAT5b in IL-15-expanded L-chain-specific CD8+ T cells resulted in significant downregulation of CD27 and CD28 (Fig. 3B). These data indicate that IL-15 regulates the senescence delay of antigen-specific T cells through the JAK3-STAT5 signaling pathway.
Figure 3.
IL-15 regulates senescence delay through the JAK3-STAT5 signaling pathway in Id-specific Tcells. (A) Id L-chain-specific T cells were expanded with IL-15 (50 ng/mL) by rapid expansion protocol (REP) for 12 d before the addition of the signaling pathway inhibitors shown. The effect of signaling pathway inhibitors on CD27 and CD28 expression in Id L-chain-specific (P28) memory CD8+ T cells were analyzed by flow cytometry on day 14. (Detailed information on signaling pathway inhibitors is listed in Table S1.) (B) IL-15-expanded Id L-chain-specific (P28) T cells on day 14 were activated by plate-bound anti-CD3 antibody for 72 h and transfected with one of two (ShRNA1 or 2) STAT5b ShRNA-containing a lentivirus or vector alone for 12 h. 48 h later, the expression of STAT5, CD27, and CD28 was analyzed by real-time PCR or flow cytometry. MFI: Mean fluorescence intensity. p < 0.05.
IL-15 strongly activates STAT5 and inhibits the expression of DNA damage genes in human CD8+ T-cells
In order to see how IL-15 activates the STATs signaling pathway, we treated the antigen-specific (Id, L-chain) CD8+ T-cell line with IL-15 or IL-2 at different concentrations for multiple time points, and analyzed the cell extracts for pSTAT5 activity through Western blotting. We observed that IL-15 treatment led to a dramatic increase of pSTAT5 signaling, compared with IL-2 treatment, in idiotype-specific CD8+ T-cell populations at all conditions, indicating that IL-15 treatment strongly activates pSTAT5 signaling in CD8+ T cells (Fig. 4A). By contrast, we found that treatment with IL-15 or IL-2 has little effect on the activation of pSTAT3 in the CD8+ T-cell population (Fig. 4A and Fig. S2A). As previous studies reported,32-34 we also found that IL-15 treatment activated pAKT signaling and resulted in higher perforin expression in CD8+ T cells (Fig. S2B and D). IL-2 treatment led to higher Phospho-S6 Ribosomal Protein expression and low pAKT activation in CD8+ T cells (Fig. S2C).
Figure 4.
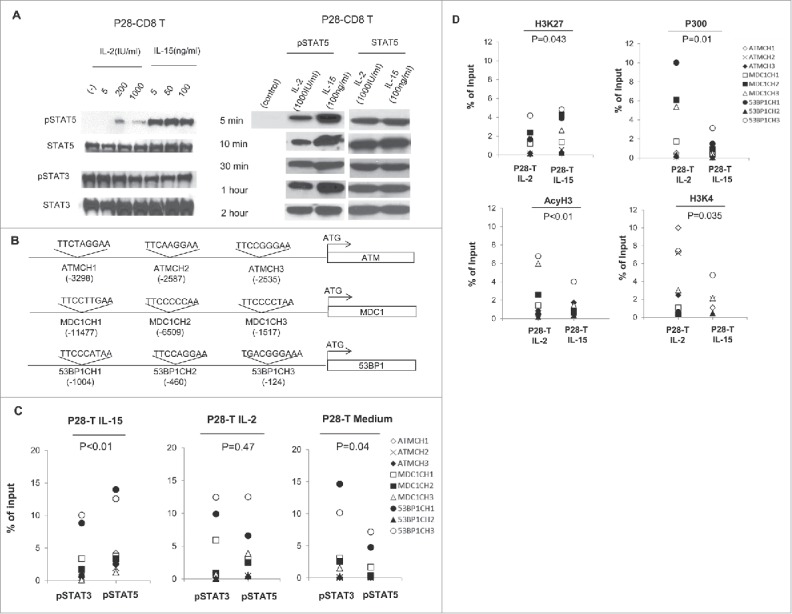
IL-15 strongly activates STAT5 signaling and changes the ratio of pSTAT5/3 signaling in CD8+ T cells. (A) Id-specific T cells starved of cytokines for 24 h were treated with IL-2 or IL-15 at different concentrations for 15 min. Total protein was extracted from the cytokine-treated T cells and equal amounts of protein were loaded into each lane. Anti-pSTAT3, anti-pSTAT5, anti-total STAT3, and anti-total STAT5 antibodies were used in Western blotting. (B) Schema for the nine potential STAT binding sites in the promoter regions of ATM, MDC1, and 53BP1. The prediction was carried out with TFSEARCH online program, and the potential STAT binding sequences and relative locations are indicated. (C) ChIP-PCR analysis of pSTAT5 and pSTAT3 binding to the STAT sites on the promoters of ATM, MDC1, and 53BP1 genes in cytokine-stimulated, day 14 IL-2- or IL-15-expanded Id L-chain-specific (P28) T cells, or unstimulated idiptype-specific CD8+ T cells. Shown are pooled data for nine STAT binding sites on the promoters of DNA damage genes. Isotype-matched antibodies were used as negative controls for all experiments (data not shown). (D) ChIP-PCR analysis of histones binding to the STAT sites on the promoters of ATM, MDC1, 53BP1 genes in day 14, IL-2- or IL-15-expanded idiotype L-chain specific T cells. H3K27: tri-methy-H3 (Lys27); P300: Histone acetyltransferase p300; H3K4: Histone tri methyl lysine 4; AcyH3: acetyl- Histone H3. MFI: Mean fluorescence intensity. p < 0.05.
In our previous data, we found 85% of senescence biomarker genes are downregulated in IL-15-expanded T cells. To confirm that the expression of these genes was regulated by IL-15, we used an online TFSEARCH program (http://www.cbrc.jp/research/db/TFSEARCH.html) and identified nine STAT consensus binding sites on the promoters of ATM, 53BP1, and MDC1 genes located between position -11477 and -124 (Fig. 4B). Through ChIP-PCR assay, we observed that there was significantly more pSTAT5 than pSTAT3 binding to the nine STAT sites in IL-15-expanded T cells (Fig. 4C, p <0.01, Paired t-test). The binding ratio of pSTAT5/pSTAT3 to the sites is not significant in IL-2-expanded T cells (p = 0.38). In unstimulated idiotype-specific CD8+ T cells, there is significant more pSTAT3 than pSTAT5 binding to the nine STATs sites (p = 0.04). Moreover, we found significantly more binding of transcriptionally repressive histones [H3K27: tri-methy-H3 (Lys27), p = 0.043] and less binding of transcriptionally active histones (P300: Histone acetyltransferase p300, p = 0.01; H3K4: Histone tri methyl lysine 4, p = 0.035; AcyH3: acetyl-Histone H3, p < 0.01) to these nine STAT sites in IL-15, compared to IL-2-expanded T cells, (Fig. 4D). Altogether, these data indicate that IL-15 can strongly activate the STAT5 signaling pathway, which inhibited the expression DNA damage genes in CD8+ T cells.
Discussion
Adoptive T cell transfer has emerged as an effective immunotherapy for both solid and hematologic cancers in a variety of clinical trials.4,5, 35 Recent studies of adoptive transfer with autologous T cells generated from patients have focused on generation of genetically modified memory CD8+ T cells with chimeric antigen receptors or T-cell receptors with a particular focus on improving the proliferation and persistence of T cells after transfer.36-40 Traditionally, IL-2 has been a central component of T-cell expansion protocols.41-43 However, IL-2-expanded T cells have significant limitations in adoptive therapy, including susceptibility to T-cell activation-induced cell death (AICD), Treg proliferation, and T-cell differentiation.44,45 Hence, there is an urgent need to find new cytokines for the growth of T cells. In this study, we found IL-15-expanded T cells mediate superior protection against tumor cells in vivo and mechanism of IL-15 is through the senescence delay/reversal of human CD8+ T cells. Specifically, we found IL-15 can strongly activate STAT5 signaling, which changed the ratio of pSTAT5/3 signaling in the CD8+ T cells and decreased the expression of DNA damage molecules. Although CD4+ T-cell senescence delay/reversal have been reported before,46,47 our results are the first to demonstrate senescence delay/reversal in CD8+ T cells.
Cellular senescence is a specific cell cycle status in which the cells permanently withdraw from the cell cycle.24 Replicative senescence (telomere-dependent) usually occurs in T cells with shorter telomere length as a process of aging isolated in elderly people.48-51 Premature senescence (telomere-independent), on the other hand, has many causes, such as DNA damage, oxygen stress, chromatin perturbation, and oncogene perturbation.52-55 Extended in vitro culturing can cause senescence.56 Human T-cell senescence has been suggested as an important reason for escape from tumor surveillance.45 Unlike phenotypic biomarkers for memory T cells, there is no defined biomarker for senescent cells and the most consistent feature of senescent cells is their resistance to enter the S/G2 cell cycle stage after proliferative stimulation.24,52, 57 Other phenotypic changes associated with senescent cells include the following: increased β-galaxidase activity58, increased expression of cell cycle inhibitors and DNA damage molecules59, increased expression of senescence-associated pro-inflammatory cytokines54,55,60, and decreased expression of CD27, CD28 biomarkers on the cell surface.31,46 Senescent human CD8+ T cells have poor proliferation capacity, defective killing abilities, and defective granule exocytosis.61,62 Thus, strategies to delay/reverse the senescence of tumor antigen-specific CD8+ T cells may improve the effectiveness of adoptive T-cell therapy. In this study, we found that IL-15-expanded idiotype L-chain-specific CD8+ T cells have decreased P53, P21WAF1, and P16INK4a expression. They also have a higher percentage of cells in the S/G2 phase after proliferative stimulation; decreased senescence-associated pro-inflammatory cytokine expression (IL-8 and TNFα); decreased senescence biomarkers expression; and higher CD27 and CD28 expression, compared to IL-2-cultured T cells. All of these changes indicate that IL-15-expanded antigen-specific memory CD8+ T cells have delayed/reverse senescence.
The Signal Transducer and Activator of Transcription (STAT) family of proteins consist of seven members that play important roles in immune system regulation.63,64 STAT proteins are highly homologous in several domains, including SH2, DNA-binding, and transactivation and they can mediate their function through the mechanism of homodimers or heterodimers.64 Recent studies found that cross-regulation among the STAT family members has an important role in the maintenance of cytokine signaling specificity.63 For example, IL-6 stimulation can form three distinct dimers: STAT1–STAT1, STAT1–STAT3, and STAT3–STAT3, which can play dramatically different functions in the cells.65 The binding ratio of different STAT members to the same STAT sites can affect gene expression and cell differentiation dramatically.66-68 In our study, we found IL-15 stimulation dramatically activated STAT5 signaling and induced more pSTAT5 binding to the nine STAT sites on the promoter of ATM, 53BP1, MDC1 DNA damage genes. As a consequence of this binding, there are less pSTAT3, more transcriptionally repressive histones (H3K27) and less transcriptionally active histones (H3K4, P300, Acy 300) binding to the promoters. DNA damage molecules are known to play critical roles in the initiation and regulation of senescence and their high expression is a biomarker for cellular senescence.60,69 The downregulation of these genes in IL-15-expanded T cells confirmed the senescence process was delayed.
In summary, we found that IL-15 can delay the senescence process in memory CD8+ T cells through the strong activation of STAT5 and the changes of pSTAT5/3 signaling in CD8+ T cells. Our results are consistent with recent studies where constitutively activated STAT5 signaling mediated strong antitumor effects and the inhibition of STAT3 led to an enhanced adoptive therapy effect.70-73 The mechanisms revealed in this study provide the basis for future rational design of strategies to improve persistence of CD8+ T-cell therapy in the clinical setting.
Materials and methods
Cell lines, antibodies, and reagents
Human myeloma cell lines U266 and ARP-1 were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 10 μg/mL gentamicin at 37°C and 5% CO2. Flow antibodies for T-cell surface biomarkers and cytokine antibodies were all from BD Biosciences or eBiosciences. The following reagents were used per manufactures' instructions: anti-P53 and anti-P21WAF1 (Genescript), anti-tubulin and anti-P16INK4a (BD biosciences), ChIP grade anti-Histone H3 (tri methyl K4) (Abcam), anti-p300 (Millipore), anti-Histone 3 tri-methy-H3 (Lys27) (Millipore), anti-Histone 3 acetylated (AcyH3) (Millipore), anti-pSTAT3 (Santa Cruz), and anti-pSTAT5 (Santa Cruz), anti-hTCRαβ-PE (eBiosciences), anti-Perforin-PE (eBiosciences), anti-Phospho-S6-FITC (cell signaling), anti-EMOES-APC (eBiosciences).
Expansion of U266 myeloma Id-specific T cells
Peptide-specific T cells (P20-T, P23-T, P25-T, P26-T, P28-T) were generated from HLA-A2+ normal donors as previously reported.17 Briefly, PBMCs (1 × 105 cells/well) were incubated with 10 μg/mL Id-specific peptide (P20, P23, P25, P26, P28) in quadruplicate in 96-well U-bottom microculture plates in 200 μL of culture medium (50% AIM-V, 50% RPMI-1640, 10% human AB serum, 100 IU/mL of IL-2) and restimulated with peptide every 3 d. After five stimulations, T cells were cultured with peptide-pulsed T2 cells and interferon (IFN)-γ production was determined from the supernatants by ELISA. The IFNγ-producing T cells were purified by an IFNγ-secreting Cell Enrichment and Detection Kit and further expanded in the presence of 30 × 106 allogeneic feeder cells and 30 ng/mL anti-CD3 antibody in a T25 flask with AIM-V media including 10% human AB serum. Cytokines (IL-2 180 IU/mL or IL-15 50 ng/mL) were added the next day. The culture medium was changed with same cytokine conditions on day 5 and every 3 d subsequently for 14–18 days, as described in the REP.20,21
Adoptive T-cell therapy
Six-to-twelve-week-old NOD SCID IL-2 receptor γc chain knockout mice (Jackson Laboratory, Stock# 005557), were injected by IV with 0.2 × 106 U266 or ARP-1 human myeloma cells on day 0. Mice were irradiated (200 Cy) on day 2 and received 1 × 107 Id-specific T cells on day 3, followed by rhIL-2 at 10,000 IU with IP injection twice daily for a total of six doses. Tumor growth was monitored by an ELISA assay of tumor-specific serum-secreted Ig protein (IgE for U266 and IgA for ARP-1, Bethyl laboratories) and the survival time of the mice was recorded.
Cell cycle assay
Id-specific T cells (1 × 106) expanded with IL-2 or IL-15 for 14 d were put in a complete T-cell medium in a 24-well plate which was coated with 1 µg/mL of OKT3 antibody. Seventy-two hours later, the T cells were stained with anti-human CD8+, CD62L, and CD45RA for 30 min, washed in 1XPBS, and fixed with 70% ethanol for overnight. The next day, 5 µg/mL Propidium iodide (PI) was added for 15 min at 37°C to stain the cells. After washing, the T cells were analyzed by cytometry. The fluorescence intensity of the stained cells was used to determine the G0/G1 and S/G2 phase of T cells.
Intracellular staining of pro-inflammatory cytokines
2 × 106 idiotype-specific T cells expanded with IL-2 or IL-15 for 14 d were washed and stimulated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA) and 250 ng/mL ionomycin; after 2 h, 5 µg/mL brefeldin A was added. Five hours later the cells were stained with anti-human CD8+, CD62L, and CD45RA for 30 min, washed in 1XPBS, fixed and permeabilized (BD Cytofix/Cytoperm Plus kit). Following this procedure the cells were stained with cytokine-specific antibodies and analyzed by flow cytometry.
Western blotting
Approximately 20 μg of total cell protein was extracted from Id-specific T cells and a standard Western blot assay protocol was followed.19
Signaling pathway inhibition assay
Idiotype-specific T cells expanded with 50 ng/mL IL-15 and allogeneic feeder cells for 12 d were cultured with signaling pathway inhibitors in the presence of 50 ng/mL IL-15 in complete T cell medium. The concentrations of signaling pathway inhibitors used are listed in Table S1. The expression of CD27 and CD28 were analyzed on day 14 by flow cytometer.
STAT5 ShRNA knockdown
IL-15-expanded, day 14 idiotype-specific T cells were activated with a plate-coated in OKT3 antibody for 72 h, washed, and transfected with lentivirus containing STAT5b-ShRNA (ShRNA 1: NM_012448/TRCN0000232137. ShRNA2: NM_012448/TRCN0000232140, sigma) in the presence of 8 ug/mL of polybrene for 12 h. The cells were then washed with 1 X PBS, and incubated in cytokine-free T-cell complete medium for another 48 h. The expression of STAT5b was analyzed by real-time RT-PCR normalized with GAPDH expression and the surface expression of CD27 and CD28 were analyzed by flow cytometry.
Real-time PCR array assay
3 µg of RNA was extracted from IL-2 or IL-15-expanded idiotype-specific T cells and reverse transcribed into cDNA with the Superscript III kit (Invitrogen). The expression of 84 cellular senescence genes and MDC1 gen was analyzed with primers pre-located inside the real-time PCR array (Qiagen, Cat# PAHS-050ZC), using the Applied Biosystems StepOne™/Real-Time PCR System. The real-time PCR conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 1 min. The results were analyzed by Qiagen on-line software and the list of genes are in Table 1.
ChIP-qPCR assays
IL-2 or IL-15-expanded idiotype-specific T cells were cross-linked and lysed with the ChIP assay kit (Cat# 26156, Thermo scientific). The digested chromatin was then immune-precipitated with 2 µg of anti-human pSTAT3, pSTAT5, Histone 3 tri-methy-H3 (Lys27), Histone H3 (tri methyl K4), Histone 3 acetylated, or p300 antibodies. The recovered DNA was purified through a column and amplified by real-time PCR at the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min. The ChIP primers used are listed in Table S2. The percentage of input was calculated as: % Input = 100 × 2^ (Average Ct – Adjusted Input Ct). In all assays, only living cells were analyzed and the dead cells were removed with a dead cell removal kit (Cat# 130-090-101) from Mitenyi Biotec. Isotype-matched antibodies were used as negative control for all experiments (data not shown).
Statistical analysis
The Student t-test was used to compare various experimental groups; p values <0.05 were considered statistically significant. Unless otherwise indicated, means and standard deviations (SD) are shown.
Study approval
Animal studies were approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Frances E. Dressman for her kind assistance in editing the manuscript and we thank Stephanie S Watowich and Haiyan Li for their kind suggestions.
Funding
This study was conducted with support from the Leukemia & Lymphoma Society Specialized Center of Research Grant #7262-08 (LWK), the Multiple Myeloma SPORE Grant P50CA142509, the Brian D. Novis Research Grant from the International Myeloma Foundation (JW), the Lady Leukemia League Research Grant (JW), and the National Natural Science Foundation of China Grant No. 81570189 (JW), and Guangzhou Department of Science and Information Technology, People's Republic of China (No 2014Y2-00092).
Author Contributions
J. W. and L.W. K. designed experiments. J.W., K.M., F.C., S.K, Z.J., X.X., and B. F. performed experiments. H.J., J.Q., L.Z., J.Y., S.N., and Q.Y. provided critical reagents or suggestions. J. W. and L.W.K analyzed data and wrote the paper.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinicians 2016; 66(1):7-30; PMID:26742998; http://dx.doi.org/ 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.June CH. Adoptive T cell therapy for cancer in the clinic. J Clin Investig 2007; 117(6):1466-76; PMID:17549249; http://dx.doi.org/ 10.1172/JCI32446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.June CH. Principles of adoptive T cell cancer therapy. J Clin Investig 2007; 117(5):1204-12; PMID:17476350; http://dx.doi.org/ 10.1172/JCI31446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer 2008; 8(4):299-308; PMID:18354418; http://dx.doi.org/ 10.1038/nrc2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley ME, Rosenberg SA. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nat Rev Cancer 2003; 3(9):666-75; PMID:12951585; http://dx.doi.org/ 10.1038/nrc1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schluns KS, Lefrancois L. Cytokine control of memory T-cell development and survival. Nat Rev Immunol 2003; 3(4):269-79; PMID:12669018; http://dx.doi.org/ 10.1038/nri1052 [DOI] [PubMed] [Google Scholar]
- 7.Badovinac VP, Tvinnereim AR, Harty JT. Regulation of antigen-specific CD8+ T cell homeostasis by perforin and interferon-γ. Science 2000; 290(5495):1354-7; PMID:11082062; http://dx.doi.org/ 10.1126/science.290.5495.1354 [DOI] [PubMed] [Google Scholar]
- 8.Refaeli Y, Van Parijs L, London CA, Tschopp J, Abbas AK. Biochemical mechanisms of IL-2–regulated fas-mediated T cell apoptosis. Immunity 1998; 8(5):615-23; PMID:9620682; http://dx.doi.org/ 10.1016/S1074-7613(00)80566-X [DOI] [PubMed] [Google Scholar]
- 9.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 1998; 9(5):669-76; PMID:9846488; http://dx.doi.org/ 10.1016/S1074-7613(00)80664-0 [DOI] [PubMed] [Google Scholar]
- 10.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol 2001; 167(12):6869-76; PMID:11739504; http://dx.doi.org/ 10.4049/jimmunol.167.12.6869 [DOI] [PubMed] [Google Scholar]
- 11.Tan JT, Dudl E, LeRoy E, Murray R, Sprent J, Weinberg KI, Surh CD. IL-7 is critical for homeostatic proliferation and survival of naïve T cells. Proc Natl Acad Sci 2001; 98(15):8732-7; PMID:11447288; http://dx.doi.org/24751819 10.1073/pnas.161126098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spolski R, Leonard WJ. Interleukin-21: a double-edged sword with therapeutic potential. Nat Rev Drug Discov 2014; 13(5):379-95; PMID:24751819; http://dx.doi.org/ 10.1038/nrd4296 [DOI] [PubMed] [Google Scholar]
- 13.Cousens LP, Orange JS, Biron CA. Endogenous IL-2 contributes to T cell expansion and IFN-gamma production during lymphocytic choriomeningitis virus infection. J Immunol 1995; 155(12):5690-9; PMID:7499855 [PubMed] [Google Scholar]
- 14.Khoruts A, Mondino A, Pape KA, Reiner SL, Jenkins MK. A natural immunological adjuvant enhances T cell clonal expansion through a CD28-dependent, interleukin (IL)-2-independent mechanism. J Exp Med 1998; 187(2):225-36; PMID:9432980; http://dx.doi.org/ 10.1084/jem.187.2.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li XC, Demirci G, Ferrari-Lacraz S, Groves C, Coyle A, Malek TR, Strom TB. IL-15 and IL-2: a matter of life and death for T cells in vivo. Nat Med 2001; 7(1):114-8; PMID:11135625; http://dx.doi.org/ 10.1038/83253 [DOI] [PubMed] [Google Scholar]
- 16.Klebanoff CA, Finkelstein SE, Surman DR, Lichtman MK, Gattinoni L, Theoret MR, Grewal N, Spiess PJ, Antony PA, Palmer DC et al.. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc Natl Acad Sci U S A 2004; 101(7):1969-74; PMID:14762166; http://dx.doi.org/ 10.1073/pnas.0307298101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weng J, Cha SC, Matsueda S, Alatrash G, Popescu MS, Yi Q, Molldrem JJ, Wang M, Neelapu SS, Kwak LW. Targeting human B-cell malignancies through Ig light chain-specific cytotoxic T lymphocytes. Clin Cancer Res 2011; 17(18):5945-52; PMID:21813633; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-0970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng J, Neelapu SS, Woo AF, Kwak LW. Identification of human idiotype-specific T cells in lymphoma and myeloma. Curr Top Microbiol Immunol 2011; 344:193-210; PMID:20549471; http://dx.doi.org/ 10.1007/82_2010_70 [DOI] [PubMed] [Google Scholar]
- 19.Weng J, Rawal S, Chu F, Park HJ, Sharma R, Delgado DA, Fayad L, Fanale M, Romaguera J, Luong A et al.. TCL1: a shared tumor-associated antigen for immunotherapy against B-cell lymphomas. Blood 2012; 120(8):1613-23; PMID:22645177; http://dx.doi.org/ 10.1182/blood-2011-09-382838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Topalian SL, Muul LM, Solomon D, Rosenberg SA. Expansion of human tumor infiltrating lymphocytes for use in immunotherapy trials. J Immunol Methods 1987; 102(1):127-41; PMID:3305708; http://dx.doi.org/ 10.1016/S0022-1759(87)80018-2 [DOI] [PubMed] [Google Scholar]
- 21.Riddell SR, Greenberg PD. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J Immunol Methods 1990; 128(2):189-201; PMID:1691237; http://dx.doi.org/ 10.1016/0022-1759(90)90210-M [DOI] [PubMed] [Google Scholar]
- 22.Miyakawa Y, Ohnishi Y, Tomisawa M, Monnai M, Kohmura K, Ueyama Y, Ito M, Ikeda Y, Kizaki M, Nakamura M. Establishment of a new model of human multiple myeloma using NOD/SCID/gammac(null) (NOG) mice. Biochem Biophys Res Commun 2004; 313(2):258-62; PMID:14684154; http://dx.doi.org/ 10.1016/j.bbrc.2003.11.120 [DOI] [PubMed] [Google Scholar]
- 23.Mirandola L, Yu Y, Chui K, Jenkins MR, Cobos E, John CM, Chiriva-Internati M. Galectin-3C inhibits tumor growth and increases the anticancer activity of bortezomib in a murine model of human multiple myeloma. PLoS One 2011; 6(7):e21811; PMID:21765917; http://dx.doi.org/ 10.1371/journal.pone.0021811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passos JF, Simillion C, Hallinan J, Wipat A, von Zglinicki T. Cellular senescence: unravelling complexity. Age 2009; 31(4):353-63; PMID:19618294; http://dx.doi.org/ 10.1007/s11357-009-9108-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campisi J. senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 2005; 120(4):513-22; PMID:15734683; http://dx.doi.org/ 10.1016/j.cell.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 26.Ye J, Huang X, Hsueh EC, Zhang Q, Ma C, Zhang Y, Varvares MA, Hoft DF, Peng G. Human regulatory T cells induce T-lymphocyte senescence. Blood 2012; 120(10):2021-31; PMID:22723548; http://dx.doi.org/ 10.1182/blood-2012-03-416040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol 2005; 37(5):961-76; PMID:15743671; http://dx.doi.org/ 10.1016/j.biocel.2004.10.013 [DOI] [PubMed] [Google Scholar]
- 28.Zhang H. Molecular signaling and genetic pathways of senescence: Its role in tumorigenesis and aging. J Cell Physiol 2007; 210(3):567-74; PMID:17133363; http://dx.doi.org/ 10.1002/jcp.20919 [DOI] [PubMed] [Google Scholar]
- 29.Fridman AL, Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene 2008; 27(46):5975-87; PMID:18711403; http://dx.doi.org/ 10.1038/onc.2008.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akbar AN, Henson SM. Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nat Rev Immunol 11(4):289-95; PMID:21436838; http://dx.doi.org/ 10.1038/nri2959 [DOI] [PubMed] [Google Scholar]
- 31.Montes CL, Chapoval AI, Nelson J, Orhue V, Zhang X, Schulze DH, Strome SE, Gastman BR. Tumor-induced senescent T cells with suppressor function: A potential form of tumor immune evasion. Cancer Research 2008; 68(3):870-9; PMID:18245489; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2282 [DOI] [PubMed] [Google Scholar]
- 32.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 2010; 32(1):79-90; PMID:20096607; http://dx.doi.org/ 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malamut G, El Machhour R, Montcuquet N, Martin-Lannerée S, Dusanter-Fourt I, Verkarre V, Mention JJ, Rahmi G, Kiyono H, Butz EA et al.. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease–associated inflammation and lymphomagenesis. J Clin Investig 120(6):2131-43; PMID:20440074; http://dx.doi.org/ 10.1172/JCI41344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornish GH, Sinclair LV, Cantrell DA. Differential regulation of T-cell growth by IL-2 and IL-15. Blood 2006; 108(2):600-8; PMID:16569767; http://dx.doi.org/ 10.1182/blood-2005-12-4827 [DOI] [PubMed] [Google Scholar]
- 35.Gattinoni L, Powell DJ, Rosenberg SA, Restifo NP. Adoptive immunotherapy for cancer: building on success. Nat Rev Immunol 2006; 6(5):383-93; PMID:16622476; http://dx.doi.org/ 10.1038/nri1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Besser MJ, Shapira-Frommer R, Treves AJ, Zippel D, Itzhaki O, Hershkovitz L, Levy D, Kubi A, Hovav E, Chermoshniuk N et al.. Clinical responses in a phase II study using adoptive transfer of short-term cultured tumor infiltration lymphocytes in metastatic melanoma patients. Clin Cancer Res 2010; 16(9):2646-55; PMID:20406835; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-0041 [DOI] [PubMed] [Google Scholar]
- 37.Schmitt TM, Ragnarsson GB, Greenberg PD. T cell receptor gene therapy for cancer. Hum Gene Ther 2009; 20(11):1240-8; PMID:19702439; http://dx.doi.org/ 10.1089/hum.2009.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM et al.. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 2002; 298(5594):850-4; PMID:12242449; http://dx.doi.org/ 10.1126/science.1076514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardy NM, Fellowes V, Rose JJ, Odom J, Pittaluga S, Steinberg SM, Blacklock-Schuver B, Avila DN, Memon S, Kurlander RJ et al.. Costimulated tumor-infiltrating lymphocytes are a feasible and safe alternative donor cell therapy for relapse after allogeneic stem cell transplantation. Blood 2012; 119(12):2956-9; PMID:22289893; http://dx.doi.org/ 10.1182/blood-2011-09-378398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liddy N, Bossi G, Adams KJ, Lissina A, Mahon TM, Hassan NJ, Gavarret J, Bianchi FC, Pumphrey NJ, Ladell K et al.. Monoclonal TCR-redirected tumor cell killing. Nat Med 2012; 18(6):980-7; PMID:22561687; http://dx.doi.org/ 10.1038/nm.2764 [DOI] [PubMed] [Google Scholar]
- 41.Cantrell D, Smith K. The interleukin-2 T-cell system: a new cell growth model. Science 1984; 224(4655):1312-6; PMID:6427923; http://dx.doi.org/ 10.1126/science.6427923 [DOI] [PubMed] [Google Scholar]
- 42.Gillis S, Smith KA. Long term culture of tumour-specific cytotoxic T cells. Nature 1977; 268(5616):154-6; PMID:145543; http://dx.doi.org/ 10.1038/268154a0 [DOI] [PubMed] [Google Scholar]
- 43.Morgan D, Ruscetti F, Gallo R. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science 1976; 193(4257):1007-8; PMID:181845; http://dx.doi.org/ 10.1126/science.181845 [DOI] [PubMed] [Google Scholar]
- 44.Crompton JG, Sukumar M, Restifo NP. Uncoupling T-cell expansion from effector differentiation in cell-based immunotherapy. Immunological Rev 2014; 257(1):264-76; PMID:24329803; http://dx.doi.org/ 10.1111/imr.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crespo J, Sun H, Welling TH, Tian Z, Zou W. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr Opin Immunol 2013; 25(2):214-21; PMID:23298609; http://dx.doi.org/ 10.1016/j.coi.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Mitri D, Azevedo RI, Henson SM, Libri V, Riddell NE, Macaulay R, Kipling D, Soares MV, Battistini L, Akbar AN. Reversible senescence in human CD4+CD45RA+CD27- memory T cells. J Immunol 187(5):2093-100; PMID:21788446; http://dx.doi.org/ 10.4049/jimmunol.1100978 [DOI] [PubMed] [Google Scholar]
- 47.Warrington KJ, Vallejo AN, Weyand CM, Goronzy J Jr. CD28 loss in senescent CD4+ T cells: reversal by interleukin-12 stimulation. Blood 2003; 101(9):3543-9; PMID:12506015; http://dx.doi.org/ 10.1182/blood-2002-08-2574 [DOI] [PubMed] [Google Scholar]
- 48.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 2004; 14(4):501-13; PMID:15149599; http://dx.doi.org/ 10.1016/S1097-2765(04)00256-4 [DOI] [PubMed] [Google Scholar]
- 49.Fagagna FdAd, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003; 426(6963):194-8; PMID:14608368; http://dx.doi.org/ 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- 50.Adams PD. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell 2009; 36(1):2-14; PMID:19818705; http://dx.doi.org/ 10.1016/j.molcel.2009.09.021 [DOI] [PubMed] [Google Scholar]
- 51.Chen WH, Kozlovsky BF, Effros RB, Grubeck-Loebenstein B, Edelman R, Sztein MB. Vaccination in the elderly: an immunological perspective. Trends Immunol 2009; 30(7):351-9; PMID:19540808; http://dx.doi.org/ 10.1016/j.it.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8(9):729-40; PMID:17667954; http://dx.doi.org/ 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- 53.Lleonart ME, Artero-Castro A, Kondoh H. Senescence induction; a possible cancer therapy. Mol Cancer 2009; 8:3; PMID:19133111; http://dx.doi.org/ 10.1186/1476-4598-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N et al.. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 2008; 133(6):1006-18; PMID:18555777; http://dx.doi.org/ 10.1016/j.cell.2008.03.038 [DOI] [PubMed] [Google Scholar]
- 55.Kuilman T, Michaloglou C, Vredeveld LCW, Douma S, van Doorn R, Desmet CJ et al.. Oncogene-Induced Senescence Relayed by an Interleukin-Dependent Inflammatory Network. Cell 2008; 133(6):1019-31; PMID:18555778; http://dx.doi.org/ 10.1016/j.cell.2008.03.039 [DOI] [PubMed] [Google Scholar]
- 56.Kassem M, Ankersen L, Eriksen EF, Clark BF, Rattan SI. Demonstration of cellular aging and senescence in serially passaged long-term cultures of human trabecular osteoblasts. Osteoporos Int 1997; 7(6):514-24; PMID:9604046; http://dx.doi.org/ 10.1007/BF02652556 [DOI] [PubMed] [Google Scholar]
- 57.d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer 2008; 8(7):512-22; PMID:18574463; http://dx.doi.org/ 10.1038/nrc2440 [DOI] [PubMed] [Google Scholar]
- 58.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci 1995; 92(20):9363-7; PMID:7568133; http://dx.doi.org/22698404 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, Tavana O, Yang P, Manshouri T, Li Y et al.. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell 2012; 21(6):793-806; PMID:22698404; http://dx.doi.org/ 10.1016/j.ccr.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodier F, Coppe J-P, Patil CK, Hoeijmakers WAM, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 2009; 11(8):973-9; PMID:19597488; http://dx.doi.org/ 10.1038/ncb1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Appay V, Nixon DF, Donahoe SM, Gillespie GM, Dong T, King A, Ogg GS, Spiegel HM, Conlon C, Spina CA et al.. HIV-specific CD8(+) T cells produce antiviral cytokines but are impaired in cytolytic function. J Exp Med 2000; 192(1):63-75; PMID:10880527; http://dx.doi.org/ 10.1084/jem.192.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vallejo AN. CD28 extinction in human T cells: altered functions and the program of T-cell senescence. Immunol Rev 2005; 205:158-69; PMID:15882352; http://dx.doi.org/ 10.1111/j.0105-2896.2005.00256.x [DOI] [PubMed] [Google Scholar]
- 63.Darnell JE. STATs and Gene Regulation. Science 1997; 277(5332):1630-5; PMID:9287210; http://dx.doi.org/ 10.1126/science.277.5332.1630 [DOI] [PubMed] [Google Scholar]
- 64.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 2003; 3(11):900-11; PMID:14668806; http://dx.doi.org/ 10.1038/nri1226 [DOI] [PubMed] [Google Scholar]
- 65.Costa-Pereira AP, Tininini S, Strobl B, Alonzi T, Schlaak JF, Is'harc H, Gesualdo I, Newman SJ, Kerr IM, Poli V. Mutational switch of an IL-6 response to an interferon-γ-like response. Proc Natl Acad Sci 2002; 99(12):8043-7; PMID:12060750; http://dx.doi.org/24058752 10.1073/pnas.122236099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avalle L, Pensa S, Regis G, Novelli F, Poli V. STAT1 and STAT3 in tumorigenesis: A matter of balance. JAKSTAT 2012; 1(2):65-72; PMID:24058752; http://dx.doi.org/ 10.4161/jkst.20045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cohen PA, Koski GK, Czerniecki BJ, Bunting KD, Fu X-Y, Wang Z, Zhang WJ, Carter CS, Awad M, Distel CA et al.. STAT3- and STAT5-dependent pathways competitively regulate the pan-differentiation of CD34pos cells into tumor-competent dendritic cells. Blood 2008; 112(5):1832-43; PMID:18577706; http://dx.doi.org/ 10.1182/blood-2007-12-130138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang X-P, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G et al.. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol 2011; 12(3):247-54; PMID:21278738; http://dx.doi.org/ 10.1038/ni.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003; 426(6963):194-8; PMID:14608368; http://dx.doi.org/ 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- 70.Grange M, Buferne M, Verdeil G, Leserman L, Schmitt-Verhulst AM, Auphan-Anezin N. Activated STAT5 promotes long-lived cytotoxic CD8+ T cells that induce regression of autochthonous melanoma. Cancer Res 2012; 72(1):76-87; PMID:22065720; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-2187 [DOI] [PubMed] [Google Scholar]
- 71.Kujawski M, Zhang C, Herrmann A, Reckamp K, Scuto A, Jensen M, Deng J, Forman S, Figlin R, Yu H. Targeting STAT3 in Adoptively Transferred T Cells Promotes Their In Vivo Expansion and Antitumor Effects. Cancer Res 2010; 70(23):9599-610; PMID:21118964; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mulé J, Kerr WG et al.. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med 2005; 11(12):1314-21; PMID:16288283; http://dx.doi.org/ 10.1038/nm1325 [DOI] [PubMed] [Google Scholar]
- 73.Yang H, Yamazaki T, Pietrocola F, Zhou H, Zitvogel L, Ma Y, Kroemer G. Improvement of immunogenic chemotherapy by STAT3 inhibition. OncoImmunology 2016; 5(2):e1078061; PMID:27057456; http://dx.doi.org/ 10.1080/2162402X.2015.1078061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.