Abstract
Acute ischemic stroke (AIS) accounts for more than 80% of the approximately 610,000 new stroke cases worldwide every year. Both ischemia and reperfusion can cause death, damage, and functional changes of affected nerve cells, and these alterations can result in high rates of disability and mortality. Therefore, therapies aimed at increasing neuroprotection and neurorepair would make significant contributions to AIS management. However, with regard to AIS therapies, there is currently a large gap between experimental achievements and practical clinical solutions (EC-GAP-AIS). Here, by integrating curated disease-gene associations and interactome network known to be related to AIS, we investigated the molecular network mechanisms of multi-module structures underlying AIS, which might be relevant to the time frame subtypes of AIS. In addition, the EC-GAP-AIS phenomenon was confirmed and elucidated by the shortest path lengths and the inconsistencies in the molecular functionalities and overlapping pathways between AIS-related genes and drug targets. Furthermore, we identified 23 potential targets (e.g. ADORA3, which is involved in the regulation of cellular reprogramming and the extracellular matrix) and 46 candidate drugs (e.g. felbamate, methylphenobarbital and memantine) that may have value for the treatment of AIS.
Acute ischemic stroke (AIS) is a disease that is characterized by neuronal dysfunction and apoptosis induced by the interruption of blood supply resulting from the occlusion or rupture of blood vessels1. It is the most common cause of death and a major cause of disability worldwide2. Each year, 795,000 people experience a new or recurrent stroke. Approximately 610,000 of these strokes are first attacks, of which 87% are ischemic3. 5 years after a stroke, approximately 47% of patients died, and more than one-third of all survivors are left disabled4. In the United States, the costs associated with treatment for ischemic stroke are large financial burden, totaling more than $70 billion5. The high rates of stroke-associated mortality and disability result from neuronal injury6. However, the mechanisms underlying neuronal injury in AIS are poorly described. Previous studies have shown that ischemic stroke initiates a generalized series of events that occur at the onset of cerebral ischemia7. These include cellular bioenergetic failure, oxidative stress, microvascular injury, inflammation, and the eventual necrosis of neuronal, glial and endothelial cells. The time points at which these events occur could be specifically targeted by therapies. However, a number of drugs that have been shown to confer neuroprotective effects on preclinical experiments have failed in a clinical setting8. This might be owing to complicated factors involving in treatment of heterogeneous patients9. It is widely accepted that this heterogeneity might be the consequence of treatments outside the time frame of efficacy in a real-world AIS clinical setting10. Hence, effective drugs are rarely shown to promote neuroprotection and neurorepair of AIS, and the underlying molecular mechanisms of the gap between experimental achievements and clinical solutions remain to be fully explored.
Recently, a new trend in drug development has been to translate the research mode from a single molecule to multiple molecules combined with biological pathways and networks that provides a new method of drug development for complex diseases11. The latest evidence shows that different neuropathologies share important commonalities12. N-methyl-d-aspartate (NMDA) receptors play specific roles in pathological roles that are shared across various neurological and psychiatric disorders13. For instance, DJ-1, a Parkinson’s disease gene, is also a key regulator of stroke14. Hence, many investigations now focus on identifying new drugs for AIS therapy using therapies that have been proven effective in other diseases.
Network medicine has become increasingly important for identifying novel disease mechanisms and predicting drugs15. It provides a network-based approach to elucidate the underlying molecular mechanisms, mainly in terms of disease modules, of disease phenotypes and disease-disease associations16,17,18. To utilize incomplete interactome data for investigating a diseasome (i.e., a disease-disease relationship), Menche et al. proposed novel shortest path-based measurement to evaluate overlap between disease modules19. Moreover, a novel algorithm was proposed to detect disease modules using incomplete interactome data by taking advantage of the network expansion of disease-related seed genes20. A number of studies21,22 have investigated the disease modules associated with specific disease phenotypes, such as asthma, diabetes and cancer, for which a single disease module would mainly be detected. Recent work has integrated both large-scale electronic medical records and genomic data to detect thousands of novel associations between Mendelian and complex diseases. These studies have revealed that a nondegenerate, phenotypic code links each complex disorder (e.g. cancer, stroke and type 2 diabetes) to a unique collection of Mendelian loci. In addition, recent studies of the molecular subtypes of complex diseases, such as cancer23, type 2 diabetes24 and psychiatric disorders25, have indicated that incorporating the molecular profiles of disease phenotype subtypes that have previously been considered as single disorder would substantially improve our understanding of disease pathophysiology and the outcomes of treatments. Therefore, investigating the disease modules corresponding to the time-frame subtypes of AIS would be a promising research avenue which increase our understanding of the mechanisms underlying the gap between experimental achievements and clinical solutions for AIS.
Here, according to the underlying molecular subtypes of complex diseases and the fragmentation phenomena of disease modules that were extracted from incomplete interactome data19, we used a novel disease module detection strategy to identify multi-module structures that might correspond to the disease subtypes of AIS. We first partitioned the entire PPI network into hundreds of topological modules, and then we detected the significant relevant modules of AIS using correlation analysis between the modules and curated AIS-related genes. The curated AIS-related genes were extracted from three phenotype-genotype associations, including the Coremine literature database26, OMIM27 and DiseaseConnect28. The human interactome data were filtered from the String 9.1 PPI database29. We used a widely used community detection method (i.e., BGLL) to identify topological modules from the entire human PPI network. After the disease modules associated with AIS were identified by the correlation analysis, we performed a functional analysis of the AIS-related disease modules using gene ontology and pathway enrichment analyses. The key biological functional features of each AIS-related disease module were then mapped to the temporal pathophysiological events that were proposed by Saenger et al.7. To identify potential drug targets for early time frame subtypes of AIS, we calculated the shortest path distance between AIS disease genes and drug targets. Finally, we confirmed the molecular mechanisms associated with the gap between experimental achievements and clinical solutions for AIS therapies and identified promising novel targets and related drugs for AIS that might confer neuroprotective efficacy.
Results
AIS-related disease–gene relationships
To identify AIS-related disease terms, we searched the Medical Subject Headings (MeSH, 2014 version) terminology database on the MeSH Browser website (https://www.nlm.nih.gov/mesh/MBrowser.html) using the key words “stroke” and “infarction”. Using this approach, we ultimately identified (confirmed by the neurobiologists in our author list) 12 AIS-related MeSH headings (Table 1, Table S1).
Table 1. Acute ischemic stroke–related MeSH headings.
| ID | MeSH headings | Scope Note |
|---|---|---|
| D002544 | Cerebral Infarction | The formation of an area of necrosis in the cerebrum caused by an insufficiency of arterial or venous blood flow. Infarcts of the cerebrum are generally classified by hemisphere, lobe, arterial distribution, and etiology. |
| D020767 | Intracranial Thrombosis | Formation or presence of a blood clot in a blood vessel within the SKULL. Intracranial thrombosis can lead to thrombotic occlusions and brain infarction. |
| D020766 | Intracranial Embolism | Blocking of a blood vessel in the skull by an embolus which can be a blood clot or other undissolved material in the blood stream. |
| D002542 | Intracranial Embolism and Thrombosis | Embolism or thrombosis involving blood vessels which supply intracranial structures. Emboli may originate from extracranial or intracranial sources. |
| D020243 | Infarction, Anterior Cerebral Artery | Necrosis occurring in the anterior cerebral artery system, including branches such as Heubner’s artery. Infarction in anterior cerebral artery usually results in sensory and motor impairment in the lower body. |
| D020244 | Infarction, Middle Cerebral Artery | Necrosis occurring in the middle cerebral artery distribution system which brings blood to the entire lateral aspects of each cerebral hemisphere. Clinical signs include impaired cognition; aphasia; agraphia; weak and numbness in the face and arms, contralaterally or bilaterally depending on the infarction. |
| D020762 | Infarction, Posterior Cerebral Artery | Necrosis induced by ischemia in posterior cerebral artery distribution system which supplies portions of the brain stem, thalamus, temporal and occipital lobe. Clinical features include olfaction disorders and visual problems. |
| D002546 | Ischemic Attack, Transient | Brief reversible episodes of focal, nonconvulsive ischemic dysfunction of the brain having a duration of less than 24 hours, and usually less than 1 hour, caused by transient thrombotic or embolic vessel occlusion or stenosis. |
| D046589 | CADASIL | CADASIL is an acronym for Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy. |
| D020925 | Hypoxia-Ischemia, Brain | A disorder characterized by a reduction of oxygen in the blood combined with reduced blood flow to brain from a localized obstruction of a cerebral artery or from systemic hypoperfusion. Prolonged hypoxia-ischemia is associated with TIA; brain infarction and other conditions. |
| D059409 | Stroke, Lacunar | Stroke caused by lacunar infarction or other small vessel diseases of the brain. It features hemiparesis, hemisensory, or hemisensory motor loss. |
| D002545 | Brain Ischemia | Localized reduction of blood flow to brain tissue due to arterial obstruction or systemic hypoperfusion. This frequently occurs in conjunction with brain hypoxia. Prolonged ischemia is associated with brain infarction. |
Using these 12 MeSH headings as disease keywords, we downloaded 1425 significant disease-gene associations (p < 0.05) in batch mode from the Coremine database26. To obtain reliable associations, we performed an extensive check of the PubMed literatures related to the 1425 disease-gene associations, and we confirmed the existence of 1042 (1042/1425 = 73.12%) AIS disease-gene associations involving 606 distinct genes. When we mapped these genes to the human PPI network (see the Supplementary text for our preprocessing methods), only 537 (537/606, 88.61%) genes were kept in the analysis. We visualized the PPI network of the remained AIS-related genes as shown in Fig. 1. In this network, 433 genes (433/606, 71.45%) were included in the giant component (over 12 folds density of the global human PPI network), whereas 69 (69/606, 11.4%) genes were not included, and 104 (104/606, 17.2) genes were scattered in the network. This result is consistent with both the agglomeration of disease proteins in the neighborhood of the observable module and the fragmentation of disease modules20 in the context of incomplete interactome data.
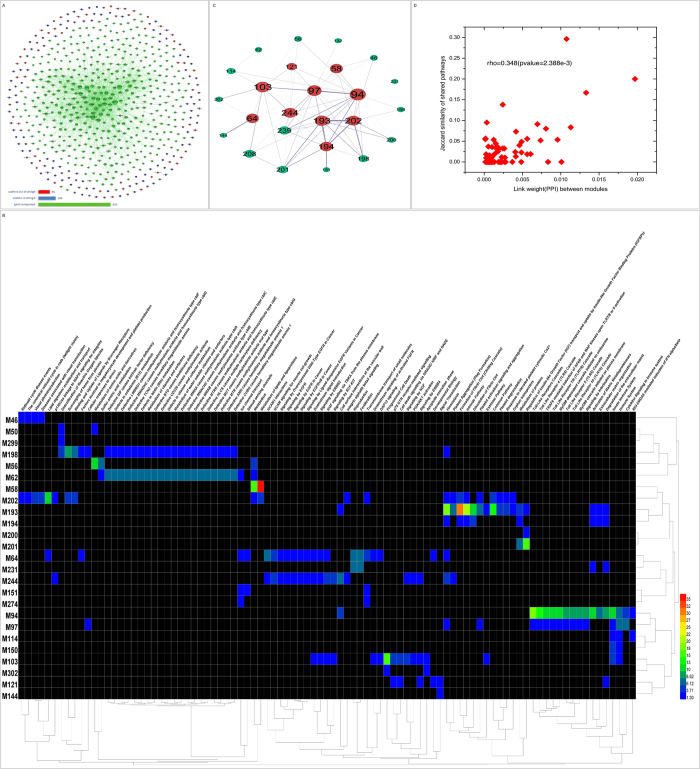
Figure 1. AIS PPI network, intra-module connectivity and heat map of the enriched pathways of the modules.
(A) AIS PPI network. There were a total of 606 AIS genes. Of these, there were 537 genes (~88.61%) in String 9.1 and 433 genes (~71.45%) in the giant component. (B) Heat map of the enriched pathways of the modules with an OR > 2. Pathways are aligned along the x-axis, while modules are aligned along the y-axis. Color code: red, small corrected p value and high enrichment; and green, large corrected p value and low enrichment. (C) Intra-module connectivity of the modules with an OR > 2. The red nodes represent the 10 with a large size and higher OR. The green nodes are other modules with an OR > 2. The thickness of the edge is proportional to its weight. Node sizes correspond to the number of edges that cross the node. (D) Connection between the edge weight and the pathways shared between the modules. There is a positive correlation (calculated by Spearman correlation) between the edge weight in (C) and shared pathways between modules.
To identify the general functional characteristics of AIS-related genes, we performed GO and pathway enrichment analyses of these genes and detected the significant biological processes in which these genes participate (Table S3-S6). In an ischemic area, as a result of a lack of energy, a cascade of neurochemical events occur, ranging from disrupted ion homoeostasis to necrotic and apoptotic cell death2. The enrichment analysis results indicated specific biological process features for AIS. For instance, the enriched pathway “Unblocking of NMDA receptor, glutamate binding and activation” (corrected p-value = 2.94 × 10−4) is related to glutamate release and calcium transport30, whereas the biological processes “Toll-Like Receptor 2 (TLR2) Cascade,” (corrected p-value = 1.759 × 10−3) and “Toll-Like Receptor TLR1:TLR2 Cascade” (corrected p-value = 1.759 × 10−3) are involved in oxidative stress and cellular necrosis31.
Significant AIS topological modules
To identify the significant disease modules that might correspond to the temporal subtypes of AIS, we partitioned the human PPI network using a widely used community detection algorithm. We confirmed 314 modules of the whole PPI network (see Methods and supplementary text). We obtained 131 modules, including at least one gene from the 537 AIS-related gene list, in which there were 29 modules with significant (Odds Ratio (OR) > 2.0) number of AIS-related genes. To elucidate the inter-module connectivity, we constructed a network using the 29 modules as nodes and links that represented the shared interactions between modules and a heat map that clustered similar modules based on their shared enriched pathways (see Methods, Fig. 1B and C). We found that there were positive correlations (rho = 0.384, p-value = 2.388e-3) between the link weights and shared pathways of modules (Fig. 1D), indicating that the closer two modules are linked, the higher the degree of shared biological processes between them. For example, we found strong interactions between M94 and M97 and many enriched pathways, such as the Toll-Like Receptor Cascades, the Toll-Like Receptor 4 (TLR4) Cascade, and Signaling by Interleukins, are shared between these two modules. We have listed the top 10 topological modules with the highest ORs. These are the modules that were the focus of our further investigations.
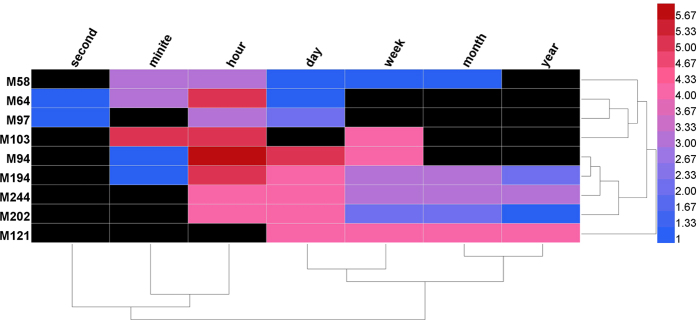
Next, we performed a functional analysis of the 10 modules using GO and pathway perspectives, and we found that each of these 10 modules were associated with highly enriched processes. For example, for module M64, the highly enriched GO processes were “ion transport” and “calcium ion transport”, and the enriched pathways were closely relates to the regulation of NMDA receptors, in which Ca2+ plays a pivotal role in regulating NMDA receptor activity32 (Table S8). Another module, M103, was associated with processes involved in necrosis and programmed cell death. To define the temporal subtypes of AIS, we annotated the pathophysiological events that were involved in the enriched pathways according to the relevant previous studies7,10,33,34 (Fig. 2 and Table S2). Our results showed that several modules, such as M64 and M103, contained distinct features that were associated with early molecular events (i.e., occurred within minutes to hours) in AIS. These modules would therefore be appropriate targets for early-stage neuroprotective interventions.
Figure 2. The heatmap of the time frame information of enriched Reactome pathways of ten significant disease modules.
The different color types correspond to the number of enriched pathways of the module in the time frame.
The molecular network characteristics of AIS-related drug targets
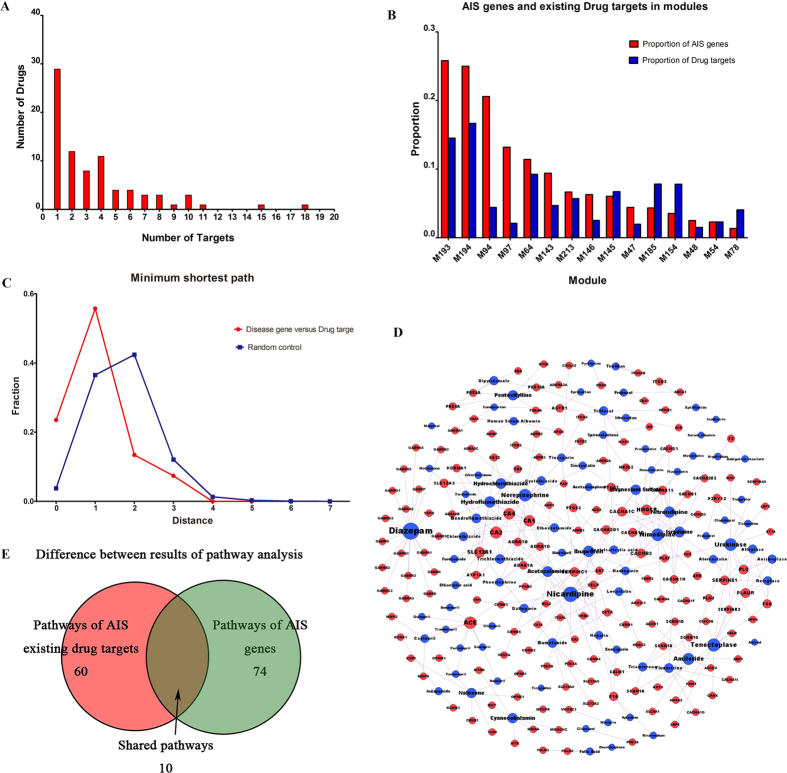
We identified 87 drugs from AIS guidelines and their 161 drug targets from the DrugBank database (see Methods, Fig. 3D, Table S2). Only 29 (29/87, 33.3%) of the drugs have a single target (Fig. 3A). A total of 149 (149/161, 92.55%) targets were scattered in the topological modules, and of these, 51 (51/161, 31.68%) targets were included in the 29 significant AIS modules. To measure the molecular interactions between AIS drug targets and genes, we calculated the minimum shortest path lengths between them. This approach was used in a similar previous study35(see the Methods section). The results showed that the distances were more enriched at the lower range of distances (i.e., < = 1, Fig. 3C) than the random control. This means that existing AIS drugs regulate diseases by directly targeting disease genes or the neighbors of AIS genes, which might explain the success of current therapies to treat AIS. We also calculated the degrees of AIS drug targets and found that more than 45% of the targets had large degrees (>50) that were statistically higher than those of the entire PPI network (Table S7). These data indicate that shorter distances might be the consequence of the hub effects of AIS-related drug targets.
Figure 3. Network of existing AIS drug-targets, statistical analysis of drug targets and analysis of minimum shortest path analysis.
(A) Analysis of drug-target interactions. Most drugs have several targets. (B) Graph showing the statistical analysis of AIS genes and drug targets in modules. For M193, M194 and M64, the proportions of AIS genes and targets are much higher than those of other modules. (C) The distribution of minimum shortest paths for the disease data (red) and random control (blue) groups. Enrichment occurs at distances 0 and 1. (D) AIS existing drug-target network. The blue nodes represent existing AIS drugs, and the red nodes represent their targets. (E) In the pathway analysis showing the results for AIS genes and their existing drug targets, there were 10 overlapping pathways.
To further investigate the molecular mechanisms underlying the gap between experimental achievements and clinical solutions with regard to neuroprotection effects, we measured the overlapping pathways both AIS drug targets and genes participating. We identified 84 pathways that were enriched in AIS-related disease genes and 70 pathways that were enriched in AIS drug targets. Only 10 pathways overlapped (P = 2.2 × e−16), and no neuroprotection-associated pathways were included in the overlapping pathways (Fig. 3E). This result indicates that although current AIS drug targets tend to intervene directly with AIS genes, they are not capable of regulating the neuroprotection-associated pathways that are involved in the early stage pathophysiological events of AIS. These findings suggest that specific and definable molecular network mechanisms underlie the huge gap between experimental achievements and practical clinical solutions (EC-GAP-AIS). A variety of neuroprotective agents have been used in experiments, but these have failed to achieve efficacy in clinical applications8. This might be because these agents do not target the correct temporal subtype (i.e. stages) of AIS.
Novel potential targets and drugs for neuroprotection in AIS
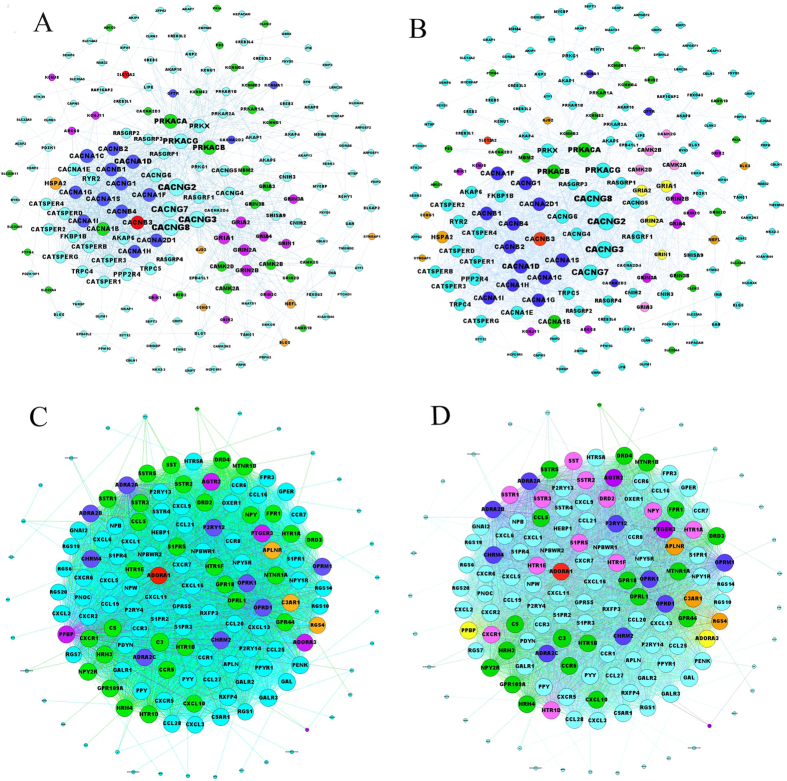
Based on the significant disease modules (in particular the modules incorporating early stage pathways) of AIS, we would be able to detect novel drug targets to promote neuroprotection in AIS. Here, we used DrugBank to filter the novel drug targets (those that have not been previously recognized as AIS drug targets) that might act via direct intervention (shortest path length< = 1) with AIS disease genes through the early-stage pathways. First, we identified 5 candidate modules from the significant identified modules that had high ratios of both AIS genes and drug targets in DrugBank. These were M58, M64, M94, M193 and M145. According to the identified functional characteristics (according to GO and pathway analysis), we found that 3 of these modules (i.e., M58, M94 and M193) were not associated with any neuroprotective pathways. These pathways were therefore excluded from further analysis. For example, M58 is closely related to lipid metabolism, and M94 is highly involved in immune responses. It has previously been established that the immune system contributes to the brain damage that is produced by ischemia and that the immune response is involved in all stages of the ischemic cascade36. M193 has been shown to be involved in the blood coagulation process. Therefore, these two modules, M64 and M145, have been confirmed as final candidate modules for neuroprotection-promoting drug target discovery. The M64 module (Fig. 4A) includes 184 genes, 21 (21/184, 11.4%) of which are associated with AIS and 57 (57/184 = 31.0%) of which are identified as drug targets in the DrugBank database. We considered the 27 drug targets, which directly or adjacently interact (i.e. the shortest path length < = 1) with the AIS genes, as preliminary candidate drug targets. The results showed that the M64 module is a particular type of molecular module that is incorporated in early-stage AIS events that are associated with neuroprotective processes (Table 2, Table S9). The highly enriched specific pathways, including “Unblocking of NMDA Receptor, Glutamate Binding and Activation”, of M64 (Fig. S2) are also enriched pathways of AIS genes. For example, “Unblocking of NMDA Receptor, Glutamate Binding and Activation” is involved in the initial stage (events occurring within minutes to hours) of AIS. A total of 13 (13/27, 48.1%) preliminary candidate drug targets were shown to be involved in this neuroprotection-related pathway. To further investigate the potential biological functions of these drug targets, we used a recently available proteomics database (called The Human Protein Atlas)37 to determine the tissues in which the genes of these targets are significantly expressed. Finally, we confirmed that 9 (9/13, 69.2%) of the targets are expressed in the nervous system, including 4 direct targets, GRIA1, GRIA2, GRIN1and GRIN2A (all of which belong to the glutamate-gated ion channel family), and 5 neighbor targets, CAMK2A, CAMK2B, CAMK2D, CAMK2G and GRIA3 (the first 4 of which belong to the calcium-dependent protein kinase (CAMK) family). These 9 drug targets are potential targets for inducing neuroprotective activity in AIS. Many reports have supported the importance of NMDA receptor subtypes in both neuronal survival and neuronal death38, and switches between NMDA receptor subtypes may contribute to reduced plasticity by influencing the binding of active CAMK family members39. We obtained 21 clinical drugs, including Primidone and Pethidine, that have targets among the 9 proteins formerly identified in DrugBank (Table S11). Although these 21 drugs are currently used primarily for epilepsy, pain relief and neurodegenerative diseases, searches for the latest drug clinical trials in ClinicalTrials.gov40 reveal that recent trials have involved using one or a combination of these drugs (e.g.., Primidone and Pethidine) as treatments for AIS.
Figure 4. Visualization of M64 and M145.
(A) and (B), visualization of M64. (C) and (D), visualization of M145. The light blue nodes are not the AIS genes or drug targets in the DrugBank database. The green nodes are the targets of drugs that were not found to be associated with AIS in the DrugBank database. The blue nodes are the targets of AIS drugs that were not associated with AIS genes. The orange-yellow nodes are AIS genes that were not drug targets in the DrugBank database. The purple nodes indicate AIS genes that were targets of drugs that were not associated with AIS in the DrugBank database. The red nodes indicate AIS genes and their drug targets. The pink nodes shown in (B) and (D) indicate potential targets that were not AIS genes. The yellow nodes in B indicate potential targets that were found to be AIS genes.
Table 2. Pathways enriched for M64.
| Pathway | PV | CPV | Time frame |
|---|---|---|---|
| Activation of NMDA receptor upon glutamate binding and postsynaptic events | 1.73E-20 | 1.36E-18 | mh |
| Glutamate Binding, Activation of AMPA Receptors and Synaptic Plasticity | 3.54E-20 | 2.63E-18 | smh |
| Trafficking of AMPA receptors | 3.54E-20 | 2.63E-18 | hd |
| Unblocking of NMDA receptor, glutamate binding and activation | 4.17E-19 | 2.99E-17 | mh |
| Depolarization of the Presynaptic Terminal Triggers the Opening of Calcium Channels | 1.22E-15 | 6.96E-14 | mh |
| Post-NMDA receptor activation events | 1.62E-15 | 8.97E-14 | mh |
| Ras activation upon Ca2+ influx through NMDA receptor | 4.55E-14 | 2.27E-12 | mh |
| CREB phosphorylation through the activation of CaMKII | 5.85E-13 | 2.78E-11 | mh |
| CREB phosphorylation through the activation of Ras | 1.72E-12 | 7.69E-11 | mh |
| Integration of energy metabolism | 2.59E-11 | 1.01E-09 | m |
| NCAM1 interactions | 8.29E-10 | 2.84E-08 | h |
| Rap1 signaling | 6.49E-09 | 1.97E-07 | hd |
| NCAM signaling for neurite out-growth | 7.11E-08 | 1.84E-06 | dwMy |
| PKA activation | 2.28E-07 | 5.62E-06 | mh |
| PKA activation in glucagon signaling | 3.06E-07 | 7.43E-06 | mh |
Here, we describe 15 highly enriched pathways of M64. Most of these pathways are induced during the early stage of ischemic stroke (within minutes to hours (mh)). Two pathways are involved from hours to days (hd), and the pathway “NCAM signaling for neurite out-growth” can last for days, weeks, months or years (dwMy) (Table S6).
Based on our identification of potential targets, we extracted 21 approved drugs, all of which were either antagonists of NMDA receptors or inhibitors of CAMK2. The drugs are used to treat neurological diseases (e.g., seizures, Alzheimer’s disease, and Parkinson’s disease), psychosis (e.g., bipolar disorder, schizophrenia, and delirium), and chronic myelogenous leukemia and to induce and maintain general anesthesia during surgery (Fig. 4A and B).
The M145 module included 149 nodes and was associated with 47 existing drug targets and 9 AIS genes. Of these, 8 (8/47, 17.02%) targets were involved in the highest M145-enriched pathway, “G Alpha (i) Signaling Events,” which is associated with the G-protein coupled receptor (GPCR) (Table S10). The classical signaling mechanism in this pathway involves the inhibition of cyclic adenosine monophosphate (cAMP)-dependent pathways, which are implicated in neuronal cell death41. Studies have shown that astrocytes mediate Ca2+ signaling by stimulating GPCRs42 and that this activity could occur within minutes to hours following stroke33. Finally, we identified 14 drug targets for nervous system (Table 3), such as ADORA3, PPBP and CXCR1, in the DrugBank database, in which only 6 targets (i.e., ADORA3, DRD2, HTR1A, HTR1D, HTR1E, and HTR1F) were associated with 25 drugs (i.e. agonists) (Table S11). Of these 25 agonists, several are dopamine receptor agonists or 5-hydroxytryptamine receptors agonists (Fig. 4C and D). Several studies have demonstrated that dopamine receptor agonists can protect against ischemia-induced neurodegeneration43 and that 5-hydroxytryptamine receptor agonists can reduce infarct volumes44.
Table 3. Spatial localization of potential acute ischemic stroke targets in M64 and M145.
| Tissue | Cerebral Cortex |
Hippocampus |
||||
|---|---|---|---|---|---|---|
| Cell | Endothelial cells | Glial cells | Neuronal cells | Neuropil | Glial cells | Neuronal cells |
| M64 | GRIA2 | CAMK2D | CAMK2A | CAMK2A | GRIA3 | CAMK2A |
| CAMK2A | CAMK2G | CAMK2B | GRIN1 | CAMK2B | ||
| GRIN1 | CAMK2D | CAMK2B | CAMK2D | |||
| GRIA3 | CAMK2G | CAMK2D | CAMK2G | |||
| GRIN2A | GRIN2A | CAMK2G | GRIA1 | |||
| GRIA1 | GRIA2 | |||||
| GRIA2 | GRIA3 | GRIA2 | ||||
| GRIA3 | GRIN1 | GRIN1 | ||||
| GRIN1 | GRIN2A | |||||
| GRIN2A | ||||||
| M145 | DRD2 | HTR1D | CXCR1 | ADORA3 | DRD2 | CXCR1 |
| HTR1E | HTR1D | |||||
| HTR1A | HTR1A | DRD2 | HTR1D | |||
| HTR1E | HTR1D | HTR1A | HTR1E | HTR1E | ||
| HTR1E | HTR1D | |||||
| NPY | HTR1E | NPY | ||||
| PPBP | HTR1F NPY | |||||
| SST | S1PR5 | SSTR3 | ||||
| SSTR2 | SSTR1 | |||||
| SSTR3 | SSTR2 | |||||
| SSTR3 | ||||||
We identified 9 potential targets in M64 and 14 potential targets in M145.
Discussion
Ischemic stroke is a heterogeneous disorder with a variety of clinical symptoms and causes and a diversity of disease subtypes, including CADASIL, lacunar stroke and middle cerebral artery infarction. Early stage intervention has an important impact on prognosis in ischemic stroke patients. However, few such interventions are currently available for use in a clinical setting. Investigations of the molecular network mechanisms (in term of disease modules) that contribute to ischemic stroke are key to identifying the manifestation subtypes and temporal stages of this disorder, and knowledge gained in both of these areas will increase our ability to generate tailored early-stage therapies. We identified over 1000 high-quality AIS disease-gene associations in the PubMed literature database and 29 significant disease modules in the human PPI network. We confirmed the importance of several disease modules, such as M64 and M145, which correspond to the temporal subtypes of AIS and would be highly valuable for early-stage therapies (mainly involving neuroprotection). Furthermore, we identified molecular mechanisms that underlie the gap between experimental achievements and clinical solutions for AIS treatment by measuring the difference between the pathways involving AIS genes and existing drug targets. Based on this investigation, a total of 23 potential targets and dozens of FDA approved drugs (for other disease conditions) were identified. Although the ultimate validation of these novel results will require systematic experimental and clinical studies, we searched the recent literature and found isolated studies that partially validated our results, supporting the reliability of these data. For example, both NMDA receptors and GPCR- related pathways have been identified in M64 disease modules, and related studies have shown that NMDA receptor blockers exert a neuroprotective effect while GPCR agonists are associated with cell apoptosis45,46,47. Otherwise, the activation of NMDA receptors and GPCRs contributes to Ca2+ overload10,42, which occurs within minutes of stroke onset33.
NMDA receptors are involved in a variety of neurological and psychiatric diseases48,49. For instance, NMDA receptors initiate neuronal death and the neurodegenerative processes in Alzheimer’s disease50. These antagonists can be used to treat cognitive dysfunctions, such as dementia51. Many successful experiments have demonstrated that NMDA antagonists confer a neuroprotective effect against the progression of ischemic stroke52,53, and GRIN2A is increased immediately after the onset of stroke symptoms7. Due to the shared molecular and phenotype features associated with various neurological diseases, the drugs used to treat the other similar neurological diseases may also create an effective neuroprotective effect against AIS. Of the candidate drugs identified in this study, felbamate and methylphenobarbital, which are used to treat epilepsy, and memantine, which is used to treat Alzheimer’s disease, may also be effectively used to treat AIS.
The clinical failure of NMDA receptor antagonists is partly because of the time frame in which they are applied. Whereas in experimental models, the onset of ischemia and reperfusion can be precisely defined, this is not possible in a clinical setting, and treatment might therefore be delayed before the presence of the disease is realized10. In a clinical setting, the timing of treatment might be delayed until the patient becomes aware of the cerebral ischemia. The time points at which these clinical drugs are used are often outside the window of opportunity to act as an effective neuroprotective treatment7.
In addition, the significant pathways associated with M145 and G alpha (i) signaling events exert a neuroprotective effect by inhibiting the cAMP-dependent pathway, which inhibits apoptosis41. This pathway also participates in the initial stage of stroke by regulating Ca2+signaling. This pathway is also related to stem cell functions. The role played by G alpha (i) signaling in pluripotent stem cells is largely unknown, but it involves the maintenance of pluripotency and the directed differentiation of human embryonic stem cells54. Histone modifications are thought to play certain key roles in cell reprogramming55, which plays a crucial role in establishing nuclear totipotency during normal development56. In the current study, the candidate ADORA3 is targeted by the antagonist aminophylline. The description of aminophylline in the DrugBank database indicates that aminophylline can modify histones by activating histone deacetylase. Hence, ADORA3 could be involved at a specific point during reprogramming. In addition, reprogramming may represent an endogenous process that protects the brain against further injury57. Therefore, regulating G alpha (i) signaling events might be a new avenue for further studies of AIS treatments.
Moreover, ADORA3 agonists can regulate the extracellular matrix (ECM) to protect against neuronal death58. During the initial stage of stroke, the upregulation of matrix metalloproteinases (MMPs) damages the blood brain barrier (BBB) by degrading the neurovascular matrix and thereby contributing to neuronal death. However, MMPs also promote angiogenesis during neurovascular repair phases34. Published evidence has shown that ADORA3 agonists can increase the secretion of MMPs59.These agonists may therefore promote effective neuroprotection and neurorepair in AIS.
Methods
Disease-gene associations
To identify AIS-related disease terms, we searched the Medical Subject Headings (MeSH, 2014 version) terminology database using the key words “stroke” and “infarction” at the MeSH Browser website (https://www.nlm.nih.gov/mesh/MBrowser.html). Following this search, the neurobiologists in our author list ultimately confirmed 12 AIS-related MeSH headings. Using these 12 MeSH headings as disease keywords, we downloaded 1425 significant disease-gene associations (p < 0.05) in batch mode from the Coremine database.
We searched the PubMed literature database for these disease-gene associations, and we confirmed 1042 disease-gene associations by reading more than 4500 PubMed articles.
AIS existing drugs-targets associations
In recent years, the American College of Cardiology (ACC) and the American Heart Association (AHA) have classified ischemic stroke as a atherosclerotic cardiovascular disease events60. We obtained known AIS drugs from the guideline designed by AHA and American Stroke Association (ASA) professionals61,62. Comprehensive information was obtained from the DrugBank database about these drugs63.
Detection of PPI topological modules
Community structures are widely distributed in complex networks. Each community is comprised of nodes that densely connect its members and are sparsely connected with the nodes in other modules64. A variety of community detection algorithms can be used to identify the topological modules in a large-scale network. We used a widely used algorithm called BGLL65, which is based on modularity evaluation, to obtain the topological modules of the full PPI network. Because BGLL yielded some modules with very large (e.g. several thousand) member nodes, we iteratively divided the modules using BGLL, resulting in topological modules that contain between 5 and 400 member nodes. However, different community detection algorithms would exactly obtain different results. Therefore, to validate the potential influence of our results, we performed another analysis using a network partition method based on the NMF66 algorithm to obtain a similar number of communities for comparison (see Supplementary text).
GO enrichment analysis
GO enrichment analysis defines gene groups based on the categories in the Gene Ontology database67. Using the plugin BiNGO 2.44 of Cytoscape 2.8.2 68,69, an open-source software platform for visualizing molecular interaction networks, we analyzed the disease genes, existing drug targets, and proteins in the modules that we previously characterized.
Reactome pathway analysis
Pathway analysis has become an important method for gaining insight into the biological functions underlying genes and proteins70. The Reactome database71 is a manually curated, open-source and open-data resource of human pathways. We obtained the enriched Reactome pathways using the online software program KOBAS2.072.
Intra-module connectivity
Next, we extracted the outgoing edges between the nodes in the different modules with an OR > 2. A link was documented when two modules were found to have PPI interactions between each other. For example, if there are c nodes in module M1, d nodes in M2, and e edges between M1 and M2, the weight of the edge M1-M2 would be:
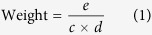 |
The heavier the weight, the closer the interaction between the two modules. Using the topological connectivity, we were able identify biological connections using the functional analysis73.
The shortest paths between drug targets and seed genes
Shortest paths are significant topological and statistical quantities that are used to analyze social and biological networks. The most outstanding example of the use of these quantities is the well-known small world property of many complex networks18. We used Dijkstra’s algorithm to identify the shortest path lengths between AIS drug targets and the genes of interest confirmed in this study74. To obtain random controls for the target-genes, we generated 100 independent randomized samples using the PPI network. Significant differences were calculated using t-tests (see supplementary text).
Additional Information
How to cite this article: Wang, Y. et al. Network-Based Approach to Identify Potential Targets and Drugs that Promote Neuroprotection and Neurorepair in Acute Ischemic Stroke. Sci. Rep. 7, 40137; doi: 10.1038/srep40137 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The study was supported by the National S&T Major Project of China (2012ZX09503-001-003) and NSFC project (61105055).
Footnotes
Author Contributions C.S. and X.Z. conceived and designed the research. X.Z., C.S., J.M., Y.W., H.L., Y.L., G.L. and P.Z. performed the following research projects: curation of the AIS disease-gene associations (X.Z., C.S., Y.W., H.L., Y.L., H.C., X.Y., T.Z. and M.F.), data analyses (Y.W., G.L. and P.Z.) and results validation (C.S. and Y.W.). X.Z., C.S., Y.W., J.M., H.L. and Y.L. wrote the manuscript. All authors have reviewed and revised the manuscript.
References
- González R. G. et al. Of referencing in Acute ischemic stroke (ed. González R. G. et al.). (Springer-Verlag Berlin Heidelberg, 2011). [Google Scholar]
- Donnan G. A. et al. Stroke. Lancet. 371, 1612–1623 (2008). [DOI] [PubMed] [Google Scholar]
- Mozaffarian D. et al. Heart disease and stroke statistics–2015 update: a report from the American Heart Association. Circulation. 131, e29–322, 10.1161/CIR.0000000000000152 (2014). [DOI] [PubMed] [Google Scholar]
- Luengo-Fernandez R. et al. Population-based study of disability and institutionalization after transient ischemic attack and stroke: 10-year results of the Oxford Vascular Study. Stroke. 44, 2854–2861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakaran S., Ruff I. & Bernstein R. A. Acute stroke intervention: a systematic review. Jama. 313, 1451–1462 (2015). [DOI] [PubMed] [Google Scholar]
- Xian Y. et al. Association between stroke center hospitalization for acute ischemic stroke and mortality. Jama. 305, 373–380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger A. K. & Christenson R. H. Stroke biomarkers: progress and challenges for diagnosis, prognosis, differentiation, and treatment. Clin Chem. 56, 21–33 (2010). [DOI] [PubMed] [Google Scholar]
- Cook D. J., Teves L. & Tymianski M. Treatment of stroke with a PSD-95 inhibitor in the gyrencephalic primate brain. Nature. 483, 213–217 (2012). [DOI] [PubMed] [Google Scholar]
- Xu S. Y. & Pan S. Y. The failure of animal models of neuroprotection in acute ischemic stroke to translate to clinical efficacy. Med Sci Monit Basic Res. 19, 37–45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirnagl U., Iadecola C. & Moskowitz M. A. Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci. 22, 391–397 (1999). [DOI] [PubMed] [Google Scholar]
- Anighoro A., Bajorath J. & Rastelli G. Polypharmacology: challenges and opportunities in drug discovery. J Med Chem. 57, 7874–7887 (2014). [DOI] [PubMed] [Google Scholar]
- Chen M. J. et al. Gene profiling reveals hydrogen sulphide recruits death signaling via the N-methyl-D-aspartate receptor identifying commonalities with excitotoxicity. J Cell Physiol. 226, 1308–1322 (2011). [DOI] [PubMed] [Google Scholar]
- Kalia L. V., Kalia S. K. & Salter M. W. NMDA receptors in clinical neurology: excitatory times ahead. Lancet Neurol. 7, 742–755 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleyasin H. et al. The Parkinson’s disease gene DJ-1 is also a key regulator of stroke-induced damage. Proc Natl Acad Sci USA. 104, 18748–18753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman E. K. & Loscalzo J. Network medicine approaches to the genetics of complex diseases. Discov Med. 14, 143–152 (2012). [PMC free article] [PubMed] [Google Scholar]
- Barabasi A. L., Gulbahce N. & Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 12, 56–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. & Butte A. J. Network medicine in disease analysis and therapeutics. Clin Pharmacol Ther. 94, 627–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. Human symptoms-disease network. Nat Commun. 5, 4212, 10.1038/ncomms5212 (2014). [DOI] [PubMed] [Google Scholar]
- Menche J. et al. Uncovering disease-disease relationships through the incomplete interactome. Science. 347, 1257601, 10.1126/science.1257601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiassian S. D., Menche J. & Barabasi A. L. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput Biol. 11, e1004120, 10.1371/journal.pcbi.1004120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. et al. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes in asthma. Hum Mol Genet. 24, 3005–3020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. P. et al. Isoquercetin protects cortical neurons from oxygen-glucose deprivation-reperfusion induced injury via suppression of TLR4-NF-small ka, CyrillicB signal pathway. Neurochem Int. 63, 741–749 (2013). [DOI] [PubMed] [Google Scholar]
- Bailey P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med. 7, 311ra174, 10.1126/scitranslmed.aaa9364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H. et al. Examining the phenotypic heterogeneity of early Autism Spectrum Disorder: subtypes and short-term outcomes. J Child Psychol Psychiatry. 71, 10.1111/jcpp.12448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenssen T. K. et al. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 28, 21–28 (2001). [DOI] [PubMed] [Google Scholar]
- Amberger J. S. et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43, D789–798, 10.1093/nar/gku1205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C. et al. DiseaseConnect: a comprehensive web server for mechanism-based disease-disease connections. Nucleic Acids Res. 42, W137–146, 10.1093/nar/gku412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–815, 10.1093/nar/gks1094 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth L. P., Kuner T. & Sakmann B. Adjacent asparagines in the NR2-subunit of the NMDA receptor channel control the voltage-dependent block by extracellular Mg2+. J Physiol. 506, 13–32 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M. et al. Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J Immunol. 164, 5998–6004 (2000). [DOI] [PubMed] [Google Scholar]
- Xin W. K. et al. A functional interaction of sodium and calcium in the regulation of NMDA receptor activity by remote NMDA receptors. J Neurosci. 25, 139–148 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone F. C. & Feuerstein G. Z. Inflammatory mediators and stroke: new opportunities for novel therapeutics. J Cereb Blood Flow Metab. 19, 819–834 (1999). [DOI] [PubMed] [Google Scholar]
- Liu Z. & Chopp M. Astrocytes, therapeutic targets for neuroprotection and neurorestoration in ischemic stroke. Prog Neurobiol. 15, 10.1016/j.pneurobio.2015.09.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim M. A. et al. Drug-target network. Nat Biotechnol. 25, 1119–1126 (2007). [DOI] [PubMed] [Google Scholar]
- Iadecola C. & Anrather J., The immunology of stroke: from mechanisms to translation. Nat Med. 17, 796–808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M. et al. Proteomics. Tissue-based map of the human proteome. Science. 347, 1260419, 10.1126/science.1260419 (2015). [DOI] [PubMed] [Google Scholar]
- Spaethling J., Le L. & Meaney D. F. NMDA receptor mediated phosphorylation of GluR1 subunits contributes to the appearance of calcium-permeable AMPA receptors after mechanical stretch injury. Neurobiol Dis. 46, 646–654 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria A. & Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 48, 289–301 (2005). [DOI] [PubMed] [Google Scholar]
- Zarin D. A. et al. The ClinicalTrials.gov results database–update and key issues. N Engl J Med. 364, 852–860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domin H. et al. Neuroprotective potential of the group III mGlu receptor agonist ACPT-I in animal models of ischemic stroke: In vitro and in vivo studies. Neuropharmacology. 102, 276–294 (2016). [DOI] [PubMed] [Google Scholar]
- Choudhury G. R. & Ding S. Reactive astrocytes and therapeutic potential in focal ischemic stroke. Neurobiol Dis. 85, 234–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck J. H. et al. De novo expression of dopamine D2 receptors on microglia after stroke. J Cereb Blood Flow Metab. 35, 1804–1811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X. et al. Effect of treadmill exercise on 5-HT, 5-HT1A receptor and brain derived neurophic factor in rats after permanent middle cerebral artery occlusion. Neurol Sci. 35, 761–766 (2014). [DOI] [PubMed] [Google Scholar]
- Yuan H. et al. Context-dependent GluN2B-selective inhibitors of NMDA receptor function are neuroprotective with minimal side effects. Neuron. 85, 1305–1318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. & Templeton D. M. Cadmium activates CaMK-II and initiates CaMK-II-dependent apoptosis in mesangial cells. FEBS Lett. 581, 1481–1486 (2007). [DOI] [PubMed] [Google Scholar]
- Rickhag M. et al. Comprehensive regional and temporal gene expression profiling of the rat brain during the first 24 h after experimental stroke identifies dynamic ischemia-induced gene expression patterns, and reveals a biphasic activation of genes in surviving tissue. J Neurochem. 96, 14–29 (2006). [DOI] [PubMed] [Google Scholar]
- Paoletti P., Bellone C. & Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 14, 383–400 (2013). [DOI] [PubMed] [Google Scholar]
- Hardingham G. E. & Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 11, 682–696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R. New developments on the role of NMDA receptors in Alzheimer’s disease. Curr Opin Neurobiol. 22, 559–563 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordji K., Becerril-Ortega J. & Buisson A. Synapses, NMDA receptor activity and neuronal Abeta production in Alzheimer’s disease. Rev Neurosci. 22, 285–294 (2011). [DOI] [PubMed] [Google Scholar]
- Sun M. et al. Isoflurane preconditioning provides neuroprotection against stroke by regulating the expression of the TLR4 signalling pathway to alleviate microglial activation. Sci Rep. 5, 11445, 10.1038/srep11445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H. et al. A neuroprotective role of the NMDA receptor subunit GluN3A (NR3A) in ischemic stroke of the adult mouse. Am J Physiol Cell Physiol. 308, C570–577, 10.1152/ajpcell.00353.2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doze V. A. & Perez D. M. GPCRs in stem cell function. Prog Mol Biol Transl Sci. 115, 175–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. et al. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol Cell Biol. 24, 5710–5720 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Dean W. & Walter J. Epigenetic reprogramming in mammalian development. Science. 293, 1089–1093 (2001). [DOI] [PubMed] [Google Scholar]
- Marsh B. et al. Systemic lipopolysaccharide protects the brain from ischemic injury by reprogramming the response of the brain to stroke: a critical role for IRF3. J Neurosci. 29, 9839–9849 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain S., Shearer T. W. & Crosson C. E. Mechanisms linking adenosine A1 receptors and extracellular signal-regulated kinase 1/2 activation in human trabecular meshwork cells. J Pharmacol Exp Ther. 320, 258–265 (2007). [DOI] [PubMed] [Google Scholar]
- Jian Liu K. & Rosenberg G. A. Matrix metalloproteinases and free radicals in cerebral ischemia. Free Radic Biol Med. 39, 71–80 (2005). [DOI] [PubMed] [Google Scholar]
- Andrus B. & Lacaille D. 2013 ACC/AHA guideline on the assessment of cardiovascular risk. J Am Coll Cardiol. 63, 2886, 10.1016/j.jacc.2014.02.606 (2013). [DOI] [PubMed] [Google Scholar]
- Jauch E. C. et al. Guidelines for the early management of patients with acute ischemic stroke: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 44, 870–947 (2013). [DOI] [PubMed] [Google Scholar]
- Kernan W. N. et al. Guidelines for the prevention of stroke in patients with stroke and transient ischemic attack: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 45, 2160–2236 (2014). [DOI] [PubMed] [Google Scholar]
- Law V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42, D1091–1097, 10.1093/nar/gkt1068 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich M. T. et al. Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics. 24, i223–231, 10.1093/bioinformatics/btn161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel V. D. G. J. L., Lambiotte R. et al. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment 10, P10008 (2008). [Google Scholar]
- Lee Daniel D. & Learning H. S. S. The parts of objects by non-negative matrix factorization. Nature. 401, 788–791 (1999). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. NOA: a novel Network Ontology Analysis method. Nucleic Acids Res. 39, e87, 10.1093/nar/gkr251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M. E. et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 27, 431–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K. & Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 21, 3448–3449 (2005). [DOI] [PubMed] [Google Scholar]
- Khatri P., Sirota M. & Butte A. J. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 8, e1002375, 10.1371/journal.pcbi.1002375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D. et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–477, 10.1093/nar/gkv1351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C. et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–322, 10.1093/nar/gkr483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Liu Z. P. & Chen L. Identification of dysfunctional modules and disease genes in congenital heart disease by a network-based approach. BMC Genomics. 12, 592, 10.1186/1471-2164-12-592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormen T. H. Of referencing in Introduction to algorithms (ed. Cormen T. H.) 658–662 (MIT press, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.