Abstract
The conserved cAMP-dependent protein kinase (PKA) is composed of the regulatory and catalytic subunits and acts as the central component of the cAMP signaling pathway. In the human fungal pathogen Candida albicans, the PKA regulatory subunit Bcy1 plays a critical role in the regulation of cell differentiation and death. It has long been considered that Bcy1 is essential for cell viability in C. albicans. In the current study, surprisingly, we found that Bcy1 is not required for cell growth, and we successfully generated a bcy1/bcy1 null mutant in C. albicans. Deletion of BCY1 leads to multiple cellular morphologies and promotes the development of filaments. Filamentous and smooth colonies are two typical morphological types of the bcy1/bcy1 mutant, which can undergo spontaneous switching between the two types. Cells of filamentous colonies grow better on a number of different culture media and have a higher survival rate than cells of smooth colonies. In addition, deletion of BCY1 significantly increased the frequency of white-to-opaque switching on N-acetylglucosamine (GlcNAc)-containing medium. The bcy1/bcy1 null mutant generated herein provides the field a new resource to study the biological functions of the cAMP signaling pathway in C. albicans.
Keywords: Candida albicans, PKA regulatory subunit, Bcy1, filamentation, white-opaque switching, cAMP signaling pathway
Introduction
The cAMP signaling pathway regulates a plethora of biological processes in eukaryotic organisms (Wang and Heitman, 1999; Pan et al., 2000; Gancedo, 2001; Chin et al., 2002; Chiaradonna et al., 2008). In the human fungal pathogen Candida albicans, this pathway plays a central role in the regulation of morphological transitions, carbon source utilization, quorum sensing, cell death, and virulence (Leberer et al., 2001; Rocha et al., 2001; Phillips et al., 2006; Biswas et al., 2007; Huang et al., 2010; Huang, 2012; Du et al., 2015). In response to environmental stimulation [such as increased CO2 levels, N-acetylglucosamine (GlcNAc), serum, and elevated temperatures], cells of C. albicans activate the cAMP signaling pathway and undergo morphological changes (Leberer et al., 2001; Phillips et al., 2006; Biswas et al., 2007; Huang et al., 2010; Huang, 2012).
Morphological plasticity is a striking feature of pathogenic Candida species and is tightly linked to virulence (Biswas et al., 2007; Whiteway and Bachewich, 2007; Huang, 2012). C. albicans can exist in a number of morphological forms, such as the yeast form, filaments (hyphae and pseudohyphae), and white, gray, and opaque cell types (Biswas et al., 2007; Whiteway and Bachewich, 2007; Huang, 2012). Different cell types of C. albicans play distinct roles in its life cycle. For example, the yeast cell can be easily disseminated to different tissues through the host circulatory system, while filamentous cells are better at invading tissue and initiating infections (Gow et al., 2012). White cells are more virulent in systemic infections, while opaque and gray cells have an enhanced ability to colonize cutaneous tissues (Tao et al., 2014b). The cAMP-PKA pathway is the major pathway involved in the regulation of morphological transitions and virulence in C. albicans (Biswas et al., 2007; Huang, 2012). Ras1 is upstream of the cAMP signaling pathway and is required for serum-induced true hyphal formation, but it is not essential for cell growth and the development of pseudohyphae in C. albicans (Feng et al., 1999). In response to extracellular stimuli, the activated Ras protein (Ras1) signals the adenylyl cyclase Cyr1 (also named Cdc35) to increase the synthesis of cAMP in C. albicans (Feng et al., 1999; Rocha et al., 2001). Deletion of CYR1 in C. albicans results in slow cell growth and serious defects in filamentation (Rocha et al., 2001). Cyr1 functions as a sensor for multiple extracellular signals including CO2, quorum sensing molecules, GlcNAc, and bacterial peptidoglycan (Wang, 2013). The alterations of cAMP levels modulate the activity of the cAMP-dependent protein kinase (PKA).
The PKA kinase is composed of two catalytic and two regulatory subunits in C. albicans (Biswas et al., 2007). Tpk1 and Tpk2 are two isoforms of the PKA catalytic subunit, which physically interacts with and is regulated by Bcy1, the regulatory subunit in C. albicans (Bockmühl et al., 2001; Cassola et al., 2004; Giacometti et al., 2012; Schaekel et al., 2013). The binding of cAMP to the regulatory subunit leads to the release and activation of the catalytic subunits.Tpk1 and Tpk2 play distinct and redundant roles in the regulation of filamentation, the stress response, and glycogen storage (Bockmühl et al., 2001; Giacometti et al., 2009). Bockmühl et al. (2001) have shown that Tpk1 is required for the formation of filaments on solid media, while Tpk2 is required for filamentation in liquid media. The authors further conclude that the two isoforms of the catalytic subunit are essential for cell viability because they were unable to generate the tpk1/tpk1 tpk2/tpk2 double mutant (Bockmühl et al., 2001; Giacometti et al., 2009). The PKA regulatory subunit Bcy1 plays a negative role in the regulation of the cAMP signaling pathway in fungal species (Cassola et al., 2004; Giacometti et al., 2006, 2012; Schaekel et al., 2013). Cassola et al. (2004) demonstrated that it is not possible to generate a BCY1 null mutant in a WT strain of C. albicans, since inactivation of BCY1 leads to constitutive activation of the cAMP/PKA pathway (Cassola et al., 2004). Alternatively, they generated a bcy1/bcy1 tpk2/tpk2 double mutant by deletion of BCY1 in a tpk2/tpk2 background strain. This double mutant exhibits a defect in the development of filaments in response to GlcNAc and serum (Cassola et al., 2004). And deletion of BCY1 affects the nuclear localization of Tpk1, suggesting that Bcy1 may also regulate the activity of the catalytic subunit by controlling its subcellular localization.
In the present study, surprisingly, we found that Bcy1 is not essential for cell growth of C. albicans. We successfully deleted both alleles of BCY1 and generated a bcy1/bcy1 null mutant in a laboratory wild type strain of C. albicans. This mutant provides an opportunity to revisit the biological roles of the PKA regulatory subunit in this important fungal pathogen. Deletion of BCY1 in C. albicans leads to multiple cellular morphologies and hyperfilamentation in certain media. We further show that Bcy1 plays an important role in the regulation of carbon source utilization and in white-opaque switching.
Materials and methods
Culture conditions, strains, and plasmids
The strains used in this study are listed in Table S1. YPD medium (2% glucose, 2% peptone, 1% yeast extract) and modified Lee's glucose medium (Huang et al., 2010) were used for routine culture of C. albicans. The red dye phloxine B (5 μg/mL) was added to the solid medium for the filamentation and white-opaque switching assays. Media used for spot serial dilution growth assays: YPD, Lee's (Lee's medium without sugar) (Lee et al., 1975), Lee's media with different carbon sources (replacement of glucose with 1.25% fructose or 3% ethanol plus 2% glycerol), YNB media [yeast nitrogen base with 0.5% ammonium sulfate and 3 amino acids (0.13 g/L leucine, 0.03 g/L histidine, and 0.04 g/L arginine) and different carbon sources (2% glucose, 2% fructose, 2% mannitol, or 4% glycerol)].
BCY1 were deleted in two WT strains (SN152 and SN152a) using the same strategy as described below. The two alleles of BCY1 were deleted using the fusion PCR strategy (Noble and Johnson, 2005). The first allele of BCY1 was replaced with the fusion PCR products of the CdHIS1 marker amplified from plasmid pSN52. The second allele of BCY1 was deleted with the fusion PCR products of the CmLEU2 marker amplified from pSN40. The primers used for the PCR reactions are listed in Table S2. To construct the BCY1-reconstituted strain, the fusion PCR product of three fragments (the CdARG4 marker amplified from plasmid pSN69, and fragments of BCY1 3′-UTR and BCY1 ORF plus 5′-UTR) was used for transformation of the bcy1/bcy1 mutant. The two BCY1-related fragments were amplified from genomic DNA of C. albicans (SC5314) with primer pairs (BCY1-5F-COM plus BCY1-5R-COM and BCY1-3R-COM plus BCY1-3R-COM, respectively).
Plasmid pACT1 was used to construct the TPK1 and TPK2-overexpressing plasmids (Huang et al., 2010). The PCR products of TPK1 were digested with EcoRV and HindIII and inserted into the EcoRV/HindIII site of pACT1 to generate plasmid pACT-TPK1. The PCR products of TPK2 were digested with StuI and HindIII and inserted into the EcoRI/HindIII site of pACT1, to generate plasmid pACT-TPK2. The AscI-linearized overexpressing plasmids were used for transformation of the WT and bcy1/bcy1 mutant.
White-opaque switching assay
White-opaque switching assays were performed as described previously (Xie et al., 2013). Lee's glucose and Lee's GlcNAc media were used for the quantitative switching assays. The cells were cultured on the plates at 25°C for 5 days. The cell identity was assessed by cellular morphology and verified by cell type-specific genes (data not shown).
Filamentation assays
Lee's glucose, Lee's GlcNAc, and YPD media were used for the filamentation assays. The cells were cultured at 25° and 37°C as indicated in the figure legends. For quantitative filamentous-smooth colony type switching assays (Figure 4), cells from a filamentous or smooth colony grown on Lee's glucose medium for 3 days were resuspended and cultured in liquid Lee's glucose for 24–96 h at 25°C. A small aliquot of cells was collected at the time point indicated and replated on YPD plates. After 5 days of growth at 30°C, colonies of the smooth and filamentous types were counted. The switching frequency = (number of colonies with alternative phenotype/total colony number) × 100%.
PI and DAPI staining assays
The cells were grown in liquid Lee's glucose medium for 48 h at 25°C and collected for propidium iodide (PI) and 4′-6-diamidino-2-phenylindole (DAPI) staining assays as described previously (Du et al., 2015). The cells were washed with 1 × phosphate-buffered saline (PBS) and resuspended in 1 × PBS. PI was added to the cells at a concentration of 2 μg/mL. The cells were stained for 15 min at room temperature in the dark with slight shaking and used for microscopy assays. For the DAPI staining assays, cells collected from liquid Lee's glucose medium were first fixed in 70% ethanol for 20 min and then stained with 1 μg/mL of DAPI. The cells were then washed with 1 × PBS and used for microscopy assays.
Quantitative real-time PCR (qRT-PCR) assay
Cells were grown Lee's GlcNAc plates at 25°C for 3 days and collected for total RNA extraction. The qRT-PCR assay was performed as described in our previous report (Tao et al., 2014a). Total RNA was used to synthesize cDNA with RevertAid H Minus reverse transcriptase (Thermo Scientific). Quantification of transcripts was performed in Bio-Rad CFX96 real-time PCR detection system using SYBR green. The relative expression level of each gene was normalized to that of C. albicans ACT1.
Results
Generation of the bcy1/bcy1 null mutant in C. albicans
Although the PKA regulatory subunit plays critical roles in a variety of biological processes in fungal species, the gene encoding this subunit is not essential for cell growth in many fungi including the model organisms, Saccharomyces cerevisiae (Cannon and Tatchell, 1987; Toda et al., 1987), Schizosaccharomyces pombe (DeVoti et al., 1991), and Neurospora crassa (Bruno et al., 1996), and the human fungal pathogens, Aspergillus fumigates (Zhao et al., 2006) and Cryptococcus neoformans (D'Souza et al., 2001). However, it has been thought that BCY1, the sole gene encoding the PKA regulatory subunit in C. albicans, is an essential gene (Cassola et al., 2004). Considering the conserved feature of the cAMP signaling pathway, we suspected that BCY1 might not be an essential gene in C. albicans, and thus the failure to obtain its null mutant in a previous report (Cassola et al., 2004) could be due to technical reasons. Using a fusion PCR deletion and prototrophic selection strategy (Noble and Johnson, 2005), we successfully deleted the two alleles of BCY1 in a WT strain of C. albicans (SN152, Figure 1). Correct integration of the CdHIS1 and CmLEU2 markers into the BCY1 locus was confirmed using PCR with two sets of flanking checking primers (Figure 1B, lanes 1–4). Moreover, one set of ORF primers was used to verify the absence of the BCY1 ORF region in the genome (Figure 1B, lanes 5 and 6). These results indicate that the two alleles of BCY1 were successfully deleted and replaced by the CdHIS1 and CmLEU2 markers, respectively. Therefore, as in other previously described fungi (Cannon and Tatchell, 1987; Toda et al., 1987; DeVoti et al., 1991; Bruno et al., 1996; D'Souza et al., 2001 and Zhao et al., 2006), the conserved PKA regulatory subunit Bcy1 is also not required for cell viability in C. albicans.
Figure 1.
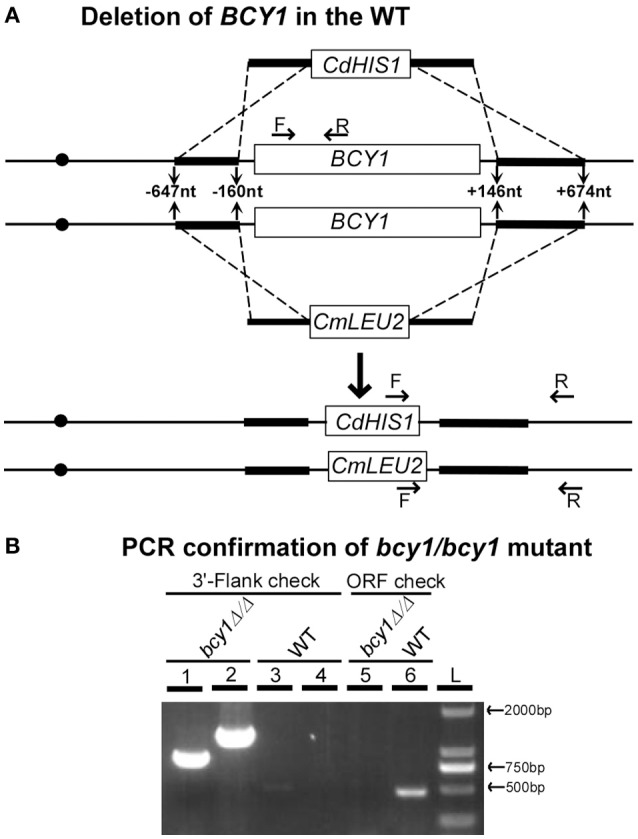
Deletion of both alleles of BCY1 in C. albicans. (A) Strategy for BCY1 deletion. Fusion PCR deletion assays were used to delete the two alleles of BCY1 as described in the Methods Section. CdHIS1 and CmLEU2 were used as selective markers for BCY1 deletion. (B) PCR confirmation of BCY1 deletion in the WT strain SN152. The primers used are indicated in (A). Lanes 1 and 3: 3′-flanking checking primers for CdHIS1 integration; lanes 2 and 4: 3′-flanking checking primers for CmLUE2 integration; lanes 5 and 6: BCY1 ORF checking primers. L, DNA ladder.
Multiple morphologies of the bcy1/bcy1 null mutant
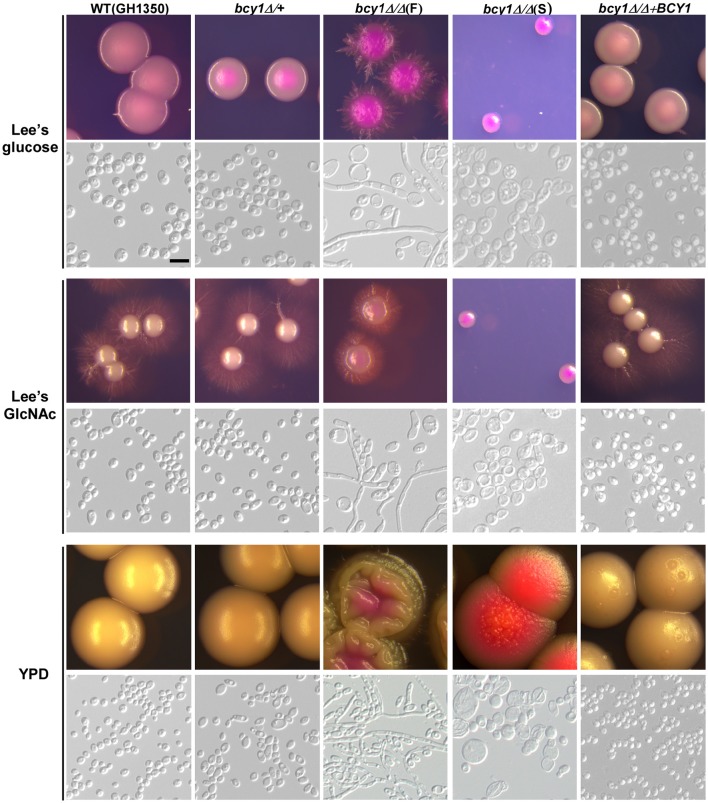
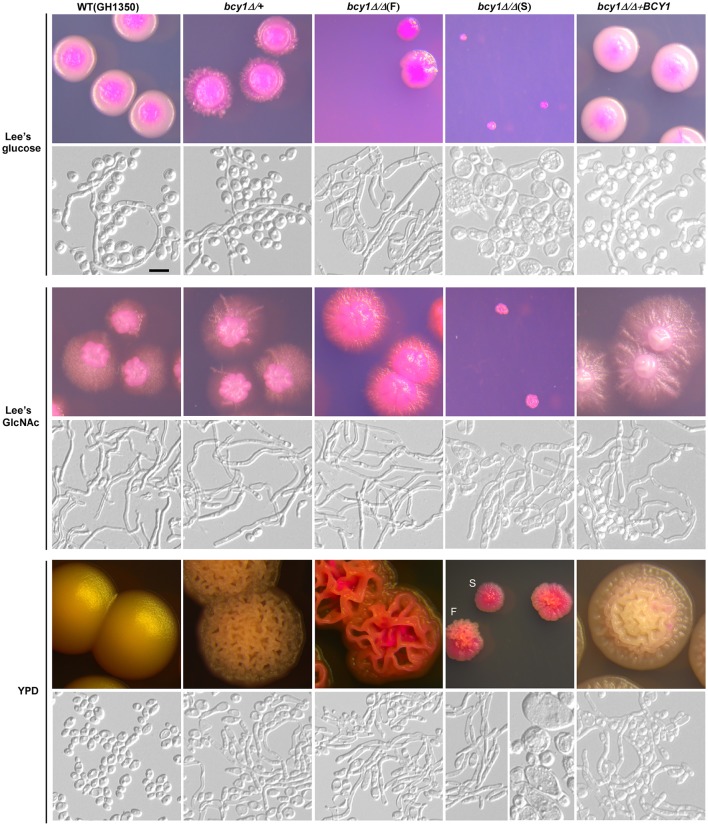
To evaluate the function of Bcy1 during filamentous development of C. albicans, we cultured the cells of WT, BCY1/bcy1, bcy1/bcy1, and BCY1-reconstituted strains on three different media (Lee's glucose, Lee's GlcNAc, and YPD) at two temperatures (25 and 37°C). These culture conditions were used because the three media exhibit different levels of filamentation induction in C. albicans. A high temperature (37°C) promotes filamentation, whereas a low temperature (25°C) favors yeast cell growth. Therefore, a combination of these conditions would facilitate the discrimination of the filamentation ability of different strains. At 25°C (Figure 2), deletion of one allele of BCY1 had no obvious effect on filamentous growth. Consistently, the BCY1-reconstituted strain also exhibited a similar phenotype to that of the WT control. However, the bcy1/bcy1 null mutant displayed a serious growth defect and had two distinct colony phenotypes (smooth and filamentous) on all three media. Cells of the smooth colonies were swollen and looked unhealthy, whereas filamentous cells had a much healthier appearance. On Lee's GlcNAc medium, a portion of the bcy1/bcy1 mutant cells exhibited an opaque-like phenotype (Figure 2). At 37°C (Figure 3), two colony types of the bcy1/bcy1 mutant were observed on three media. On Lee's glucose medium, filamentous colonies of the bcy1/bcy1 mutant underwent more robust filamentation than the WT, BCY1/bcy1, and BCY1-reconstituted strains, whereas on Lee's GlcNAc medium, all four strains exhibited strong filamentation at 37°C. These results are consistent with previous reports showing that GlcNAc is a potent yeast-to-filamentous growth inducer (Simonetti et al., 1974). The bcy1/bcy1 mutant showed the most robust filametation on YPD medium, while the BCY1/bcy1 and BCY1-reconstituted strains exhibited an intermediate level of filamentation at 37°C. The WT control maintained the yeast form on YPD medium at 37°C. These results suggest that Bcy1 plays a critical role in the regulation of filamentation and that the dosage of Bcy1 also affects this biological process.
Figure 2.
Cellular and colony morphologies of the WT, BCY1/bcy1, and bcy1/bcy1 mutants, and the BCY1-reconstituted strain on Lee's glucose, Lee's GlcNAc, and YPD media at 25°C. The cells were cultured on Lee's glucose and Lee's GlcNAc medium plates for 5 days or on YPD plates for 3 days. The bcy1/bcy1 mutant consists of two colony phenotypes: smooth (S) and filamentous (F). Scale bar, 10 μm.
Figure 3.
Cellular and colony morphologies of the WT, BCY1/bcy1, and bcy1/bcy1 mutants, and BCY1-reconstituted strain on Lee's glucose, Lee's GlcNAc, and YPD media at 37°C. The cells were cultured on Lee's glucose and Lee's GlcNAc medium plates for 5 days or on YPD plates for 3 days. At 37°C, all colonies of the bcy1/bcy1 mutant underwent hyper-filamentation on Lee's GlcNAc medium. Scale bar, 10 μm.
The bcy1/bcy1 null mutant can switch between the smooth and filamentous phenotypes
The bcy1/bcy1 mutant has two colony phenotypes: smooth and filamentous (Figures 2, 3, 4A). To test whether the two phenotypes could switch between each other, we cultured the two cell types on YPD medium and calculated the switching frequencies of the original to alternative cell type after 3, 5, and 8 days at 30°C (Figure 4B). Extension of incubation on solid medium promoted the filamentous phenotype. To further characterize the switching feature of the bcy1/bcy1 mutant, the smooth and filamentous colonies were cultured in liquid Lee's glucose medium for 0–96 h at 25°C (Figure 4C). The cells were then replated onto YPD medium and cultured at 30°C for 5 days. As shown in Figure 4C, the filamentous-to-smooth (F-to-S) switching frequencies at different time points were similar (20–40%), whereas the smooth-to-filamentous (S-to-F) switching frequencies increased dramatically with extension of the initial culture time.
Figure 4.
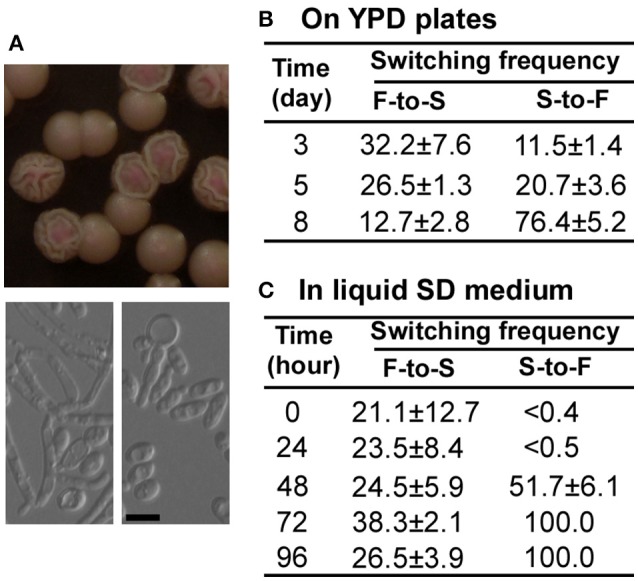
Switching between smooth (S) and filamentous (F) forms of the bcy1/bcy1 mutant at 30°C. (A) Cellular and colony morphologies of the bcy1/bcy1 mutant. The cells were cultured on YPD medium for 4 days. Scale bar, 10 μm. (B) Switching frequencies of S-to-F and F-to-S on YPD medium. Smooth and filamentous cells were plated on YPD medium and cultured for 3–8 days. (C) Switching frequencies of S-to-F and F-to-S in liquid SD medium. Cells were incubated in liquid SD medium for 0–96 h and then replated on YPD medium plates for 5 days.
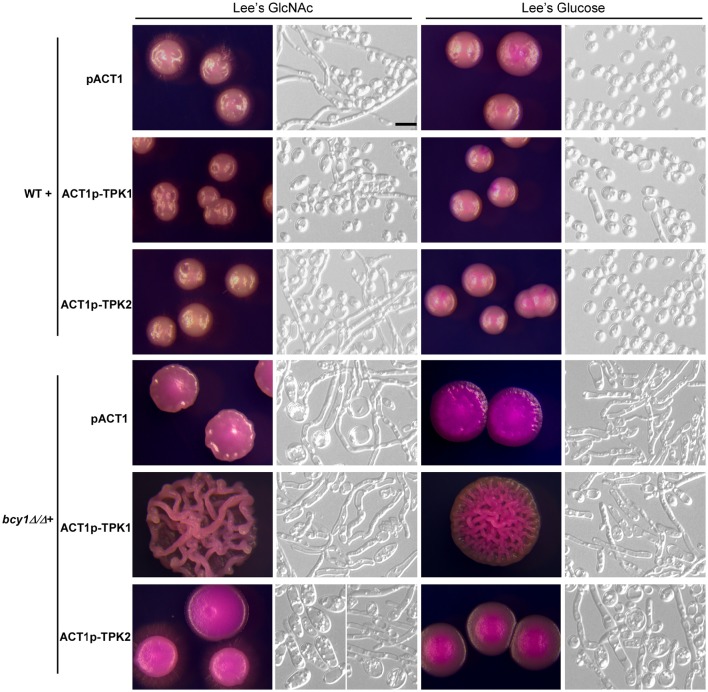
Effect of overexpression of TPK1 and TPK2 in the bcy1/bcy1 null mutant
Deletion of BCY1 causes constitutive activation of the PKA kinase. We examined the effect of overexpression of TPK1 and TPK2, which encode the two isoforms of the catalytic subunit, in the bcy1/bcy1 null mutant. As shown in Figure 5, overexpression of TPK1 in the bcy1/bcy1 null mutant resulted in hyper-filamentation at 25°C, while overexpression of TPK2 did not promote filamentation but led to the formation of two types of colonies: filamentous and opaque-like. On Lee's GlcNAc medium, one colony type was similar to the opaque phenotype, while the other underwent hyper filamentation. Of note, it was very difficult to obtain TPK2-overexpressing transformants in the bcy1/bcy1 null mutant, suggesting that overexpression of TPK2 may cause rapid cell death.
Figure 5.
Overexpression of TPK1 and TPK2 in the WT and bcy1/bcy1 mutant. Cells were cultured on Lee's glucose and Lee's GlcNAc media at 25°C for 5 days. Scale bar, 10 μm.
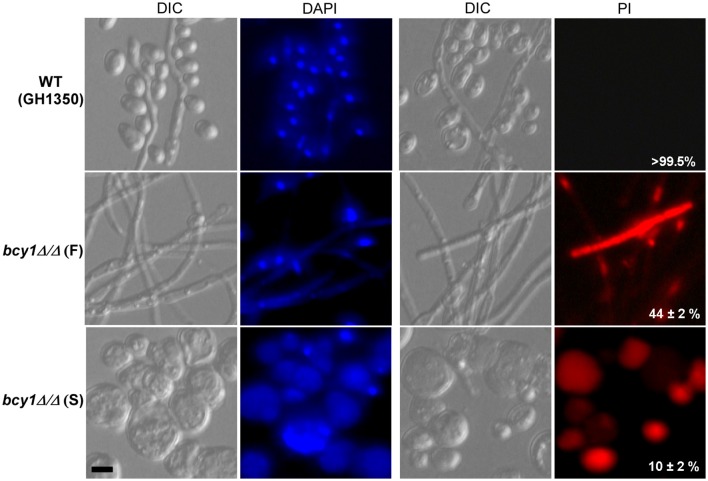
Deletion of BCY1 promotes cell death
The cAMP signaling pathway regulates cell death in C. albicans (Phillips et al., 2006). Because the deletion of BCY1 results in constitutive activation of this pathway, we next tested the viability of bcy1/bcy1 mutant cells during incubation in Lee's glucose medium. Cells of the WT, smooth, and filamentous types of the bcy1/bcy1 null mutant were grown in liquid Lee's glucose medium for 48 h at 25°C. The cells were then collected and stained with DAPI and PI. As shown in Figure 6, cells of the WT and filamentous form of the bcy1/bcy1 mutant had intact DAPI-stained nuclei, while most cells of the smooth type of the bcy1/bcy1 mutant had a fragmented nucleus or had no intact nuclei. PI staining verified that most cells of the smooth form of the bcy1/bcy1 mutant underwent cell death. Quantitative assays demonstrated that more than 95% of the smooth cells and approximately 70% of filamentous cells of the bcy1/bcy1 mutant were dead after 48 h of incubation in Lee's glucose medium at 25°C. Of note, more than 99% the WT control cells remained viable under the same culture conditions.
Figure 6.
Deletion of BCY1 in C. albicans promotes cell death, especially in the bcy1/bcy1 smooth (S) form. Cells of the WT, bcy1/bcy1 filamentous (F), and bcy1/bcy1 smooth (S) forms were incubated in Lee's glucose liquid medium for 48 h with shaking, and then used for DAPI and PI staining assays. The percentages (on PI-images) represent the survival rates using plating assays. DIC, differential interference contrast. Scale bar, 10 μm.
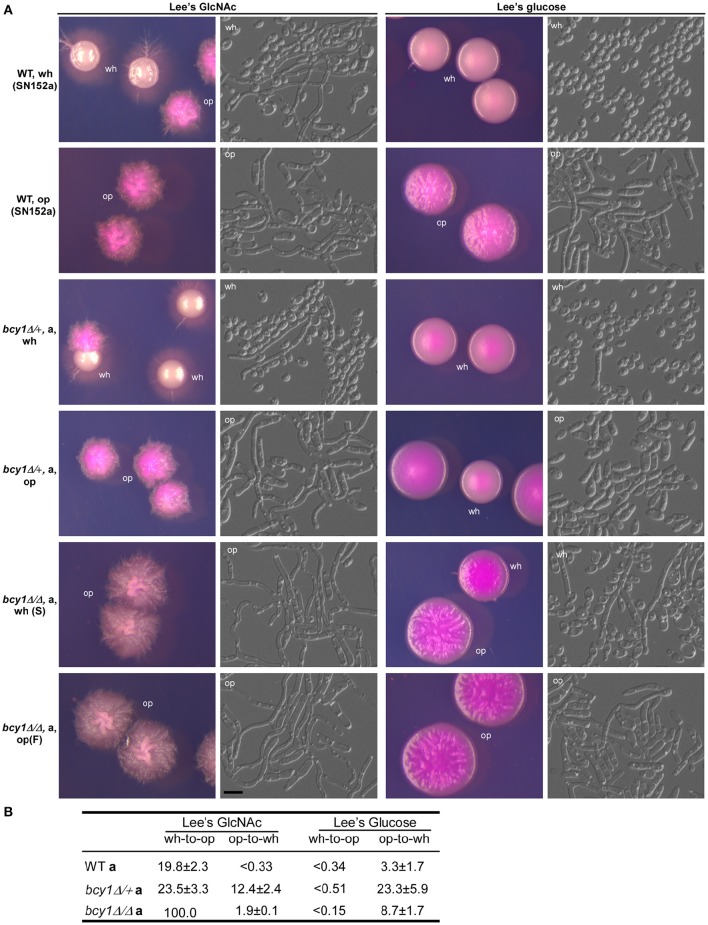
Role of Bcy1 in the regulation of white-opaque switching
Huang et al. (2010) have demonstrated that activation of the cAMP signaling pathway promotes white-opaque switching in C. albicans (Huang et al., 2010). As shown in Figure 2, deletion of BCY1 in the WT strain (MTLa/α) induced the formation of an opaque-like cell type on Lee's GlcNAc medium. Therefore, we deleted BCY1 in a white-opaque switchable MTLa/Δ strain (SN152a Tao et al., 2014a) to generate the BCY1/bcy1 and bcy1/bcy1 mutants (MTLa/Δ). The colony and cellular morphologies of the WT, BCY1/bcy1, and bcy1/bcy1 mutants (MTLa/Δ) are shown in Figure 7A. On both Lee's GlcNAc and Lee's glucose media, the BCY1/bcy1 mutant exhibited similar colony and cellular phenotypes to those of the WT strain. Both the smooth and filamentous types of the bcy1/bcy1 mutant could undergo white-opaque switching on Lee's glucose medium. However, both types underwent hyperfilamentation on Lee's GlcNAc medium. Replating and cell type-specific gene expression assays indicated that on Lee's GlcNAc medium, the filamentous cells had an opaque identity (Figure S1). Quantitative switching assays demonstrated that deletion of both alleles of BCY1 caused a mass conversion to the opaque phenotype on Lee's GlcNAc medium (Figure 7B). These results suggest that Bcy1 plays a negative role in the regulation of the white-to-opaque transition under this culture condition.
Figure 7.
Deletion of BCY1 affects white-opaque switching in C. albicans. Cells were cultured on Lee's glucose and Lee's GlcNAc media at 25°C for 5 days. Scale bar, 10 μm. (A) Cellular and colony morphologies of the WT, BCY1/bcy1, and bcy1/bcy1 mutants. On Lee's GlcNAc medium, no white colonies were observed since white cells switched to the opaque phenotype under this culture condition. Wh, white; op, opaque. (B) White-to-opaque and opaque-to-white switching frequencies of the WT, BCY1/bcy1, and bcy1/bcy1 mutants.
Role of Bcy1 in the regulation of carbon source utilization
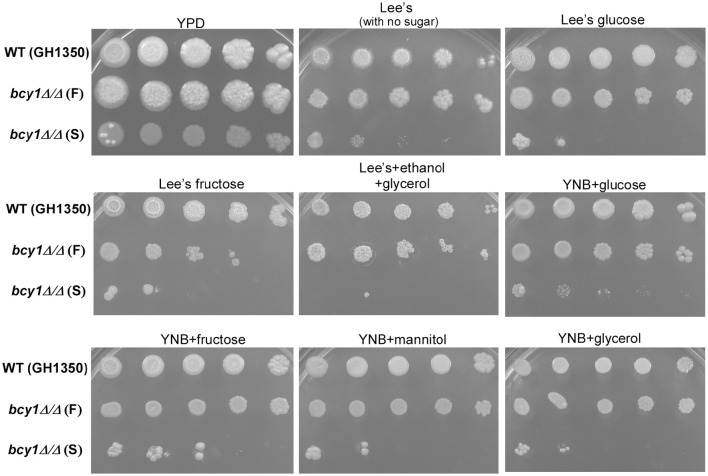
Next, we tested whether the regulatory subunit Bcy1 was involved in the regulation of carbon source utilization in C. albicans. As shown in Figure 8, nine media (including rich YPD medium, four Lee's media, and four YNB media containing different types of carbon sources) were used for this assay. The filamentous form of the bcy1/bcy1 mutant grew well on all media, although its growth rate was slower than that of the WT. The smooth form of the bcy1/bcy1 mutant grew well on YPD medium. However, the cells of this form showed a serious growth defect on both Lee's and YNB media. This defect did not appear to be related to the fermentative or non-fermentative features of the carbon sources.
Figure 8.
Bcy1 regulates carbon source utilization. Cells of the WT, bcy1/bcy1 filamentous (F), and bcy1/bcy1 smooth (S) forms were adjusted to 5 × 105 cells/mL, and then spotted onto nine different medium plates at 10-fold serial dilutions. The plates were incubated at 37°C for 4 days. Media used: rich medium YPD; Lee's, Lee's medium containing no sugar; Lee's glucose, Lee's medium containing 1.25% glucose; Lee's fructose, Lee's medium containing 1.25% fructose; Lee's+ethanol+glycerol, Lee's medium containing 3% ethanol and 2% glycerol; YNB+glucose (fructose, mannitol, glycerol), YNB plus 2% glucose (2% fructose, 2% mannitol, or 4% glycerol).
Discussion
The cAMP/PKA pathway plays a critical role in the regulation of a number of features of the human fungal pathogen C. albicans (Hogan and Sundstrom, 2009; Huang, 2012). The regulatory subunit Bcy1 has been considered to be essential in this fungus (Cassola et al., 2004). In this study, we demonstrate that the two alleles of BCY1 could be deleted in C. albicans. Given the conserved feature of the cAMP signaling pathway in fungal species, our finding is reasonable because the orthologs of Bcy1 in several fungal species (such as Bcy1 in S. cerevisiae, Csg1 in S. pombe, and pkaR in Aspergillus fumigatus and Cryptococcus neoformans) are not essential for cell viability (Cannon and Tatchell, 1987; Toda et al., 1987; DeVoti et al., 1991; Bruno et al., 1996; D'Souza et al., 2001; Zhao et al., 2006). Based on the bcy1/bcy1 null mutant generated in C. albicans, we re-evaluated the roles of Bcy1 in the regulation of filamentation, cell growth, and carbon source utilization. We also found that Bcy1 regulates white-opaque switching in C. albicans.
Deletion of CYR1, the sole gene encoding the adenylyl cyclase in C. albicans, completely blocked filamentation in response to several potent inducers including serum, CO2, and bacterial peptidoglycan (Rocha et al., 2001; Klengel et al., 2005; Xu et al., 2008). Activation of the cAMP-PKA pathway by ectopic expression of the activating form of Ras1 (Ras1V13) or deletion of the high affinity cyclic nucleotide phosphodiesterase-encoding gene PDE2 results in hyperfilamentation in C. albicans (Feng et al., 1999; Jung and Stateva, 2003). As expected, deletion of BCY1 in C. albicans promotes filamentation under conditions favoring yeast cell growth (such as at low temperature and in rich media, Figures 2, 3). Similar to the pde2/pde2 mutant (Jung and Stateva, 2003), the PKA catalytic subunit could be constitutively activated in the bcy1/bcy1 mutant. Moreover, the phenotypes of the bcy1/bcy1 mutant are highly similar to the hyperactive CYR1 mutant (Bai et al., 2011). The activated cAMP-PKA pathway then modulates downstream transcription factors (such as Efg1 and Flo8), which regulate filament-specific gene expression and promote filamentation (Bockmühl and Ernst, 2001; Cao et al., 2006).
Mutation of the PKA regulatory subunit in S. cerevisiae causes a variety of phenotypes (Cannon et al., 1990). In C. albicans, we found that deletion of BCY1 also resulted in multiple colony and cellular phenotypes including yeast, filamentous, and opaque-like forms (Figure 2). Interestingly, different cell types of the bcy1/bcy1 mutant exhibited different cell growth and carbon nutrient utilization abilities (Figure 8). Filamentous cells grew much better than cells of the smooth (yeast) form on all media, suggesting that both fermentative and non-fermentative carbon sources can be utilized by filamentous cells of the mutant. Switching between the filamentous and yeast cell forms can occur (Figure 4). Filamentous cells are healthier and display a better survival rate than cells of the smooth form when grown in regular media. Moreover, an extended culture time (which may represent a stressful condition) appeared to promote yeast-to-filamentous cell growth in the bcy1/bcy1 mutant. These results suggest that deletion of BCY1 in C. albicans promotes cell death, potentially due to constitutive activation of the cAMP-PKA pathway. Consistent with this idea, Phillips et al. (2006) demonstrated that the protein level of Bcy1 declined significantly during acetic acid-induced programmed cell death (Phillips et al., 2006). Filamentation in the bcy1/bcy1 mutant could be a strategy of cells to avoid cell death or to improve their anti-stress abilities. Consistent with our study, Laprade et al. (2016) recently reported that filamentation of C. albicans provides protection against antifungal-induced programmed cell death (Laprade et al., 2016).
White-opaque switching is another important feature of C. albicans and is involved in the regulation of virulence, sexual mating, and stress responses (Slutsky et al., 1987; Lohse and Johnson, 2009; Soll, 2009). Inactivation of the cAMP signaling pathway by the deletion of CYR1 suppresses GlcNAc and CO2-induced white-to-opaque switching, whereas activation of this pathway by ectopic expression of the activating form of RAS1, RAS1V13, or deletion of PDE2 promotes the opaque phenotype (Huang et al., 2009, 2010). Consistent with these observations, deletion of BCY1 promotes white-to-opaque switching in C. albicans (Figure 7). This promoting effect is dosage-dependent because the deletion of one allele of BCY1 leads to a moderate increase in this switch and deletion of both alleles causes a mass conversion on GlcNAc-containing media. GlcNAc is a potent inducer of the opaque phenotype. Consistent with the phenotype in the bcy1/bcy1 mutant, Huang et al. (2010) reported that the deletion of PDE2 also results in a mass white-to-opaque conversion on GlcNAc-containing media (Huang et al., 2010).
In summary, we successfully generated a bcy1/bcy1 mutant in C. albicans, which clarifies the essential role of this PKA regulatory subunit and provides a new avenue to study the cAMP-PKA pathway in this medically important pathogen. Our study also provides new insights into the functional roles of Bcy1 in the regulation of filamentation, carbon source utilization, and white-opaque switching. The results reported herein further confirm the conserved features and central role of the cAMP-PKA pathway in the regulation of a variety of biological features of C. albicans.
Author contributions
XD and GH designed the study. XD, CC, and QZ performed experiments. GH, XD, CC, and QZ analyzed data. GH, XD, CC, and QZ wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Suzanne Noble for the generous gifts of plasmids and strains. This work was supported by grants from the Chinese National Natural Science Foundation (31370175, 31170086, and 81322026) and the “100 Talent Program” grant from the Chinese Academy of Sciences (to GH).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02127/full#supplementary-material
Relative gene expression levels in white and opaque cells. Cells collected from Lee's GlcNAc medium plates (three days, at 25°C) were used for qRT-PCR assays. The values of the expression level of each gene in white cells of the WT strain were set as “1.” White and opaque cells of the WT served as controls. bcy1/bcy1 op, opaque cells plated on Lee's GlcNAc medium; bcy1/bcy1 original wh, white cells plated on Lee's GlcNAc medium. ACT1 served as the reference gene for normalization.
Strains used in this study.
Primers used in this study.
References
- Bai C., Xu X. L., Wang H. S., Wang Y. M., Chan F. Y., Wang Y. (2011). Characterization of a hyperactive Cyr1 mutant reveals new regulatory mechanisms for cellular cAMP levels in Candida albicans. Mol. Microbiol. 82, 879–893. 10.1111/j.1365-2958.2011.07859.x [DOI] [PubMed] [Google Scholar]
- Biswas S., Van Dijck P., Datta A. (2007). Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71, 348–376. 10.1128/MMBR.00009-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmühl D. P., Ernst J. F. (2001). A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmühl D. P., Krishnamurthy S., Gerads M., Sonneborn A., Ernst J. F. (2001). Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42, 1243–1257. 10.1046/j.1365-2958.2001.02688.x [DOI] [PubMed] [Google Scholar]
- Bruno K. S., Aramayo R., Minke P. F., Metzenberg R. L., Plamann M. (1996). Loss of growth polarity and mislocalization of septa in a Neurospora mutant altered in the regulatory subunit of cAMP-dependent protein kinase. EMBO J. 15, 5772–5782. [PMC free article] [PubMed] [Google Scholar]
- Cannon J. F., Gitan R., Tatchell K. (1990). Yeast cAMP-dependent protein kinase regulatory subunit mutations display a variety of phenotypes. J. Biol. Chem. 265, 11897–11904. [PubMed] [Google Scholar]
- Cannon J. F., Tatchell K. (1987). Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7, 2653–2663. 10.1128/MCB.7.8.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Lane S., Raniga P. P., Lu Y., Zhou Z., Ramon K., et al. (2006). The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17, 295–307. 10.1091/mbc.E05-06-0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassola A., Parrot M., Silberstein S., Magee B. B., Passeron S., Giasson L., et al. (2004). Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot. Cell 3, 190–199. 10.1128/EC.3.1.190-199.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaradonna F., Balestrieri C., Gaglio D., Vanoni M. (2008). RAS and PKA pathways in cancer: new insight from transcriptional analysis. Front. Biosci. 13:5257–5278. 10.2741/3079 [DOI] [PubMed] [Google Scholar]
- Chin K. V., Yang W. L., Ravatn R., Kita T., Reitman E., Vettori D., et al. (2002). Reinventing the wheel of cyclic AMP: novel mechanisms of cAMP signaling. Ann. N.Y. Acad. Sci. 968, 49–64. 10.1111/j.1749-6632.2002.tb04326.x [DOI] [PubMed] [Google Scholar]
- DeVoti J., Seydoux G., Beach D., McLeod M. (1991). Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 10, 3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza C. A., Alspaugh J. A., Yue C., Harashima T., Cox G. M., Perfect J. R., et al. (2001). Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21, 3179–3191. 10.1128/MCB.21.9.3179-3191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Guan G., Li X., Gulati M., Tao L., Cao C., et al. (2015). N-acetylglucosamine-induced cell death in Candida albicans and its implications for adaptive mechanisms of nutrient sensing in yeasts. MBio 6, e01376–e01315. 10.1128/mBio.01376-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Summers E., Guo B., Fink G. (1999). Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181, 6339–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M. (2001). Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25, 107–123. 10.1111/j.1574-6976.2001.tb00573.x [DOI] [PubMed] [Google Scholar]
- Giacometti R., Kronberg F., Biondi R. M., Hernández A. I., Passeron S. (2012). Cross regulation between Candida albicans catalytic and regulatory subunits of protein kinase A. Fungal Genet. Biol. 49, 74–85. 10.1016/j.fgb.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Giacometti R., Kronberg F., Biondi R. M., Passeron S. (2009). Catalytic isoforms Tpk1 and Tpk2 of Candida albicans PKA have non-redundant roles in stress response and glycogen storage. Yeast 26, 273–285. 10.1002/yea.1665 [DOI] [PubMed] [Google Scholar]
- Giacometti R., Souto G., Silberstein S., Giasson L., Cantore M. L., Passeron S. (2006). Expression levels and subcellular localization of Bcy1p in Candida albicans mutant strains devoid of one BCY1 allele results in a defective morphogenetic behavior. Biochim. Biophys. Acta 1763, 64–72. 10.1016/j.bbamcr.2005.11.016 [DOI] [PubMed] [Google Scholar]
- Gow N. A., van de Veerdonk F. L., Brown A. J., Netea M. G. (2012). Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10, 112–122. 10.1038/nrmicro2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D. A., Sundstrom P. (2009). The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 4, 1263–1270. 10.2217/fmb.09.106 [DOI] [PubMed] [Google Scholar]
- Huang G. (2012). Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence 3, 251–261. 10.4161/viru.20010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Srikantha T., Sahni N., Yi S., Soll D. R. (2009). CO(2) regulates white-to-opaque switching in Candida albicans. Curr. Biol. 19, 330–334. 10.1016/j.cub.2009.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Yi S., Sahni N., Daniels K. J., Srikantha T., Soll D. R. (2010). N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6:e1000806. 10.1371/journal.ppat.1000806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Stateva L. I. (2003). The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology 149, 2961–2976. 10.1099/mic.0.26517-0 [DOI] [PubMed] [Google Scholar]
- Klengel T., Liang W. J., Chaloupka J., Ruoff C., Schröppel K., Naglik J. R., et al. (2005). Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 15, 2021–2026. 10.1016/j.cub.2005.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade D. J., Brown M. S., McCarthy M. L., Ritch J. J., Austriaco N. (2016). Filamentation protects Candida albicans from amphotericin B-induced programmed cell death via a mechanism involving the yeast metacaspase, MCA1. Microb. Cell 3, 285–292. 10.15698/mic2016.07.512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Harcus D., Dignard D., Johnson L., Ushinsky S., Thomas D. Y., et al. (2001). Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42, 673–687. 10.1046/j.1365-2958.2001.02672.x [DOI] [PubMed] [Google Scholar]
- Lee K. L., Buckley H. R., Campbell C. C. (1975). An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13, 148–153. 10.1080/00362177585190271 [DOI] [PubMed] [Google Scholar]
- Lohse M. B., Johnson A. D. (2009). White-opaque switching in Candida albicans. Curr. Opin. Microbiol. 12, 650–654. 10.1016/j.mib.2009.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S. M., Johnson A. D. (2005). Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4, 298–309. 10.1128/EC.4.2.298-309.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Harashima T., Heitman J. (2000). Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3, 567–572. 10.1016/S1369-5274(00)00142-9 [DOI] [PubMed] [Google Scholar]
- Phillips A. J., Crowe J. D., Ramsdale M. (2006). Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 103, 726–731. 10.1073/pnas.0506405103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha C. R., Schröppel K., Harcus D., Marcil A., Dignard D., Taylor B. N., et al. (2001). Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12, 3631–3643. 10.1091/mbc.12.11.3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaekel A., Desai P. R., Ernst J. F. (2013). Morphogenesis-regulated localization of protein kinase A to genomic sites in Candida albicans. BMC Genomics 14:842. 10.1186/1471-2164-14-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti N., Strippoli V., Cassone A. (1974). Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 250, 344–346. 10.1038/250344a0 [DOI] [PubMed] [Google Scholar]
- Slutsky B., Staebell M., Anderson J., Risen L., Pfaller M., Soll D. R. (1987). “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169, 189–197. 10.1128/jb.169.1.189-197.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R. (2009). Why does Candida albicans switch? FEMS Yeast Res. 9, 973–989. 10.1111/j.1567-1364.2009.00562.x [DOI] [PubMed] [Google Scholar]
- Tao L., Cao C., Liang W., Guan G., Zhang Q., Nobile C. J., et al. (2014a). White cells facilitate opposite- and same-sex mating of opaque cells in Candida albicans. PLoS Genet. 10:e1004737. 10.1371/journal.pgen.1004737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Du H., Guan G., Dai Y., Nobile C. J., Liang W., et al. (2014b). Discovery of a “white-gray-opaque” tristable phenotypic switching system in candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol. 12:e1001830. 10.1371/journal.pbio.1001830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., et al. (1987). Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 1371–1377. 10.1128/MCB.7.4.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Heitman J. (1999). Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2, 358–362. 10.1016/S1369-5274(99)80063-0 [DOI] [PubMed] [Google Scholar]
- Wang Y. (2013). Fungal adenylyl cyclase acts as a signal sensor and integrator and plays a central role in interaction with bacteria. PLoS Pathog. 9:e1003612. 10.1371/journal.ppat.1003612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Bachewich C. (2007). Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61, 529–553. 10.1146/annurev.micro.61.080706.093341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Tao L., Nobile C. J., Tong Y., Guan G., Sun Y., et al. (2013). White-opaque switching in natural MTLa/alpha isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 11:e1001525. 10.1371/journal.pbio.1001525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. L., Lee R. T., Fang H. M., Wang Y. M., Li R., Zou H., et al. (2008). Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 4, 28–39. 10.1016/j.chom.2008.05.014 [DOI] [PubMed] [Google Scholar]
- Zhao W., Panepinto J. C., Fortwendel J. R., Fox L., Oliver B. G., Askew D. S., et al. (2006). Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect. Immun. 74, 4865–4874. 10.1128/IAI.00565-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative gene expression levels in white and opaque cells. Cells collected from Lee's GlcNAc medium plates (three days, at 25°C) were used for qRT-PCR assays. The values of the expression level of each gene in white cells of the WT strain were set as “1.” White and opaque cells of the WT served as controls. bcy1/bcy1 op, opaque cells plated on Lee's GlcNAc medium; bcy1/bcy1 original wh, white cells plated on Lee's GlcNAc medium. ACT1 served as the reference gene for normalization.
Strains used in this study.
Primers used in this study.