ABSTRACT
Herpes simplex virus 1 (HSV-1) latency entails the repression of productive (“lytic”) gene expression. An attractive hypothesis to explain some of this repression involves inhibition of the expression of ICP0, a lytic gene activator, by a viral microRNA, miR-H2, which is completely complementary to ICP0 mRNA. To test this hypothesis, we engineered mutations that disrupt miR-H2 without affecting ICP0 in HSV-1. The mutant virus exhibited drastically reduced expression of miR-H2 but showed wild-type levels of infectious virus production and no increase in ICP0 expression in lytically infected cells, which is consistent with the weak expression of miR-H2 relative to the level of ICP0 mRNA in that setting. Following corneal inoculation of mice, the mutant was not significantly different from wild-type virus in terms of infectious virus production in the trigeminal ganglia during acute infection, mouse mortality, or the rate of reactivation from explanted latently infected ganglia. Critically, the mutant was indistinguishable from wild-type virus for the expression of ICP0 and other lytic genes in acutely and latently infected mouse trigeminal ganglia. The latter result may be related to miR-H2 being less effective in inhibiting ICP0 expression in transfection assays than a host microRNA, miR-138, which has previously been shown to inhibit lytic gene expression in infected ganglia by targeting ICP0 mRNA. Additionally, transfected miR-138 reduced lytic gene expression in infected cells more effectively than miR-H2. While this study provides little support for the hypothesis that miR-H2 promotes latency by inhibiting ICP0 expression, the possibility remains that miR-H2 might target other genes during latency.
IMPORTANCE Herpes simplex virus 1 (HSV-1), which causes a variety of diseases, can establish lifelong latent infections from which virus can reactivate to cause recurrent disease. Latency is the most biologically interesting and clinically vexing feature of the virus. Ever since miR-H2's discovery as a viral microRNA bearing complete sequence complementarity to the mRNA for the important viral gene activator ICP0, inhibition of ICP0 expression by miR-H2 has been a major hypothesis to help explain the repression of lytic gene expression during latency. However, this hypothesis remained untested in latently infected animals. Using a miR-H2-deficient mutant virus, we found no evidence that miR-H2 represses the expression of ICP0 or other lytic genes in cells or mice infected with HSV-1. Although miR-H2 can repress ICP0 expression in transfection assays, such repression is weak. The results suggest that other mechanisms for miR-H2 activity and for the repression of lytic gene expression during latency deserve investigation.
KEYWORDS: herpes simplex virus, latency, miRNA
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a ubiquitous human pathogen that mainly infects the facial mucosal surfaces. Productive (“lytic”) infection of HSV-1 in these peripheral tissues is followed by the establishment of a latent infection in the trigeminal ganglion (TG), from which the virus can periodically reactivate to cause recurrent disease (reviewed in reference 1). During latency, the virus does not replicate, and lytic viral gene expression is strongly repressed, with the only abundantly expressed viral genes contained within a locus that encodes the latency-associated transcripts (LATs) (1, 2) and several HSV-1 microRNAs (miRNAs) (3–5). In mouse models of HSV-1 latency, one or more products from the LAT locus can promote the establishment and maintenance of latency by repressing lytic gene expression (6–8). There is also evidence for these gene products protecting neurons during acute infection (9–14) and playing a role in reactivation from latency (8, 15–19), chromatin modifications (20–22), and immune evasion (23).
miRNAs are small RNAs that repress gene expression by binding to their target mRNAs in the RNA-induced silencing complex (RISC) and reducing mRNA stability and/or translation. The seed region of an miRNA (nucleotides 2 to 8 starting from the 5′ end) is critical for its functions, although other parts of the miRNA can also contribute (24). Among the HSV-1 miRNAs encoded in the LAT locus, miR-H2 is one of the most highly expressed during latent infection of mouse TGs (3, 4, 25) and in a cell culture model of neuronal latency (26). Its expression during latency depends on a 200-bp DNA segment that includes the LAT promoter (25), and it can be expressed in cell culture following transfection of a plasmid encoding the LAT transcription unit (3). Thus, miR-H2 may perform one or more functions of the LAT locus. miR-H2 is completely complementary to coding sequences within the mRNA that encodes infected cell protein 0 (ICP0). ICP0 is an E3 ubiquitin ligase that counteracts cellular repressive functions, including intrinsic and innate immunity (27–31), and is important for viral replication and gene expression in many cell types and for reactivation from latency (32–36). The complementarity of miR-H2 and ICP0 mRNA, prior results showing that one or more products of the LAT locus repress lytic gene expression in mouse neurons (6–8), and transient-transfection experiments showing that miR-H2 can repress ICP0 expression in cell culture (3, 37, 38) led to the attractive hypothesis that miR-H2 could promote latency by repressing ICP0 expression, thereby globally repressing lytic gene expression (3, 37–39). HSV-2, which is closely related to HSV-1, also encodes a miR-H2 (alternatively known as miR-III) that is completely complementary to coding sequences in the HSV-2 ICP0 mRNA and can similarly repress ICP0 expression in transfection assays (40). In further support of the hypothesis, although miR-H2 is expressed rather weakly during lytic infection (3, 4, 25), it can be found bound to ICP0 mRNA in photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) assays (38, 41). Interestingly, a host miRNA, miR-138, can downregulate ICP0 by targeting the 3′ untranslated region (UTR) of the ICP0 transcript, thereby promoting latency by repressing lytic gene expression and increasing host survival (41). This raises the possibility that the virus might be equipped with multiple mechanisms of repressing ICP0 expression to promote latency.
Some mutational studies investigating miR-H2 have been published (37–39). Two miR-H2 mutant viruses derived from strain McKrae were reported to exhibit increased ICP0 protein expression 4 and 6 h after infection of rabbit skin (RS) cells and increased viral virulence and reactivation rates in a mouse model of ocular inoculation (37, 39). A mutant virus in a strain 17 background was reported to show increased ICP0 expression at both the mRNA and protein level 8 h after infection of 293T cells but reduced viral DNA replication in Neuro-2A cells following infection at a low but not a high multiplicity of infection (MOI) (38). Regardless, the critical question of whether miR-H2 can downregulate the expression of ICP0 and other lytic genes during the establishment or maintenance of latency has not been tested. To investigate this and related questions, we generated a mutant HSV-1 in a strain KOS background that is drastically impaired for miR-H2 expression, as well as a rescued derivative, and characterized their phenotypes in cell culture and a mouse model. Our results provide little or no support for the hypothesis that miR-H2 represses the expression of ICP0 during infection.
RESULTS
miR-H2 can repress ICP0 protein and ICP0 mRNA expression in transfected cells.
To test whether we could confirm previous results that HSV-1 miR-H2 can repress ICP0 protein expression during transfection (3, 37, 38), we cotransfected 293T cells with an ICP0-expressing plasmid (pRS-1) and either a miR-H2 mimic or a control mimic with an irrelevant sequence, followed by Western blotting of ICP0 and, as a loading control, actin. As expected, transfection of the miR-H2 mimic reduced the level of ICP0 protein relative to the level seen following transfection of the control mimic (Fig. 1A). When transfected together with pRS-1 and the miR-H2 mimic, a miR-H2 inhibitor, compared to a control inhibitor with an unrelated sequence, led to increased ICP0 protein levels (Fig. 1B). Since a previous experiment that observed miR-H2 inhibition of ICP0 protein expression failed to show a concomitant reduction in ICP0 mRNA (3), we were interested in whether this could be confirmed. However, measuring ICP0 mRNA levels by quantitative reverse transcription PCR (qRT-PCR) in a transfection experiment performed in the same manner as the experiment whose results are shown in Fig. 1A revealed that ICP0 mRNA expression was reduced 2.5-fold by miR-H2 (P = 0.022, two-tailed paired t test) (Fig. 1C). Thus, miR-H2 can repress ICP0 expression at both the protein and mRNA level in transfected cells.
FIG 1.
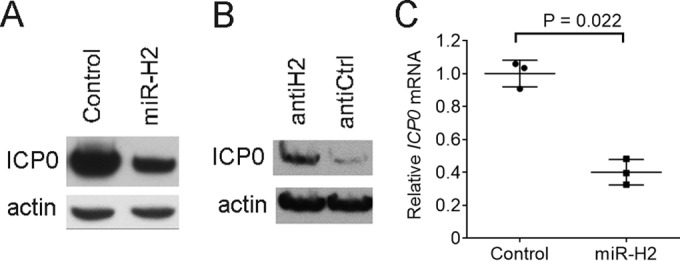
Effects of miR-H2 on ICP0 protein and ICP0 mRNA expression in transfected cells. (A) 293T cells were cotransfected with 75 ng of pRS-1 and 16 nM either irrelevant control mimic or miR-H2 mimic. At 30 h posttransfection (hpt), ICP0 and, as a loading control, actin levels were analyzed by Western blotting. (B) 293T cells were cotransfected with pRS-1, 8 nM miR-H2 mimic, and 20 nM either miR-H2 inhibitor (antiH2) or control inhibitor (antiCtrl). At 30 hpt, ICP0 and actin levels were analyzed by Western blotting. (C) Transfection was performed as described in the legend to panel A in triplicate, with ICP0 mRNA levels, normalized to the levels of human GAPDH mRNA, measured by qRT-PCR at 30 hpt. Horizontal bars and error bars represent mean values and standard deviations, respectively.
Design and validation of mutations that disrupt miR-H2 expression.
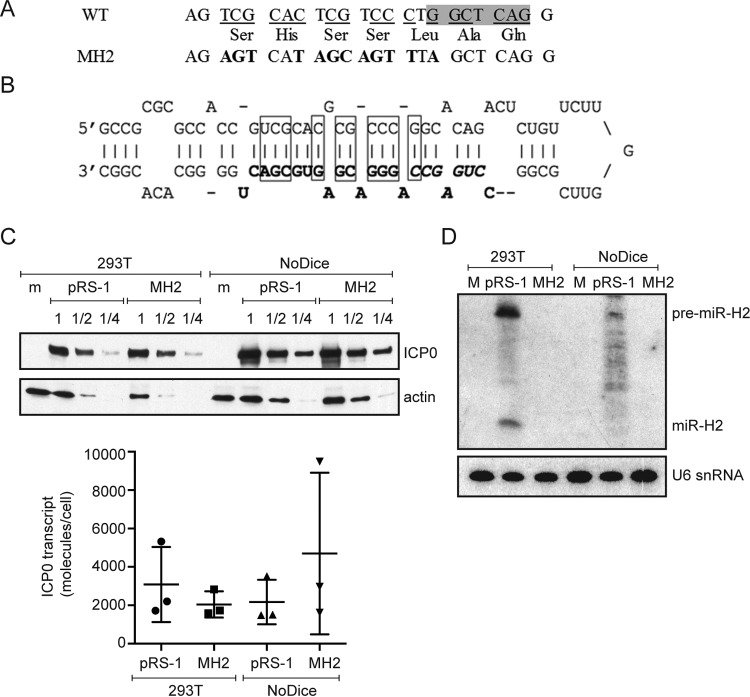
We then sought to design mutations that would disrupt miR-H2 expression. Our design took into account two considerations. First, as miR-H2 is encoded in the strand antisense to the ICP0 open reading frame, we would need to maintain the ICP0 amino acid sequence using codon redundancy. Second, given that mutations are introduced into both strands of the DNA, any mutant form of miR-H2, were it to be expressed, would still be completely complementary to the mutated ICP0 transcript expressed from the opposite strand and could target the mutated ICP0 transcript. Therefore, to prevent a mutant form of miR-H2 from being expressed, sufficient nucleotide substitutions would be needed to disrupt base pairs in the pre-miR-H2 hairpin so that the stem-loop structure required for miRNA biogenesis would no longer be formed. Based on these considerations, we designed a mutated sequence with 12 nucleotide substitutions (Fig. 2A). The mutations are expected to disrupt 10 bp within the hairpin (Fig. 2B). The resulting sequence was not predicted to form any stem-loop structure by the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). To test whether the mutations affect ICP0 protein or ICP0 mRNA expression, we introduced the mutations into the pRS-1 plasmid, resulting in the pRS-1MH2 plasmid, which expressed amounts of ICP0 protein (determined by Western blotting of a dilution series of the samples) and ICP0 mRNA (determined by qRT-PCR) similar to the amounts expressed by pRS-1 when transfected into 293T cells (Fig. 2C). To eliminate the concern that miR-H2 might be expressed from the wild-type (WT) plasmid, which could affect ICP0 levels, we measured miR-H2 levels from pRS-1 plasmid-transfected cells by qRT-PCR and Northern blot analyses. Remarkably, a strong signal was detected by qRT-PCR (data not shown), and we could easily detect both pre-miR-H2 and mature miR-H2 by Northern blot analysis despite there being no known promoter in the plasmid that would drive miR-H2 expression (Fig. 2D). To eliminate any effect on ICP0 expression due to miR-H2 expression, we employed NoDice cells, which lack Dicer and are derived from 293T cells (42). Transfection of the pRS-1 plasmid into these cells resulted in the expression of pre-miR-H2 and a ladder of RNA products on the Northern blot. Some very faint bands were roughly the size of miR-H2, but none precisely comigrated with miR-H2 (Fig. 2D). Following transfection of pRS-1 or pRS-1MH2 into the NoDice cells, the two plasmids expressed similar amounts of ICP0 mRNA and ICP0 protein (Fig. 2C), suggesting that the MH2 mutations did not affect ICP0 expression at the level of either mRNA or protein in the absence of miR-H2. We also observed no expression of any miRNA-sized species hybridizing to the probe for miR-H2 when pRS-1MH2 was transfected into either 293T or NoDice cells (Fig. 2D). The effect of the MH2 mutations on miR-H2 expression was examined further after the construction of a mutant virus (see below).
FIG 2.
miR-H2 mutations that do not affect ICP0 expression. (A) Mutations introduced into miR-H2. The top row shows the WT ICP0 gene sequence antisense to miR-H2, with the shaded area representing the sequence complementary to the seed region. The bottom row gives the corresponding mutant sequence with the altered nucleotides in bold letters. The middle row shows the predicted amino acid sequence encoded by both DNA sequences. (B) Sequence and hairpin structure of pre-miR-H2 showing mature miR-H2 in boldface, its seed region in boldface italics, and the base pairs that would be disrupted by the MH2 mutations in rectangles. (C) (Top) HEK293T wild-type and NoDice cells were either mock transfected (m) or transfected with the ICP0-expressing plasmid pRS-1 or the corresponding plasmid containing the miR-H2 mutations, pRS-1MH2. Protein samples were harvested at 24 hpt and serially diluted in twofold steps. Western blot analysis was conducted to compare the ICP0 protein levels in the different samples with the level of actin as an internal control. (Bottom) Total RNA samples were prepared at 24 hpt and analyzed for ICP0 mRNA levels by qRT-PCR. Horizontal bars and error bars represent mean values and standard deviations of the means, respectively, for three independent experiments. Differences were not statistically significant in either cell type (paired Student's t test). (D) 293T or NoDice cells were either mock transfected (M), or transfected with pRS-1 or pRS-1MH2 as indicated. At 24 hpt, <200-nucleotide RNA was isolated from transfected cells and analyzed by Northern blot hybridization, first being probed for miR-H2 and then stripped and probed with the U6 snRNA probe as an internal loading control.
Construction of miR-H2 mutant virus.
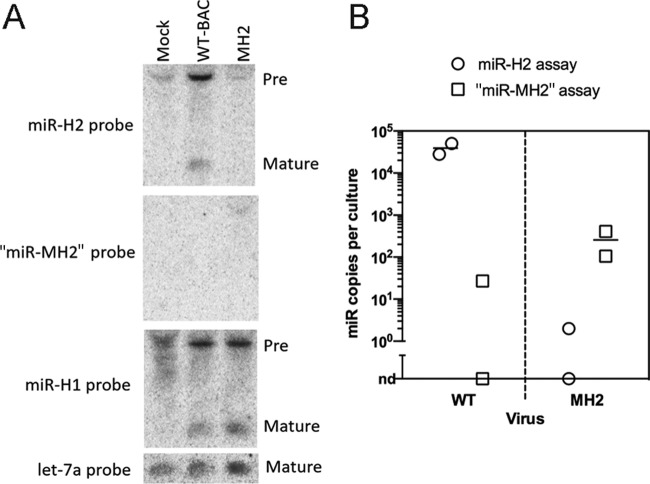
Using bacterial artificial chromosome (BAC) technology, we introduced the MH2 mutations into a BAC containing the genome of HSV-1 strain KOS (referred to hereinafter as WT), resulting in a mutant BAC and virus named MH2. Starting with the MH2 BAC, we restored the wild-type sequence to generate a rescued BAC and virus named RH2. Northern blot hybridization confirmed that, whereas mature miR-H2 was readily detectable in WT virus-infected cells, it was undetectable in MH2 mutant-virus-infected cells (Fig. 3A, top). Moreover, when we stripped and reprobed the membrane for “miR-MH2” (the putative mutant version of miR-H2 expressed from the MH2 virus) using the same conditions, no mature miRNA signal was detected in any sample, although a larger species was detected in RNA from MH2-infected cells (but not in the other samples) (Fig. 3A). The MH2 virus did, however, express miR-H1, similarly to the WT (Fig. 3A). To more sensitively evaluate the amount of “miR-MH2” possibly expressed from the mutant virus, we developed a stem-loop qRT-PCR assay that could quantify “miR-MH2” by comparing the signal from tested samples to that from a dilution series of synthetic miRNA. Using this assay and a previously developed assay for miR-H2 (3), we quantified miR-H2 and “miR-MH2” in WT- and MH2 virus-infected 293T cells, respectively, normalizing to the let-7a levels. As we have found that longer RNA species, such as pre-miRNAs, can sometimes meaningfully contribute to the signal in stem-loop qRT-PCR assays (M. F. Kramer and J. M. Pesola, unpublished results), we size fractionated the RNAs on an SDS-PAGE urea gel and extracted RNA with lengths of approximately 18 to 28 nucleotides. qRT-PCR assays using this size-fractionated RNA revealed that the “miR-MH2” levels in MH2 virus-infected cells were 150-fold lower than the miR-H2 levels in WT virus-infected cells (Fig. 3B). As miR-H2 is expressed to a maximum of 35 molecules/cell (4), this fold reduction corresponds to the MH2 virus expressing <1 “miR-MH2” molecule for every four cells. In a separate experiment, 293T cells infected with the RH2 virus showed levels of miR-H2 similar to the levels in 293T cells infected with WT virus as measured by qRT-PCR (data not shown). Thus, the MH2 mutations drastically reduce miR-H2 expression.
FIG 3.
Mutant HSV-1 with drastically reduced miR-H2 expression. (A) 293T cells were either mock infected or infected with WT (WT-BAC) or MH2 virus at an MOI of 10 for 18 h, followed by Northern blotting. The probes used for hybridization are shown to the left, and the positions of miRNA species (pre- and mature) are indicated to the right. (B) “miR-MH2” expressed in MH2-infected cells is present at levels 150-fold below the level of miR-H2 in WT-infected cells. qRT-PCR assays for miR-H2 (circles) and “miR-MH2” (squares) were performed on duplicate gel-extracted short RNA samples from 293T cells infected with WT (left) or MH2 (right) virus. Signals detected in WT-infected cells by the “miR-MH2” assay and in MH2-infected cells by the miR-H2 assay were either very low or not detectable (nd). Expression levels were normalized to that of host miRNA let-7a. Bars represent mean values.
MH2 shows WT levels of virus production and no increase in ICP0 gene expression in lytically infected cells.
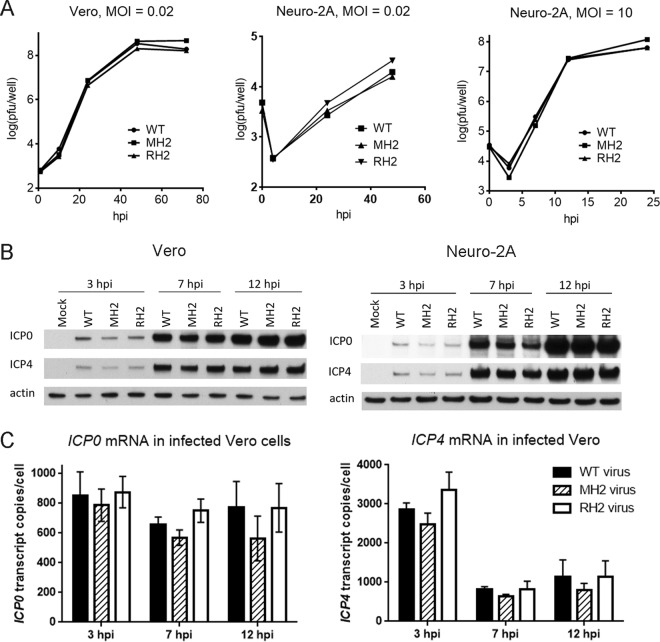
To investigate the effects of the MH2 mutations on viral replication and ICP0 gene expression in lytically infected cells, we infected Vero or Neuro-2A cells with WT, MH2, and RH2 viruses at different multiplicities of infection (MOIs). These viruses exhibited similar replication kinetics in Vero cells at an MOI of 0.02 and in Neuro-2A cells at MOIs of 0.02 and 10 (Fig. 4A). They expressed similar levels of ICP0 and, as a control, ICP4 proteins at 3, 7, and 12 h postinfection (hpi) when Vero or Neuro-2A cells were infected at an MOI of 10, as analyzed by Western blotting, and there certainly were no increases in ICP0 expression in MH2-infected cells (Fig. 4B). (If anything, the ICP0 and ICP4 levels were lower in MH2-infected cells than in WT- or RH2-infected cells in this experiment, but this was not the case in other experiments [data not shown].) They also expressed similar ICP0 and ICP4 (Fig. 4C) and ICP27 (data not shown) mRNA levels at these time points when Vero cells were infected at an MOI of 10, as measured by qRT-PCR. Notably, the qRT-PCR results also revealed that ICP0 mRNA reached levels of 800 molecules/cell as early as 3 hpi and remained above 500 molecules/cell throughout the rest of the infection cycle (Fig. 4C). Under the same conditions, we have previously shown that in lytically infected Vero cells, miR-H2 accumulates gradually during infection, to only 10 molecules/cell at 6 hpi and 35 molecules/cell at 18 hpi (4). Thus, the MH2 mutations did not affect virus production and did not increase ICP0 gene expression in lytically infected cells, and ICP0 mRNA is much more abundant than miR-H2 in this setting.
FIG 4.
The MH2 mutations have little effect on viral replication and ICP0 expression during lytic infection. (A) Replication kinetics in Vero cells at an MOI of 0.02 (left), in Neuro-2A cells at an MOI of 0.02 (middle) and in Neuro-2A cells at an MOI of 10 (right). Shown are viral titers over time. Each point represents the average value of triplicates. (B) Vero (left) or Neuro-2A (right) cells were infected with WT, MH2, or RH2 virus at an MOI of 10, followed by Western blotting for ICP0, ICP4, and actin levels. Viruses used for infection are indicated above each gel lane, and the infection times are shown on top. (C) Vero cells were infected with WT (black bar), MH2 (hatched bar), or RH2 (white bar) at an MOI of 10 for 3, 7, or 12 h, and then ICP0 (left) and ICP4 (right) transcript levels were measured by qRT-PCR. The error bars represent the standard deviations of duplicates. No difference was significant according to one-way ANOVA tests.
The MH2 mutations have little effect on viral replication or gene expression in acutely and latently infected mouse TG.
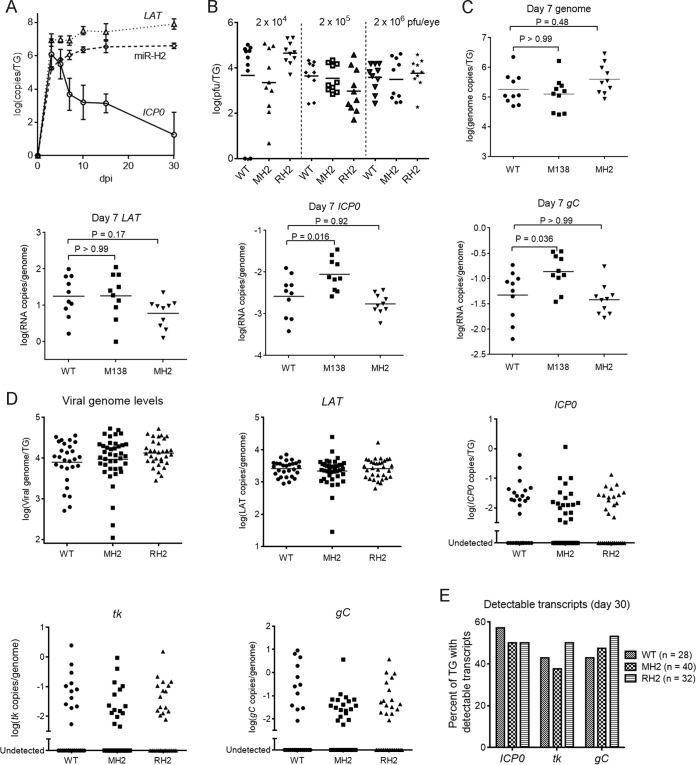
To characterize the mutant virus phenotype in vivo, we employed a mouse model in which, following corneal inoculation, virus replicates productively in the eye and TG and then establishes latent infection in TG neurons (43). Prior to testing the mutant, we used qRT-PCR to assess the expression of stable LAT, miR-H2, and ICP0 mRNA expression in TGs at different time points after the mice were inoculated on the cornea with KOS strain at a dose of 2 × 105 PFU/eye. Consistent with previous results for the expression of LAT and HSV-1 miRNAs using strains KOS and F (25, 44, 45), LAT and miR-H2 expression increased between 3 and 30 days postinfection (dpi), with both plateauing by 15 dpi (Fig. 5A); however, these results differed from a report using strain F that LAT and HSV-1 miRNAs decreased substantially between 14 and 28 dpi (46). As expected, the levels of ICP0 transcripts declined after 3 dpi (Fig. 5A). The levels of miR-H2 were similar to those of ICP0 transcripts at 5 dpi and greatly exceeded those of ICP0 transcripts thereafter (Fig. 5A). Our subsequent studies therefore focused on 5 dpi and later.
FIG 5.
The MH2 mutations have little effect on viral replication and transcript levels in murine TGs. (A) Time course of miR-H2, LAT, and ICP0 mRNA expression in murine TGs. Mice were inoculated on the cornea with 2 × 105 PFU of KOS per eye. At different dpi, 3 or 4 mice were euthanized and TGs were harvested and analyzed for ICP0 transcripts, stable LAT, and miR-H2 by qRT-PCR. RNA copy numbers were obtained by comparing the signals with standard curves generated using in vitro-transcribed ICP0 mRNA, LAT, or synthetic miR-H2. (B) Viral titers in TGs at 5 dpi following inoculation, with doses indicated at the top and virus names at the bottom. Data for different doses are separated by vertical dashed lines. Each point represents a value from one TG. The horizontal lines represent the geometric mean values (undetectable values were included in the calculations as 0). One-way ANOVA tests did not detect any significant differences between titers of viruses at each dose (P ≥ 0.16). (C to E) Mice were inoculated on the cornea with 2 × 105 PFU/eye of the indicated virus. (C) Viral genome and transcript levels in TGs at 7 dpi. Each point represents a value from one TG. The names of the transcripts assayed are indicated at the top of each graph. Vertical axes show logarithmic values, and RNA levels were normalized to viral genome levels. The horizontal lines represent the geometric mean values. P values shown above the brackets were obtained by one-way ANOVA tests with Bonferroni's multiple comparison test. (D) Viral genome and transcript levels at 30 dpi as described in the legend to panel C, except that mean values are not indicated in the plots for viral lytic transcripts due to the large numbers of undetectable values. One-way ANOVA (for viral genome and LAT) and Kruskal-Wallis tests (for ICP0, tk, and gC transcripts) did not detect any significant differences (P ≥ 0.48). (E) Percentages of TGs exhibiting detectable lytic transcripts. Each vertical bar represents the value for each virus (indicated in the key) and each RNA (below the graph). Fisher's exact tests did not detect any significant differences for each transcript (P ≥ 0.34).
We next compared WT, MH2, and RH2 viruses for viral replication in TGs after inoculation on the cornea using three different doses. Despite their 100-fold range, the three doses resulted in similar viral titers in TG homogenates at 5 dpi. We note that in a previous report (43), TG virus titers at 3 dpi increased substantially with inoculum dose through this range. Differences in the time points of analysis (dpi) and experimental methods may account for the different results. Regardless, we found no significant differences in titers detected among the viruses with any given dose (P ≥ 0.16) (Fig. 5B). Next, we compared WT and MH2 viruses for the expression of ICP0 and other viral genes at 7 dpi by measuring transcript levels using qRT-PCR. The M138 mutant virus, in which miR-138 binding sites in ICP0 mRNA are inactivated, was included as a positive control because it expresses increased lytic transcript levels at this time point (41). With a dose of 2 × 105 PFU/eye, WT and MH2 viruses expressed similar levels of ICP0 and gC transcripts per viral genome, while the levels of these transcripts per genome were significantly higher with the M138 virus (Fig. 5C). The level of LAT expression per genome appeared lower in MH2-infected ganglia than in WT-infected ganglia; however, that difference was not significant (P = 0.17) (Fig. 5C).
To assess the role of miR-H2 during latency, we infected mice with WT, MH2, or RH2 virus with a dose of 2 × 105 PFU/eye. At 30 dpi, we harvested TGs for DNA and RNA analyses using quantitative PCR (qPCR) and qRT-PCR, respectively. The viral genome and LAT intron levels were similar for the three viruses (Fig. 5D), suggesting that the MH2 virus was able to establish latency normally in TGs. Lytic transcripts were only detectable in some ganglia, as reported before (7, 41, 44, 47). The three viruses produced similar levels of ICP0, tk, and gC transcripts, as well as similar percentages of TGs with detectable ICP0, tk, or gC transcripts (Fig. 5E).
Thus, the MH2 mutations did not detectably affect the expression of ICP0 or other lytic genes tested during the establishment of latency (7 dpi) or while latency was maintained (30 dpi).
The MH2 mutations have no significant effect on virulence or reactivation.
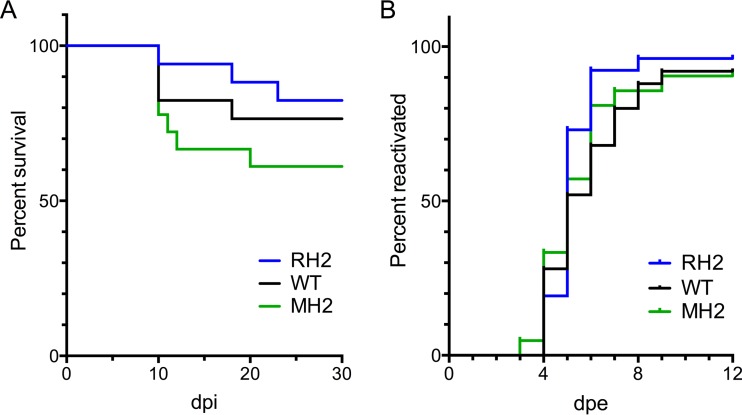
We next assessed the impact of the MH2 mutations on viral virulence and reactivation from latency. We initially examined virulence in the experiment examining transcript levels described above. In that experiment, after infection for 30 days, the mortality rates were 5/20, 4/24, and 4/20 (number of dead mice/total number in group) for WT, MH2, and RH2 viruses, respectively; these rates were not significantly different from each other (P = 0.71 for WT versus MH2, and P = 1 for MH2 versus RH2, Fisher's exact tests). To examine virulence further and assess the role of miR-H2 during reactivation from latency, mice were infected on the cornea at a dose of 1 × 104 PFU/eye. We employed this lower inoculum dose because we thought that it might reveal differences in reactivation more readily. Mice were infected for 30 days before explant of ganglia to induce reactivation. We also assessed the mortality of mice over this time frame (Fig. 6A). Overall, we saw mortalities in the same range as seen with 2 × 105 PFU/eye (see above), which is consistent with the similar amounts of viral replication at 5 dpi with doses ranging from 2 × 104 to 2 × 106 PFU/eye (Fig. 5B). Although by 30 dpi, mice inoculated with MH2 virus showed a roughly twofold-higher level of mortality than mice inoculated with WT and RH2 viruses (Fig. 6A), the differences were not statistically significant. The kinetics of explant reactivation were also similar for the three viruses (Fig. 6B). Thus, the MH2 mutations did not significantly affect virulence or reactivation from latency.
FIG 6.
Viral virulence and reactivation from latency. Mice were corneally inoculated with 1 × 104 PFU/eye of the indicated virus (n = 17 to 18 mice per virus group). The three viruses showed similar back titers of inocula, 1-dpi-eye-swab titers, and 3-dpi-TG titers (not shown). (A) Percentages of mice that died or were euthanized due to severe morbidity over time. Differences were not statistically significant (log-rank test, P > 0.1). (B) At 30 dpi, surviving mice were euthanized and their TGs were explanted onto a monolayer of Vero cells in tissue culture medium for induction of reactivation. Reactivation was assayed by the presence of infectious virus in the medium. Plotted are percentages of cultures (n = 22 to 28 per virus group) producing infectious virus at the indicated days after explantation (dpe). Differences were not statistically significant (log-rank test, P > 0.5).
miR-H2 is less potent than miR-138 as a repressor of ICP0 expression.
Like miR-H2, miR-138 was previously shown to repress ICP0 expression in transfection assays. However, unlike the MH2 mutations, ICP0 mutations that prevent miR-138 repression result in increased lytic gene expression in TGs (41). We therefore compared miR-H2 and miR-138 in an experiment where different amounts of miR-H2 or miR-138 were added during cotransfection with pRS-1 into 293T cells. While both miR-H2 and miR-138 could repress ICP0 mRNA and protein expression in a dose-dependent manner, miR-H2 was about twofold less effective than miR-138 in repressing ICP0 mRNA expression at all doses tested (Fig. 7A). It was also less potent in repressing ICP0 protein expression, as it took ∼4 times as much miR-H2 as miR-138 to obtain the same reduction in expression (Fig. 7B). Additionally when a mixture of miR-138 and miR-H2 with a 1:1 ratio was transfected, the effects were similar to the results of adding miR-138 alone (Fig. 7A and B), suggesting that in the presence of miR-138, additional miR-H2 could not reduce ICP0 expression much further. Next, cells were infected with WT virus after being transfected with miR-H2, miR-138, or a negative-control mimic, followed by analysis of ICP0, ICP27, TK, and gC protein expression by Western blotting. miR-138 decreased the levels of all four proteins more effectively than miR-H2 (Fig. 7C). These results suggest that, even when present at high levels, miR-H2 is not as effective a repressor of ICP0 expression as miR-138 under the conditions tested.
FIG 7.
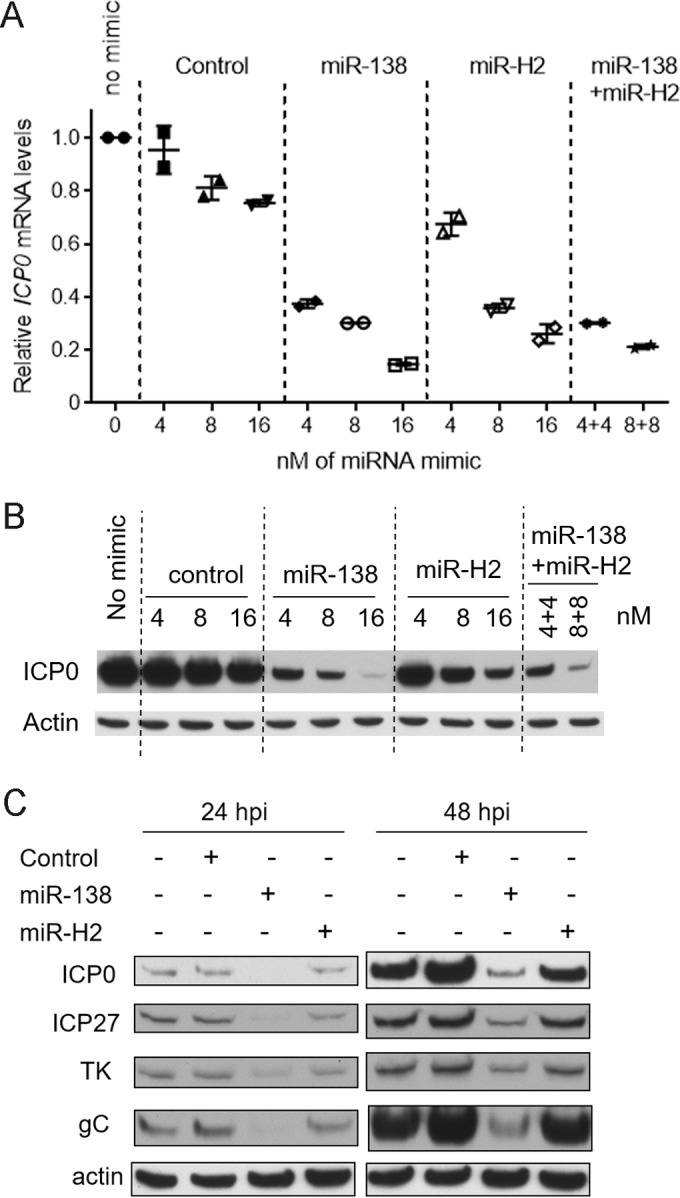
Weak repression by miR-H2. (A) 293T cells were cotransfected with 75 ng of pRS-1 together with the concentrations indicated on the x axis of an irrelevant control mimic or an miR-138 or miR-H2 mimic or both, as indicated above the graph. In the cells analyzed in the last two lanes, miR-138 and miR-H2 were both transfected. At 30 hpt, ICP0 and GAPDH mRNA levels were analyzed by qRT-PCR. Shown are ICP0 mRNA levels normalized by GAPDH mRNA levels, with the bars and error bars representing mean values and standard deviations of duplicates. (B) Transfection was performed as described in the legend to panel A. ICP0 and actin protein levels were analyzed by Western blotting. The mimics used and the concentrations are indicated above the gel images. (C) 293T cells were mock transfected or transfected with 16 nM irrelevant control mimic, miR-138 mimic, or miR-H2 mimic. At 24 hpt, the cells were infected with WT virus for 24 or 48 h at an MOI of 0.1. The cells were then harvested and analyzed for the different proteins listed on the left using Western blotting.
DISCUSSION
Since its discovery (3), miR-H2 has been hypothesized to repress ICP0 expression, thereby repressing lytic gene expression during latent infections. However, this hypothesis had not been tested. Here, we show that miR-H2 can reduce ICP0 mRNA and protein expression in transfected cells. We designed and validated mutations that do not affect ICP0 expression but drastically reduce miR-H2 expression. We find that a mutant virus containing these mutations exhibits WT levels of viral replication and no increase in ICP0 expression in lytically infected cells. In a mouse model, the mutant virus does not differ significantly from the WT in terms of virulence and reactivation. Importantly, the mutant exhibits WT levels of expression of ICP0 and other lytic genes in acutely and latently infected mouse TGs, arguing against the hypothesis that miR-H2 represses lytic gene expression by targeting ICP0 mRNA in vivo. Finally we show that miR-H2 is a relatively weak repressor of ICP0. We discuss each of these results below.
miR-H2 can downregulate ICP0 mRNA, as well as protein, expressed during transfection.
Our results confirm previous findings that HSV-1 miR-H2 can downregulate ICP0 protein levels (3, 37, 38) and additionally show that miR-H2 can reduce ICP0 mRNA levels, contrary to a previous report (3) but consistent with other previous results in infected cells transfected with an HSV-1 miR-H2 mimic (38) or an HSV-2 miR-H2 (miR-III) duplex (40). Different conditions, including different time points taken and different reagents for expressing miRNAs, may have led to the discrepant results. However, our results are more in line with what would be expected, as mammalian miRNAs have been shown to predominantly function to reduce mRNA levels (48).
Problems and solutions related to mutating miRNAs located within mRNA-coding regions.
Altering miRNAs located within mRNA-coding regions by utilizing codon redundancy runs the risk of influencing the expression of the protein independent of effects on the miRNA. In checking whether our MH2 mutations affect ICP0 expression using a transfection assay, we were surprised to find that miR-H2 is expressed by the ICP0-expressing plasmid, which could influence the outcomes of the test. This problem was solved by using a Dicer knockout cell line, which prevented miR-H2 expression and showed that ICP0 expression is indeed unaffected by the MH2 mutations in the absence of miR-H2 expression. A second issue concerns the potential expression of a mutant form of miR-H2, which could target the mutant ICP0 mRNA encoded by the opposite strand. Our analyses confirm that any potential “miR-MH2” expression from MH2 virus is negligible compared to the level of miR-H2 expression from WT BAC virus. Moreover, such “miR-MH2” expression as was observed seems more likely to be due to degradation products of longer transcripts or contamination by longer products rather than authentic miRNA, especially as the MH2 mutations prevent stem-loop formation. Previous miR-H2 mutational studies took similar approaches in mutation design (37–39). The concern about miR-H2-independent mutational effects on ICP0 expression was noted in the study with a miR-H2 mutant virus generated in a strain McKrae background (McK-ΔH2) (37). These authors stated that they did not observe any miR-H2 expression following transfection of their ICP0-expressing plasmid; however, the type and sensitivity of the assay(s) used were not specified. None of the previous studies experimentally addressed the concern of mutant miR-H2 expression (37–39).
miR-H2 does not repress ICP0 expression or viral replication during lytic infection of cells.
We find no evidence that miR-H2 downregulates ICP0 expression during lytic infection, as in lytically infected Vero and Neuro-2A cells, ICP0 expression was not increased by the MH2 mutations. In comparison, mutations altering miR-138 binding sites on the ICP0 mRNA result in increased ICP0 expression during lytic infection of cells containing high levels of miR-138 (41). Weak repression of ICP0 expression (see below) might be one explanation for miR-H2's ineffectiveness. Another explanation comes from our finding that, at least in Vero cells, vastly less miR-H2 is expressed than ICP0 mRNA, making it unlikely that miR-H2 can have a robust impact on ICP0 expression during lytic infection. These results contrast with previous reports that concluded there was increased ICP0 expression in 293 cells infected with the strain 17 miR-H2 mutant virus (17syn+mH2) (38), as well as in RS cells infected with McK-ΔH2 (37). While viral strain and cell line differences might bear on the discrepancies, we note that the previously reported increases in ICP0 expression were either very modest (≤1.3-fold [38]) or not quantified (37). Thus, it seems possible that our results and these previous results may not be so different after all.
Our observation that MH2 exhibits WT levels of virus production was not surprising, given the low abundance of miR-H2 during lytic infection and given that we observed no increase in ICP0 expression. Our findings of WT levels of virus production in Neuro-2A cells did not mirror the curious decreases in viral DNA synthesis exhibited by the 17syn+mH2 viral isolates in these cells (38). More work is needed to understand whether differences in mutations and viral strains are solely responsible for the different results reported.
miR-H2 does not detectably repress lytic gene expression in acutely or latently infected mouse TGs.
In mice, the MH2 mutant and WT viruses expressed ICP0 and other viral transcripts at levels that were indistinguishable at both 7 and 30 dpi. Notably, the results for ICP0 expression are consistent with previous results showing that a LAT deletion mutation, which leads to 2 to 3 orders of magnitude lower miR-H2 levels during latent infection, so that they are indistinguishable from background levels (25), also did not appreciably affect spliced ICP0 transcript levels at 30 dpi (49). We note, however, that the LAT deletion did result in increases in other lytic transcripts (7). Thus, a product of the LAT locus other than miR-H2 is likely to be responsible for the repression of these lytic transcripts.
Our results cannot be readily explained in terms of miR-H2/ICP0 ratios, because miR-H2 is highly expressed at these time points. However, miR-H2's weak repression of ICP0 expression, even when overexpressed in infected 293T cells, might prevent us from detecting a difference between the mutant virus and the WT in comparison to the strong downregulation resulting from miR-138 overexpression. Although a mutant virus with altered miR-138 target sites on the ICP0 mRNA showed significantly increased ICP0 transcript levels at 7 dpi (41; this study), the increases were only about threefold. Therefore, it would not be surprising for a weaker repressor to display no detectable effect.
The miR-H2-deficient mutant virus is similar to the WT for virulence and reactivation rates.
No statistically significant difference was detected between the MH2 mutant virus and the WT in terms of mortality or the rate of reactivation. This is consistent with the lack of any effect on viral gene expression or viral replication in our studies but inconsistent with the significantly increased mortality and rate of reactivation exhibited by the McK-ΔH2 mutant (37). Based on our sample sizes, our studies of virulence and reactivation had 75% and 80% power, respectively, to detect differences as large as those found in the Mck-ΔH2 studies. Other differences between the Mck-ΔH2 studies and ours include mortality being followed for only 10 days in the Mck-ΔH2 studies, differences in statistical methods used to analyze reactivation data, and the Mck-ΔH2 rescue virus not being directly compared to the WT McKrae strain. Regardless, the different results could be due to different viral and mouse strains. Interestingly, a mutant with the Mck-ΔH2 miR-H2 mutations introduced into an LAT deletion background (dLAT-ΔH2), which severely decreases miR-H2 expression during latency (25) but has little or no effect on miR-H2 levels during acute infection or in cell culture (25, 39), exhibited similar phenotypic changes in virulence and an even more apparent effect on reactivation compared to the mutant in the WT McKrae background (39). Regardless, phenotypes exhibited by miR-H2 mutant viruses would be unlikely to be due to derepression of ICP0, as our studies have found no evidence that miR-H2 represses ICP0 expression during infection.
miR-H2 is a weak repressor of ICP0 expression.
Despite the perfect complementarity between miR-H2 and ICP0 mRNA, our current results clearly show that, at least relative to miR-138, miR-H2 is a weak repressor of ICP0. Localization of the miR-H2 target site within the ICP0 coding region might account for its weak repression. Although there is evidence for miRNA binding sites in coding regions (50), in animals, most sites demonstrated to play important roles are located within 3′ UTRs (reviewed in reference 24). This is thought to be related to hindered accessibility of the RISC to miRNA binding sites in coding regions when active translation is occurring (51). In support of this, it has been shown that most miRNA binding sites located in coding regions only marginally reduce mRNA levels (52) and that converting 3′ UTRs to coding regions by modifying stop codons ablates or diminishes translation inhibition (51, 53).
A previous result that might initially seem inconsistent with our findings of low effectiveness of miR-H2 repression of ICP0 expression is that the miR-H2 target site exhibited more reads than the miR-138 target sites in a PAR-CLIP assay of lytically infected cells (41). However, the cross-linking efficiency in the PAR-CLIP assay is affected by the number of 4-thiouracil molecules incorporated into the RNA, and the two miR-138 target sites have only one uracil between them, whereas the miR-H2 target site has five. Also, the outcomes of CLIP analysis do not always correlate with repression strength, as many CLIP-identified miRNA binding sites have been found not to mediate repression (54).
What role can miR-H2 play? Although our results failed to detect a biological function for miR-H2, we note that the miR-H2 seed region (but not the entire sequence) is conserved between HSV-1 and HSV-2 (4, 55) and that miR-H2 is highly expressed during latency (3, 4, 25), suggesting that it may have a role to play, more likely during latency and, perhaps, reactivation than during lytic infection, including acute infection in vivo. We acknowledge that our mouse model does not fully recapitulate HSV-1 infection and disease in humans, so it might not capture important roles for miR-H2. That said, it is possible that as a fine tuner of ICP0 expression, miR-H2 may serve as a backup latency stabilizing factor, and its loss may be inconsequential in the presence of redundant mechanisms or its effects are too small to be detected in the context of highly variable gene expression data. Another possibility is that miR-H2 could act by targeting genes other than ICP0 that affect aspects of latency other than repression of lytic gene expression, such as maintaining neuronal survival. For example, the defects in DNA synthesis reported for the 17syn+mH2 mutant might be due to targets other than ICP0, as suggested previously (38). Future efforts focusing on targets of miR-H2 other than ICP0 and on how products of the LAT locus other than miR-H2 repress lytic gene expression may be fruitful.
MATERIALS AND METHODS
Cells and viruses.
Vero, 293T, and Neuro-2A cells were maintained as previously described (41). NoDice cells (clone number 4-25) (42) were a generous gift of Bryan Cullen (Duke University) and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. Bacterial artificial chromosome (BAC)-derived WT, MH2, and RH2 viruses (described below) were used for all infections, except that the miR-H2 expression time course analysis used HSV-1 strain KOS. Standard protocols were used to prepare and propagate viruses and for infection and plaque assays (56).
Plasmids.
The ICP0-expressing plasmid (pRS-1) was described previously (41). The pRS-1MH2 plasmid was created using a QuikChange site-directed mutagenesis kit (Agilent) and two primers (primer 1, CGCCGTGTGCCCCAGAGTCATAGCAGTTTAGCTCAGGCCGCGAACC, and primer 2, GGTTCGCGGCCTGAGCTAAACTGCTATGACTCTGGGGCACACGGCG; mutated sequences are in boldface), following the manufacturer's protocol.
Transfections.
All transfections were performed in 24-well plates using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer's instructions. For transfection of pRS-1 or pRS-1MH2 alone into 293T or NoDice cells, 150 ng of a plasmid was added per well. For cotransfection, 75 ng of pRS-1 plasmid was transfected into 293T cells in each well together with the indicated concentration of miScript miR-H2, miR-138, or an irrelevant control mimic (from Qiagen). The control mimic has the following sequence: GGCACGUCAGUAGCGGUUAGGUU. In the experiment whose results are shown in Fig. 1B, 20 nM miR-H2 inhibitor or control inhibitor (kindly provided by Nelson Chau of Regulus Therapeutics, Inc., San Diego, CA) was also added. The miR-H2 inhibitor was completely complementary to miR-H2-3p, while the control was a version of an inhibitor of miR-155 with six base mutations. Transfected cells were harvested 24 or 30 h posttransfection (hpt), as indicated in the figure legends, for Western blot and RNA analyses. For the transfection-infection experiment, 293T cells in each well of a 24-well plate were transfected with 16 nM control, miR-H2, or miR-138 mimic. Twenty-four hours later, the cells were infected with WT virus at an MOI of 0.1 for 24 or 48 h before being harvested for Western blot analyses.
Western blot analyses.
Western blotting was performed as described previously (57). The following antibodies and dilutions were used: ICP0 mouse monoclonal antibody (Abcam), 1:5,000; ICP27 mouse monoclonal antibody (Virusys), 1:1,000; TK mouse monoclonal antibody (a generous gift from Bill Summers, Yale University), 1:1,000; gC mouse monoclonal antibody (Fitzgerald), 1:1,000; β-actin monoclonal antibody (Sigma), 1:10,000; and horseradish peroxidase (HRP)-conjugated goat anti-mouse antiserum (SouthernBiotech), 1:2,000.
Quantitative PCR and RT-PCR.
To quantify mRNA in transfected 293T or NoDice cells or in infected Vero cells (MOI of 10), RNA was isolated using an RNeasy minikit (Qiagen). To quantify viral genomes and transcripts in mouse TGs, DNA and RNA were isolated using an AllPrep DNA/RNA minikit (Qiagen). Reverse transcription (RT) was performed using a QuantiTect RT kit (Qiagen) and specific primers described previously (41). The manufacturer's instructions were followed in all these cases. Previously described SYBR green PCR assays were used to quantify viral genomic DNA, the LAT intron, and ICP0, ICP27, ICP4, tk, and gC transcripts, as well as mouse and human GAPDH (encoding glyceraldehyde-3-phosphate dehydrogenase) transcripts (41). Viral transcript levels were normalized to GAPDH transcript levels. For miRNA quantification, total RNA from cells or tissues was purified using an RNeasyPlus minikit (Qiagen), following the protocol to retain small RNAs provided by the manufacturer. miR-H2, “miR-MH2,” and let-7a levels were evaluated by qRT-PCR using the TaqMan microRNA reverse transcription kit and TaqMan gene expression master mix (Applied Biosystems) according to the manufacturer's instructions. The primers and probes used for the miR-H2 and let-7a assays have been described previously (3). The “miR-MH2” assay utilized the same reverse primer as those assays along with the following oligomers: TCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGAGTC (stem-loop primer), TCATAACCTGAGCTAAACTGCT (forward primer), and 6-FAM-TACGACAGAGTCATA-MGB-NFQ (TaqMan probe [6-FAM is 6-carboxyfluorescein]; Applied Biosystems). The viral miRNA levels were normalized to the let-7a levels. For all qRT-PCR assays, RNAs were quantified using standard curves with serially diluted RNA (synthetic mimics for miRNA, in vitro transcripts for mRNA, and total RNA for GAPDH).
Northern blot analyses.
From 293T or NoDice cells transfected with 150 ng of pRS-1 or pRS-1-MH2 plasmid as described above or from 293T cells infected with WT or MH2 viruses at an MOI of 10 for 18 h, RNA less than 200 nucleotides long was isolated using a mirVana miRNA isolation kit (Ambion) according to the manufacturer's instructions. Northern blot analyses of RNA from infected cells were performed as described previously (25). For transfected cells, we used a nonradioactive method, following a protocol (58) for the design of locked-nucleic acid (LNA) probes, denaturing polyacrylamide gel electrophoresis, transfer to a nylon membrane, and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide cross-linking. The LNA probes were synthesized and labeled with digoxigenin (DIG) at both ends by Exiqon. The membrane was prehybridized in 10 ml of ULTRAhyb ultrasensitive hybridization buffer (Ambion) at 60°C for 30 min, hybridized with the labeled probe at 60°C overnight, and then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% SDS for 15 min each and three times with 0.1× SSC plus 0.1% SDS for 15 min each at 60°C. Block, wash, and development buffers for the following steps were provided in a DIG wash and block buffer set (Roche). The membrane was blocked using block buffer for 1 h, incubated with anti-digoxigenin-alkaline phosphatase (AP) antibody (1:15,000, Roche) in block buffer for 30 min, and washed 4 times (10 min each) with wash buffer at room temperature. Images were visualized after applying the alkaline phosphatase substrate CDP-Star (Roche) diluted 100-fold in development buffer and exposed to X-ray films. For multiple hybridizations of the same membranes, membranes were stripped by two incubations (10 min each) at 100°C with 0.1% SDS and 5 mM EDTA before being probed with another digoxigenin-labeled LNA probe. The probes used for Northern blot hybridizations are listed in Table 1.
TABLE 1.
Sequences of Northern blot probes
| Probe targeta | Sequenceb |
|---|---|
| miR-H2 DNA | 5′ AGTCGCACTCGTCCCTGGCTCAGG 3′ |
| “miR-MH2” DNA | 5′ AGAGTCATAGCAGTTTAGCTCAGG 3′ |
| miR-H1 DNA | 5′ TCCACTTCCCGTCCTTCCATC 3′ |
| let-7a DNA | 5′ AACTATACAACCTACTACCTCA 3′ |
| miR-H2 LNA | 5′ DIG-AGTCGC+AC+T+CG+T+CCCTGGCTCAGG-DIG 3′ |
| U6 LNA | 5′ CGAAT+TTG+CGT+GT+CAT+CCTTGC-dig 3′ |
LNA, locked nucleic acid.
DIG, digoxigenin labeling; +N, modified residue in LNA.
Construction of MH2 and RH2 viruses.
A BAC containing WT HSV-1 strain KOS, referred to herein as WT BAC was described previously (57). Using the procedures summarized below, the MH2 mutations (same as in pRS-1MH2) were introduced into WT BAC to generate MH2 BAC. Then, WT sequences were introduced back into the MH2 BAC to generate RH2 BAC. Procedures to introduce mutations into the repeat regions of the viral genome in the BACs have been described elsewhere (41). Briefly, the sequence to be mutated in one copy of ICP0 (copy 1) was replaced with a zeocin resistance (Zeo) cassette and that in the other copy (copy 2) was replaced with a kanamycin resistance (Kan) cassette designed to introduce the mutations in a scarless manner using two-step Red-mediated recombination (59). After copy 2 was mutated, Zeo in copy 1 was replaced with the Kan cassette, permitting the introduction of the same mutations into copy 1. The primers used to amplify and incorporate the Zeo and Kan cassettes and the accompanying MH2 and RH2 sequence changes into the HSV-1 genome are listed in Table 2. The BACs resulting from each step were verified by sequencing using primers spanning the mutation site (forward, GACCCCATAGTGATCAGCGACTCC, and reverse, TTTTCCTGGCCCGACCCGCGCCTCTT), as well as by restriction fragment length polymorphism using restriction endonucleases EcoRI and HindIII. A unique restriction product pattern is seen whenever Zeo or Kan is inserted into the genome, due to the existence of a HindIII restriction site in Kan and an EcoRI restriction site inserted into the 5′ end of Zeo. To generate viruses, the WT, MH2, and RH2 BACs were separately transfected into Vero cells using Lipofectamine 2000 (ThermoFisher) following the manufacturer's instructions. The sequences surrounding the mutation site in the virus stocks were confirmed by sequencing the PCR products generated by the above-described primers.
TABLE 2.
Primers used to construct MH2 and RH2 BACs
| Change | Step | Sequencea |
|
|---|---|---|---|
| Forward | Reverse | ||
| From WT to MH2 | Amplify and incorporate Zeo cassette | GCCGGCCGCCCGCCCCGCGGACGCGCGCCGTGTGCCCCAGAGTCATAGCAGTTTAGCTCAGGAATTCtgttgacaattaatcatcggcat | GCGCCCGGCACAGACTCTGTTCTTGGTTCGCGGCCTGAGCTAAACTGCTATGACTCTGGGGCACACtcagtcctgctcctcggcca |
| Amplify and incorporate Kan cassette | GCCGGCCGCCCGCCCCGCGGACGCGCGCCGTGTGCCCCAGAGTCATAGCAGTTTAGCTCAGGCCGCGAtagggataacagggtaatcgattt | GCGCCCGGCACAGACTCTGTTCTTGGTTCGCGGCCTGAGCTAAACTGCTATGACTCTGGGGCACACGGgccagtgttacaaccaattaacc | |
| From MH2 to RH2 | Amplify and incorporate Zeo cassette | GCCGGCCGCCCGCCCCGCGGACGCGCGCCGTGTGCCCCAGTCGCACTCGTCCCTGGCTCAGGAATTCtgttgacaattaatcatcggcat | GCGCCCGGCACAGACTCTGTTCTTGGTTCGCGGCCTGAGCCAGGGACGAGTGCGACTGGGGCACACtcagtcctgctcctcggcca |
| Amplify and incorporate Kan cassette | CCGGCCGCCCGCCCCGCGGACGCGCGCCGTGTGCCCCAGTCGCACTCGTCCCTGGCTCAGGCCGCGtagggataacagggtaatcgattt | GCGCCCGGCACAGACTCTGTTCTTGGTTCGCGGCCTGAGCCAGGGACGAGTGCGACTGGGGCACACGgccagtgttacaaccaattaacc | |
Lowercase letters are the sequences overlapping the Zeo or Kan gene. Letters in italics are the introduced EcoRI restriction sites. Letters in boldface indicate the mutated regions.
Gel fractionation to isolate miRNA.
We previously isolated mature miRNA using the flashPAGE fractionator and disposable gels (Ambion); however, those products have been discontinued. As we have found that certain stem-loop qRT-PCR assays efficiently detect larger species, such as pre-miRNAs, that contain the miRNA sequences (M. F. Kramer and J. M. Pesola, unpublished results), we modified a published protocol (60) as described below in order to isolate miRNA for the experiment to measure the effects of the MH2 mutations on miRNA expression by qRT-PCR. Samples of total RNA (purified as described above for miRNA quantification) and DynaMarker prestained RNA marker (DiagnoCine) were resolved on 15% Tris-borate-EDTA (TBE)-urea polyacrylamide gels (Invitrogen) in nonadjacent lanes. The portion of each gel lane corresponding to RNA lengths of approximately 18 to 28 bases was excised with a clean scalpel and sliced in half lengthwise. To elute the RNA, gel pieces were sieved through a gel breaker tube (IST Engineering), mixed with 320 μl of 0.3 M NaCl, frozen on dry ice, thawed, and left to shake at room temperature for 7 h. The mixture was spun through a 5-μm filter tube (IST Engineering), and RNA in the filtrate was precipitated in 75% ethanol with 5-μg/ml linear acrylamide (Ambion) as the carrier, washed with 80% ethanol, air dried for 8 min, and resuspended in 12 μl of water.
Animal procedures.
The animal housing and experimental procedures were approved by the Institutional Animal Care and Use Committee of Harvard Medical School in accordance with federal guidelines. Seven-week-old male CD-1 mice (Charles River Laboratories) were anesthetized by intraperitoneal injection of 100 mg/kg of body weight of ketamine (Fort Dodge Animal Health) and 10 mg/kg of xylazine (Lloyd Laboratories). Three microliters of virus, corresponding to one of the doses indicated in the legend to Fig. 5 or Fig. 6, was dropped onto each scarified cornea. Mortality figures include mice that had died and those that were moribund and therefore euthanized. For TG acquisition, mice were sacrificed by cervical dislocation while anesthetized with 3% isoflurane in oxygen with a flow rate of 0.5 ml/min using an anesthesia machine (Colonial Medical Supply). For nucleic acid extraction, TGs were rapidly removed and placed on dry ice before storing at −80°C.
Reactivation assays.
TGs removed from mice at 30 dpi were placed immediately in DMEM supplemented with 10% FBS on ice. After finishing dissection of all TGs, the medium with TGs was transferred onto monolayers of Vero cells in 12-well plates and incubated at 37°C for 12 days. Each day until cytopathic effects in the reactivating wells were obvious, 50-μl aliquots of medium were harvested from each well and stored at −80°C. The presence of infectious virus in the daily aliquots was determined by adding the medium to confluent 24-well plates containing Vero cells and monitoring for cytopathic effects over 4 days.
Statistical analyses.
Statistical analyses were performed using Prism 6 and StatMate 2.0 (GraphPad). The mRNA levels in transfected cells were analyzed by two-tailed paired t test for comparisons of two groups or by one-way analysis of variance (ANOVA) with Bonferroni's multiple-comparison test for comparisons of three groups. The viral titers and levels of DNA and RNA were analyzed by one-way ANOVA with Bonferroni's multiple-comparison test, except that lytic transcript levels in 30-dpi TG samples were analyzed by Kruskal-Wallis test. The percentages of TGs exhibiting detectable lytic transcripts were analyzed by Fisher's exact test. The numbers of mice infected for evaluation of mortality and reactivation rates were chosen to provide 75% to 85% statistical power to detect differences at least as large as those reported in another research group's study (37). Survival curves and reactivation kinetics were analyzed by the log-rank (Mantel-Cox) test.
ACKNOWLEDGMENTS
We thank the following investigators for generous provision of reagents: Bryan Cullen (Duke University) for the NoDice cell line, Bill Summers (Yale University) for anti-TK antibody, and Nelson Chau (Regulus Therapeutics, Inc.) for the miRNA inhibitors.
This work was supported by grants from the National Institutes of Health (grants R21 AI105896 and P01 AI098681) to D.M.C. and startup funds from Zhejiang University to D.P. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
D.P. and D.C. conceived the study and drafted the manuscript; D.P., J.M.P., G.L., and S.M. acquired the data; and all authors analyzed and interpreted data, critically revised the manuscript, approved it, and agreed to be accountable for it.
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex virus, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racianello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–1059. [DOI] [PubMed] [Google Scholar]
- 3.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol 84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol 83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garber DA, Schaffer PA, Knipe DM. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol 71:5885–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen SH, Kramer MF, Schaffer PA, Coen DM. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol 71:5878–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicoll MP, Hann W, Shivkumar M, Harman LE, Connor V, Coleman HM, Proenca JT, Efstathiou S. 2016. The HSV-1 latency-associated transcript functions to repress latent phase lytic gene expression and suppress virus reactivation from latently infected neurons. PLoS Pathog 12:e1005539. doi: 10.1371/journal.ppat.1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 10.Thompson RL, Sawtell NM. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J Virol 75:6660–6675. doi: 10.1128/JVI.75.14.6660-6675.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inman M, Perng GC, Henderson G, Ghiasi H, Nesburn AB, Wechsler SL, Jones C. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J Virol 75:3636–3646. doi: 10.1128/JVI.75.8.3636-3646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed M, Lock M, Miller CG, Fraser NW. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J Virol 76:717–729. doi: 10.1128/JVI.76.2.717-729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branco FJ, Fraser NW. 2005. Herpes simplex virus type 1 latency-associated transcript expression protects trigeminal ganglion neurons from apoptosis. J Virol 79:9019–9025. doi: 10.1128/JVI.79.14.9019-9025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Chentoufi AA, Hsiang C, Carpenter D, Osorio N, BenMohamed L, Fraser NW, Jones C, Wechsler SL. 2011. The herpes simplex virus type 1 latency-associated transcript can protect neuron-derived C1300 and Neuro2A cells from granzyme B-induced apoptosis and CD8 T-cell killing. J Virol 85:2325–2332. doi: 10.1128/JVI.01791-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leib DA, Bogard CL, Kosz-Vnenchak M, Hicks KA, Coen DM, Knipe DM, Schaffer PA. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol 63:2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson RL, Sawtell NM. 2011. The herpes simplex virus type 1 latency associated transcript locus is required for the maintenance of reactivation competent latent infections. J Neurovirol 17:552–558. doi: 10.1007/s13365-011-0071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perng GC, Dunkel EC, Geary PA, Slanina SM, Ghiasi H, Kaiwar R, Nesburn AB, Wechsler SL. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol 68:8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trousdale MD, Steiner I, Spivack JG, Deshmane SL, Brown SM, MacLean AR, Subak-Sharpe JH, Fraser NW. 1991. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J Virol 65:6989–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Block TM, Deshmane S, Masonis J, Maggioncalda J, Valyi-Nagi T, Fraser NW. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192:618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- 20.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giordani NV, Neumann DM, Kwiatkowski DL, Bhattacharjee PS, McAnany PK, Hill JM, Bloom DC. 2008. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J Virol 82:6056–6060. doi: 10.1128/JVI.02661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava R, Dervillez X, Khan AA, Chentoufi AA, Chilukuri S, Shukr N, Fazli Y, Ong NN, Afifi RE, Osorio N, Geertsema R, Nesburn AB, Wechsler SL, BenMohamed L. 2016. The herpes simplex virus latency-associated transcript gene is associated with a broader repertoire of virus-specific exhausted CD8+ T cells retained within the trigeminal ganglia of latently infected HLA transgenic rabbits. J Virol 90:3913–3928. doi: 10.1128/JVI.02450-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer MF, Jurak I, Pesola JM, Boissel S, Knipe DM, Coen DM. 2011. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology 417:239–247. doi: 10.1016/j.virol.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurak I, Hackenberg M, Kim JY, Pesola JM, Everett RD, Preston CM, Wilson AC, Coen DM. 2014. Expression of herpes simplex virus 1 microRNAs in cell culture models of quiescent and latent infection. J Virol 88:2337–2339. doi: 10.1128/JVI.03486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanfranca MP, Mostafa HH, Davido DJ. 2014. HSV-1 ICP0: an E3 ubiquitin ligase that counteracts host intrinsic and innate immunity. Cells 3:438–454. doi: 10.3390/cells3020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukashchuk V, Everett RD. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol 84:4026–4040. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roizman B, Zhou G, Du T. 2011. Checkpoints in productive and latent infections with herpes simplex virus 1: conceptualization of the issues. J Neurovirol 17:512–517. doi: 10.1007/s13365-011-0058-x. [DOI] [PubMed] [Google Scholar]
- 30.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurak I, Silverstein LB, Sharma M, Coen DM. 2012. Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J Virol 86:10093–10102. doi: 10.1128/JVI.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai W, Astor TL, Liptak LM, Cho C, Coen DM, Schaffer PA. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol 67:7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai W, Schaffer PA. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol 66:2904–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Silverstein S. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J Virol 66:2916–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halford WP, Schaffer PA. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J Virol 75:3240–3249. doi: 10.1128/JVI.75.7.3240-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson RL, Sawtell NM. 2006. Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J Virol 80:10919–10930. doi: 10.1128/JVI.01253-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang X, Brown D, Osorio N, Hsiang C, Li L, Chan L, BenMohamed L, Wechsler SL. 2015. A herpes simplex virus type 1 mutant disrupted for microRNA H2 with increased neurovirulence and rate of reactivation. J Neurovirol 21:199–209. doi: 10.1007/s13365-015-0319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flores O, Nakayama S, Whisnant AW, Javanbakht H, Cullen BR, Bloom DC. 2013. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol 87:6589–6603. doi: 10.1128/JVI.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang X, Brown D, Osorio N, Hsiang C, BenMohamed L, Wechsler SL. 2016. Increased neurovirulence and reactivation of the herpes simplex virus type 1 latency-associated transcript (LAT)-negative mutant dLAT2903 with a disrupted LAT miR-H2. J Neurovirol 22:38–49. doi: 10.1007/s13365-015-0362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang S, Patel A, Krause PR. 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol 83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. 2014. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe 15:446–456. doi: 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. 2014. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leib DA, Coen DM, Bogard CL, Hicks KA, Yager DR, Knipe DM, Tyler KL, Schaffer PA. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol 63:759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramer MF, Chen SH, Knipe DM, Coen DM. 1998. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J Virol 72:1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du T, Zhou G, Roizman B. 2013. Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc Natl Acad Sci U S A 110:E2621–E2628. doi: 10.1073/pnas.1309906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du T, Zhou G, Roizman B. 2011. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci U S A 108:18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raja P, Lee JS, Pan D, Pesola JM, Coen DM, Knipe DM. 2016. A herpesviral lytic protein regulates the structure of latent viral chromatin. mBio 7:e00633-16. doi: 10.1128/mBio.00633-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen SH, Lee LY, Garber DA, Schaffer PA, Knipe DM, Coen DM. 2002. Neither LAT nor open reading frame P mutations increase expression of spliced or intron-containing ICP0 transcripts in mouse ganglia latently infected with herpes simplex virus. J Virol 76:4764–4772. doi: 10.1128/JVI.76.10.4764-4772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnall-Levin M, Zhao Y, Perrimon N, Berger B. 2010. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc Natl Acad Sci U S A 107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. 2009. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol 16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin HR, Ganem D. 2011. Viral microRNA target allows insight into the role of translation in governing microRNA target accessibility. Proc Natl Acad Sci U S A 108:5148–5153. doi: 10.1073/pnas.1102033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umbach JL, Cullen BR. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coen DM, Fleming HE Jr, Leslie LK, Retondo MJ. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J Virol 53:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan D, Coen DM. 2012. Quantification and analysis of thymidine kinase expression from acyclovir-resistant G-string insertion and deletion mutants in herpes simplex virus-infected cells. J Virol 86:4518–4526. doi: 10.1128/JVI.06995-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu TT, Li Z, John B. 2014. Enhanced detection of small RNAs using a nonradioactive approach. Methods Mol Biol 1173:123–133. doi: 10.1007/978-1-4939-0931-5_11. [DOI] [PubMed] [Google Scholar]
- 59.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 60.Pfeffer S, Lagos-Quintana M, Tuschl T. 2005. Cloning of small RNA molecules. Curr Protoc Mol Biol Chapter 26:Unit 26.4. doi: 10.1002/0471142727.mb2604s72. [DOI] [PubMed] [Google Scholar]