ABSTRACT
Uncoating of a virus particle to expose its nucleic acid is a critical aspect of the viral multiplication cycle, as it is essential for the establishment of infection. In the present study, we investigated the role of plant HSP70 homologs in the uncoating process of Cucumber necrosis virus (CNV), a nonenveloped positive-sense single-stranded RNA [(+)ssRNA] virus having a T=3 icosahedral capsid. We have found through Western blot analysis and mass spectrometry that the HSP70 homolog Hsc70-2 copurifies with CNV particles. Virus overlay and immunogold labeling assays suggest that Hsc70-2 is physically bound to virions. Furthermore, trypsin digestion profiles suggest that the bound Hsc70-2 is partially protected by the virus, indicating an intimate association with particles. In investigating a possible role of Hsc70-2 in particle disassembly, we showed that particles incubated with Hsp70/Hsc70 antibody produce fewer local lesions than those incubated with prebleed control antibody on Chenopodium quinoa. In conjunction, CNV virions purified using CsCl and having undetectable amounts of Hsc70-2 produce fewer local lesions. We also have found that plants with elevated levels of HSP70/Hsc70 produce higher numbers of local lesions following CNV inoculation. Finally, incubation of recombinant Nicotiana benthamiana Hsc70-2 with virus particles in vitro leads to conformational changes or partial disassembly of capsids as determined by transmission electron microscopy, and particles are more sensitive to chymotrypsin digestion. This is the first report suggesting that a cellular Hsc70 chaperone is involved in disassembly of a plant virus.
IMPORTANCE Virus particles must disassemble and release their nucleic acid in order to establish infection in a cell. Despite the importance of disassembly in the ability of a virus to infect its host, little is known about this process, especially in the case of nonenveloped spherical RNA viruses. Previous work has shown that host HSP70 homologs play multiple roles in the CNV infection cycle. We therefore examined the potential role of these cellular components in the CNV disassembly process. We show that the HSP70 family member Hsc70-2 is physically associated with CNV virions and that HSP70 antibody reduces the ability of CNV to establish infection. Statistically significantly fewer lesions are produced when virions having undetectable HSc70-2 are used as an inoculum. Finally incubation of Hsc70-2 with CNV particles results in conformational changes in particles. Taken together, our data point to an important role of the host factor Hsc70-2 in CNV disassembly.
KEYWORDS: Hsp70, heat shock 70-kDa protein, Hsc70, heat shock cognate 70-kDa protein, Cucumber necrosis virus, coat protein, Tombusvirus, virus uncoating, virus disassembly, Tombusviridae
INTRODUCTION
Nonenveloped viruses are made up of a nucleic acid genome and a protein coat that plays an essential role in protection of the genome from nuclease attack in host cells. The nucleic acid is enclosed in the capsid, which is composed of repeating virus-encoded polypeptide units stabilized by divalent cations, cementing proteins, or disulfide bonds (1). The stable proteinaceous coat must be at least partially shed in order to release the encapsidated genome into the host cell to initiate an infection. The metastability associated with viruses allows structural rearrangements within capsids (1, 2) and is also important to prevent premature disassembly of capsids, which will not only expose the genome to host nucleases but will also compromise the targeting of the genome to its site of replication. Enveloped viruses mostly enter and uncoat by membrane fusion followed by receptor-mediated endocytosis (3). On the other hand, nonenveloped viruses mainly disassemble by initiating a conformational change in their capsids either by using a receptor or mechanical or chemical cues or via cotranslational disassembly (1, 4–6). Structural transformations of viral capsids are also triggered through interaction with chaperones such as members of the heat shock protein 70 (HSP70) family, including the constitutively expressed isoform Hsc70 (heat shock cognate 70-kDa protein) and the stress-inducible isoform Hsp70 (heat shock 70-kDa protein) (4, 7). Although constitutively expressed, Hsc70 can also be induced under specific conditions (8, 9) and can synergistically complement the functions of Hsp70 to preserve cellular integrity during metabolic challenges (10). Hsc70 is one of the most abundant cytosolic HSP70 isoforms and has many distinct functions, both in preventing or reversing protein aggregation and in disassembling protein complexes (7, 11). Hsc70s have been found to play an important role in the disassembly of large macromolecular complexes (7, 11, 12) such as the DNA-replication origin complexes (13) and clathrin-coated vesicles (14–17). Recent studies have revealed that Hsc70 functions by trapping the conformational fluctuations in the clathrin lattice, causing local strains to accumulate (17). This destabilizes the lattice, causing it to disintegrate in the cytoplasm. Due to the ability of Hsc70 homologs to disaggregate stable oligomeric complexes (18), viruses have evolved to recruit these cellular disassembling machines to uncoat their capsids for the establishment of infection. Hsc70 homologs have been found to promote the uncoating processes of several animal viruses, such as polyomaviruses, papillomaviruses, reoviruses, adenoviruses, nodaviruses, rotaviruses, and Nervous necrosis virus (19–25). However, there are no reports regarding the involvement of cellular Hsc70 in the disassembly of plant viruses, although the virally encoded HSP70 homolog of Beet yellows virus has been hypothesized to play a role in disassembly of the helical capsid (26). We wished to determine whether Nicotiana benthamiana HSP70 family homologs play a role in the disassembly of the plant virus Cucumber necrosis virus (CNV). This would extend our previous observation that HSP70 homologs play multiple roles in the CNV multiplication cycle (27).
CNV is a nonenveloped positive-sense single-stranded RNA [(+)ssRNA] virus that belongs to the Tombusvirus genus in the Tombusviridae family (28). The CNV capsid is a T=3 icosahedron having a diameter of 34 nm and consists of 180 identical copies of a 41-kDa coat protein (CP), that encapsidates a 4.7-kb monopartite RNA molecule. Each of the CP subunits folds into three major domains: an RNA binding domain (R), the shell domain (S), and the protruding domain (P). The P and the S domains are connected by a short hinge (h) region while the arm (a) region flexibly tethers the R and S domains, both allowing for T=3 icosahedral symmetry and quasiequivalent subunit interactions (29, 30). To meet the requirements of quasiequivalence, the CP adopts three different conformations (A, B, and C). The arms are disordered in the A and B subunits, while C subunits are characterized by ordered arms that extend to form the β-annulus at the particle 3-fold axis (29, 30). In analogy to Tomato bushy stunt virus (TBSV), Ca2+ ions stabilize the interactions between adjacent A, B, and C subunits in the shell domain by bringing together aspartate residues at the particle quasi-3-fold axis (Q3) (31, 32). The isoelectric point of the CNV virion is pH 5.2; hence, the particles are stable at acidic pH. However, particles expand above pH 7, with this being enhanced in the presence of a Ca2+ chelator (32, 33). The internally located R domain and arm region externalize during particle expansion, and a hole is created at the particle Q3 axis (32), which is believed to allow for exit of virion RNA (34, 35). The expanded state of CNV as well as that of other members of the Tombusviridae is believed to represent an uncoating intermediate (30, 34, 36).
CNV particles are transmitted in nature through the recognition of specific amino acids in the shell and the protruding domains of the CP by glycoprotein receptors present on the surface of zoospores of the fungus Olpidium bornovanus (37, 38). Receptor binding initiates a conformational change that is essential for vector transmission (33, 38–40). CNV particles have been found to interact with Hsp70 of fungal zoospores as determined by Western blotting and virus overlay assays (unpublished observations). It is possible that the Hsp70 associated with the vector zoospores is responsible for bringing about the conformational change required for cellular entry of CNV particles, as has been reported for rotavirus entry into cells (21). This, and our recent finding that HSP70 homologs play multiple roles in the CNV infection cycle (27), raised the possibility of the involvement of plant Hsp70 or Hsc70 homologs in the disassembly of CNV. Here we report several observations that suggest that CNV particles co-opt Hsc70-2 and/or Hsp70 for uncoating. CNV particle preparations were found to contain Hsc70, which is associated with virions as determined by its comigration with CNV particles in an agarose gel and by immunogold labeling of CNV particles using an HSP70 antibody. The bound Hsc70-2 is intimately associated with CNV particles, as it is relatively resistant to trypsin digestion. Heat-shocked plants that express HSP70 at a high level or plants overexpressing Hsc70-2 produce a significantly higher number of local lesions when inoculated with CNV particles than do untreated plants. This is not observed when such treated plants are inoculated with viral RNA, suggesting that the increased numbers of local lesions are due to increased viral disassembly rather than an increase in the efficiency of some other aspect of the initiation of infection. In addition, CNV particles incubated with Hsc70 partially disassemble or are conformationally altered and are more sensitive to chymotrypsin digestion. Taken together, our data strongly support that CNV co-opts cellular Hsc70 for disassembly.
RESULTS
Hsc70-2 is present in CNV particle preparations.
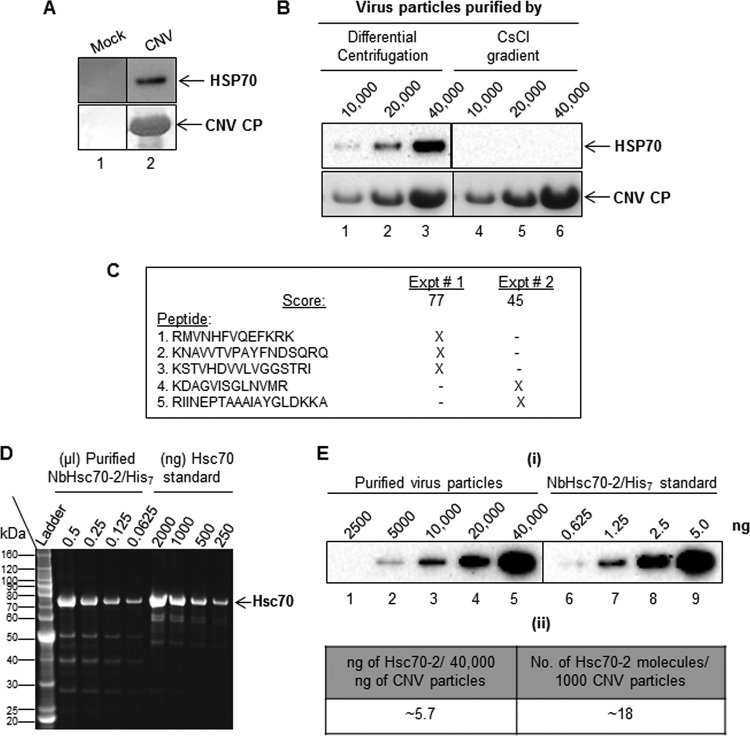
We have previously found that HSP70 homologs play multiple roles in the CNV infection cycle and that Hsc70-2 coimmunoprecipitates with CNV CP (27). This prompted us to determine whether Hsc70-2 is present in CNV particle preparations. To do this, we purified CNV particles from equal masses of mock- or CNV-inoculated N. benthamiana leaf material using polyethylene glycol (PEG) precipitation followed by differential centrifugation (see Materials and Methods). After resuspending the pellets in equal volumes, we conducted Western blot analyses using equal volumes of purified material and a monoclonal antibody that detects both Hsp70 and Hsc70 (HSP70 antibody). Figure 1A shows that HSP70 can be readily detected in virion preparations extracted from CNV-infected plants (lane 2) when a large amount (40 μg) of denatured CNV particles is loaded onto the gel. However, no HSP70 signal was detected when an equal volume of material obtained from a mock virion preparation (lane 1) was loaded on the gel, suggesting that HSP70 copurifies with CNV virions.
FIG 1.
Hsc70-2 is present in CNV virion preparations. (A) Western blot analysis of CNV virion preparations. Virion extraction was from equal masses of mock (lane 1)- and CNV (lane 2)-inoculated leaves. Pellets were resuspended in an equal volume of 10 mM sodium acetate (pH 5.8), and equal volumes of material (40,000 ng of CNV) were loaded onto a denaturing NuPAGE gel, followed by blotting to PVDF membranes. The upper panel was probed with an antibody that reacts to both Hsp70 and Hsc70 (here referred to as HSP70 antibody), and the lower panel was stained with Ponceau S to visualize CNV CP. (B) CNV virions (10,000, 20,000 and 40,000 ng) extracted by either differential centrifugation (lanes 1 to 3) or CsCl gradient centrifugation (lanes 4 to 6) were subjected to SDS-PAGE followed by Western blotting analyses. The upper panel shows a Western blot probed with an HSP70 antibody, and the lower panel shows the blot stained with Ponceau S to visualize CNV CP. (C) Summary of Hsc70-2 peptides detected by mass spectrometric analysis of proteins present in two independently purified CNV virion preparations (experiments [Expt] 1 and 2). The score shown for each experiment is based on a MASCOT search. X indicates the presence of the peptide in the mass spectrometric analysis. A BLAST analysis of the 5 detected peptides in the taxid Nicotianoideae identified 3 proteins that showed 100% identity to all five peptides (accession numbers AAP04522, AAR17080 and XP_009620324.1). All three proteins were most similar to Hsc70-2-like proteins as determined by BLAST analysis. The nucleotide sequences of these genes are most similar (95 to 96%) to that of N. benthamiana Hsc70-2 (Nbv5tr6412958; University of Sydney, Australia [http://benthgenome.qut.edu.au/]). These peptides were not detected in mock virus preparations extracted in a similar manner from the same amount of leaf tissue used to extract virions. (D) Denaturing gel electrophoresis of several dilutions (shown as microliters) of recombinant N. benthamiana Hsc70-2/His7 protein purified from bacterial cells using a Talon metal affinity matrix (Clontech). Bovine Hsc70 was used as the mass and size standard (shown in nanograms). The first lane shows a protein molecular mass standard (ladder) in kilodaltons. (E) Estimate of amounts of Hsc70-2 present in CNV virion preparations. Panel I, Western blot of CNV virion preparations probed with an HSP70 antibody. CNV particles (2,500, 5000, 10,000, 20,000, and 40,000 ng, as indicated) were electrophoresed under denaturing conditions and blotted to PVDF membranes prior to incubating with antibody. Dilutions of bacterially expressed N. benthamiana Hsc70-2/His7 (0.625, 1.25, 2.5, and 5.0 ng) probed with HSP70 antibody were used to estimate the amounts of Hsc70-2 present in CNV virion preparations. Panel ii, summary of amounts of Hsc70-2 present in virion preparations as determined by densitometry using Image Lab software version 5.1 (Bio-Rad). In an independent experiment, we found approximately 3.5 ng of Hsc70-2/His7 per 40 μg of virus (i.e., approximately 11 molecules of Hsc70-2 in 1,000 CNV particles).
HSP70 was also detected in virion preparations extracted from CNV-infected plants using a miniprep procedure (41) or following sucrose density gradient purification (reference 42 and unpublished observations). However, when virus purification from CNV-infected plants was performed under stringent conditions using cesium chloride (CsCl) gradients (29), HSP70 was not detectable. Figure 1B shows that when equal amount of virions extracted by either differential centrifugation (lanes 1 to 3) or CsCl gradients (lanes 4 to 6) were subjected to SDS-PAGE followed by Western blotting analyses, HSP70 was not detectable in the latter, suggesting that HSP70 may not be firmly attached to CNV virions. This is consistent with our observation that levels of HSP70 in CNV preparations decline after long-term storage of the virus (unpublished observations), possibly due to release from virions and/or degradation.
To determine which isoform of the HSP70 protein family is present in purified CNV virion preparations, mass spectrometry (MS) was conducted in 2 independent experiments. In each experiment, equal volumes of material obtained from equal masses of mock- and CNV-inoculated leaf material were subjected to CNV purification procedures. In total, 5 Hsc70 peptides were specifically identified in CNV virion preparations, each showing 100% identity to the deduced amino acid sequence of N. benthamiana Hsc70-2 (Gene ID Nbv5tr6412958 in the N. benthamiana database available through the University of Sydney, Australia [http://benthgenome.qut.edu.au/]) (Fig. 1C). These peptides were not detected in preparations purified in a similar manner from equal masses of mock-inoculated leaves, which indicates that Hsc70-2 copurifies with CNV virions possibly due to an interaction between the virion and Hsc70-2.
To quantify the amount of Hsc70-2 present in CNV preparations, we purified bacterially expressed recombinant N. benthamiana Hsc70-2 containing 7 C-terminal histidine (His) residues (NbHsc70-2/His7) (Fig. 1D) as described previously (27). Increasing masses of CNV particles along with several different dilutions of recombinant NbHsc70-2/His7 were electrophoresed through an SDS-polyacrylamide gel, and then Western blot analysis was conducted using an HSP70 antibody (which detects both Hsp70 and Hsc70) (Fig. 1E). From these experiments, we estimate that approximately 5.7 ng of N. benthamiana Hsc70-2 is present in 40 μg of CNV, which corresponds to approximately 18 molecules of Hsc70-2 per 1,000 CNV particles. In an independent experiment, a value of approximately 11 molecules of Hsc70-2 per 1,000 virions was calculated. Taken together, the results strongly suggest that N. benthamiana Hsc70-2 is present in CNV virion preparations.
We also assessed whether another member of the Tombusviridae family, the Aureusvirus Cucumber leaf spot virus (CLSV) is associated with HSP70 by using differential centrifugation as described above. Figure 2 shows that CLSV is associated with HSP70 (panel i) and that approximately 4.5 ng per 40,000 ng of CLSV is present (panel ii), which is similar to that observed with CNV. This observation shows that another virus in the Tombusviridae family is associated with HSP70 homologs, reinforcing the notion that this association has biological significance.
FIG 2.
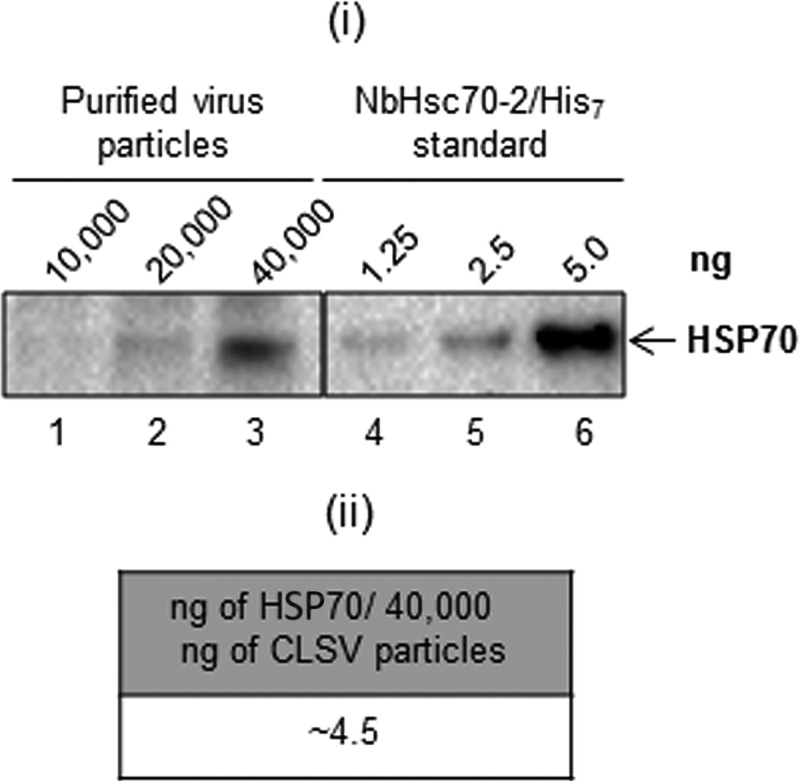
HSP70 copurifies with CLSV. (i) Western blot of CLSV virion preparations (extracted by differential centrifugation as described for CNV) probed with an HSP70 antibody. CLSV particles (10,000, 20,000, and 40,000 ng, as indicated) were electrophoresed under denaturing conditions and blotted to PVDF membranes prior to incubating with antibody. Dilutions of N. benthamiana Hsc70-2/His7 (1.25, 2.5, and 5.0 ng) probed with HSP70 antibody were used to estimate the amount of HSP70 present in CLSV virion preparations. (ii) Summary of amounts of HSP70 present in virion preparations as determined by densitometry using Image Lab software version 5.1 (Bio-Rad).
CNV particles bind Hsc70 in vitro.
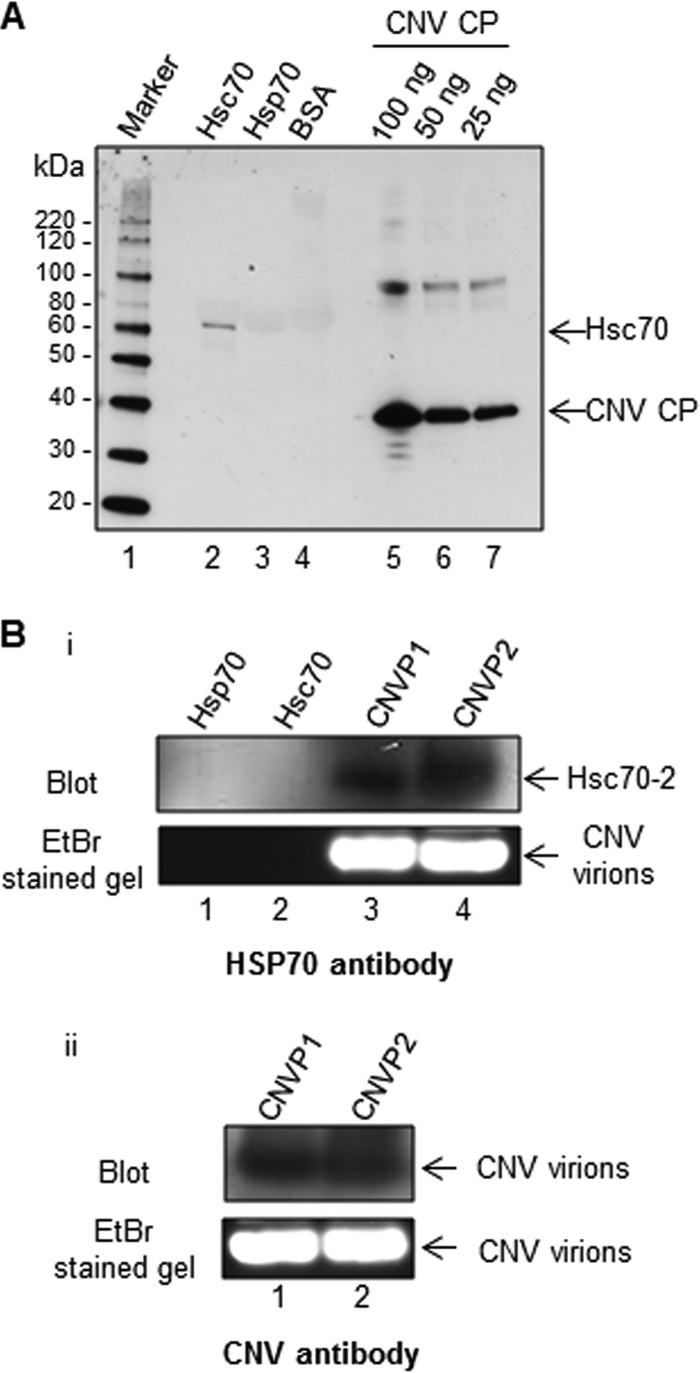
To determine if CNV particles can bind HSP70 homologs in vitro, we performed a virus overlay assay as described previously (38). Ten micrograms of recombinant Hsc70 (bovine), Hsp70 (human), and bovine serum albumin (BSA) as a negative control along with CNV CP controls was subjected to SDS-PAGE and blotted onto a nitrocellulose membrane. The proteins were allowed to renature in situ overnight. The membrane was then incubated with CNV particles for 3 h, washed, and then probed with a CNV CP-specific antibody. As shown in Fig. 3A, the antibody bound to protein in the lane loaded with Hsc70 but not to that in the lanes containing Hsp70 or BSA, suggesting that CNV virions can bind Hsc70 in vitro. This reinforces our finding that Hsc70 can interact with and therefore copurify with CNV during virus extraction.
FIG 3.
Purified CNV particles bind to Hsc70 in vitro and in vivo. (A) Virus overlay assay showing that CNV particles bind to Hsc70 in vitro. Equal masses (10 μg) of bovine Hsc70, human Hsp70, and BSA as a negative control along with CNV CP standards (100, 50, and 25 ng, as indicated) were subjected to SDS-PAGE. The proteins were blotted to a nitrocellulose membrane and allowed to renature overnight. The membrane was then incubated with CNV particles, washed, and then probed with a CNV CP-specific antibody. The first lane shows a protein molecular size marker with sizes of protein bands indicated to the left of the blot. (B) HSP70 antibody binds to CNV particles subjected to agarose gel electrophoresis, suggesting a physical interaction between Hsc70 and CNV particles in vivo. Twenty micrograms of two independently purified CNV particle preparations (CNVP1 and CNVP2) were electrophoresed through 1% agarose gel, blotted, and probed with either an HSP70 antibody (panel i) or a CNV antibody made to bacterially expressed S and P domains of the CP as a positive control (panel ii. The upper panel shows the blots, and the lower panel shows the ethidium bromide (EtBr)-stained gels. In panel I, 100 ng of human Hsp70 and bovine Hsc70 was also included in the gel to rule out the possibility that these proteins might comigrate with CNV particles and thus artifactually indicate that CNV particles are bound to Hsc70.
Hsc70-2 is bound to virions in purified CNV preparations.
To determine if Hsc70-2 is bound to CNV virions in purified particle preparations, 20 μg of CNV particles from two independent purifications was electrophoresed through 1% (wt/vol) agarose gels. Recombinant Hsp70 and Hsc70 were also loaded on the gel. Proteins were blotted to polyvinylidene difluoride (PVDF) membranes, followed by Western blotting analyses using an HSP70 (Fig. 3B, panel i) or CNV (Fig. 3B, panel ii) polyclonal antibody. It can be seen that a signal was obtained using the HSP70 antibody and that this signal is present at a position on the blot that is similar to that of CNV particles probed with a CNV polyclonal antibody (compare blots in Fig. 3B, panel i, lanes 3 and 4, with those in Fig. 3B, panel ii, lanes 1 and 2). Such binding was not apparent at a similar position in the lanes containing purified Hsp70 and Hsc70 (Fig. 3B, panel i, lanes 1 and 2, respectively), showing that the signal to CNV particles is not due to fortuitous comigration of CNV particles with free HSP70 homologs. These results indicate that Hsc70-2 detected by mass spectrometry in CNV preparations is bound to virions.
Hsc70-2 is associated with CNV virions as determined by immunogold labeling.
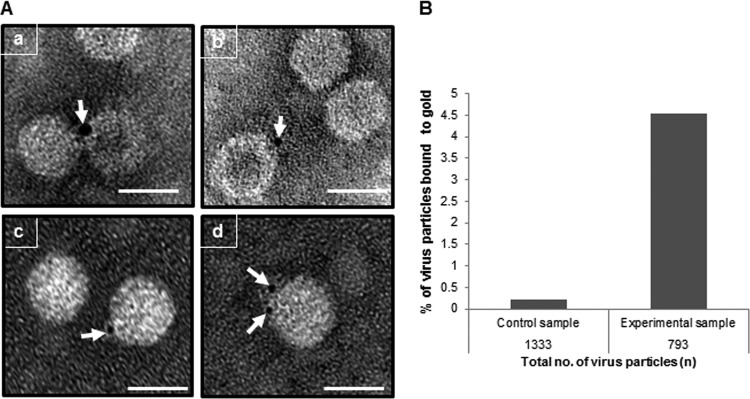
The results shown so far provide evidence that N. benthamiana Hsc70-2 is bound to CNV virions. To more directly assess this, we performed immunogold labeling experiments. One microgram of CNV particles was adsorbed onto Formvar-carbon-coated nickel grids, followed by incubation with HSP70 antibody. After washing, the grids were incubated with a goat anti-mouse secondary antibody conjugated to 4-nm colloidal gold particles to label Hsc70-2 molecules present in CNV virion preparations. After gold labeling, the virus particles were negatively stained and analyzed by transmission electron microscopy (TEM). A control experiment was performed in a similar manner except that no primary antibody was used. Figure 4A shows that CNV virions are labeled with gold, as indicated by the arrows. From three independent experiments it was found that, when labeled, the majority of the virions are labeled with 1 (∼53%) or 2 (∼19%) gold particles (representative images are shown in Fig. 4A). The remaining virus particles were labeled with 3 (∼8.8%), 4 (∼5.8%), 5 (∼7.3%), or >5 (∼5.8%) gold particles (data not shown). These data indicate that Hsc70-2 is bound to CNV particles and that some particles are bound to more than 1 Hsc70-2 molecule.
FIG 4.
Immunogold labeling experiment showing that purified CNV particles are bound to Hsc70. (A) One microgram of purified CNV particles was adsorbed onto Formvar-carbon-coated nickel grids, followed by incubation with 1% BSA–1× PBS as blocking agent. The grids were then incubated with HSP70 antibody at 4°C overnight. After incubation, the grids were washed and treated for 1 h at RT with a goat anti-mouse antibody conjugated to 4-nm gold particles in 1% BSA–1× PBS. After washing, virus particles were negatively stained with 2% uranyl acetate. A negative-control experiment was performed in a similar fashion except the particles were incubated with buffer instead of the primary antibody. The grids were analyzed by TEM at 100 kV, and the images were taken at a magnification of ×80,000. Panels a, b, and c, virus particles bound to 1 gold particle, as shown by the arrow; panel d, a virus particle bound to 2 gold particles. The scale bar represents 34 nm. The experiment was repeated 3 times. (B) A graphical representation of the percentage of virus particles bound to gold in the control (without primary antibody) and experimental (with primary antibody) samples. Immunogold labeling was conducted as described for panel A, and 100 randomly selected fields were photographed for subsequent analysis. The total numbers of virus particles were counted in the control (1,333) and experimental (793) samples along with the numbers of particles bound to gold.
To assess whether binding to CNV particles is specific, 100 random TEM images were taken of both the experimental samples and the control (with no primary antibody) prepared as described above. The total numbers of virions, as well as the number of labeled virions, were counted, and the percentage of virions bound to gold was determined and is shown graphically in Fig. 4B. As can be seen, approximately 20 times more particles are gold labeled in the experimental sample compared to the control sample. These observations suggest that in the experimental treatment, gold-labeled antibody is bound predominantly to CNV particles containing Hsc70-2 rather than being randomly distributed on the grid. Moreover, the bound Hsc70-2 is accessible to antibody, suggesting that the Hsc70-2 epitope is on the particle surface rather than being present within virions. It is also noted that about 4.5% of CNV particles were bound to gold, which extrapolates to approximately 70 particles of gold per 1,000 CNV particles given that many of the labeled virions contain more than 1 bound antibody (see above) (Fig. 4). This ratio of Hsc70-2 particles to CNV particles is slightly higher than but similar to that observed using Western blot analyses, which suggested that approximately 11 to 18 molecules of Hsc70-2 are present in 1,000 CNV particles (Fig. 1E). Taken together, the presented data strongly support that a subset of CNV particles are bound to Hsc70-2, with approximately 1.1 to 7% of the particles being bound as determined by Western blotting and immunogold labeling.
The Hsc70-2 bound to CNV in purified virion preparations is partially protected from trypsin digestion.
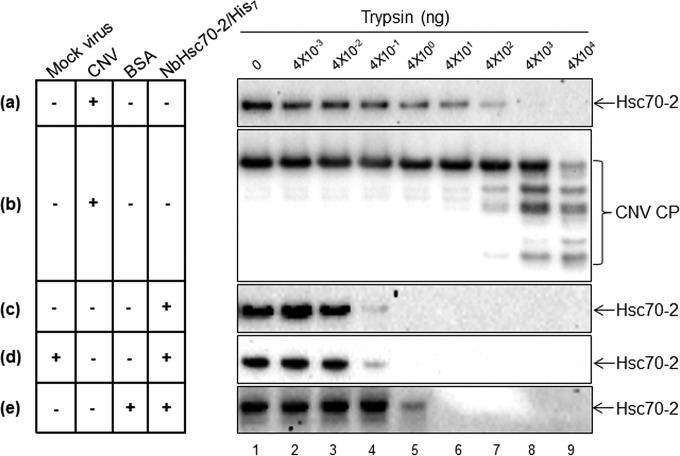
Previous work from other labs has shown that HSP70 homologs can be incorporated into the interior of lentivirus virions (43). However, our immunogold labeling results indicate that Hsc70-2 is at least partially present on the surface of CNV particles. To further assess whether the bound Hsc70-2 is exposed or partially exposed on the exterior of CNV virions, we performed a trypsin digestion assay. CNV virions (40 μg) were treated with increasing amounts of trypsin, and the presence of Hsc70-2 was assessed by Western blotting analyses (Fig. 5). HSP70 antibody was used in the blot in Fig. 5a, and CNV CP antibodies SP and RAD were utilized for Fig. 5b. It can be seen that Hsc70-2 is sensitive to trypsin, since levels decline noticeably when 4 ng of trypsin is used (Fig. 5a, lane 5) and no Hsc70-2 is apparent when approximately 40,000 ng of trypsin is used (Fig. 5a, lane 8). This indicates that Hsc70-2 is at least partially exposed on the particle surface rather than being present entirely within virions, consistent with the results of immunogold labeling experiments. Also, while levels of Hsc70-2 start to degrade partially at 4 ng of trypsin, the CP in virions is still intact (Fig. 5b, lane 5). However, Hsc70-2 is efficiently degraded only when the virions start to degrade (Fig. 5a and b, compare lanes 7 to 9), suggesting that although Hsc70-2 is exposed on the exterior of CNV, it is partially protected by virions. The conclusion that CNV virions partially protect Hsc70-2 from trypsin degradation is reinforced by the observation that free bacterially expressed NbHsc70-2/His7 is susceptible to digestion with significantly smaller amounts of trypsin. For example, free NbHsc70-2/His7 is completely digested using 4 ng of trypsin (Fig. 5c, lane 5), whereas Hsc70-2 present in CNV preparations requires approximately 40,000 ng of trypsin for complete digestion (Fig. 5a, lane 9). However, it is possible that the decreased sensitivity of CNV-bound Hsc70-2 to trypsin could simply be due to the presence of a large amount of CNV particles in the digestion mixture For Fig. 5a, which could competitively protect the Hsc70-2 from degradation. To rule out this possibility, NbHsc70-2/His7 was incubated with 40 μg of BSA (which is equivalent to the amount of CNV particles used for Fig. 5a), followed by trypsin digestion. It can be seen in Fig. 5e that the presence of BSA does partially interfere with digestion of NbHsc70-2/His7 as would be expected. However, unlike the case for Hsc70-2 in CNV virions, only 10-fold more trypsin (rather than the 1,000-fold seen with CNV virions in Fig. 5a) was required to completely digest NbHsc70-2/His7 in the presence of BSA. Thus, these data indicate that the reduced digestion of Hsc70-2 observed in the presence of CNV is due to partial protection by CNV virions. In order to rule out the possibility that components of the CNV virus preparation interfere with the trypsin sensitivity of CNV-bound Hsc70-2 seen in Fig. 5a, we incubated NbHsc70-2/His7 with the same volume of “virus” extracted from an equivalent mass of mock-inoculated leaves that was used for CNV extraction from infected leaves for Fig. 5a (Fig. 5d). We found that the trypsin digestion pattern of NbHsc70-2/His7 in the presence of the mock “virus” preparation was similar to that of free NbHsc70-2/His7 (compare Fig. 5c and d). This observation reinforces our hypothesis that CNV particles partially protect the bound Hsc70-2 from trypsin digestion and that this protection is not due to interference by components of the virus preparation. The implied intimate interaction between CNV particles and Hsc70-2 suggests that bound Hsc70-2 may be involved in some aspect of the establishment of infection, such as disassembly, or that it may assist in CNV particle stability.
FIG 5.
The Hsc70-2 bound to virions is partially protected from protease digestion. (a and b) A trypsin protection assay was performed by incubating 40 μg of purified CNV virions with increasing amounts of trypsin (0 to 4 × 104 ng, as indicated) at RT for 30 min. Samples were then electrophoresed through a NuPAGE gel, blotted, and then probed with either an HSP70 antibody (a) or a mixture of two CNV CP antibodies that react to the CNV R and arm domains (RAD) or shell and protruding domains (SP) (b). (c) A similar experiment was performed using 6.5 ng of recombinant NbHsc70-2/His7 (which corresponds to the approximate amount present in 40 μg of CNV virions) to compare the trypsin sensitivity of free Hsc70-2 with that of Hsc70-2 present in CNV virion preparations (a). (d) NbHsc70-2/His7 (6.5 ng) was mixed with mock virus extracted from a mass of leaf material equal to that used for CNV for panel a to determine if components of the virus preparation interfere with trypsin sensitivity of Hsc70-2. (e) Recombinant NbHsc70-2/His7 (6.5 ng) was mixed with 40 μg of BSA prior to trypsin treatment. The blots in panels c, d, and e were each probed with HSP70 antibody under conditions identical to those for panel a. Note that trypsin-treated CNV has numerous degradation products (shown by brackets in panel b). The blots in panels a and c to e were each exposed for 39 min.
HSP70 antibody-incubated virus particles or CsCl-purified CNV virions having undetectable amounts of HSP70 produce a reduced number of local lesions on C. quinoa.
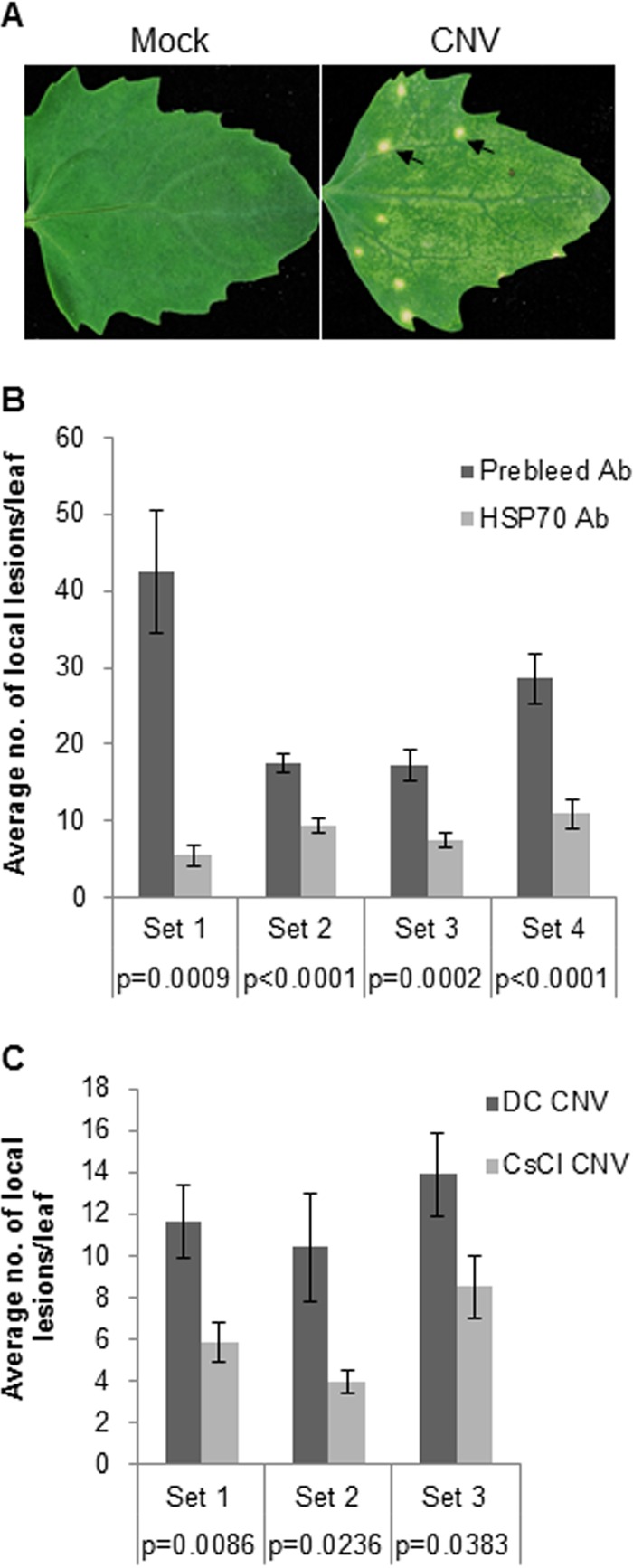
Local lesion assays provide a means for measuring infection by virus particles or viral RNA inocula where each lesion is believed to result from a single particle (or single uncoating event) or a single infectious RNA molecule (44). Viral RNA replicates in the inoculated cell, and in the case of CNV, newly replicated viral RNA (rather than virions) moves to adjacent cells where replication is established once again, with further viral RNA cell-to-cell movement eventually resulting in the formation of a visible local lesion (Fig. 6A). To assess the possibility that the Hsc70-2 associated with CNV particles assists in disassembly during the establishment of infection, we incubated CNV particles with HSP70 antibody or with control prebleed antibody and then conducted a local lesion assay using the local lesion host Chenopodium quinoa. For this experiment, 400 pg of virus particles (∼0.04 fmol) was incubated with HSP70 antibody (∼6 fmol) at 37°C for 1 h to bind the Hsc70-2 molecules associated with particles. An equivalent amount of prebleed antibody was used in control experiments in place of HSP70 antibody. After incubation, 40 pg of virions was used to inoculate C. quinoa leaves, the average number of local lesions per leaf was determined, and Student's t test was conducted. Figure 6B shows that in 4 independent experiments, significantly lower numbers of local lesions per leaf were obtained using virions incubated with HSP70 antibody than using virions incubated with prebleed antibody. These observations are consistent with the notion that the Hsc70-2 present in virion preparations plays a role in the establishment of CNV infection, possibly by enhancing disassembly in inoculated cells.
FIG 6.
CNV particles incubated with HSP70 antibody or having undetectable amounts of HSP70 produce fewer local lesions on the local lesion host Chenopodium quinoa. (A) Locals lesions produced on C. quinoa leaves following inoculation with CNV virions. The leaf in the left panel was mock inoculated with buffer only, and the leaf in the right panel was inoculated with CNV virions. Arrows point to the local necrotic lesions observed at 5 dpi. (B) CNV particles (400 pg) were incubated with an equal mass of prebleed IgG or HSP70 antibody at 37°C for 1 h, and then 40 pg of virus was used to inoculate individual C. quinoa leaves. The numbers of local lesions per leaf were counted at 5 dpi. A graphical representation of the average number of local lesions per leaf obtained from 4 independent experiments (set 1, n = 8; set 2, n = 10; set 3, n = 20; set 4, n = 23) is shown (n represents the number of leaves counted for each treatment in an experimental set). (C) Graphical representation of 3 independent local lesion assays (set 1, n = 22; set 2, n = 14; set 3, n = 22) showing the average numbers of local lesions per leaf on C. quinoa plants inoculated with 40 pg/leaf of CNV virions extracted by either differential centrifugation (DC CNV) or CsCl gradient (CsCl. CNV). The lesions were counted at 5 dpi. The P values for each experimental set as determined by Student's t test are shown and indicate that the differences are statistically significant (i.e., P < 0.05). The standard error is shown by the brackets above the bars.
Also, we wanted to determine if particles extracted via differential centrifugation, which have detectable amounts of HSP70 (Fig. 1B), would produce a greater number of local lesions on C. quinoa than CsCl-purified virions, which have undetectable amounts of HSP70. Therefore, equal amounts of virions (40 pg) extracted by differential centrifugation (DC CNV) or by CsCl gradients (CsCl CNV) were inoculated onto C. quinoa leaves. Figure 6C shows that in 3 independent experiments, significantly greater numbers of local lesions were observed in DC CNV virions than in CsCl CNV virions. These results suggest that Hsc70-2 bound to virions plays an important role in the establishment of infection, possibly via enhancing disassembly in inoculated cells.
Overexpression of Hsc70-2 or Hsp70 leads to enhanced CNV disassembly efficiency.
To further examine the possibility that N. benthamiana Hsc70-2 may play a role in CNV disassembly during the establishment of infection, we agroinfiltrated plants either with Hsc70-2 cloned as previously described into the binary vector pBin(+) [pNbHsc70-2/pBin(+)] (25) or with empty pBin(+) (EV) as a control (27). We then inoculated infiltrated leaves with CNV to determine if overexpression of Hsc70-2 would lead to enhanced local lesion production. To confirm that Hsc70-2 was overexpressed, total leaf protein samples were collected at 1 and 3 days postagroinfiltration (dpai) and analyzed through Western blot analysis using an HSP70 antibody. We observed that plants agroinfiltrated with pNbHsc70-2/pBin(+) contained significantly elevated levels of Hsc70-2 at 3 dpai compared those in to the EV control (Fig. 7A, compare lanes 3 and 4). We therefore inoculated agroinfiltrated leaves at 3 dpai, when the expression levels of Hsc70-2 were high, with 100 ng of CNV and then counted the number of local lesions at 4 to 6 days postinoculation (dpi). Three independent experiments were conducted, and in each experiment, plants overexpressing Hsc70-2 were found to develop significantly (P < 0.05) greater numbers of local lesions than EV-infiltrated plants (Fig. 7B). We suggest that overexpression of N. benthamiana Hsc70-2 leads to enhanced disassembly efficiency of CNV, leading to a greater number of local lesions. It is noted here that CNV does not normally produce discrete, countable, local lesions on inoculated leaves of N. benthamiana; rather, the lesions are broad and spreading (unpublished observations). However, we have found that discrete lesions are produced when plants are agroinfiltrated. This is likely due to the combined immune response of N. benthamiana to CNV infection and the pattern-triggered immunity induced by Agrobacterium tumefaciens (45).
FIG 7.
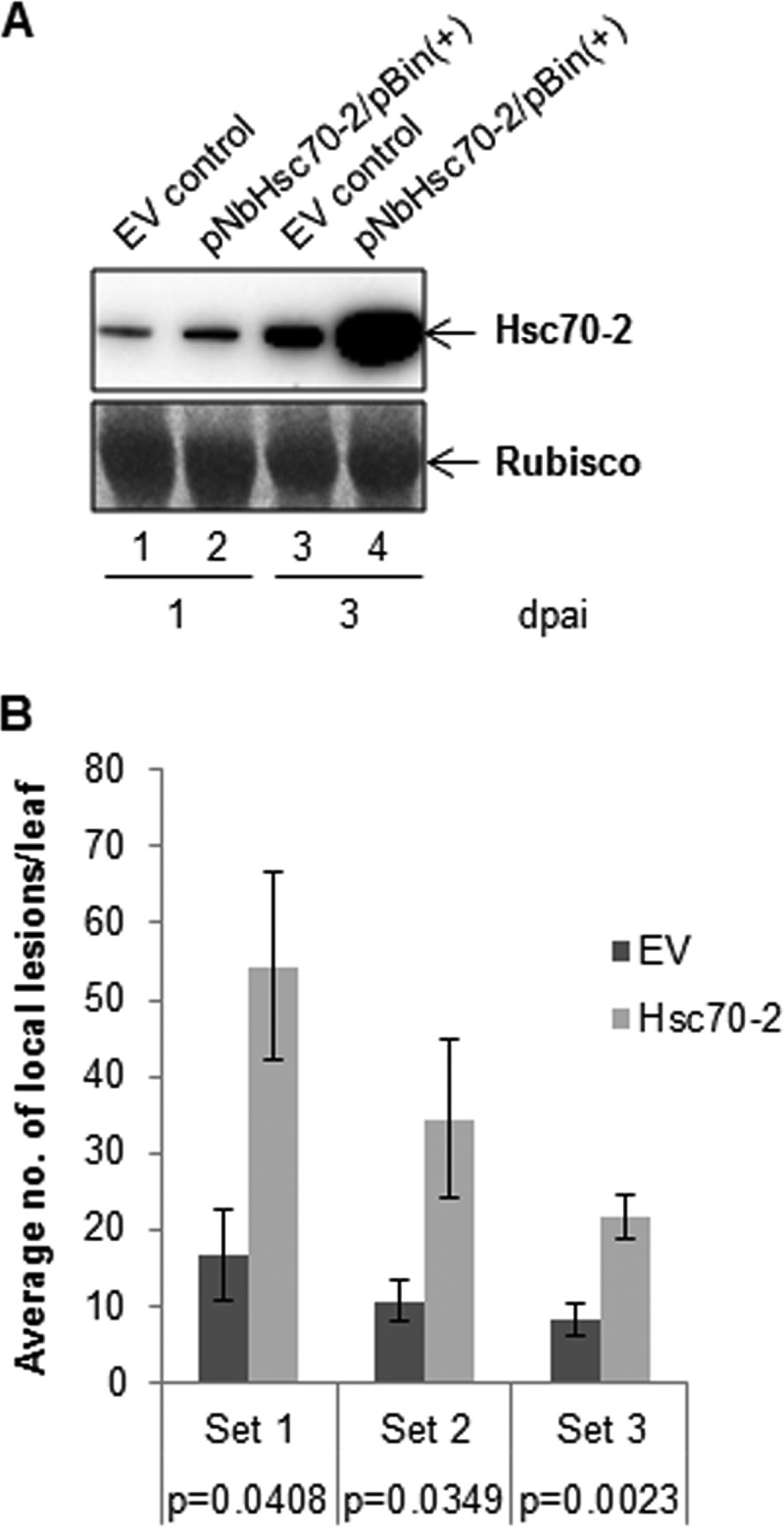
N. benthamiana leaves agroinfiltrated with Hsc70-2 show an increase in the number of local lesions produced by CNV in comparison to plants agroinfiltrated with empty vector (EV). (A) Western blot analysis of leaf extracts from N. benthamiana plants agroinfiltrated with EV or pNbHsc70-2/pBin(+) at 1 and 3 dpai, showing overexpression of Hsc70-2. Two or three leaves from two plants were collected and ground in liquid nitrogen, and 100 mg of the ground leaf material was added to 350 μl of LDS protein extraction buffer. Equal volumes were electrophoresed using SDS-PAGE and probed with an HSP70-specific antibody. A Ponceau S-stained image of the blot showing levels of RubisCO used as a loading control is in the lower panel. (B) Local lesion assay on N. benthamiana plants that were agroinfiltrated with EV or pNbHsc70-2/pBin(+) as indicated and then 3 days later were inoculated with 100 ng of CNV virions. Note that at the time plants are inoculated, the levels of NbHsc70-2 are elevated (panel A, compare lanes 3 and 4). A graphical representation of the average number of local lesions per leaf on empty vector (EV)- or Hsc70-2-agroinfiltrated plants from 3 independent local lesion assays (set 1, n = 6; set 2, n = 8; set 3, n = 16) is shown (n represents the number of leaves counted for each treatment in an experimental set). The P values for each experimental set as determined by Student's t test are shown and indicate that the differences are statistically significant (i.e., P < 0.05). The standard error is shown by the brackets above the bars. It is noted that CNV normally produces a spreading-necrosis phenotype on N. benthamiana leaves, where local lesions cannot be counted. However, following agroinfiltration, discrete local lesions are produced (unpublished observations), presumably due to agroinfiltrated plants having already established a defense response to Agrobacterium tumefaciens.
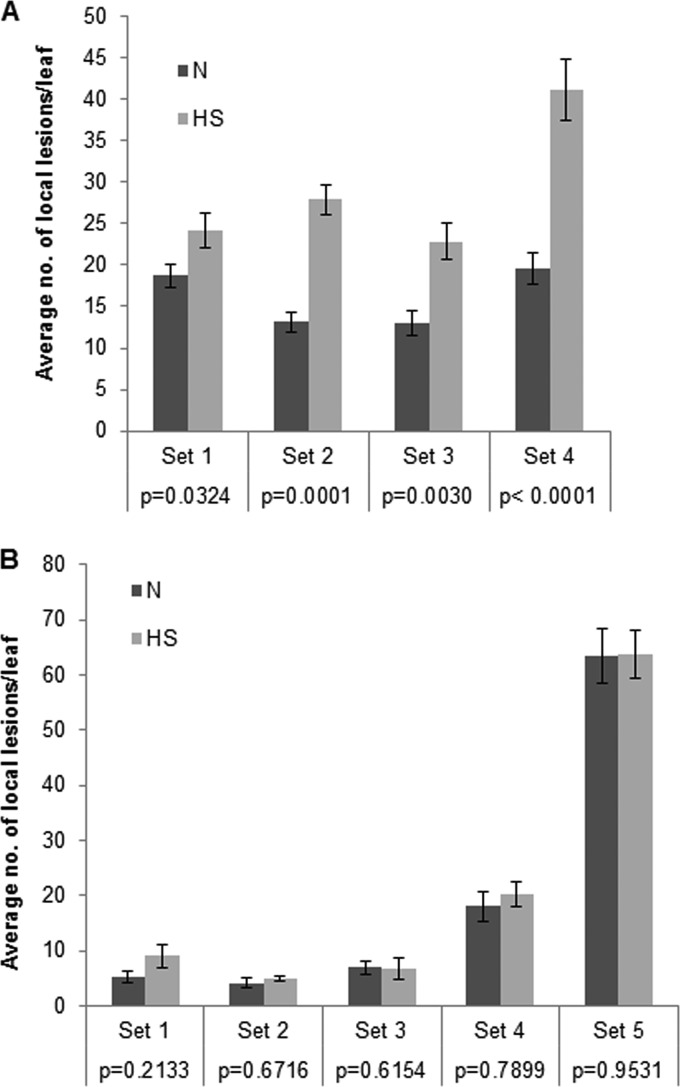
We also wished to determine whether overexpression of HSP70 homologs would lead to increased disassembly efficiency in the CNV local lesion host C. quinoa. Since C. quinoa is a poor host for transient protein expression via agroinfiltration (unpublished findings), we utilized heat shock treatment to increase the levels of HSP70 homologs. Previously we have found that heat shock of C. quinoa plants at 48°C for 30 min followed by a 2-h recovery period leads to overexpression of Hsp70, and possibly Hsc70, at 2 h post-heat treatment and that high levels of expression remains for up to 3 days post-heat shock (27). Hence, we inoculated the heat-shocked (HS) or non-heat-shocked (N) plants at 2 h after treatment with 40 pg of CNV particles and examined the number of local lesions produced per leaf in both sets of plants. Figure 8A shows a graphical representation of the average number of local lesions per leaf on HS and N plants. Four independent experiments were conducted, and in each experiment it was shown that HS plants developed a significantly (P < 0.05) greater number of local lesions per leaf than N plants, suggesting that Hsc70 and/or Hsp70 may enhance CNV disassembly efficiency. To distinguish between greater disassembly efficiency and the possibility that heat-shocked plants might simply support CNV replication and accumulation to a higher level, as reported previously (27, 46), we conducted a similar local lesion analysis with CNV virion RNA as the inoculum. We found that in 5 independent experiments the average numbers of local lesions per leaf on the HS and N plants were not statistically significantly different (P > 0.05) (Fig. 8B), suggesting that the greater number of local lesions found in heat-shocked plants using CNV particles could be due to enhanced disassembly efficiency of CNV virions rather than increased capacity of heat-shocked C. quinoa plants to support CNV infection. Our results are therefore consistent with the hypothesis that HSP70 homologs assist in the CNV uncoating process.
FIG 8.
Heat-shocked C. quinoa leaves show an increase in the number of local lesions in comparison to untreated plants when virions are used as the inoculum but not when virion RNA is used. (A) Graphical representation of the average number of local lesions per leaf on heat-shocked (HS) and non-heat-shocked (N) C. quinoa plants from 4 independent local lesion assays (set 1, n = 20; set 2, n = 38, set 3, n = 27; set 4, n = 19) using virions as the inoculum (40 pg/leaf) (n represents the number of leaves counted for each treatment in an experimental set). The P values for each experimental set as determined by Student's t test are shown and indicate that the differences are statistically significant (i.e., P < 0.05). (B) Graphical representation of 5 independent local lesion assays (set 1, n = 12; set 2, n = 15; set 3, n = 17; set 4, n = 18; set 5, n = 32) conducted as for panel A except that virion RNA (1 ng/leaf) was used as an inoculum. Note that P values are greater than 0.05, indicating that the differences in local lesion numbers are not statistically significant.
Incubation of virus particles with N. benthamiana Hsc70-2 at pH 7.5 leads to partial disassembly and/or conformational change of CNV virions.
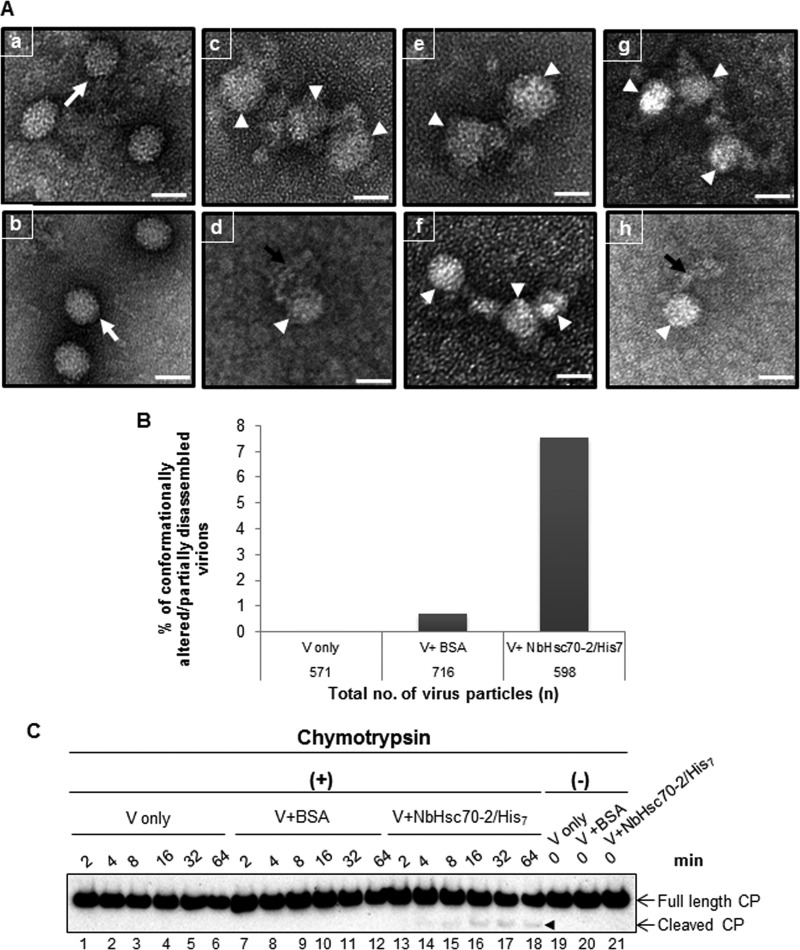
Studies have shown that Hsc70 can initiate the disassembly of oligomeric protein complexes (18). Also as described above, Hsp70 and Hsc70 have been found to have roles in inducing conformational change in viral capsids which is then followed by entry and uncoating in cells (21, 47). We hypothesize that Hsc70-2 induces a conformational change in CNV capsids which may contribute to disassembly of particles in the cytoplasm. To assess this, we performed an in vitro binding assay using 600 ng of CNV particles and 1,800 ng of NbHsc70-2/His7 or recombinant BSA as a control under optimized Hsc70-2 binding conditions at pH 7.5 in the presence of 1× EDTA-free protease inhibitor (see Materials and Methods). After incubation for 3.5 h, the particles were stained with 2% uranyl acetate and analyzed by TEM. It can be seen in Fig. 9A that incubation of virus particles with NbHsc70-2/His7 results in conformationally altered or partially dissociated virions (panels c to h, white arrowheads), which was not observed following incubation of particles in binding buffer (panel a). In some cases it appeared that viral RNA could be seen extruding from particles or perhaps instead that “strings” of CP subunits were found associated with particles (panels d and h, black arrows). These findings are consistent with the hypothesis that N. benthamiana Hsc70-2 plays a role in the disassembly of CNV particles at pH 7.5.
FIG 9.
Incubation of CNV with NbHsc70-2/His7 at pH 7.5 leads to partial disassembly of virions. (A) Six hundred nanograms of CNV virions was incubated with NbHsc70-2/His7 binding buffer (panel a),1,800 ng of BSA (panel b), or 1,800 ng of NbHsc70-2/His7 (panels c to h) under optimized binding conditions at pH 7.5 in the presence of 1× EDTA-free protease inhibitor for 3.5 h. Particles were adsorbed to Formvar-coated nickel grids and subjected to negative staining with 2% uranyl acetate. Intact virus particles are shown by white arrows in panels a and b. The partially disassembled virions are shown by white arrowheads in panels c to h. The black arrow represents possible viral RNA extruding from a partially disassembled virion or coat protein subunits (panels d and h). The scale bar represents 34 nm. The experiment was repeated 3 times, and representative results are shown. (B) A graphical representation of the percentage of conformationally altered or partially disassembled virions in samples prepared as for panel A. One hundred fields were randomly selected and photographed for subsequent analysis. The total numbers of virus particles were counted in the controls (either virus treated with NbHsc70-2/His7 binding buffer [V only; n = 571] or virus treated with BSA [V+BSA; n = 716]) or the experimental sample (virus treated with NbHsc70-2/His7 [V+Hsc70; n = 598]), along with the number of particles that were conformationally altered or partially disassembled. An approximately 11-fold increase in the percentage of conformationally altered/partially disassembled virions was observed in V+Hsc70-2 compared to V+BSA samples. Approximately 7.5% of CNV virions were found to be conformationally altered or partially disassembled in the presence of NbHsc70-2/His7. (C) Six hundred nanograms of virions was treated with binding buffer only (lanes 1 to 6), BSA (lanes 7 to 12), or NbHsc70-2/His7 (lanes 13 to 18) as for panel A but in the absence of any protease inhibitor. After incubation for 3.5 h, the virions were treated with 10 ng of chymotrypsin at RT, and equal volumes of samples were collected and mixed with the reaction stop buffer at indicated time points, followed by SDS-PAGE and Western blot analyses using the CNV CP antibody RAD. Equal amounts of samples from each treatment were collected and processed as described above before the addition of chymotrypsin (lanes 19 to 21). The chymotrypsin cleavage products in V+NbHsc70-2/His7 samples are shown by the arrowhead.
To further assess the validity of the observed phenomena, an in vitro binding assay was performed as described above, and 100 random images were photographed. The total number of CNV particles was counted along with the number of virions that were conformationally altered or partially disassembled. Figure 9B shows that an approximately 11-fold increase in the percentage of conformationally altered or partially disassembled virions was found when CNV was incubated with NbHsc70-2/His7 (7.5%) compared to the BSA control (0.69%). These results are consistent with our hypothesis that Hsc70-2 may play a role in CNV disassembly in inoculated cells.
To further show that some virions undergo structural changes in the presence of exogenously added NbHsc70-2/His7, chymotrypsin digestion assays on virions incubated in the presence or absence of N. benthamiana Hsc70-2 were conducted. It can be seen in Fig. 9C, lanes 13 to 18, that a low proportion of CP associated with CNV virions was digested by chymotrypsin, whereas buffer- or BSA-treated virions were insensitive to chymotrypsin (Fig. 9C, lanes 1 to 12). The sensitivity of Hsc70-2-treated CNV particles to chymotrypsin suggests that Hsc70-2 induces a conformational change in particles that renders them susceptible to protease treatment. We note that only a low level of digestion by chymotrypsin is observed. This could be due to inefficient binding of Hsc70-2 to CNV particles in the in vitro assay used, as was observed in TEM analysis of Hsc70-2-treated CNV (see above), and a resulting low level of conformational change and thus chymotrypsin sensitivity.
DISCUSSION
Little is known about the means by which nonenveloped virus particles disassemble to establish infection in a host. We provide evidence that Hsc70-2 is associated with CNV particles and that Hsc70-2 and/or HSP70 homologs can assist in the CNV disassembly process. Previous work has demonstrated that HSP70 homologs are highly upregulated during CNV infection and that they contribute to multiple aspects of the CNV infection process, such as in accumulation of viral RNA, accumulation of CP, pseudovirion assembly, and targeting of CNV CP to chloroplasts (27). HSP70 homologs have been found to promote the uncoating of several animal icosahedral viruses, such as rotaviruses, polyomaviruses, reoviruses, and papillomaviruses (19–21, 47). Our studies show for the first time that a plant HSP70 homolog, Hsc70-2, may be involved in the disassembly of an icosahedral plant virus, CNV.
Hsc70 and Hsp70 are molecular chaperones that play important roles in protein folding and preventing aggregate formation (7). They also have been found to promote the disassembly of macromolecular complexes such as clathrin-coated vesicles (14–16). Studies have shown that Hsc70 not only uncoats clathrin but also primes it to reform clathrin-coated pits (48). Studies on human polyomavirus suggest that Hsp70 also plays dual roles during virus infection by facilitating both capsid assembly (49) and disassembly (19). Recently we have found that Hsc70-2 plays a role in the assembly of pseudovirions that accumulate during agroinfiltration of plants with CNV CP (27). Our observation that Hsc70-2 is likely involved in the uncoating of CNV capsids is consistent with the notion that Hsc70-2 might be involved in both CNV assembly and disassembly, as in polyomaviruses and papillomaviruses (19, 49).
As shown in Fig. 1, Hsc70-2 copurifies with CNV, suggesting an association between CNV particles and Hsc70-2. Virus overlay assays (Fig. 3A) show that CNV particles can bind bovine Hsc70. Moreover, Fig. 3B shows that CNV particles remain associated with Hsc70-2 following agarose gel electrophoresis, reinforcing our notion that Hsc70-2 is physically bound to CNV particles. Immunogold labeling experiments provide further evidence that Hsc70-2 is bound to particles, with the majority of particles containing 1 or 2 bound Hsc70-2 molecules (Fig. 4). The nature of the association between CNV particles and Hsc70-2 was examined using trypsin sensitivity assays, and it was found that Hsc70-2 present in CNV preparations is less sensitive to trypsin treatment than “free” Hsc70-2, requiring between 1,000- and 10,000-fold more trypsin for complete digestion (Fig. 5). This suggests that Hsc70-2 is intimately associated with CNV particles.
We have also examined whether CLSV is associated with HSP70 homologs and found that approximately 4.5 ng of HSP70 per 40,000 ng of CLSV is present (Fig. 2). This is similar to the level of Hsc70-2 detected in CNV preparations (3.5 to 5.7 ng/40,000 ng of CNV) (Fig. 1). Additionally, in a preliminary experiment we have found that N. benthamiana plants agroinfiltrated with Hsc70-2 and inoculated with CLSV as described for CNV develop visibly greater numbers of local lesions (unpublished observations). The finding that CLSV is associated with HSP70 homologs suggests that other members of the Tombusviridae may similarly be associated with HSP70. Further experiments are required to evaluate this possibility and its significance to the disassembly process.
Although our data show that Hsc70-2 is intimately associated with CNV virions, we do not know details of its association from a structural point of view. However, in analogy to the maturation protein of MS2 bacteriophage, which replaces 1 CP dimer in the T=3 structure, it is possible that Hsc70-2 (∼70 kDa) may a replace a CNV CP dimer (∼82 kDa).
How, then, does Hsc70-2 become associated with CNV particles? HSP70 homologs are known to require a short region of hydrophobic residues flanked by basic amino acids for efficient binding of substrates (12, 50, 51). Interestingly, the 18-amino acid (aa) β-region of the CNV CP arm is highly hydrophobic, and this is flanked by highly basic regions in the adjacent R domain and ε region of the arm. It is possible that this represents the site of interaction of Hsc70-2. This is a likely possibility, since it is known that the CNV arm region acts as a transit peptide targeting CNV CP to chloroplasts (52), and many transit peptides are believed to interact with HSP70 homologs (53–55). Moreover, we have shown that HSP70 homologs are required for CNV entry into chloroplasts and that Hsc70-2 increases the accumulation of CNV virus-like particles (27). In addition, we have found that Hsc70-2 interacts with CNV CP in vivo (27). We propose that Hsc70-2 bound to the CNV arm region of the CP becomes associated with CNV particles during virion assembly. Alternatively, Hsc70-2 may associate with the arm region following assembly during the CNV breathing process when the arm becomes externalized. Consistent with these suggestions, we have found that mutant CNV particles that lack the β-region of the arm do not associate with HSP70 homologs to a detectable level as determined by Western blotting (unpublished observations). Additionally, unlike WT CNV, mutant virions do not produce a significantly greater number of local lesions on heat-shocked than on non-heat-shocked C. quinoa (unpublished observations). Further experiments are required to address the site on CNV CP or particles where Hsc70-2 is bound and how Hsc70-2 becomes associated with virions.
In Fig. 6B and C, respectively, we found that CNV particles incubated with HSP70 antibody or CsCl-purified virions having undetectable amounts of HSP70 produce fewer local lesions on C. quinoa. This finding suggests that the Hsc70-2 associated with CNV particles enhances the establishment of infection. This could be due to a role for Hsc70-2 in initiating virus disassembly. On the other hand, it is known that HSP70 homologs play a role in the formation of the replication complexes of CNV and TBSV (56, 57). Therefore, it is possible that the associated Hsc70-2 assists in establishing infection by providing a source of Hsc70 for replication complex formation. It is also noted that since CsCl purified virions produce local lesions on C. quinoa, it is possible that virion preparations might contain a very low level of Hsc70-2 which is beyond the capacity of detection by Western blotting analyses. Alternatively, Hsc70-2 may enhance infectivity of CNV virions but may not be essential. Enhancement of infection could be due to a role in facilitating disassembly or perhaps by supplying the HSP70 homolog required for formation of the initial replication complex.
It is possible that many CNV virions may not be able to efficiently establish an infection in a susceptible host due to the absence of Hsc70-2 in the particle. This is consistent with the great number of virions required to establish an infection. For example, in C. quinoa, 40 pg of virus (∼2.7 × 106 molecules) produces an average of only approximately 20 to 40 local lesions. Although numerous factors certainly play a role in the ability of a particle to establish an infection, it is possible that the association of Hsc70-2 with CNV may comprise one contribution to the establishment of infection.
To assess our hypothesis that Hsc70-2 assists CNV in disassembly, we examined particles incubated with an excess of bacterially expressed Hsc70-2 by TEM analysis and found that approximately 7.5% of treated virions showed observable changes in morphology (see Fig. 9B), We also examined Hsc70-2-treated particles for conformational changes by conducting chymotrypsin sensitivity assays, where we found that a small proportion of the CP of these particles becomes accessible to chymotrypsin treatment (Fig. 9C). Together, these observations suggest that exogenously added Hsc70-2 changes the conformation of CNV particles, consistent with a role in virus disassembly.
We note that only a small proportion of CNV particles are altered following incubation with bacterially expressed Hsc70-2 (Fig. 9B). One likely explanation for this is that HSP70 homologs are known to have a lower affinity for natively folded polypeptides than for misfolded or alternatively folded polypeptides (11). Also, the in vitro reaction conditions likely do not sufficiently mimic the complex intracellular environment where CNV disassembly occurs.
We conducted local lesion assays using either CNV virions (Fig. 8A) or CNV RNA (Fig. 8B) on heat-shocked C. quinoa to distinguish between the possibilities that HSP70 homologs assist in CNV disassembly or assist in some other aspect of the initiation of infection, such as supplying HSP70 homologs needed for formation of the replication complex. We found that although the number of local lesions did not change in heat-shocked versus non-heat-shocked plants when viral RNA was used as the inoculum, the number of local lesions was significantly higher when virions were used as the inoculum. This reinforces our conclusion that HSP70 homologs, or more specifically Hsc70-2, assists in the efficiency of the establishment of infection by assisting in particle disassembly. This finding also suggests that cytoplasmic HSP70 homologs may further assist in CNV disassembly beyond that which is facilitated by CNV-bound Hsc70-2.
Conformational change of viral capsids during cellular entry and uncoating has been suggested or documented for several animal and plant viruses, including human rhinovirus 2, poliovirus, BK polyomavirus, TBSV, Tobacco mosaic virus (TMV), Turnip crinkle virus (TCV), Carnation mottle virus (CarMV), Southern bean mosaic virus (SBMV), Turnip yellow mosaic virus, Brome mosaic virus (BMV), Cowpea chlorotic mottle virus (CCMV), Alfalfa mosaic virus (AMV), Red clover necrotic mosaic virus (RCNMV), and Flock House virus (FHV) (5, 6, 34, 58–62). In the case of TBSV, TCV, RCNMV, and SBMV, it has been suggested that particles swell in the higher-pH, low-calcium environment of the cytoplasm, leaving a hole at the particle 3-fold axis where the RNA can exit (34–36, 63–65). Additionally, for AMV, TMV, TCV, CCMV, FHV, and BMV (5, 6, 34, 61) it has been suggested that the particles undergo cotranslational disassembly whereupon the exposed virion RNA of partially disassembled particles binds ribosomes and subsequent translation assists in further uncoating of particles. We note that following numerous attempts to observe cotranslational disassembly of CNV in wheat germ extracts and rabbit reticulocyte lysates, translation products were not observed even when particles were swollen prior to addition to the cell-free translation systems or when particles were pretreated with Hsc70-2 (unpublished observations). Thus, it is possible that CNV differs from other similar nonenveloped viruses and TMV in that disassembly does not occur cotranslationally. Alternatively, it may be that the cell-free systems are lacking the components normally present in a cell that are required for cotranslational disassembly of CNV.
The data described in this article support the notion that CNV CP can co-opt HSP70 homologs for virion disassembly. Like most spherical viruses, CNV persists in a metastable state where it can exist in two distinct conformations: a compact closed state and a relaxed (expanded) open conformation (2, 30, 32, 34, 35). Virions can undergo a breathing phenomenon during which they change from a closed to an open conformation. These two states of the particle could be influenced by conditions within the host cell such as high pH or low calcium ions, where the particle would be in the expanded conformation. In analogy to TBSV, the R and arm region wholly or partially externalize during particle expansion, and an opening is created at the particle quasi-3-fold axis (31). It is possible that the viral RNA comes out of the hole from the 3-fold axis that is created during particle swelling, as has been suggested for Carnation mottle virus and other viruses (35). It is possible that CNV-bound Hsc70-2 assists in this breathing phenomenon, promoting expansion under favorable cytoplasmic conditions such as elevated pH and low calcium. Experiments were conducted in which CNV particles were incubated with Hsc70-2 at pH 7.5 and then analyzed for a change in conformation by agarose gel electrophoresis, a procedure that can readily identify swollen CNV particles (33); however, no difference could be discerned using this method, possibly due to a low number of particles that may be affected by Hsc70-2 as deduced from the TEM and chymotrypsin experiments conducted for Fig. 9.
We provide the following hypothetical model for CNV virion disassembly facilitated by Hsc70-2. According to this model, CNV acquires Hsc70-2 during the assembly process, possibly due to Hsc70-2 interaction with the transit peptide-like region of the arm of the CNV CP. The disassembly process is facilitated by the bound Hsc70-2 along with the high-pH environment of the cytoplasm, resulting in CNV expansion which creates an opening in the particle at the quasi-3-fold axis. As the arm and R domains externalize through this opening, virion RNA may be concomitantly externalized. Recently, we have shown that the R domain of CNV contains a highly basic KGKKGK region which likely binds viral RNA within the capsid (66). Also, the arm domain contains a KGRKPR region which may similarly bind virion RNA (unpublished observations). Thus, it seems possible that RNA bound to these basic sequences may exit the virion in association with R and arm domain externalization. The RNA may then be accessible for translation of the p33 and p92 open reading frames (ORFs), which encode the auxiliary replicase protein and RNA-dependent RNA polymerase, respectively. As we have found that CNV produces a greater number of local lesions on plants overexpressing HSP70 homologs (Fig. 6 and 7), it is possible that cytoplasmic HSP70 homologs may further assist in CNV disassembly beyond that facilitated by the virion-associated Hsc70-2. This may allow for a more complete disassembly and release of viral RNA, which may be required to “untangle” the partially disassembled virion. Finally, it is possible that the Hsc70-2 associated with the particle or the sequestered HSP70 homologs may assist in the formation of the replication complex.
In summary, we have provided evidence that CNV particles are associated with Hsc70-2 and that this association may contribute to particle disassembly. We believe that this finding may provide fruitful insight into the multifaceted aspects of the disassembly processes of other nonenveloped viruses as well.
MATERIALS AND METHODS
Virus purification.
Approximately 1.5 μg (in 110 μl) of T7 polymerase runoff transcripts of an infectious wild-type (WT) CNV cDNA clone (pK2/M5) was used to inoculate 3 or 4 leaves of 4- to 6-week-old Nicotiana benthamiana as previously described (67). At 5 dpi, infected leaves were collected and macerated in 100 ml of 25 mM potassium phosphate (KPO4) buffer (pH 6.8) and used for inoculation using carborundum as an abrasive and a sterile sponge. For mock inoculation, 3 or 4 leaves of 4- to 6-week-old healthy N. benthamiana were ground as described above and the resulting ground material was used for inoculation. At 6 dpi, approximately 60 to 100 g of mock-infected or infected leaf tissue was collected and homogenized in a blender using at least 5 volumes of 100 mM sodium acetate (NaOAc) (pH 5.0) containing 10 mM β-mercaptoethanol. The homogenate was filtered through 2 layers of Miracloth (Calbiochem), and the filtrate was gently rotated at 4°C for at least 1 h and then centrifuged at 8,000 × g for 15 min at 4°C to remove plant debris. The solution was adjusted to 8% polyethylene glycol 8000 (PEG 8000) (Sigma-Aldrich) and incubated for at least 2 h at 4°C with constant stirring. The virus was pelleted at 8,000 × g for 20 min at 4°C and then resuspended in 10 ml of 10 mM NaOAc, pH 5.0. The suspension was gently rotated at 4°C overnight and then subjected to centrifugation at 8,000 × g for 20 min at 4°C. The supernatant (containing virions) was transferred to polycarbonate tubes (Beckman polycarbonate centrifuge tubes, 26.3 ml) and then subjected to ultracentrifugation (Beckman L8-70M ultracentrifuge) at 40,000 rpm for 4 h at 4°C in a Beckman type 50.2 Ti rotor. The supernatant was discarded, and the pellet was resuspended in 500 to 1000 μl of 10 mM NaOAc, pH 5.0. The suspension was gently rotated at 4°C overnight and then centrifuged again at 20,000 × g for 15 min at 4°C. The supernatant (containing virions) was collected and stored at 4°C until further use. The virus concentration was determined spectrophotometrically (the absorbance at 260 nm of a 1-mg/ml suspension of CNV is 4.5). CsCl purification of CNV was performed as previously described (29).
SDS-PAGE and Western blot analysis.
Protein samples used for electrophoresis were prepared as described previously (27). Samples were electrophoresed through NuPAGE 4 to 12% Bis-Tris gels (Thermo Fisher Scientific), blotted onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad), and probed with either a monoclonal antibody that detects both Hsc70 and Hsp70 (ADI-SPA-820-F [Enzo Life Sciences]) (here referred to as HSP70 antibody), a rabbit polyclonal antibody specific to bacterially expressed CNV S and P domain sequences (SP antibody) or R and arm domains (RAD antibody), or a rabbit polyclonal antibody raised against CNV particles (CNV polyclonal antibody). Antigen-antibody complexes were detected with peroxidase-labeled goat anti-mouse or goat anti-rabbit antibodies as appropriate (Sigma).
Ponceau S staining.
Ponceau S (Sigma-Aldrich) staining of blots was conducted as described previously (27). CNV CP or ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) was used as the loading control to standardize the mass of total protein loaded onto the gel prior to blotting.
Mass spectrometry.
Two independent virus purification experiments were performed with equal amounts of CNV- and mock-inoculated leaf tissue using the differential centrifugation technique described above. Preparations were resuspended in identical volumes, and equal amounts of the two samples (mock and CNV infected) were analyzed for the presence of Hsc70-2 by mass spectrometry (MS) using the Proteomics Core Facility at the University of British Columbia Center for High-Throughput Biology. Samples were subjected to denaturing gel electrophoresis, and bands greater and less than the size of CNV CP (41 kDa) were extracted, digested with trypsin, and then differentially labeled with formaldehyde isotopologues. Samples were pooled and subjected to liquid chromatography-tandem MS (LC-MS/MS) analysis. MASCOT was used to identify peptides using the Swiss-Prot database (68).
Purification of bacterially expressed N. benthamiana Hsc70-2 (pNbHsc70-2/His7).
The previously described N. benthamiana Hsc70-2 cloned into the bacterial expression vector pET24D(+) with a 7× His tag (27) was purified using the Talon Superflow metal affinity resin (Clontech Laboratories) as recommended by the manufacturer. Isopropylthio-β-galactoside (IPTG) (1 mM) (Thermo Fisher Scientific) was used to induce 1,000 ml of log-phase (optical density at 600 nm [OD600] = 0.6 to 0.8) bacterial culture. Hsc70-2 was purified under denaturing conditions from the total (soluble and insoluble) lysate as described previously (27). The purified recombinant protein (NbHsc70-2/His7) was concentrated using an Amicon Ultra-2ml centrifugal filter with a 50-kDa nominal molecular mass limit and was quantified by running several dilutions on an SDS-polyacrylamide gel with bovine Hsc70 (ADI-SPP-751-D; Enzo Life Sciences) as the mass standard. The purified protein was stored at 4°C.
Quantification of Hsc70-2 present in CNV virion preparations.
Increasing amounts (from 2.5 μg to 40 μg) of purified virus particles were mixed with protein denaturation buffer (LDS; Thermo Fisher Scientific) in the presence of sample reducing agent. The mixture was heated at 70°C for 10 min and subjected to denaturing acrylamide gel electrophoresis followed by Western blotting as described above. Several dilutions of NbHsc70-2/His7 (from 0.625 ng to 10 ng) were also electrophoresed for quantifying the amount of Hsc70-2 present in virion preparations. For detection of Hsc70-2, HSP70 antibody was used at a 1/2,000 dilution (1-mg/ml stock). The antigen-antibody complex was detected using a goat anti-mouse antibody (Sigma). The amount of Hsc70-2 present in virion preparations was determined by densitometric analysis utilizing Image Lab software version 5.1 (Bio-Rad Laboratories). Two independent Western blot analyses at two different amounts (20 μg and 40 μg) of virus were conducted, followed by quantification as described above, and an average was taken to estimate the mass of N. benthamiana Hsc70-2 present in CNV virion preparations.
Agarose gel electrophoresis of purified particles followed by Western blotting.
Twenty micrograms of purified virus particles from two independent purifications was electrophoresed through 1% (wt/vol) agarose gels in TB buffer (45 mM Tris, 45 mM boric acid, pH 8.3) as described previously (37). One hundred nanograms of bovine Hsc70/Hsp73 (ADI-SPP-751-D; Enzo Life Sciences) (here referred to as bovine Hsc70) and human Hsp70/Hsp72 (ADI-NSP-555-D, Enzo Life Sciences) (here referred to as human Hsp70) was also loaded onto the gel as controls. The particles were stained with ethidium bromide (EtBr) as described previously (27). After visualization, electrophoresed particles and proteins were blotted onto PVDF membranes in TB buffer using capillary transfer (69). After overnight transfer, the membrane was divided into two halves and each of the two blots was blocked with 5% skim milk in 1× Tris-buffered saline (TBS) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2.5 mM KCl) containing 0.4% Triton X-100 for 1 h at room temperature (RT). After blocking, one of the membranes was probed with a CNV polyclonal antibody, while the other was incubated with HSP70 antibody. Antigen-antibody complexes were detected using peroxidase-labeled goat anti-rabbit or goat anti-mouse antibody (Sigma) for detecting CNV particles and HSP70, respectively. The Amersham ECL Prime Western blotting detection reagent (GE Healthcare Life Sciences) was used for detection of bound antibodies.
Virus overlay assay.
Ten micrograms of recombinant human Hsp70, bovine Hsc70, or BSA (Sigma) was mixed with the protein denaturation LDS buffer (Thermo Fisher Scientific) and electrophoresed along with CNV CP mass standards through a NuPAGE 12% Bis-Tris gel using morpholinepropanesulfonic acid (MOPS) buffer (Thermo Fisher Scientific). The proteins were transferred to a nitrocellulose membrane (0.45-μm pore size; Bio-Rad) and allowed to renature overnight at 4°C in the presence of 4% BSA in 1× phosphate-buffered saline (PBS) as described previously (38). After overnight incubation with BSA, the membrane was washed 3 times with 1× PBS and then blocked with 1× PBS containing 5% skim milk for 1 h at room temperature (RT). After being washed 3 times with 1× PBS, the membrane was incubated with 100 μg of CNV particles in 10 ml of sodium phosphate (NaPO4) buffer (pH 7.6) for 3 h at RT. The membrane was then washed and probed with a CNV CP-specific antibody (SP).
Immunogold labeling assay.
One microgram of CNV virions (in 10 μl) was adsorbed onto Formvar-carbon coated nickel grids for 15 min. The solution was gently removed with Whatman filter paper no. 1 and then placed on 50-μl drops of blocking agent (1% BSA–1× PBS) for 10 min. Excess blocking agent was removed as described above, and grids were placed on 10-μl drops of either HSP70 antibody (1/5 dilution of a 1-mg/ml stock) or no antibody (negative control) in 1% BSA–1× PBS. After an overnight incubation at 4°C, the grids were washed 3 times in 50-μl drops of 1× PBS for 5 min each at room temperature (RT). The grids were then incubated with 4-nm colloidal gold–AffiniPure goat anti-mouse IgG(H+L) (Jackson ImmunoResearch Laboratories, Inc.) at a 1/20 dilution in 1% BSA–1× PBS (10 μl) for 1 h at RT. The grids were washed again in 1× PBS as described above, followed by 4 additional washes in distilled water (10 μl) for 1 min each. The grids were then stained with 2% uranyl acetate for 30 s and analyzed by transmission electron microscopy (TEM) using a Hitachi H-7100 TEM. The images were captured at 100 kV at a magnification of ×80,000 using an Orius charge-coupled device (CCD) camera.
Trypsin digestion of CNV preparations.
Forty micrograms of virus particles (in 5 μl 10 mM NaPO4 buffer, pH 7.6) was subjected to different concentrations of trypsin (Sigma) at RT (see the legend to Fig. 4) for 30 min as described previously with some modifications (33, 34). The reactions were stopped by adding LDS sample buffer (Thermo Fisher Scientific) (to a final concentration of 1× LDS buffer) containing sample reducing agent and 5× cOmplete protease inhibitor (Roche) and heating the samples at 70°C for 10 min. The samples were electrophoresed through a 4 to 12% Bis-Tris NuPAGE gel and analyzed by Western blotting using either HSP70 antibody or a mixture of CNV CP antibodies SP and RAD. Control reaction mixtures included 6.5 ng of NbHsc70-2/His7 (the approximate amount associated with 40 μg of CNV particles as determined from Fig. 1) alone or in conjunction with 40 μg of BSA and were treated with trypsin as described above. In a further control, 6.5 ng of NbHsc70-2/His7 was incubated with an equivalent amount of extract of “virus” obtained from mock-inoculated leaf tissue (followed by trypsin digestion as described above) in order to rule out the possibility that components in the CNV virion extract interfere with the trypsin digestion assay.
Local lesion analysis following incubation with HSP70 antibody.
CNV virions (400 pg) were incubated with an equivalent mass of either HSP70 antibody or prebleed IgG antibody in 25 mM potassium phosphate (KPO4) buffer (pH 6.8) at 37°C for 1 h in a 10-μl reaction volume. After incubation, the volume was increased to 100 μl with 25 mM KPO4 (pH 6.8), and 10 μl (containing 40 pg of virions) was used to inoculate Chenopodium quinoa, a CNV local lesion host. Three to five plants of the same age and 3 or 4 leaves at identical developmental positions were rubbed with carborundum and then immediately inoculated. Lesions were counted at 4 to 7 dpi. The number of leaves counted (n) for each experimental set is given in the figure legends. Student's t test using GraphPad (GraphPad Software, Inc.) was conducted to determine if statistically significant differences existed between the two treatments. P values of less than 0.05 were considered to indicate statistically significant differences.
Local lesion assay on N. benthamiana expressing Hsc70-2.
N. benthamiana Hsc70-2 cloned into the binary vector pBin(+) as previously described [pNbHsc70-2/pBin(+)] (27), was used for agroinfiltration of N. benthamiana. Two leaves at identical developmental positions, each from 3 to 8 N. benthamiana plants (4 to 6 weeks old) were agroinfiltrated with either empty pBin(+) vector (EV) or pNbHsc70-2/pBin(+). To ascertain that the levels of Hsc70-2 increased in agroinfiltrated leaves, total-leaf-protein (TLP) samples were collected as described previously (27) at the time points indicated in the figure legends and subjected to Western blot analysis using HSP70 antibody. For local lesion assays, N. benthamiana plants were agroinfiltrated, and at 3 days postagroinfiltration (dpai), leaves were inoculated with 100 ng of CNV particles. Lesions were counted at 4 to 6 dpi. The number of leaves counted (n) for each experimental set is given in the figure legends. Student's t test was conducted to determine if differences between treatments were statistically significant using GraphPad software. P values of less than 0.05 were considered to be statistically significant.
Local lesion assays on HS C. quinoa.
The CNV local lesion host C. quinoa was heat shocked (HS) at 48°C for 30 min, followed by a 2-h recovery period at 26°C, as has been described for Arabidopsis thaliana (70). Untreated plants (no treatment) of the same age were kept at 26°C. Western blot analysis was utilized to confirm induction of HSP70. As a means for estimating the disassembly efficiency of CNV, C. quinoa plants of identical age were either heat shocked (HS) or not heat shocked (untreated) as described above. At 2 h posttreatment, 3 or 5 leaves each from 3 to 8 plants at identical developmental positions in HS or untreated plants were rubbed with carborundum and then inoculated with CNV. A pilot experiment was conducted to assess the concentration range in which the number of local lesions produced by CNV virions or virion RNA was linearly proportional to the inoculum concentration, as described previously (52). A concentration of 40 pg virus/leaf or 1 ng virion RNA/leaf was used for virus or virion RNA inoculations, respectively, on HS or untreated plants. Four to six leaves per plant on 3 to 6 plants per experiment were inoculated with CNV virion RNA. Lesions were counted at 5 to 7 dpi. The number of leaves counted (n) for each experimental set is given in the figure legends. Student's t test was used to evaluate statistical differences between the number of local lesions in HS and untreated plants. P values of less than 0.05 were considered to indicate statistically significant differences.
In vitro CNV/Hsc70-2 binding assay.
CNV particles (600 ng) were treated with Hsc70 binding buffer or with 1,800 ng BSA (Sigma) or 1,800 ng NbHsc70-2/His7 under binding conditions (10 mM Tris-HCl [pH 7.5], 50 mM potassium chloride [KCl], 5 mM magnesium chloride [MgCl2], 1 mM dithiothreitol [DTT], 1 mM ATP) in the presence of 1× cOmplete EDTA-free protease inhibitor as previously established for Hsc70 (71, 72) with some modifications. The mixture was incubated at 25°C for 3.5 h. The particles were then adsorbed onto Formvar-carbon-coated copper grids for 10 min, followed by negative staining with 2% uranyl acetate for 30 s. Images were taken at 100 kV and ×80,000 as described above (73).
Chymotrypsin sensitivity assay.
In vitro binding assays were performed as described above except that no protease inhibitor was added in the reaction mixture. After incubating for 3.5 h at 25°C, 10 ng of chymotrypsin (Sigma) was added to the reaction tube, and samples were collected at different times from 2 to 64 min as indicated in the legend to Fig. 9C and mixed with the reaction stop buffer (containing LDS with 5× cOmplete Protease Inhibitor [Roche] and sample reducing agent). A control representative sample was collected from each tube, i.e., virus treated with binding buffer only or virus treated with BSA or NbHsc70-2/His7, at 0 min before the addition of chymotrypsin and processed similarly. The collected samples were heated at 70°C for 10 min, followed by SDS-PAGE and Western blot analyses. A CNV CP antibody RAD (see above) was utilized for the detection of CP.
ACKNOWLEDGMENTS
This work was supported by NSERC Discovery Grant RGPIN 43840-11.
We thank Michael Weis for help with electron microscopy. We also thank Ron Reade and Jane Theilmann for excellent technical support and helpful discussions.
REFERENCES
- 1.Suomalainen M, Greber UF. 2013. Uncoating of non-enveloped viruses. Curr Opin Virol 3:27–33. doi: 10.1016/j.coviro.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Mateu MG. 2013. Assembly, stability and dynamics of virus capsids. Arch Biochem Biophys 531:65–79. doi: 10.1016/j.abb.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Lanzrein M, Schlegel A, Kempf C. 1994. Entry and uncoating of enveloped viruses. Biochem J 302:313–320. doi: 10.1042/bj3020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai B. 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu Rev Cell Dev Biol 23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- 5.Shaw JG, Plaskitt KA, Wilson TM. 1986. Evidence that tobacco mosaic virus particles disassemble contranslationally in vivo. Virology 148:326–336. doi: 10.1016/0042-6822(86)90329-6. [DOI] [PubMed] [Google Scholar]
- 6.Hiscox JA, Ball LA. 1997. Cotranslational disassembly of flock house virus in a cell-free system. J Virol 71:7974–7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer MP, Bukau B. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo S, Ahola V, Shu C, Xu C, Wang R. 2015. Heat shock protein 70 gene family in the Glanville fritillary butterfly and their response to thermal stress. Gene 556:132–141. doi: 10.1016/j.gene.2014.11.043. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Zhao J, Sheng Y, Xiao YF, Zhang YJ, Bai LX, Tan Y, Xiao LB, Xu GC. 2016. Identification of heat shock cognate protein 70 gene (Alhsc70) of Apolygus lucorum and its expression in response to different temperature and pesticide stresses. Insect Sci 23:37–49. doi: 10.1111/1744-7917.12193. [DOI] [PubMed] [Google Scholar]
- 10.Chong KY, Lai CC, Su CY. 2013. Inducible and constitutive HSP70s confer synergistic resistance against metabolic challenges. Biochem Biophys Res Commun 430:774–779. doi: 10.1016/j.bbrc.2012.11.072. [DOI] [PubMed] [Google Scholar]
- 11.Finka A, Sharma SK, Goloubinoff P. 2015. Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones. Front Mol Biosci 2:29. doi: 10.3389/fmolb.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer MP. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol 153:1–46. [DOI] [PubMed] [Google Scholar]
- 13.Dodson M, Roberts J, McMacken R, Echols H. 1985. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc Natl Acad Sci U S A 82:4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenberg E, Greene LE. 2007. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic (Copenhagen, Denmark) 8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 15.Pishvaee B, Costaguta G, Yeung BG, Ryazantsev S, Greener T, Greene LE, Eisenberg E, McCaffery JM, Payne GS. 2000. A yeast DNA J protein required for uncoating of clathrin-coated vesicles in vivo. Nat Cell Biol 2:958–963. doi: 10.1038/35046619. [DOI] [PubMed] [Google Scholar]
- 16.Newmyer SL, Schmid SL. 2001. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J Cell Biol 152:607–620. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bocking T, Aguet F, Harrison SC, Kirchhausen T. 2011. Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat Struct Mol Biol 18:295–301. doi: 10.1038/nsmb.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sousa RJ. 2014. Structural mechanisms of chaperone mediated protein disaggregation. Front Mol Biosci 1:12. doi: 10.3389/fmolb.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chromy LR, Oltman A, Estes PA, Garcea RL. 2006. Chaperone-mediated in vitro disassembly of polyoma- and papillomaviruses. J Virol 80:5086–5091. doi: 10.1128/JVI.80.10.5086-5091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanovic T, Agosto MA, Chandran K, Nibert ML. 2007. A role for molecular chaperone Hsc70 in reovirus outer capsid disassembly. J Biol Chem 282:12210–12219. doi: 10.1074/jbc.M610258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero CA, Bouyssounade D, Zarate S, Isa P, Lopez T, Espinosa R, Romero P, Mendez E, Lopez S, Arias CF. 2002. Heat shock cognate protein 70 is involved in rotavirus cell entry. J Virol 76:4096–4102. doi: 10.1128/JVI.76.8.4096-4102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saphire AC, Guan T, Schirmer EC, Nemerow GR, Gerace L. 2000. Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J Biol Chem 275:4298–4304. doi: 10.1074/jbc.275.6.4298. [DOI] [PubMed] [Google Scholar]
- 23.Niewiarowska J, D'Halluin JC, Belin MT. 1992. Adenovirus capsid proteins interact with HSP70 proteins after penetration in human or rodent cells. Exp Cell Res 201:408–416. doi: 10.1016/0014-4827(92)90290-O. [DOI] [PubMed] [Google Scholar]
- 24.Chroboczek J, Gout E, Favier AL, Galinier R. 2003. Novel partner proteins of adenovirus penton. Curr Top Microbiol Immunol 272:37–55. [DOI] [PubMed] [Google Scholar]
- 25.Chang JS, Chi SC. 2015. GHSC70 is involved in the cellular entry of nervous necrosis virus. J Virol 89:61–70. doi: 10.1128/JVI.02523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Napuli AJ, Falk BW, Dolja VV. 2000. Interaction between HSP70 homolog and filamentous virions of the beet yellows virus. Virology 274:232–239. doi: 10.1006/viro.2000.0475. [DOI] [PubMed] [Google Scholar]
- 27.Alam SB, Rochon D. 2016. Cucumber necrosis virus recruits cellular heat shock protein 70 homologs at several stages of infection. J Virol 90:3302–3317. doi: 10.1128/JVI.02833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rochon D, Lommel S, Martelli GP, Rubino L, Russo M. 2012. Family Tombusviridae, p 1111–1138. In King AMQ, Adam MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: ninth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom. [Google Scholar]
- 29.Li M, Kakani K, Katpally U, Johnson S, Rochon D, Smith TJ. 2013. Atomic structure of Cucumber necrosis virus and the role of the capsid in vector transmission. J Virol 87:12166–12175. doi: 10.1128/JVI.01965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katpally U, Kakani K, Reade R, Dryden K, Rochon D, Smith TJ. 2007. Structures of T = 1 and T = 3 particles of cucumber necrosis virus: evidence of internal scaffolding. J Mol Biol 365:502–512. doi: 10.1016/j.jmb.2006.09.060. [DOI] [PubMed] [Google Scholar]
- 31.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. 1978. Tomato bushy stunt virus at 2.9 A resolution. Nature 276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 32.Robinson IK, Harrison SC. 1982. Structure of the expanded state of tomato bushy stunt virus. Nature 297:563–568. doi: 10.1038/297563a0. [DOI] [Google Scholar]
- 33.Kakani K, Reade R, Rochon D. 2004. Evidence that vector transmission of a plant virus requires conformational change in virus particles. J Mol Biol 338:507–517. doi: 10.1016/j.jmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Bakker SE, Ford RJ, Barker AM, Robottom J, Saunders K, Pearson AR, Ranson NA, Stockley PG. 2012. Isolation of an asymmetric RNA uncoating intermediate for a single-stranded RNA plant virus. J Mol Biol 417:65–78. doi: 10.1016/j.jmb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CY, Zhang QF, Gao YZ, Xie L, Li HM, Hong J, Zhang CX. 2015. Uncoating mechanism of carnation mottle virus revealed by cryo-EM single particle analysis. Sci Rep 5:14825. doi: 10.1038/srep14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA. 2006. Removal of divalent cations induces structural transitions in red clover necrotic mosaic virus, revealing a potential mechanism for RNA release. J Virol 80:10395–10406. doi: 10.1128/JVI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakani K, Sgro J-Y, Rochon DA. 2001. Identification of specific cucumber necrosis virus coat protein amino acids affecting fungus transmission and zoospore attachment. J Virol 75:5576–5583. doi: 10.1128/JVI.75.12.5576-5583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kakani K, Robbins M, Rochon D. 2003. Evidence that binding of cucumber necrosis virus to vector zoospores involves recognition of oligosaccharides. J Virol 77:3922–3928. doi: 10.1128/JVI.77.7.3922-3928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rochon D, Kakani K, Robbins M, Reade R. 2004. Molecular aspects of plant virus transmission by olpidium and plasmodiophorid vectors. Annu Rev Phytopathol 42:211–241. doi: 10.1146/annurev.phyto.42.040803.140317. [DOI] [PubMed] [Google Scholar]
- 40.Kakani K, Rochon DA. 2002. Mutation of Pro-73 in the arm region of the Cucumber necrosis virus coat protein results in particles with an altered swollen conformation and loss of fungus transmissibility, p 17–22. In Proceedings of the Fifth Symposium of the International Working Group on Plant Viruses with Fungal Vectors Institute of Plant Sciences, Swiss Federal Institute of Technology, Zurich, Switzerland. [Google Scholar]
- 41.Robbins MA, Reade RD, Rochon DM. 1997. A cucumber necrosis virus variant deficient in fungal transmissibility contains an altered coat protein shell domain. Virology 234:138–146. doi: 10.1006/virop.1997.8635. [DOI] [PubMed] [Google Scholar]
- 42.Kakani K, Reade R, Katpally U, Smith T, Rochon D. 2008. Induction of particle polymorphism by Cucumber necrosis virus coat protein mutants in vivo. J Virol 82:1547–1557. doi: 10.1128/JVI.01976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurer C, Cimarelli A, Luban J. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J Virol 76:4666–4670. doi: 10.1128/JVI.76.9.4666-4670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews REF. 1991. Plant virology, 3rd ed, p 11–52 Academic Press, San Diego, CA. [Google Scholar]
- 45.Pruss GJ, Nester EW, Vance V. 2008. Infiltration with Agrobacterium tumefaciens induces host defense and development-dependent responses in the infiltrated zone. Mol Plant Microbe Interact 21:1528–1538. doi: 10.1094/MPMI-21-12-1528. [DOI] [PubMed] [Google Scholar]
- 46.Wang RY, Stork J, Nagy PD. 2009. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol 83:3276–3287. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Vargas J, Romero P, Lopez S, Arias CF. 2006. The peptide-binding and ATPase domains of recombinant hsc70 are required to interact with rotavirus and reduce its infectivity. J Virol 80:3322–3331. doi: 10.1128/JVI.80.7.3322-3331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang R, Gao B, Prasad K, Greene LE, Eisenberg E. 2000. Hsc70 chaperones clathrin and primes it to interact with vesicle membranes. J Biol Chem 275:8439–8447. doi: 10.1074/jbc.275.12.8439. [DOI] [PubMed] [Google Scholar]
- 49.Chromy LR, Pipas JM, Garcea RL. 2003. Chaperone-mediated in vitro assembly of polyomavirus capsids. Proc Natl Acad Sci U S A 100:10477–10482. doi: 10.1073/pnas.1832245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. 1997. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J 16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudiger S, Buchberger A, Bukau B. 1997. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol 4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 52.Xiang Y, Kakani K, Reade R, Hui E, Rochon D. 2006. A 38-amino-acid sequence encompassing the arm domain of the cucumber necrosis virus coat protein functions as a chloroplast transit peptide in infected plants. J Virol 80:7952–7964. doi: 10.1128/JVI.00153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivey RA III, Subramanian C, Bruce BD. 2000. Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol 122:1289–1299. doi: 10.1104/pp.122.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang XP, Glaser E. 2002. Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci 7:14–21. [DOI] [PubMed] [Google Scholar]
- 55.Rial DV, Arakaki AK, Ceccarelli EA. 2000. Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur J Biochem 267:6239–6248. doi: 10.1046/j.1432-1327.2000.01707.x. [DOI] [PubMed] [Google Scholar]
- 56.Serva S, Nagy PD. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol 80:2162–2169. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pogany J, Stork J, Li Z, Nagy PD. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci U S A 105:19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hewat EA, Neumann E, Blaas D. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol Cell 10:317–326. doi: 10.1016/S1097-2765(02)00603-2. [DOI] [PubMed] [Google Scholar]
- 59.Racaniello VR. 1996. Early events in poliovirus infection: virus-receptor interactions. Proc Natl Acad Sci U S A 93:11378–11381. doi: 10.1073/pnas.93.21.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang M, Abend JR, Tsai B, Imperiale MJ. 2009. Early events during BK virus entry and disassembly. J Virol 83:1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brisco M, Hull R, Wilson TM. 1986. Swelling of isometric and of bacilliform plant virus nucleocapsids is required for virus-specific protein synthesis in vitro. Virology 148:210–217. doi: 10.1016/0042-6822(86)90416-2. [DOI] [PubMed] [Google Scholar]
- 62.Matthews RE, Witz J. 1985. Uncoating of turnip yellow mosaic virus RNA in vivo. Virology 144:318–327. doi: 10.1016/0042-6822(85)90274-0. [DOI] [PubMed] [Google Scholar]
- 63.Martin SL, He L, Meilleur F, Guenther RH, Sit TL, Lommel SA, Heller WT. 2013. New insight into the structure of RNA in red clover necrotic mosaic virus and the role of divalent cations revealed by small-angle neutron scattering. Arch Virol 158:1661–1669. doi: 10.1007/s00705-013-1650-6. [DOI] [PubMed] [Google Scholar]
- 64.Aramayo R, Merigoux C, Larquet E, Bron P, Perez J, Dumas C, Vachette P, Boisset N. 2005. Divalent ion-dependent swelling of tomato bushy stunt virus: a multi-approach study. Biochim Biophys Acta 1724:345–354. doi: 10.1016/j.bbagen.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 65.Brisco M, Haniff C, Hull R, Wilson TM, Sattelle DB. 1986. The kinetics of swelling of southern bean mosaic virus: a study using photon correlation spectroscopy. Virology 148:218–220. doi: 10.1016/0042-6822(86)90417-4. [DOI] [PubMed] [Google Scholar]
- 66.Reade R, Kakani K, Rochon DA. 2010. A highly basic KGKKGK sequence in the RNA-binding domain of the Cucumber necrosis virus coat protein is associated with encapsidation of full-length CNV RNA during infection. Virology 403:181–188. doi: 10.1016/j.virol.2010.03.045. [DOI] [PubMed] [Google Scholar]
- 67.Rochon DM, Johnston JC. 1991. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology 181:656–665. doi: 10.1016/0042-6822(91)90899-M. [DOI] [PubMed] [Google Scholar]
- 68.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 69.Sambrook J, Russell DW. 2006. Southern blotting: capillary transfer of DNA to membranes. CSH Protoc 2006:,zfpg>pdb.prot4040. doi: 10.1101/pdb.prot4040. [DOI] [PubMed] [Google Scholar]
- 70.Jelenska J, van Hal JA, Greenberg JT. 2010. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc Natl Acad Sci U S A 107:13177–13182. doi: 10.1073/pnas.0910943107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu Z, Cyr DM. 1998. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem 273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 72.Johnson BD, Schumacher RJ, Ross ED, Toft DO. 1998. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem 273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 73.Hui E, Rochon D. 2006. Evaluation of the roles of specific regions of the Cucumber necrosis virus coat protein arm in particle accumulation and fungus transmission. J Virol 80:5968–5975. doi: 10.1128/JVI.02485-05. [DOI] [PMC free article] [PubMed] [Google Scholar]