ABSTRACT
The severity of clinical symptoms induced by pseudorabies virus (PRV) infection of its natural host is inversely related to the age of the pig. During this study, 2- and 15-week-old pigs were inoculated with PRV strain NIA3. This resulted in important clinical disease, although the associated morbidity and mortality were lower in older pigs. Quantitative PCR analysis of viral DNA in different organs confirmed the general knowledge on PRV pathogenesis. Several new findings and potential explanations for the observed age-dependent differences in virulence, however, were determined from the study of viral and cytokine mRNA expression at important sites of neuropathogenesis. First, only limited viral and cytokine mRNA expression was detected in the nasal mucosa, suggesting that other sites may serve as the primary replication site. Second, PRV reached the trigeminal ganglion (TG) and brain stem rapidly upon infection but, compared to 2-week-old pigs, viral replication was less pronounced in 15-week-old pigs, and the decrease in viral mRNA expression was not preceded by or associated with an increased cytokine expression. Third, extensive viral replication associated with a robust expression of cytokine mRNA was detected in the olfactory bulbs of pigs from both age categories and correlated with the observed neurological disease. Our results suggest that age-dependent differences in PRV-induced clinical signs are probably due to enhanced viral replication and associated immunopathology in immature TG and the central nervous system neurons of 2-week-old pigs and that neurological disease is related with extensive viral replication and an associated immune response in the olfactory bulb.
IMPORTANCE It is well known that alphaherpesvirus infections of humans and animals result in more severe clinical disease in newborns than in older individuals and that this is probably related to differences in neuropathogenesis. The underlying mechanisms, however, remain unclear. Pseudorabies virus infection of its natural host, the pig, provides a suitable infection model to study this more profoundly. We show here that the severe neurological disease observed in 2-week-old pigs does not appear to be related to a hampered innate immune response but is more likely to reflect the immature development state of the trigeminal ganglia (TG) and central nervous system (CNS) neurons, resulting in an inefficient suppression of viral replication. In 15-week-old pigs, viral replication was efficiently suppressed in the TG and CNS without induction of an extensive immune response. Furthermore, our results provide evidence that neurological disease could, at least in part, be related to viral replication and associated immunopathology in the olfactory bulb.
KEYWORDS: pseudorabies virus, neuropathogenesis, trigeminal ganglion, olfactory bulb, virulence
INTRODUCTION
Pseudorabies virus (PRV), also called suid herpesvirus 1 or Aujeszky's disease virus, is the causative agent of the economically important Aujeszky's disease in swine. PRV belongs to the family Herpesviridae, subfamily Alphaherpesvirinae, genus Varicellovirus. Members of the family Sus scrofa are the natural host and reservoir of the virus.
Current knowledge of PRV pathogenesis in its natural host, the pig, is mostly based on studies performed in the 1970s to 1990s that mainly used virus isolations and immunohistochemistry (IHC) stainings for virus detection. It is generally accepted that PRV infection starts with local replication in epithelial cells of the nasal and oral cavities; the virus can then cross the basement membrane and invade the underlying lamina propria (1–5). Via blood and lymph, PRV can spread to internal organs, either as cell-free virus or as cell-associated virus in leukocytes (6). Infection of sensory neurons innervating the infected epithelium, i.e., the trigeminal and olfactory nerves, enables the virus to invade the central nervous system (CNS). PRV can enter the trigeminal ganglia (TG) via retrograde transport through the trigeminal nerve and spread further into the sensory trigeminal nerve nuclei in the pons and medulla oblongata. From there on, virus can invade the thalamus, cerebellum, and cerebral cortex via interneuronal transmission (2). The entry of PRV into the CNS may also occur via the olfactory route by infection of olfactory neurons located in the nasal cavity mucosa, after which the virus can reach the second-order neurons located in the olfactory bulb (7). PRV replication in the upper respiratory tract, the CNS, and the reproductive organs is responsible for the respiratory, nervous, and reproductive symptoms, respectively (8).
During a productive PRV infection, viral genes are transcribed in a temporally ordered sequence consisting of three major classes of transcripts (immediate early, early, and late genes) (9, 10). A key characteristic of PRV infection is, however, that the virus can also go into a dormant state, called latency, that is predominantly established in neurons of the trigeminal ganglion (11). Latency is characterized by the presence of the viral genome as an episome in the cell without the production of infectious virus particles. The only transcripts that are detectable during latency are the specific latency-associated transcripts (LATs) (12). Whether or not the latency of alphaherpesviruses is established in the trigeminal ganglion is generally accepted to depend on a complex interplay between the neuronal environment, the virus, and the immune system (13).
The clinical outcome of PRV infection is greatly influenced by the age of the infected pig. Morbidity and mortality associated with PRV infection are higher in younger pigs and are typically associated with CNS symptoms. Older swine mostly exhibit symptoms of respiratory and reproductive disease. The cause of high mortality in piglets has historically been attributed to viral encephalitis (10), but the underlying mechanisms involved in the age-dependent differences in virulence remain unknown.
To study the neuropathogenesis of PRV in its natural host, an in vivo infection experiment was performed wherein domestic pigs of 2- and 15-week-old pigs were inoculated with the PRV reference strain NIA3. Virus spread and replication were studied using quantitative PCR (qPCR), quantitative reverse transcription-PCR (qRT-PCR), and immunofluorescence stainings. It was further evaluated whether local cytokine production correlated with observed virus replication and spread and whether potential explanations could be found for the observed age-dependent differences in clinical symptoms upon infection.
RESULTS
Clinical symptoms.
Intranasal inoculation of 2-week-old piglets with the NIA3 reference strain resulted in important clinical disease and high mortality rapidly after inoculation. Starting from 2 days postinfection (p.i.), an increase in rectal body temperature (between 40 and 41.3°C) and a general depression of all piglets, associated with severe diarrhea and vomiting, was observed. The symptoms worsened at 3 and 4 days p.i. Piglets exhibited complete exhaustion and were unable to stand up, so respiratory (nasal discharge, coughing, and heavy breathing) and neurological (circling, scratching, trembling, and paresis) symptoms could only be observed to a limited extent. Euthanasia of two piglets was scheduled for 1, 2, 3, and 4 days p.i., and additional piglets were euthanized at 2 days (one piglet), 3 days (one piglet), and 4 days (two piglets) p.i. on ethical grounds.
Also, 15-week-old sows showed important respiratory and neurological symptoms after inoculation with the NIA3 PRV reference strain. Morbidity and mortality were, however, lower than among the young piglets. The general condition of the sows started to decrease from day 2 p.i. and was associated with a loss of appetite and lethargy. On day 3, their condition worsened, accompanied by a rise in rectal temperature, which exceeded 41°C for all pigs over the next few days. Both respiratory and neurological symptoms were observed from day 4 p.i., which became more severe over the next several days. Most pigs developed the typical neurological symptoms associated with Aujeszky's disease, such as scratching, lack of coordination, circling, seizures, and paralysis between days 4 and 7 p.i. Respiratory symptoms, including dyspnea, nasal discharge, sneezing, coughing, and respiratory distress, were also observed. As planned, two predefined sows were euthanized on 1, 2, 3, 5, and 7 days p.i. Furthermore, one pig (N09) died overnight between days 5 and 6 p.i., and one additional pig (N10) was euthanized at day 6 p.i. as a consequence of severe neurological symptoms. Therefore, only two pigs remained, which were scheduled to be euthanized on days 14 and 28 p.i. It was decided to euthanize both pigs at day 14 p.i. in order to have duplicate data at that time point.
PRV DNA detection in the nervous system and visceral organs.
Analysis of the presence of viral DNA by qPCR in the different tissues collected at the day of euthanasia indicated that the NIA3 strain quickly disseminated throughout the bodies of both 2- and 15-week-old sows after intranasal inoculation (Fig. 1).
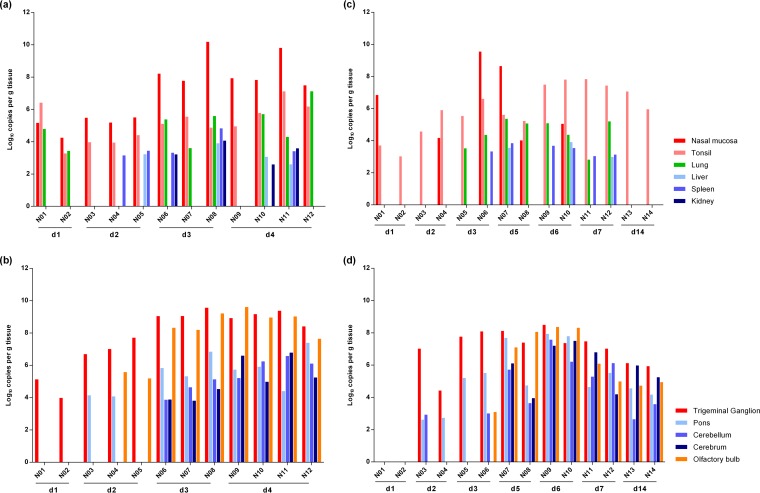
FIG 1.
Distribution of viral DNA in different tissues after NIA3 infection. Several tissues were collected from domestic 2- and 15-week-old pigs at different time points after intranasal inoculation with the NIA3 reference strain and tested by qPCR directed against gB. Viral DNA concentrations for different primary and secondary sites of replication for 2- and 15-week-old pigs (a and c, respectively) and for peripheral and central nervous system tissues for 2- and 15-week-old pigs (b and d, respectively) are shown.
At 24 h p.i., viral DNA was already detected in the nasal mucosa, tonsils, and lungs of 2-week-old piglets. Importantly, virus had also already reached the trigeminal ganglion at that time point, indicating efficient infection and retrograde transport through the trigeminal nerve in these young piglets. At 48 h p.i., viral concentrations remained similar in the nasal mucosa and tonsils. Virus was no longer detected in lung tissue but had further disseminated to other peripheral organs (liver and/or spleen) in two of three euthanized piglets. In the trigeminal ganglion, viral DNA copies had increased 300-fold, and virus had also invaded the CNS, where it was detected in the pons and/or the olfactory bulb. At days 3 and 4 p.i., an additional 100-fold increase in the concentration of viral DNA was observed in the nasal mucosa, and the peripheral organs (lung, kidney, spleen, and/or liver) of most piglets were positive for viral DNA at those time points. Furthermore, an extended invasion of the CNS was observed. The virus had by this time also reached the cerebellum and cerebrum.
A similar dissemination pattern of the NIA3 strain was found in 15-week-old pigs, except that the spread to the trigeminal ganglion, CNS, and peripheral organs showed a delay of 24 h. The most striking difference in viral dissemination between pigs of both age categories was observed at the level of the nasal mucosa. Viral DNA was detected in all inoculated 2-week-old piglets between days 1 and 4 p.i., whereas only half of the 15-week-old pigs were positive between days 1 and 3 p.i. At days 7 and 14 p.i., viral DNA was no longer detected. Despite the low replication at the nasal mucosa and the delayed spread, the tonsils of both 15-week-old sows euthanized at 24 h p.i. tested positive for viral DNA. At 48 h p.i., PRV had reached the trigeminal ganglion and had already invaded the CNS, where it was detected in the pons and/or cerebellum. At day 3, 15- and 500-fold increases in viral DNA concentrations were detected in the trigeminal ganglion and pons, respectively. Virus was also present in the olfactory bulbs of one of the two sows euthanized at that time point. However, it was only at 5 days p.i. that the virus had invaded the cerebrum. Between days 5 and 7 p.i., virus was detected in the peripheral organs. Viral DNA was, however, never detected in the kidneys. Starting from day 7 p.i., the viral DNA concentrations in all positive organs started to decrease. At 14 days p.i., virus was still detectable in the different nervous tissues, although the concentrations had decreased compared to those observed during the acute phase of infection. By that time, virus was no longer detected in peripheral organs, and only the tonsils remained positive.
Viral and cytokine gene expression at distinctive sites involved in neuropathogenesis.
Viral and cytokine mRNA expression was studied at four relevant sites involved in PRV neuropathogenesis: the nasal mucosa, the trigeminal ganglion, the brain stem (pons), and the olfactory bulb.
(i) Nasal mucosa.
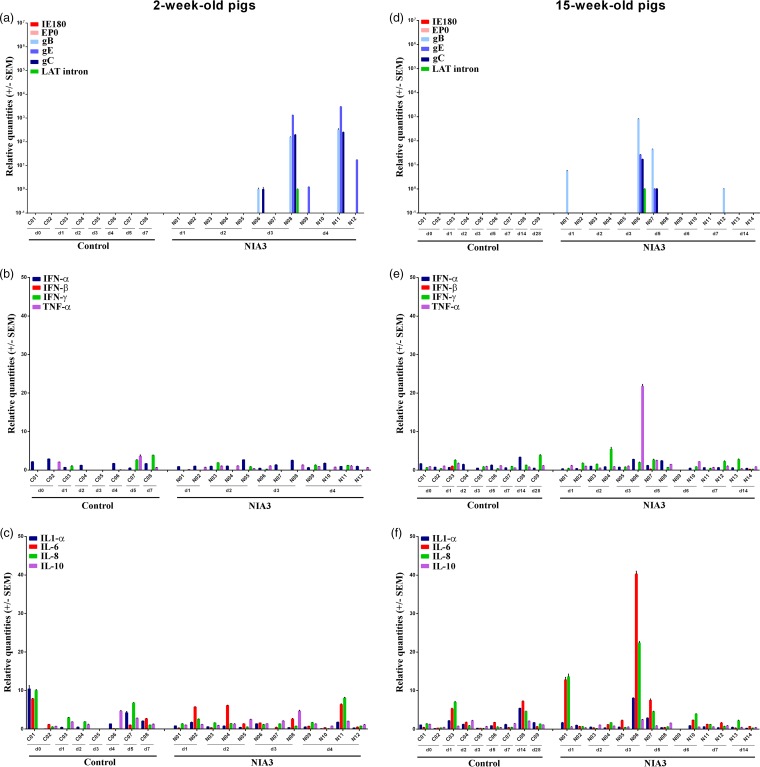
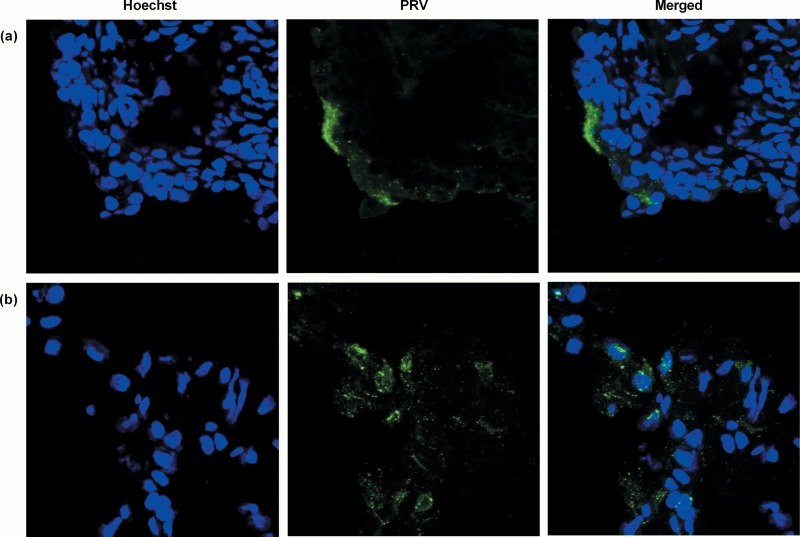
Although we showed above that PRV DNA was detected in the nasal mucosa of all 2-week-old pigs and in half of the 15-week-old pigs during the first days after NIA3 inoculation, no viral mRNA expression was detected early after infection. Viral mRNA expression was only evident from 3 days p.i. onward and to a surprisingly low extent (Fig. 2a and d). In addition, viral mRNA expression in nasal mucosa was only detected in animals that showed the highest DNA concentrations and was limited to the expression of one or more glycoproteins, except for one 2-week-old pig and one 15-week-old pig that also expressed the LAT intron. The expression of IE180 and EP0 was never detected. The low extent of viral mRNA expression was in line with the limited number of PRV plaques that we could found during analyses of immunofluorescent stainings. In 15-week-old pigs, no plaques were detected, despite the fact that 150 sections/pig were analyzed for pigs N01, N02, N03, and N04 (euthanized 1 and 2 days p.i.). Only when sections were made from 2-week-old piglet N08, which expressed the highest viral mRNA levels in nasal mucosa, was a small PRV plaque found (Fig. 3a).
FIG 2.
Nasal mucosa. Domestic 2- and 15-week-old pig were intranasally inoculated with the NIA3 strain and euthanized at different time points postinfection. mRNA expression of several PRV genes in the nasal mucosa was studied by RT-qPCR in 2- and 15-week-old pigs (a and d, respectively). Viral gene levels were expressed relative to the lowest positive sample for each age category. Furthermore, cytokine-related mRNA expression in de nasal mucosa was tested by RT-qPCR in 2- and 15-week-old pigs (panels b and c and panels e and f, respectively). Individual cytokine levels for all animals are expressed relative to the average cytokine expression in the control group (separately for 2- and 15-week-old animals).
FIG 3.
PRV immunofluorescence staining. Immunofluorescence images of the nasal mucosa (a) and olfactory bulb (b) of the NIA3-inoculated 2-week-old piglet N08 euthanized at 3 days p.i. PRV antigens (green) were stained with a combination of mouse monoclonal anti-gB and -gD antibodies (1:100 in PBS) followed by an incubation step with FITC-labeled goat anti-mouse antibodies (1:200). Nuclei (blue) were counterstained with Hoechst 33342. Images were taken with a 63× objective on a confocal microscope.
Also, only limited evidence was found for changes in cytokine expression in the nasal mucosa after the infection of pigs in both age categories with the NIA3 strain (Fig. 2b, c, e, and f). No clear upregulation of cytokines was observed in the nasal mucosa of 2-week-old piglets. Only in the 15-week-old pig, N06, euthanized at day 3 p.i., were there increases in the cytokine mRNA levels of interleukin-6 (IL-6; 40-fold), IL-8 (22-fold), and tumor necrosis factor alpha (TNF-α; 22-fold) detected. The highest viral replication was also detected in this animal.
(ii) Trigeminal ganglion.
In 2-week-old piglets, viral mRNA could be detected in the trigeminal ganglion starting from 2 days p.i.; thus, 24 h after virus had invaded the trigeminal ganglion (Fig. 4a). The highest viral mRNA levels were found on days 3 and 4 p.i., and they were comparable for all piglets euthanized at these time points. Viral mRNA of the three analyzed glycoproteins and the LAT intron was observed, but mRNA of immediate early and early genes was not detected. In 15-week-old sows, viral DNA was first detected in the trigeminal ganglion starting from day 2 p.i., correlating with the observed gC mRNA expression found in one pig at that time point (Fig. 4d). Viral replication in the trigeminal ganglion was evidenced by more extended late viral gene expression from 3 days p.i. in the trigeminal ganglia of all pigs, except for pig N05. Late viral mRNA expression was, however, less pronounced and less consistent than in 2-week-old piglets (mostly only the detection of mRNA of one or more of the glycoprotein genes). This could indicate that viral replication in the TG is at least partly inhibited in the 15-week-old pigs during the early stages of infection. At 14 days p.i., viral gene expression was no longer detected in the trigeminal ganglion, except for LAT intron expression in one of the two sows. As was the case for 2-week-old pigs, no expression of IE180 or EP0 genes was detected in the trigeminal ganglia of 15-week-old pigs. Despite the late viral mRNA expression in 2-week-old piglet N08, we did not detect any PRV-positive cells in its trigeminal ganglia by immunofluorescence staining.
FIG 4.
Trigeminal ganglion. Domestic 2- and 15-week-old pigs were intranasally inoculated with the NIA3 strain and euthanized at different time points postinfection. The mRNA expression of several PRV genes in the trigeminal ganglia was studied by RT-qPCR in 2- and 15-week-old pigs (a and d, respectively). Viral gene levels were expressed relative to the lowest positive sample for each age category. Furthermore, cytokine-related mRNA expression in the trigeminal ganglion was tested by RT-qPCR in 2- and 15-week-old pigs (panels b and c and panels e and f, respectively). Individual cytokine levels for all animals are expressed relative to the average cytokine expression in the control group (separately for 2- and 15-week-old animals).
After the inoculation of 2-week-old piglets, an upregulation of cytokine-related mRNA expression of beta interferon (IFN-β), IFN-γ, TNF-α, IL-6, and IL-10 and, to a lesser extent, IL-8 and IL-1α, was observed in the trigeminal ganglia (Fig. 4b and c). A moderate increase in TNF-α (average of 10-fold), IFN-β (average of 7-fold), IL-10 (average of 4-fold), and IL-1-α (average of 5-fold) was observed already at 2 days p.i. By the next day, the mRNA concentrations of TNF-α (average of 32-fold), IFN-β (average of 42-fold), and IL-10 (average of 14-fold) had increased notably. Furthermore, an upregulation of IFN-γ (average of 22-fold), IL-6 (average of 23-fold), and IL-8 (average of 6-fold) was also observed. From day 4 p.i. the concentrations of these upregulated cytokines started to decrease again. Interestingly, in contrast to results for 2-week-old pigs, cytokine upregulation in 15-week-old swine was clearly less pronounced and limited to a maximum 10-fold increase in IL-6, IL-8, and IL-10 between days 3 and 6 p.i. (Fig. 4e and f).
(iii) Brain stem.
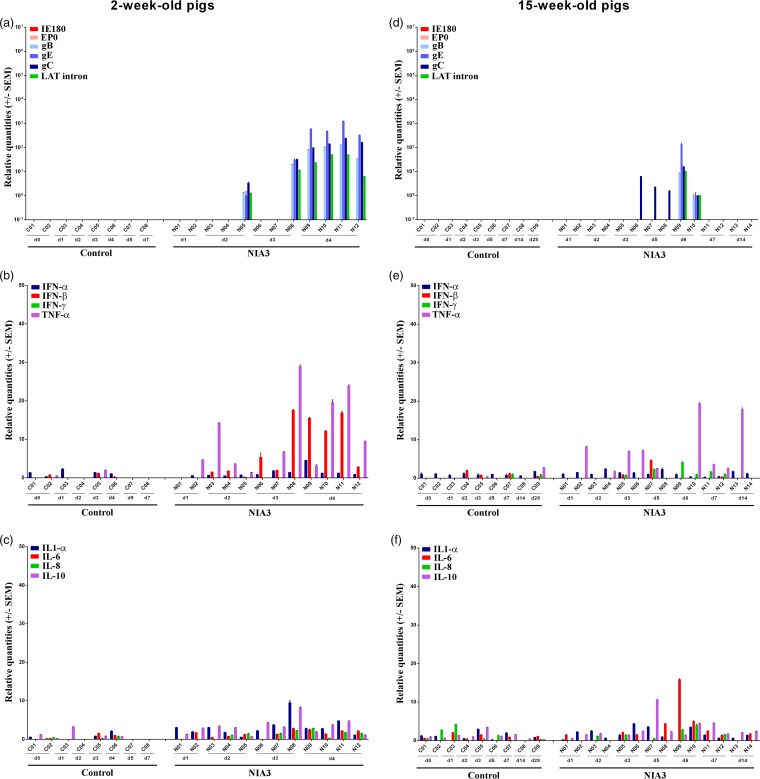
Detection of PRV DNA had shown that PRV was able to spread efficiently to the CNS and had invaded the brain stem already by day 2 p.i. in pigs of both age categories. Although viral DNA concentrations were comparable in 2- and 15-week-old pigs in the first 4 days p.i., viral mRNA expression was remarkably lower in the 15-week-old sows (Fig. 5a and d). Whereas mRNA levels of all late viral genes and LAT intron were extensively detected in all 2-week-old pigs by day 4 p.i. and already in one pig at days 2 and 3 p.i., only limited PRV-related mRNA expression was detected in five 15-week-old animals between days 3 and 6 p.i. In three of these pigs, expression was restricted to gC, indicating a suppression of late viral mRNA expression in the pons of 15-week-old pigs. As for the trigeminal ganglion, we could not confirm the presence of viral glycoproteins by immunofluorescence stainings in the pons of piglet N08 that was, however, positive for viral mRNA by qRT-PCR.
FIG 5.
Pons. Domestic 2- and 15-week-old pigs were intranasally inoculated with the NIA3 strain and euthanized at different time points postinfection. The mRNA expression of several PRV genes in the pons was studied by RT-qPCR in 2- and 15-week-old pigs (a and d, respectively). Viral gene levels were expressed relative to the lowest positive sample for each age category. Furthermore, cytokine-related mRNA expression in the pons was tested by RT-qPCR in 2- and 15-week-old pigs (panels b and c and panels e and f, respectively). Individual cytokine levels for all animals are expressed relative to the average cytokine expression in the control group (separately for 2- and 15-week-old animals).
The viral mRNA expression in the pons of 2-week-old piglets seemed to be associated with an upregulation of cytokine-related mRNA expression of IFN-β and TNF-α (Fig. 5b and c). At 2 days p.i., an increase in TNF-α (average of 6-fold) was observed. Expression continued to rise by the next day (average of 18-fold); an upregulation of IFN-β (average of 8-fold) mRNA was also observed at that time point, and the expression of both cytokines remained elevated by 4 days p.i. Cytokine upregulation in 15-week-old swine was again less clear, and no clear trend could be observed (Fig. 5e and f).
(iv) Olfactory bulb.
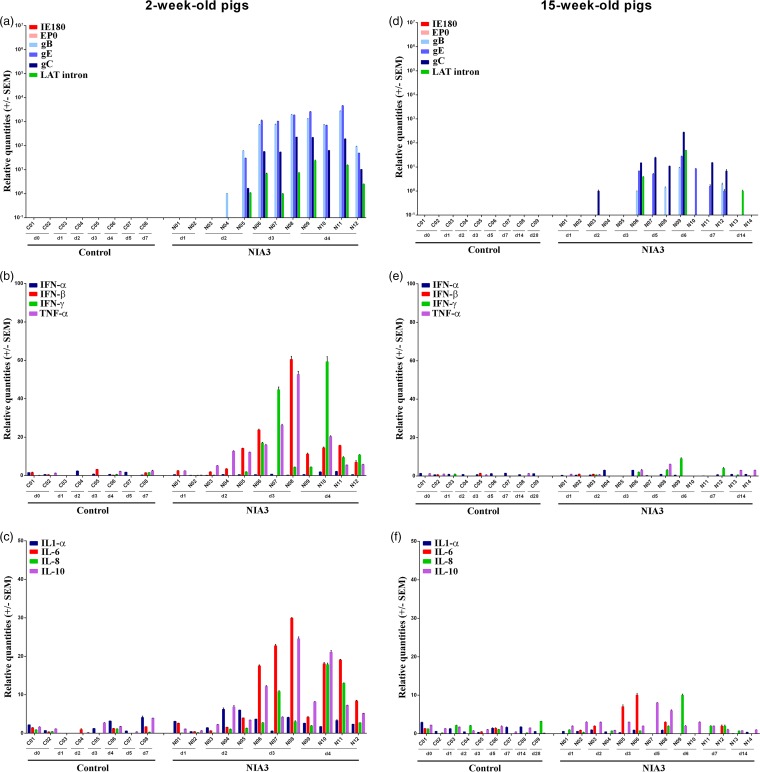
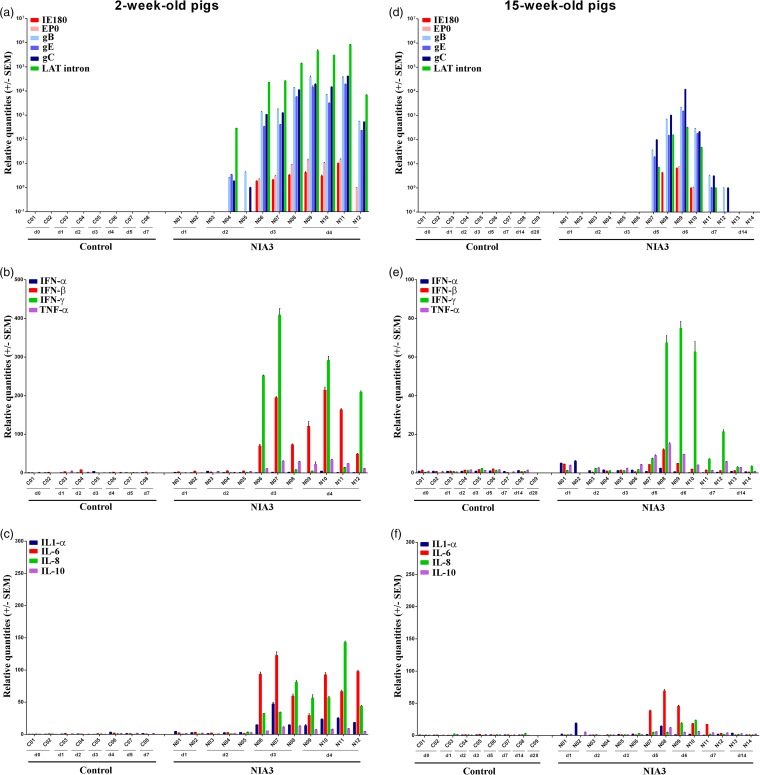
The results described above showed that immediate early and early viral gene mRNA was never detected in the nasal mucosa, trigeminal ganglia, and brain stem and late viral mRNA expression was always lower and less consistent in 15-week-old pigs than in 2-week-old pigs. The results obtained in the olfactory bulb were clearly different. Viral mRNA expression was considerably higher in the olfactory bulbs of both 2- and 15-week-old sows than in the other tissues that were analyzed, and it was the only tissue in which IE180 and EP0 mRNA expression could be detected (Fig. 6a and d). Two of three 2-week-old piglets were already positive for viral mRNA associated with glycoprotein genes and/or the LAT intron at 2 days p.i. At 3 and 4 days p.i., the mRNA concentrations had increased, associated with a consistent mRNA expression profile of immediate early, early, and late genes. Also, the LAT intron was expressed at high concentrations. For 15-week-old pigs, DNA detection had shown that PRV invaded the olfactory bulb between days 3 and 5 days p.i. Viral mRNA was only detected for the first time at day 5 p.i. By day 6, viral replication had increased, and clear expression of immediate early, early, and late genes was observed. Also, the LAT intron was detected in considerable concentrations. Interestingly, viral mRNA concentrations decreased already by day 7 p.i. By day 14 p.i., PRV-related mRNA was no longer detected. Viral replication in the olfactory bulb was confirmed by immunofluorescence stainings of the olfactory bulb of piglet N08 (Fig. 3b). Viral glycoproteins could be easily detected in the cytoplasm of cells and were visible over the entire section.
FIG 6.
Olfactory bulb. Domestic 2- and 15-week-old pigs were intranasally inoculated with the NIA3 strain and euthanized at different time points postinfection. The mRNA expression of several PRV genes in the olfactory bulb was studied by RT-qPCR in 2- and 15-week-old pigs (a and d, respectively). Viral gene levels were expressed relative to the lowest positive sample for each age category. Furthermore, cytokine-related mRNA expression in the olfactory bulb was tested by RT-qPCR in 2- and 15-week-old pigs (panels b and c and panels e and f, respectively). Individual cytokine levels for all animals are expressed relative to the average cytokine expression in the control group (separately for 2- and 15-week-old animals).
Compared to all previously discussed tissues, more pronounced cytokine mRNA expression was observed in the olfactory bulbs of both 2- and 15-week-old pigs during the first few days after NIA3 infection (Fig. 6b, c, e, and f), and the expression was higher in 2-week-old pigs than in 15-week-old pigs. The extensive upregulation of cytokine-related mRNA correlated with the expression of viral genes. At days 3 and 4 p.i. of 2-week-old pigs, a strong upregulation of IFN-β (maximum average increase of 131-fold), IFN-γ (maximum average increase of 223-fold), TNF-α (maximum average increase of 26-fold), IL-1-α (maximum average increase of 26-fold), IL-6 (maximum average increase of 92-fold), IL-8 (maximum average increase of 102-fold), and IL-10 (maximum average increase of 10-fold) was observed in the olfactory bulb. Also in 15-week-old pigs, cytokine upregulation correlated with viral replication in the olfactory bulb. An increase in cytokine-related mRNA expression was observed starting from day 5 p.i. For most upregulated cytokines, a maximum increase was observed at 6 days p.i. At this time point, increases in mRNA expression of IFN-γ (average increase of 69-fold), IL-6 (average increase of 32-fold), IL-8 (average increase of 21-fold), and TNF-α (average increase of 7-fold) were observed. Cytokine-related mRNA expression decreased again by day 7 p.i. and reached levels similar to those for the control animals by 14 days p.i.
DISCUSSION
The current knowledge on the pathogenesis of PRV in pigs mostly comes from studies performed in the last decades of the twentieth century that used virus isolations and IHC stainings for virus detection (1–5). Since more sensitive techniques such as PCR are now routinely used, we reassessed the PRV pathogenesis using those techniques. In general, our results confirmed previous observations. PRV was first detected at the nasal mucosa and tonsils, which are known places of primary replication of the virus. PRV enters the trigeminal nerve quickly after infection and reaches the TG; from there it can further invade the CNS. Viral replication at the respiratory mucosa is likely required before dissemination of PRV to the visceral organs, since virus only reached the organs several days after infection. Our results also confirmed the importance of the olfactory route as an alternative route of neuroinvasion (7, 14, 15). Despite the confirmation of the general knowledge on PRV pathogenesis, several interesting new findings resulted from the detailed study of viral and cytokine mRNA expression at important sites of neuropathogenesis.
A first observation was made at the level of the nasal mucosa, where only limited viral mRNA expression was detected in pigs of both age categories starting at 3 days p.i. These results are somewhat unexpected since previous in vivo studies with other virus strains and inoculation doses detected viral antigens in the epithelium and the underlying mucosa shortly after infection (1, 2). This indicates that the nasal mucosa only served to a limited extent as a primary replication site in our study. Most probably, primary replication occurred at another site known to be used for this purpose, such as the tonsils, oropharynx, or olfactory mucosa (reviewed in reference 16), and this difference may be due to differences in the PRV inoculation dose used. In the present study, a inoculation dose of 105 50% tissue culture infective doses (TCID50)/pig was used. This dose was chosen based on literature data showing that it is sufficient to cause important clinical disease after intranasal NIA3 infection of pigs (17, 18) and on the basis of a report by Wittmann (19) showing that 105 TCID50 is sufficient to infect pigs of all different age categories. This dose, however, is clearly lower than the inoculum doses that have been used in other studies, e.g., 1.25 × 108 PFU/pig (NIA3 strain) and 107 TCID50/pig (Kaplan strain) (1, 2). Other factors, such as differences in virulence between the viral strains used and the ages of the pigs, are also known to affect the outcome of a PRV infection (19) and may have contributed to our unexpected observations in the nasal mucosa.
More difficult to explain is the fact that despite the limited detection of viral mRNA, viral DNA was consistently detected in considerable amounts in the nasal mucosa in all of the 2-week-old pigs and in half of the 15-week-old pigs. The most plausible hypothesis appears to be that the detected DNA comes from virus that was produced at another replication site. Similar findings have already been described after intranasal inoculation of 1-week-old piglets with a gE deletion mutant of the Kaplan PRV strain (2). Although the lower stability of viral mRNA than of DNA may also affect these observations, this factor appears to be of lesser importance since viral mRNA expression was detected at later time points.
Although only limited evidence was found for viral replication in the nasal mucosa, PRV was able to quickly reach the TG in pigs of both age categories. This rapid spread to the TG suggests that PRV does not need to replicate extensively in the respiratory epithelium and may not need to breach the basement membrane to get efficient access to TG neurons. It therefore further supports different lines of evidence showing that nerve endings are present in the respiratory epithelium that can be accessed directly (2, 20, 21). The observation that PRV only reached the peripheral organs at later time points supports the need to cross the basement membrane to get access to blood and lymph.
In 2-week-old piglets, severe general and CNS symptoms were observed shortly after inoculation with the NIA3 strain. This seems to correlate with the extensive viral replication that was observed in the TG and the brain stem and the associated increase in cytokine-related mRNA expression. We found an increased expression of particularly IFN-β, TNF-α, IL-6, IL-10, and IFN-γ mRNA in the TG after PRV infection. Although speculative at this point, this suggests an infiltration of mainly macrophages and γδ TCR+ T cells (and/or NK cells), which would be in line with observations done after corneal HSV-1 infection of mice (22–24). These authors suggested that the γδ TCR+ T cells probably exert a function in suppressing HSV-1 replication by the production of IFN-γ. IFN-γ not only possesses antiviral activity but is also a potent activator of macrophages, which in turn produce proinflammatory cytokines such as TNF-α, IL-6, RANTES (“regulated on activation, normal T cell expressed and secreted”), type I IFNs, and nitric oxide (NO). Cytokine expression upon alphaherpesvirus infection of the brain stem has, however, been less well studied. We observed an upregulation of mainly TNF-α and IFN-β; this is probably the result of activation of microglia cells, the resident macrophages of the CNS. The induced immune responses at the TG and brain stem were, however, not able to block virus replication and prevent clinical disease. In line with the assumption that the high morbidity and mortality in young piglets upon PRV infection are linked to viral encephalitis, our results suggest that the observed symptoms are the result of neuronal damage caused by a combination of viral replication and immune-mediated pathology.
Despite the high expression of viral glycoprotein mRNA in the TG and brain stem of 2-week-old piglets, IE180 and EP0 mRNA expression were not detected. On the one hand, this can partly be explained by published in vitro data (25) showing that IE180 mRNA is only expressed for a limited period postinfection and is less stable than other viral mRNAs after PRV infection. We confirmed this by showing that PRV NIA3 infection of PK15 cells resulted in (i) a 10- to 100-fold-lower IE180 mRNA expression than EP0 and glycoprotein mRNA expression during the first 10 h p.i. and (ii) a significantly faster reduction in IE180 and EP0 mRNA levels (5- to 7-fold reduction) between 10 and 24 h p.i. than in glycoprotein mRNA reduction levels (1- to 2-fold reduction) (data not shown). On the other hand, the fact that the IE180 and EP0 qPCRs used were also 100 times less sensitive than the glycoprotein qPCRs contributes to this observed discrepancy in the detection of different viral mRNAs.
As indicated in the introduction, clinical symptoms observed upon PRV infection depend on the age of the pig at the moment of inoculation and tend to be less severe with increasing age of the animals. This was also observed in our study, and a comparison of viral and cytokine mRNA expression in the TG and brain stems of 2- and 15-week-old pigs provides some potential explanations. Viral mRNA expression in the TG and pons was less pronounced and less consistent in 15-week-old pigs than in 2-week-old piglets. This difference in viral replication furthermore correlated with a different expression pattern of cytokine-related mRNA in pigs of both age categories. Although a strong upregulation of especially proinflammatory cytokines and IFN-γ mRNA was observed in 2-week-old piglets, almost no changes in cytokine mRNA were detected in 15-week-old pigs. This suggests that the efficient suppression of viral replication in the TG and CNS of 15-week-old pigs may be mainly the result of intrinsic factors associated with neuronal cell biology and may be less due to an induced immune cell infiltration and associated cytokine production. Several cellular and viral properties of neurons that contribute to limiting herpesvirus replication have been described and include, among other features, a lower abundance of viral and cellular transcription factors in the nuclei of sensory neurons and the capacity of LAT transcript to repress viral lytic transcription (13, 26). Also, recent in vitro results showing that cytokine responses at the axon endings are sufficient to limit virus spread toward the cell body (27) are consistent with the hypothesis that the immune response at the level of the TG itself may be less important in limiting viral replication at that site.
Analysis of viral replication and cytokine expression at the olfactory bulb revealed some remarkable results. Extensive viral replication was detected in the olfactory bulb in pigs of both age categories; this replication was significantly higher than the viral replication in other neuronal tissues (TG and brain stem). The olfactory bulb was the only tissue where, in addition to viral glycoprotein mRNA, IE180 and EP0 mRNAs were also detected. In addition to the contributing factors mentioned above to explain the discrepancy between detection of different viral mRNAs, this suggests that tissue-dependent differences might also exist in the expression or stability of IE180 and EP0 mRNA transcripts. Extensive viral replication in the olfactory bulb further correlated with a robust expression of cytokine mRNA. Cytokine expression was most pronounced in 2-week-old pigs and included the upregulation of IFN-γ, IFN-β, IL-6, IL-8, and, to a lesser extent, TNF-α, IL-1α, and IL-10. Cytokine expression in 15-week-old pigs was somewhat less pronounced but still showed a >50-fold increase in IFN-γ and IL-6 mRNA expression. Little is currently known about the immune cells responsible for cytokine production in the olfactory bulb, but the large increase in IFN-γ suggests an infiltration of T or NK cells into the inflamed area. Although hypothetical, the high viral replication and associated immune response in the olfactory bulb seem to be linked to the neurological symptoms observed in pigs of both age categories more than the viral replication and cytokine expression in the TG and brain stem. In this respect, it is important to note that the 15-week-old pig that spontaneously died at day 5 p.i., N09, and the one that was euthanized due to the severe neurological symptoms at day 6 p.i., N10, were among those with the highest IFN-γ expression in the olfactory bulb. As such, our data suggest that infection of the olfactory bulb may play a significant role in severe PRV pathology and add to previous findings showing the importance of the olfactory bulb for neuropathogenesis and invasion of the CNS (7, 14, 15).
MATERIALS AND METHODS
Animals and viruses.
Twenty-three 15-week-old and twenty 2-week-old Belgian Landrace sows were purchased from a commercial swine herd. The sows were in good condition and tested negative in serology for PRV and classical swine fever at the beginning of the experiment. The animals were housed in BSL3 facilities on slatted floors (CODA-CERVA, Machelen). Water was available ad libitum, and the pigs were fed once each day. The animals were randomly assigned to the control or infection group. Animal experiments were performed in accordance with EU and Belgian regulations on animal welfare in experimentation. The protocol was approved by the joint ethical committee of CODA-CERVA and the Scientific Institute of Public Health Belgium (procedure agreement 121017-02). A third passage of the virulent PRV NIA3 reference strain was used.
Experimental design.
In the first experiment, nine 15-week-old sows were mock infected with PBS, and fourteen sows were intranasally inoculated (1 ml/nostril) with the PRV reference strain NIA3. A small nebulizer (1-mm spray opening) fixed on a syringe was used to drip the final infectious dose of 105 TCID50/animal into the nostrils. The sows were monitored daily for clinical symptoms and rectal body temperature. Euthanasia of one predefined pig of the control group and two predefined pigs of the NIA3-inoculated group was scheduled to occur on days 1, 2, 3, 5, 7, 14, and 28 p.i. Two additional pigs of the control group were euthanized on the day of infection (0 days p.i.). During the experiment, the animals were euthanized at the intended date or when necessary based on ethical grounds. At euthanasia, blood and several tissues (nasal mucosa, tonsils, lung, kidney, liver, spleen, olfactory bulb, trigeminal ganglion, pons, cerebellum, and cerebrum) were collected using new disposable scalpels and forceps per tissue sample to eliminate the risk of cross-contamination. Tissues were divided over multiple 2.0-ml tubes and stored at −70°C until testing.
In the second experiment, twelve female 2-week-old piglets were intranasally inoculated (0.5 ml/nostril) via a small nebulizer fixed on a syringe with a final infectious dose of 105 TCID50/animal of the NIA3 reference strain. Eight piglets were mock inoculated with phosphate-buffered saline (PBS) and held as a negative-control group. Piglets were monitored daily for clinical symptoms and rectal body temperature. Euthanasia of one predefined pig of the control group and two predefined pigs of the NIA3-inoculated group was scheduled to occur on days 1, 2, 3, 4, 5, and 7 p.i. Two additional pigs of the control group were euthanized on the day of infection (0 days p.i.). During the experiment, the animals were euthanized at the intended date or when necessary based on ethical grounds. The same samples, as for the 15-week-old pigs, were collected at the moment of euthanasia.
Sample preparation and DNA extraction.
For all collected organs except the tonsils, about 0.5 cm3 of tissue was homogenized in 1 ml of PBS by adding 10 to 15 silicon carbide beads of 1 mm (Biospec Products, Inc.) and high-speed shaking (4 min, 25 Hz) in a TissueLyser (Qiagen). For the homogenization of tonsil samples, about 0.5 cm3 of tissue was homogenized in 1 ml of PBS by adding two stainless steel beads of 5 mm (Qiagen) and high-speed shaking (10 min, 30 Hz) in a TissueLyser. For the 2-week-old piglets, the homogenization of samples was done in only 0.5 ml of PBS because of the limited amount of sample available. Genomic DNA was extracted from the homogenized tissues using a QIAamp DNA kit (Qiagen) according to the manufacturer's instructions.
RNA extraction and cDNA synthesis.
Extraction of total RNA from the homogenized tissue preparations was done by using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. Extracted total RNA was treated with Turbo DNase (Ambion) according to the manufacturer's instructions in a 50-μl reaction mixture to eliminate contaminating DNA. Subsequently, DNase-treated RNA samples were converted to cDNA using a Moloney murine leukemia virus reverse transcriptase (M-MLV RT) system (Life Technologies). For each reaction, a mix of 4 μl of RNA, 4 μl of 5× first-strand buffer, 2 μl of 0.1 M dithiothreitol, 1 μl of 10 nM dNTP mix (Roche, Basel, Switzerland), 0.2 μl of 10× hexanucleotide mix (Roche), 0.5 μl of M-MLV RT, and 8.3 μl of H2O was prepared, followed by incubation at 37°C for 60 min and then inactivation at 95°C for 10 min. For cDNA synthesis of the IE180 and EP0 templates, 1 μl of sequence-specific primers (10 μM concentrations of both reverse primers for IE180 and EP0) were used instead of hexanucleotides.
Preamplification.
A preamplification step of the cDNA was performed using a TaqMan PreAmp master mix (Applied Biosystems) according to the manufacturer's instructions to increase the amount of available material. A mix of primers (Table 1) for the selected reference genes, cytokines and viral genes (except IE180 and EP0) was made to a final concentration of 0.18 μM per primer. A separate mix containing only the reverse primers for the IE180 and EP0 gene was made since these genes mostly overlap the oppositely transcribed large latency transcript. Preamplification reaction conditions involved the amplification of 12.5 μl of cDNA in a 50-μl reaction consisting of 25 μl of TaqMan PreAmp master mix and 12.5 μl of pooled assay mix. Preamplification consisted of 14 cycles on a thermocycler (Biometra) using the following program: denaturation at 95°C for 10 min and 14 cycles of amplification (15 s at 95°C, 4 min at 60°C). The preamplified products were diluted at a ratio of 1:10 and used as the templates for the real-time qPCR analysis.
TABLE 1.
Primers and probes for seven porcine reference genes, eleven cytokines, and six PRV genes were either selected from previous studies or designed using the PrimerQuest tool
| Gene or cytokine | Description | Primer or probe sequence (5′–3′) |
Reference or source | ||
|---|---|---|---|---|---|
| Forward primer | Reverse primer | Probe | |||
| Reference genes | |||||
| ACTB | β-Actin | AGCGCAAGTACTCCGTGTG | CGGACTCGTCGTACTCCTGCTT | TCGCTGTCCACCTTCCAGCAGATGT | 29 |
| B2M | β2-Microglobulin | AAACGGAAAGCCAAATTACC | ATCCACAGCGTTAGGAGTGA | AGAAGATGAACGCGGAGCAGTCAG | 30 (primers); PrimerQuest (probe) |
| GADPH | Glyceraldehyde-3-phosphate dehydrogenase | ACATGGCCTCCAAGGAGTAAGA | GATCGAGTTGGGGCTGTGACT | CCACCAACCCCAGCAAGAGCACGC | 29 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | GTGATAGATCCATTCCTATGACTGTAGA | TGAGAGATCATCTCCACCAATTACTT | ATCGCCCGTTGACTGGTCATTACAGTAGCT | 28 |
| PPIA | Peptylpropyl isomerase A of cyclophilin A | TGCTTTCACAGAATAATTCCAGGATTTA | GACTTGCCACCAGTGCCATTA | TGCCAGGGTGGTGACTTCACACGCC | 28 |
| SDHA | Succinate dehydrogenase complex, subunit A, flavoprotein | GAACCGAAGATGGCAAGA | CAGGAGATCCAAGGCAAA | CTGCACACGTTGTACGGAAGGTCT | Primers (30); PrimerQuest (probe) |
| UBC | Ubiquitin C | GCGCACCCTGTCTGACTACA | AGATCTGCATCCCACCTCTGA | AGTCCACCCTGCACCTGGTCCTCC | 31 |
| Cytokines | |||||
| IFN-α | Interferon alpha | TGGTGCATGAGATGCTCCA | GCCGAGCCCTCTGTGCT | CAGACCTTCCAGCTCT | 29 |
| IFN-β | Interferon beta | AGCAGATCTTCGGCATTCTC | GTCATCCATCTGCCCATCAA | TAGCACTGGCTGGAATGAAACCGT | PrimerQuest |
| IFN-γ | Interferon gamma | CGATCCTAAAGGACTATTTTAATGCAA | TTTTGTCACTCTCCTCTTTCCAAT | ACCTCAGATGTACCTAATGGTGGACCTCTT | 28 |
| TNF-α | Tumor necrosis factor alpha | AACCTCAGATAAGCCCGTCG | ACCACCAGCTGGTTGTCTTT | CCAATGCCCTCCTGGCCAACG | 29 |
| IL-1α | Interleukin-1α | GTGCTCAAAACGAAGACGAACC | CATATTGCCATGCTTTTCCCAGAA | TGCTGAAGGAGCTGCCTGAGACACCC | 28 |
| IL-1β | Interleukin-1β | GTGCTGGCTGGCCCACA | GAACACCACTTCTCTCTTCA | CTCTCCACCTCCTCAAAGGG | 29 |
| IL-2 | Interleukin-2 | TGCTGATCTCTCCAGGATGC | CCTCCAGAGCTTTGAGTTCTTCTACTA | AAGCAGGCTACAGAATTGAAACACCTT | 29 |
| IL-4 | Interleukin-4 | GTCTGCTTACTGGCATGTACCA | GCTCCATGCACGAGTTCTTTCT | CCACGGACACAAGTGCGACATCACCTTAC | 29 |
| IL-6 | Interleukin-6 | CTGGCAGAAAACAACCTGAACC | TGATTCTCATCAAGCAGGTCTCC | TGGCAGAAAAAGACGGATGC | 29 |
| IL-8 | Interleukin-8 | AAGCTTGTCAATGGAAAAGAG | CTGTTGTTGTTGCTTCTCAG | TCTGCCTGGACCCCAAGGAAAAGT | 29 |
| IL-10 | Interleukin-10 | CGGCGCTGTCATCAATTTCTG | CCCCTCTCTTGGAGCTTGCTA | AGGCACTCTTCACCTCCTCCACGGC | 28 |
| Viral genes | |||||
| IE180 | Immediate early 180 | GAAGCGGGCGAGAGAAAT | GGGATTTCGCGAACTT | TCGCGAGGACCATTTGCATGC | PrimerQuest |
| EP0 | Early protein 0 | TTTGACATATCGGCGCAGT | AGAGTGTGCCTCGGACT | ATGAGCCACCAGGGTGTGAACTAT | PrimerQuest |
| gB | Glycoprotein B | ACCACCGGCGCTACTTTAAG | CCTCCAGCAGCGTCAGGTT | ACGATCAGCACGCGGGTGACC | PrimerQuest |
| gC | Glycoprotein C | CTTCGAGGTCCGCTTCTAC | TTGGCGGAGCTGAAGAG | CCGAGTACTTTGACGAGCCC | PrimerQuest |
| gE | Glycoprotein E | GCTTCCACTCGCAGCTCTTCT | GATGCAGGGCTCGTACACGTA | CCGCGCGTGGTCTCGGACA | PrimerQuest |
| LAT intron | Intron of latency-associated transcript | CTTCTTCACACGGAGGACAC | AAGCCACGCTGATACTCTTG | ATGGGCAGCAGATGGTACGGTC | PrimerQuest |
The preamplification uniformity of the different target sequences was checked according to the manufacturer's protocol. Reference samples for the different tissues (nasal mucosa, trigeminal ganglion, and CNS tissues) were tested in duplicate before and after preamplification for the different reference genes, viral genes, and cytokines. Subsequently, PreAmp uniformity values related to the reference genes ACTB, GADPH, PPIA, and UBC were calculated. Therefore, the normalized CT values were calculated for the different target genes before (cDNA) and after preamplification (PreAmp) according to the following formulas: ΔCT [cDNA] = average CT [cDNA target gene] − average CT [cDNA reference gene] and ΔCT [PreAmp] = average CT [PreAmp target gene] − average CT [PreAmp reference gene], respectively. The ΔΔCT was determined from the difference of the two ΔCTs: ΔΔCT = ΔCT [PreAmp] − ΔCT [cDNA]. A ΔΔCT close to zero indicates ideal PreAmp uniformity and a range of PreAmp uniformity values between −1.5 and +1.5 is acceptable. The mean PreAmp uniformity values for the investigated gene assays in nasal mucosa tissue related to the reference genes ACTB, GADPH, PPIA, and UBC were −0.38 ± 0.47, 0.48 ± 0.47, 0.24 ± 0.47, and 0.64 ± 0.47, respectively. The mean PreAmp uniformity values for the investigated gene assays in trigeminal ganglion tissue related to the reference genes ACTB, GADPH, PPIA, and UBC were −0.39 ± 0.96, 0.25 ± 0.65, 0.71 ± 0.96, and 0.95 ± 0.96, respectively. The mean PreAmp uniformity values for the investigated gene assays in CNS tissue related to the reference genes ACTB, GADPH, PPIA, and UBC were −0.53 ± 0.76, 0.40 ± 0.62, 0.58 ± 0.76, and 0.57 ± 0.76, respectively. Depending on the tissue tested and the reference gene used, the individual PreAmp uniformity values for the gE and IL-1α assay slightly exceeded 1.5 but were accepted for this study.
Quantitative real-time PCR (qPCR) assay design and analysis.
Primers and probes for seven porcine reference genes (ACTB, B2M, GADPH, HPRT1, PPIA, SDHA, and UBC), eleven cytokines (IFN-α, IFN-β, IFN-γ, TNF-α, IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-8, and IL-10), and six PRV genes (IE180, EP0, glycoprotein B [gB], gC, gE, and LAT intron) were either selected from previous studies (28–31) or designed using the PrimerQuest tool (Integrated DNA Technologies [IDT], Leuven, Belgium). Porcine sequences available in the NCBI databases were used to design qPCR assays, taking into account the amplicon specificities of the primers (BLAST). The synthesis of oligonucleotides was carried out by IDT. All probes were labeled with 6-carboxyfluorescein (FAM). Sequences and corresponding references are shown in Table 1. The detection limits, linear ranges, and efficiencies of the PRV qPCRs were determined by testing a 10-fold dilution series of an NIA3 PRV virus stock in PBS. This was done in triplicate, and the results are summarized in Table 2. For porcine reference genes and cytokines, a dilution of a reference sample was tested for each gene to acquire PCR efficiencies and associated standard errors using qBasePlus software (Biogazelle).
TABLE 2.
Detection limit, linear range, and efficiency of PRV qPCRs
| Target | Limit of detection (TCID50/ml) | Linear range (TCID50/ml) | Efficiency (%) |
|---|---|---|---|
| IE180 | 21 | 21–106.33 | 113 |
| EP0 | 21 | 21–106.33 | 110 |
| gB | 0.2 | 0.2–106.33 | 104 |
| gC | 0.2 | 0.2–106.33 | 93 |
| gE | 0.2 | 0.2–106.33 | 98 |
| LAT | 2 | 2–106.33 | 96 |
The qPCRs were performed in a final volume of 20 μl, containing 5 μl of DNA or cDNA, 10 μl of 2× FastStart TaqMan Probe master mix (Roche), 1 μl of probe (final concentration, 0.25 μM), 1 μl of a mix containing the forward and reverse primers (both at a final concentration of 0.9 μM), and 3 μl of H2O. The following qPCR temperature cycle was applied: denaturation by a hot start at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 45 s. All experiments were performed in duplicate on a LightCycler 480 real-time PCR system (Roche). In all qPCR analyses, negative extraction controls and negative and positive amplification controls were included, and tests were only validated when all controls were satisfactory.
Construction of a standard curve for qPCR for the gB assay.
A gB DNA standard curve was constructed for absolute quantification of the viral DNA present in the different tissues. A plasmid containing part of the gB sequence was 10-fold serially diluted resulting in a dilution series from 5 × 109 to 5 × 10−1 copies/μl. All dilutions were tested in the qPCR described above. Three independent replicates were run, the mean values of each dilution were calculated, and a standard curve was constructed by plotting the CT values against the log of the input DNA copy number.
geNorm analysis and relative quantification.
A pilot study was set up to identify the best set of reference genes to be used for normalization of qPCR data (32). Ten, fifteen, or eleven representative samples of nasal mucosa, trigeminal ganglion, or CNS tissue, respectively, were selected and analyzed for seven porcine reference genes. The obtained data were analyzed using the geNorm application available in qBasePlus software. The geNorm application ranks genes according to their stability and determines the optimal number of reference genes required for the calculation of a reliable normalization factor. This pilot study determined the most suitable and required number of reference genes for the different tissues: UBC and GADPH for nasal mucosa samples; ACTB, GADPH, and PPIA for trigeminal ganglion samples; and UBC and PPIA for CNS samples.
The quantification and normalization of results were based on the calculation of target CT values and reference gene CT values in qBasePlus software. All samples were run in duplicate for the target and reference genes. The relative expression levels were normalized with respect to the selected reference genes and to technical and experimental errors. Relative expression quantification analysis relied on the qBasePlus method (33). Individual cytokine levels for all animals were expressed relative to the average of the control group (separately for 2- and 15-week-old animals). Viral gene levels were expressed relative to the lowest positive sample for each age category.
Immunofluorescence staining.
During sample collection at the moment of euthanasia, parts of the nasal mucosa, trigeminal ganglion, pons, and olfactory bulb were embedded in Methocel (Fluka) and frozen at −70°C. Cryosections of 5 μm were made from tissues of selected animals based on the qPCR results and fixed in methanol (−20°C, 100%, 25 min). PRV-infected cells were visualized by 1 h of incubation of the cryosections with a combination of mouse monoclonal anti-gB and -gD antibodies (1:100 in PBS), followed by 1 h of incubation with fluorescein isothiocyanate (FITC)-labeled goat anti-mouse antibodies (1:200). In addition, all cryosections were incubated for 15 min with Hoechst 33342 (Invitrogen). Cryosections were washed three times with PBS after the different incubation steps. Finally, the cryosections were mounted with glycerin–Dabco 33-LV. Images of the immunofluorescence-stained cryosections were acquired using a Leica TCS SPE confocal microscope (Leica Microsystems GmbH, Heidelberg, Germany). To excite Hoechst counterstaining and FITC fluorochromes, respectively, 408- and 488-nm laser lines were used.
ACKNOWLEDGMENTS
We acknowledge veterinarians Willem Van Campe and Laurent Mostin and the animal caretakers of CODA-CERVA Machelen for their assistance in carrying out the biological sample collection, and we thank the technical personnel of the unit Enzorem for help with the analysis of the collected samples.
This study was funded by the Federal Public Service of Health, Food Chain Safety and Environment (RF11/6249), Belgium; the Federal Science Policy, BELSPO (BR/132/PI/melatin-PRV); the Concerted Research Action 01G01311 of the Research Council of Ghent University, Belgium; and the IAP (phase VII) BELVIR consortium sponsored by BELSPO.
REFERENCES
- 1.Pol JMA, Gielkens ALJ, Oirschot Van JT. 1989. Comparative pathogenesis of three strains of pseudorabies virus in pigs. Microbial 361–371. [DOI] [PubMed] [Google Scholar]
- 2.Kritas SK, Pensaert MB, Mettenleiter TC. 1994. Invasion and spread of single glycoprotein deleted mutants of Aujeszky's disease virus (Adv) in the trigeminal nervous pathway of pigs after intranasal inoculation. Vet Microbiol 40:323–334. doi: 10.1016/0378-1135(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 3.Sabó A, Rajcáni J, Blasković D. 1968. Studies on the pathogenesis of Aujeszky's disease. I. Distribution of the virulent virus in piglets after peroral infection. Acta Virol 12:214–221. [PubMed] [Google Scholar]
- 4.Sabó A, Rajcáni J, Blaskovic D. 1969. Studies on the pathogenesis of Aujeszky's disease. III. The distribution of virulent virus in piglets after intranasal infection. Acta Virol 13:407–414. [PubMed] [Google Scholar]
- 5.Wittmann G, Jakubik J, Ahl R. 1980. Multiplication and distribution of Aujeszky's disease (pseudorabies) virus in vaccinated and nonvaccinated pigs after intranasal infection. Arch Virol 66:227–240. doi: 10.1007/BF01314736. [DOI] [PubMed] [Google Scholar]
- 6.Nauwynck HJ, Pensaert MB. 1995. Cell-free and cell-associated viremia in pigs after oronasal infection with Aujeszky's disease virus. Vet Microbiol 43:307–314. doi: 10.1016/0378-1135(94)00103-4. [DOI] [PubMed] [Google Scholar]
- 7.Kritas SK, Pensaert MB, Mettenleiter TC. 1994. Role of envelope glycoproteins Gi, Gp63, and Giii in the invasion and spread of Aujeszky's disease virus in the olfactory nervous pathway of the pig. J Gen Virol 75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 8.Nauwynck H, Glorieux S, Favoreel H, Pensaert M. 2007. Cell biological and molecular characteristics of pseudorabies virus infections in cell cultures and in pigs with emphasis on the respiratory tract. Vet Res 38:229–241. doi: 10.1051/vetres:200661. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Porat T, Kaplan AS. 1985. Molecular biology of pseudorabies virus, p 105–173. In Roizman B. (ed), The herpesviruses, vol 3 Plenum Press, New York, NY. [Google Scholar]
- 10.Pomeranz LE, Reynolds AE, Hengartner CJ. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev 69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutekunst DE, Pirtle EC, Miller LD, Stewart WC. 1980. Isolation of pseudorabies virus from trigeminal ganglia of a latently infected sow. Am J Vet Res 41:1315–1316. [PubMed] [Google Scholar]
- 12.Cheung AK. 1991. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J Virol 65:5260–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Divito S, Cherpes TL, Hendricks RL. 2006. A triple entente: virus, neurons, and CD8+ T cells maintain HSV-1 latency. Immunol Res 36:119–126. doi: 10.1385/IR:36:1:119. [DOI] [PubMed] [Google Scholar]
- 14.Olander HJ, Saunders JR, Gustafson DP, Jones RK. 1966. Pathologic findings in swine affected with a virulent strain of Aujeszky's virus. Pathol Vet 3:64–82. doi: 10.1177/030098586600300104. [DOI] [PubMed] [Google Scholar]
- 15.Gerdts V, Beyer J, Lomniczi B, Mettenleiter TC. 2000. Pseudorabies virus expressing bovine herpesvirus 1 glycoprotein B exhibits altered neurotropism and increased neurovirulence. J Virol 74:817–827. doi: 10.1128/JVI.74.2.817-827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nauwynck HJ. 1997. Functional aspects of Aujeszky's disease (pseudorabies) viral proteins with relation to invasion, virulence, and immunogenicity. Vet Microbiol 55:3–11. doi: 10.1016/S0378-1135(96)01299-0. [DOI] [PubMed] [Google Scholar]
- 17.Gerdts V, Jons A, Makoschey B, Visser N, Mettenleiter TC. 1997. Protection of pigs against Aujeszky's disease by DNA vaccination. J Gen Virol 78(Pt 9):2139–2146. doi: 10.1099/0022-1317-78-9-2139. [DOI] [PubMed] [Google Scholar]
- 18.Gerdts V, Jons A, Mettenleiter TC. 1999. Potency of an experimental DNA vaccine against Aujeszky's disease in pigs. Vet Microbiol 66:1–13. doi: 10.1016/S0378-1135(98)00300-9. [DOI] [PubMed] [Google Scholar]
- 19.Wittmann G. 1991. Spread and control of Aujeszky's disease (AD). Comp Immunol Microbiol Infect Dis 14:165–173. doi: 10.1016/0147-9571(91)90129-2. [DOI] [PubMed] [Google Scholar]
- 20.Dellmann HD, Brown E. 1976. Textbook of veterinary histology. Lea & Febiger, Philadelphia, PA. [Google Scholar]
- 21.Sondersorg AC, Busse D, Kyereme J, Rothermel M, Neufang G, Gisselmann G, Hatt H, Conrad H. 2014. Chemosensory information processing between keratinocytes and trigeminal neurons. J Biol Chem 289:17529–17540. doi: 10.1074/jbc.M113.499699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan KP, Wu S, Wigdahl B, Jennings SR. 2013. Immunological control of herpes simplex virus infections. J Neurovirol 19:328–345. doi: 10.1007/s13365-013-0189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heise MT, Virgin HW. 1995. The T-cell-independent role of gamma interferon and tumor necrosis factor alpha in macrophage activation during murine cytomegalovirus and herpes simplex virus infections. J Virol 69:904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodukula P, Liu T, Rooijen NV, Jager MJ, Hendricks RL. 1999. Macrophage control of herpes simplex virus type 1 replication in the peripheral nervous system. J Immunol 162:2895–2905. [PubMed] [Google Scholar]
- 25.Feldman LT, Demarchi JM, Ben-Porat T, Kaplan AS. 1982. Control of abundance of immediate-early mRNA in herpesvirus (pseudorabies)-infected cells. Virology 116:250–263. doi: 10.1016/0042-6822(82)90417-2. [DOI] [PubMed] [Google Scholar]
- 26.Held K, Derfuss T. 2011. Control of HSV-1 latency in human trigeminal ganglia: current overview. J Neurovirol 17:518–527. doi: 10.1007/s13365-011-0063-0. [DOI] [PubMed] [Google Scholar]
- 27.Song R, Koyuncu OO, Greco TM, Diner BA, Cristea IM, Enquist LW. 2016. Two modes of the axonal interferon response limit alphaherpesvirus. mBio 7:1–13. doi: 10.3391/mbi.2016.7.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duvigneau JC, Hartl RT, Groiss S, Gemeiner M. 2005. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J Immunol Methods 306:16–27. doi: 10.1016/j.jim.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 29.Petrov A, Beer M, Blome S. 2014. Development and validation of a harmonized TaqMan-based triplex real-time RT-PCR protocol for the quantitative detection of normalized gene expression profiles of seven porcine cytokines. PLoS One 9:e108910. doi: 10.1371/journal.pone.0108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman LJ. 2006. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol 6:41. doi: 10.1186/1472-6750-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calzada-Nova G, Schnitzlein WM, Husmann RJ, Zuckermann FA. 2011. North American porcine reproductive and respiratory syndrome viruses inhibit type I interferon production by plasmacytoid dendritic cells. J Virol 85:2703–2713. doi: 10.1128/JVI.01616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellemans J, Vandesompele J. 2014. Selection of reliable reference genes for RT-qPCR analysis. Methods Mol Biol 1160:19–26. doi: 10.1007/978-1-4939-0733-5_3. [DOI] [PubMed] [Google Scholar]
- 33.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]