ABSTRACT
Since the first description of adenoviruses in bats in 2006, a number of micro- and megabat species in Europe, Africa, and Asia have been shown to carry a wide diversity of adenoviruses. Here, we report on the evolutionary, biological, and structural characterization of a novel bat adenovirus (BtAdV) recovered from a Rafinesque's big-eared bat (Corynorhinus rafinesquii) in Kentucky, USA, which is the first adenovirus isolated from North American bats. This virus (BtAdV 250-A) exhibits a close phylogenetic relationship with Canine mastadenovirus A (CAdV A), as previously observed with other BtAdVs. To further investigate the relationships between BtAdVs and CAdVs, we conducted mass spectrometric analysis and single-particle cryo-electron microscopy reconstructions of the BtAdV 250-A capsid and also analyzed the in vitro host ranges of both viruses. Our results demonstrate that BtAdV 250-A represents a new mastadenovirus species that, in contrast to CAdV, has a unique capsid morphology that contains more prominent extensions of protein IX and can replicate efficiently in a phylogenetically diverse range of species. These findings, in addition to the recognition that both the genetic diversity of BtAdVs and the number of different bat species from disparate geographic regions infected with BtAdVs appears to be extensive, tentatively suggest that bats may have served as a potential reservoir for the cross-species transfer of adenoviruses to other hosts, as theorized for CAdV.
IMPORTANCE Although many adenoviruses are host specific and likely codiverged with their hosts over millions of years, other adenoviruses appear to have emerged through successful cross-species transmission events on more recent time scales. The wide geographic distribution and genetic diversity of adenoviruses in bats and their close phylogenetic relationship to Canine mastadenovirus A (CAdV A) has raised important questions about how CAdV A, and possibly other mammalian adenoviruses, may have emerged. Although most adenoviruses tend to cause limited disease in their natural hosts, CAdV A is unusual in that it may cause high morbidity and sometimes fatal infections in immunocompetent hosts and is thus an important pathogen of carnivores. Here, we performed a comparative evolutionary and structural study of representative bat and canine adenoviruses to better understand the relationship between these two viral groups.
KEYWORDS: bat adenovirus, canine adenovirus, mastadenovirus, cross-species transmission, host range, cryo-electron microscopy, virus evolution
INTRODUCTION
The family Adenoviridae comprises a large group of double-stranded DNA (dsDNA) viruses that infect a diverse range of hosts, including mammals, birds, reptiles, amphibians, and fish (1). Although genera such as Mastadenovirus, Aviadenovirus, and Ichtadenovirus are class specific in their host range (i.e., classes Mammalia, Aves, and Pisces, respectively), other genera, such as Atadenovirus and Siadenovirus, include viruses that infect a wide range of hosts, including amphibians, reptiles, birds, placental mammals, and marsupials (2). Although it has been proposed that most adenoviruses have codiverged with their hosts over long evolutionary time scales, particularly since a number of adenoviruses appear to be host restricted to a single or few species, the wide host range of other adenoviruses (particularly members of the genus Atadenovirus) suggests that host switching has also played an important role in adenovirus evolution (2–4). Recently, an adenovirus outbreak among captive titi monkeys (Callicebus cupreus), characterized by fulminant pneumonia and hepatitis, was documented in a California research facility (5). Since the outbreak resulted in high case fatality rates, it was suggested that this newly discovered virus (titi monkey adenovirus [TMAdV]) might have jumped into titi monkeys from an unknown reservoir host, possibly cohoused macaques (5). Moreover, respiratory disease and seropositive convalescent-phase sera in both a researcher who handled the monkeys and one of his family members potentially suggested that cross-species transmission of TMAdV from monkeys to humans had occurred, followed by human-to-human transmission (5). This unique case demonstrates that cross-species transmission of adenoviruses can occur and should be regarded as a potential source of zoonotic infections.
Although bats have long been recognized as an important reservoir for a variety of different viruses such as lyssaviruses (6, 7), the number of novel viruses recovered from bats has dramatically increased in recent years due to a combination of advances in metagenomic analysis (8–10) and increased surveillance and sampling of bats in response to the discovery of their potential roles in harboring high-risk human pathogens such as severe acute respiratory syndrome (SARS)-like coronaviruses (11, 12), filoviruses (13), hantaviruses (14), and henipaviruses (15, 16). The discovery of bat adenoviruses (BtAdVs) is relatively recent, with the first, BtAdV-1 (strain FBV-1), isolated in 2006 from primary cell cultures derived from the spleen of a healthy Ryukyu flying fox (Pteropus dasymallus yayeyamae, suborder Megachiroptera) in Japan (17). Since then, a multitude of bat species, primarily those of the suborder Microchiroptera (comprising the majority of microbat families) (18, 19), have been shown to harbor adenoviruses (20–30). Formerly, only two of these viruses, BtAdV-2 (strain PPV-1), isolated from a common pipistrelle (Pipistrellus pipistrellus) in Germany (20, 23), and BtAdV-3 (strain TJM), recovered from a Rickett's big-footed bat (Myotis ricketti) in China (21), have been extensively characterized genetically. Since BtAdV-3 was the first virus in which the nearly full genome was sequenced, it has been designated the prototype bat adenovirus species, Bat mastadenovirus A, within the genus Mastadenovirus, family Adenoviridae, with BtAdV-2 being recently proposed as Bat mastadenovirus B (31). However, recent surveillance studies have identified novel adenoviruses from Chinese rufous horseshoe bats (Rhinolophus sinicus) whose genomes are considerably divergent from BtAdV A and B (32), greatly expanding the known diversity of BtAdVs. Nevertheless, with a few exceptions (33), little is currently known regarding the ecology, epizootiology, and molecular biology of BtAdVs.
Here, we characterized the complete genome of a novel bat adenovirus (BtAdV isolate 250-A) recovered from a Rafinesque's big-eared bat (Corynorhinus rafinesquii) from Kentucky, USA. Although a partial sequence of protein VIII from another bat adenovirus was previously detected in guano obtained from a roost of pallid bats (Antrozotus pallidus) in California (8), BtAdV 250-A represents the first bat adenovirus isolate in North America. We investigated the phylogenetic relationships of bat adenoviruses to other mastadenoviruses, particularly Canine mastadenovirus A (CAdV A), since CAdVs were hypothesized to have originated by interspecies transfers of adenoviruses from bats (23). In addition, we performed comparative studies on the biological properties of BtAdVs and CAdVs to determine their in vitro host ranges and how these ranges may have been shaped by their evolutionary history. Finally, we conducted mass spectrometry on purified BtAdV virions and performed single-particle cryo-electron microscopy (cryo-EM) reconstructions of the BtAdV 250-A capsid and compared the resolved structure to that of CAdV A, allowing us to examine the structural changes associated with the phylogenetic divergence between the two groups.
RESULTS
Gross and microscopic analysis of Rafinesque's big-eared bats.
Rafinesque's big-eared bats are an insectivorous species of microbat belonging to the family Vespertilonidae (commonly known as vesper or evening bats) and are a resident species in Mammoth Cave National Park (34) (Fig. 1A and B). The three bats submitted for diagnostic evaluation were juvenile males, weighing between 3.7 and 6.4 g. Due to the severe autolysis of specimen 250-C and its omission from virus isolation attempts, only bats 250-A and -B will be discussed. Gross lesions in bat 250-A included moderate, subacute, multifocal, fungal cellulitis in the patagium and moderate, acute, multifocal hemorrhage and emaciation of the carcass. Microscopically, in bat 250-A, there were focal areas of hyperkeratosis, serocellular crusts with numerous degenerate neutrophils, and perivascular accumulations of mononuclear cells in the patagium. The lungs, liver, kidneys, spleen, brain, and intestinal tract had evidence of bacterial growth that was consistent with postmortem colonization. Bat 250-B had similar gross and microscopic lesions, including malnutrition, a focus of round to oblong fungal spores mixed with keratinic debris in the patagium, and widespread postmortem colonization of bacteria in multiple tissues. Culture of samples of patagium and muzzle submitted for white nose syndrome testing revealed growth of multiple fungal contaminants, but no Pseudogymnoascus (Geomyces) species were isolated. Fluorescent antibody testing for rabies virus on brain samples was negative for both bats.
FIG 1.
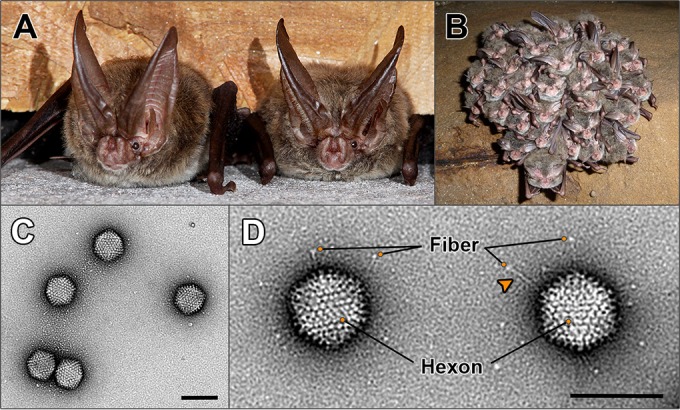
Host species from which bat adenovirus (BtAdV) 250-A was isolated and morphology of purified virions. (A) Two adult Rafinesque's big-eared bats (Corynorhinus rafinesquii) roosting in a building in Mammoth Cave National Park (MCNP), Kentucky, the site of isolation of BtAdV 250-A (photo courtesy of Tom Uhlman). (B) Rafinesque's big-eared bats hibernating in a cave in MCNP. Note the densely packed clusters formed by the bats during winter hibernation (photo courtesy of Steven Thomas, National Park Service). (C) Transmission electron micrograph of BtAdV 250-A virions negatively stained with uranyl formate, showing particles of icosahedral symmetry ∼90 nm in diameter (scale bar, 100 nm). (D) High-magnification view (52,476×) of BtAdV 250-A virions clearly showing the long flexible fiber proteins with their C-terminal knob domain visible (adjacent to the orange circles). Note an apparent kink (arrowhead) in the fiber protein, suggesting a potential hinge region imparting flexibility. Also highlighted is the hexon protein, which comprises the bulk of the adenovirus capsid, where 12 hexons form each of the 20 icosahedral facets of the particle (i.e., 240 copies per virion) (scale bar, 100 nm).
Virus isolation and initial characterization.
From gastrointestinal tissue samples taken from bat 250-A, a virus was isolated in Vero cell culture on passage 2. Brain, trachea, and heart samples from bat 250-A were negative for virus isolation. Degenerate primers based on an alignment of the DNA polymerase (Pol) sequences of various adenoviruses were used to amplify a 427-bp sequence from DNA extracted from an infected Vero cell culture. BLAST analysis of the primer-less product showed the highest nucleotide identity (74.6%) to CAdV-1, demonstrating the virus was a novel adenovirus, which was also confirmed by transmission electron microscopy (Fig. 1C and D). Due to postmortem autolysis in tissues from bat 250-A, whether the adenovirus infection was inapparent or induced pathological changes could not be conclusively determined, although the lack of virus isolation from peripheral tissues suggested the infection was likely localized (i.e., to the gastrointestinal tract) rather than systemic.
Genomic sequencing.
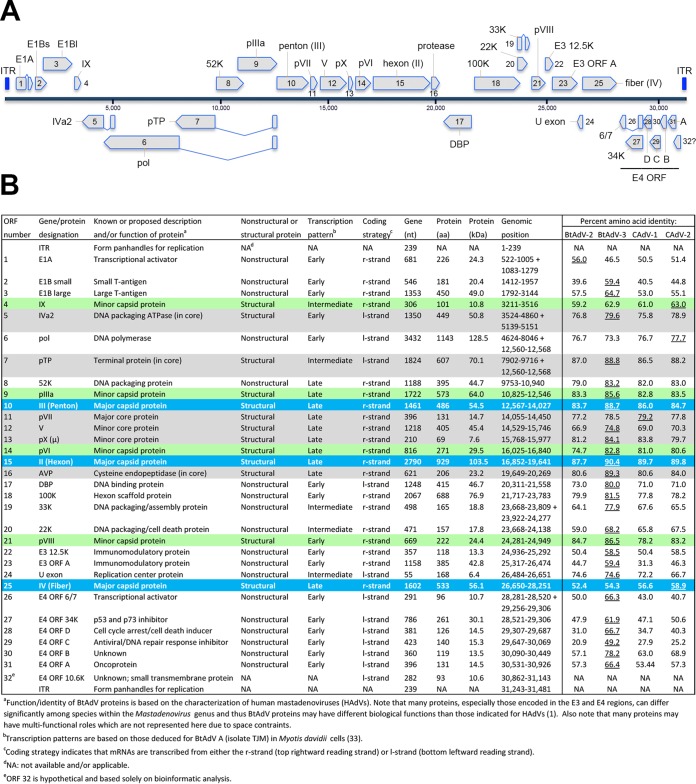
The BtAdV 250-A genome is 31,481 nucleotides (nt) in length and is thus slightly shorter than either BtAdV A (isolate TJM [31,806 nt]) or BtAdV B (isolate PPV1 [31,616 nt]). In the BtAdV 250-A genome, 31 open reading frames (ORFs) were found with recognizable similarity to known adenovirus proteins, with the inverted terminal repeats (ITRs) being 239 nt in length, within the range of other mastadenoviruses (93 to 371 nt) (1). The complete genomic organization of BtAdV 250-A, as well as its protein identities to other closely related BtAdVs and CAdV serotypes 1 and 2, is shown in Fig. 2.
FIG 2.
Genomic organization of BtAdV 250-A. (A) The 31,481-nt genome of BtAdV 250-A is shown. The inverted terminal repeats (ITRs) found at the ends of the genome are highlighted in royal blue, with genes transcribed from left to right (from the top DNA strand) shown above the blue-gray solid line, and those transcribed from right to left (from the bottom DNA strand) are indicated below. Genes that are believed to be spliced are indicated by two genetic elements that are connected by adjoining lines in a “V” or “V” pattern. (B) The 31 ORFs identified in the BtAdV 250-A genome that share homology to other BtAdVs and canine mastadenoviruses (CAdVs) are shown. All 13 tentative structural proteins are highlighted with shading: (i) the three major capsid proteins—II (hexon), III (penton), and IV (fiber)—found on the particle surface (blue), (ii) four minor capsid/cement proteins—IIIa, VI, VIII, and IX—found on or just beneath the particle surface (green), and (iii) six core proteins—IVa2, V, VII, X, TP, and the adenovirus protease (AVP)—found internally (gray). The percent amino acid identity of the BtAdV 250-A proteins to other viruses was determined using the following GenBank accession numbers: BtAdV 250-A (KX871230), BtAdV A (NC_016895), BtAdV B (JN252129), CAdV A (CAdV-1) (AC_000003), and CAdV A (CAdV-2) (AC_000020). The virus sharing the highest degree of amino acid identity to BtAdV 250-A for each protein is underlined. A lowercase “p” preceding a protein's name (i.e., pTP, pIIIa, pVI, pVII, pVIII, and pX) indicates a precursor form of the protein that is subsequently cleaved by the AVP.
The BtAdV 250-A genome shared a nucleotide identity of 72.7% to BtAdV A and 68.7% to BtAdV B. Similarly, the BtAdV 250-A genome shared nucleotide identities of 68.6 and 69.9% to CAdV-1 and CAdV-2, respectively. In contrast, comparisons of BtAdV 250-A to the recently sequenced genomes of BtAdVs from horseshoe and bent-wing bats from China (BtAdV WIV9-WIV13) exhibited only a 54.8 to 56.2% nucleotide identity (32). Comparison of the full-length Pol protein sequence of BtAdV 250-A to BtAdV A and BtAdV B demonstrated 23.3 to 26.7% amino acid divergence, thus substantiating its tentative designation as a new adenovirus species (i.e., >5 to 15% distance in Pol amino acid sequence constitutes species demarcation, in association with a lack of cross-neutralization if antisera were available) (1). In proteome comparisons between BtAdV-250, BtAdV A, BtAdV B, and the two serotypes of CAdV A, the hexon protein had the highest amino acid conservation (87.7 to 90.4% identity), whereas the E4 ORF C protein was the most divergent sequence (20.9 to 49.2% identity). Although BtAdV 250-A shared the highest amino acid identity to BtAdV A in 25 of 31 cognate proteins, it was most similar to CAdV-2 in the fiber (58.9%), IX (63.0%), and Pol (77.7%) proteins.
The BtAdV 250-A penton did not contain an Arg-Gly-Asp (RGD) motif, common in many human AdVs (HAdVs) and used to bind to the cell surface integrins αVβ3 and αVβ5 after initial coxsackievirus AdV receptor (CAR) binding by the fiber protein (35). The fiber protein of BtAdV 250-A is long (533 amino acids [aa]), although it is 9 aa shorter than the CAdV-2 fiber and is missing the 17th repeat (36) based on amino acid alignments (data not shown). However, BtAdV 250-A and CAdV-2 have recognizable identity (58.1%) at the two bends predicted in the fiber shaft of CAdV-2 at repeats 4 and 10 (36).
As previously noted for BtAdV B (23), BtAdV 250-A contains two genes in its E3 region, 12.5K and ORF A, with the latter unique to BtAdV A and B, CAdVs, skunk adenovirus (SkAdV), and equine mastadenovirus A (EAdV A) (23, 37, 38). In contrast, newly identified BtAdVs from China contain three E3 region genes: 12.5K, E3ls, and E3l, the latter being the largest adenovirus protein reported to date (32). The functional roles of the E3 proteins in these viruses are currently unknown. The E4 region, which contains six ORFs (ORFs 6/7, 34K, D, C, B, and A) in the BtAdV A and B genomes (21, 23), is found at the right end of the genome, just upstream of the ITR. A number of the genes in the E4 region are unique to mastadenoviruses (1) and, unlike other transcription units, appear to encode proteins that have dissimilar functions (35). Analysis of the BtAdV 250-A genome revealed the presence of an additional ORF (numbered as 32) in the E4 region close to the right terminal end of the genome. This ORF, capable of coding for a protein of 93 aa (putatively designated the E4 10.6K protein), partially overlaps with the 5′ end of E4 ORF A (Fig. 2A and B). Although other potentially coding ORFs were detected throughout the genome (not shown), the locality of the 10.6K ORF (between the ITR and the last gene) and its predicted topology (below) suggested that it was a good candidate for a new gene. WU-BLAST of the E4 10.6K gene product demonstrated a 34.0% amino acid identity to major facilitator superfamily (MFS) bacterial transporters, which include permeases involved in secondary active transport of small solutes, such as ions, across cellular membranes (39). Secondary structure analysis of the E4 10.6K protein predicted the presence of a central transmembrane domain, although its length differed depending upon the program used (e.g., Phyre2 [aa 24 to 53], Phobius [aa 30 to 48], and HMMTOP [aa 40 to 64]). Since the protein's short length (93 aa) and predicted central transmembrane domain are potentially suggestive of a viroporin-like or other ancillary small viral transmembrane protein (40), future work will be aimed at determining if a protein is indeed expressed from this ORF and what its potential function may be.
Mass spectrometry of purified BtAdV 250-A virions.
SDS-PAGE analysis of purified virus demonstrated at least six prominent, discrete bands ranging from approximately 10 to 100 kDa in estimated molecular mass. To conclusively determine the identity of these proteins, each band was excised from SYPRO Ruby-stained SDS gels (10% Tris-glycine gels for bands of >20 kDa and 10 to 20% Tris-Tricine gels for bands of <20 kDa) and analyzed by nano-scale high-performance liquid chromatography coupled to tandem mass spectrometry (nano-HPLC-MS/MS). The mastadenovirus particle is reported to be composed of 13 proteins: seven capsid proteins (hexon [II], penton [III], IIIa, fiber [IV], VI, VIII, and IX), and six core proteins (IVa2, V, VII, X, TP, and the adenovirus protease [AVP]) (41). The capsid is formed by three major proteins: 240 copies of the trimeric hexon which forms the bulk of the particle, and 12 pentons and their associated 12 fibers (extending from the pentons) which are found at the 5-fold vertices (35). The remaining four capsid proteins (IIIa, VI, VIII, and IX) are referred to as minor or cement capsid proteins, since they stabilize the capsid through their interactions with the hexons and pentons (35).
Mass spectrometric analysis of purified BtAdV 250-A virions identified all seven capsid proteins. In infectious virions, three of these proteins—IIIa, VI, and VIII—are cleaved from precursor (p) forms by the AVP, such that their molecular masses would be lower than expected for intact preproteins. The estimated molecular masses for the cleaved products of the BtAdV 250-A pIIIa, pVI, and pVIII, along with the core pVII protein (shown below), were based on the conserved protease cleavage sites identified in HAdVs (i.e., [M/I/L]XGX-G and/or [M/I/L]XGG-X) (42, 43), which were also present in BtAdV 250-A. However, of note, the pIIIa protein does not contain the N-terminal LXGX-G site found in HAdVs (42) but rather a homologous FXGX-G motif; thus, it is currently uncertain if this site is cleaved.
The largest and most abundant band, migrating at ∼110 kDa, was the hexon protein (with an estimated molecular mass of 103.5 kDa) (Fig. 3). Between the 50- and 75-kDa protein markers, at least three closely migrating bands were observed. The majority proteins identified in these three bands (after further separation) were protein V (ca. 45.4 kDa; upper band), protein IIIa (ca. 61.9 or 64.0 kDa for the cleaved or intact precursor, respectively; middle band), and a mixture of fiber and penton proteins (ca. 56.1 and 54.5 kDa, respectively; bottom band) (Fig. 3). Thus, protein V migrated more slowly than would be expected, possibly owing to its basic nature (pI = 10.6). Protein VI (ca. 1.4, 3.4, and 24.7 kDa for the cleavage products), a multifunctional cement protein (35), migrated between the 25- and 37-kDa markers (Fig. 3). As expected, only peptides corresponding to the 24.7-kDa fragment were detected in the band; in addition, the predicted terminal 256-IVGV-G-261 cleavage site in protein VI (42) was confirmed by peptide sequencing. The lowest band (migrating at approximately ≤10 kDa) contained peptides corresponding to the minor capsid proteins VIII (ca. 4.5, 7.7, and 12.2 kDa for the cleavage products) and IX (ca. 10.8 kDa), along with the core protein VII (ca. 2.4 and 12.3 kDa for the cleavage products) (not shown). Four core proteins—IVa2, TP, X, and AVP—were not detected in any bands; proteins IVa2, TP, and AVP may have been present in the gel but were faint due to their very low abundance (<10 copies in the case of HAdVs) within the particle (42). No cellular proteins (more than two identified peptides) were detected within BtAdV 250-A particles.
FIG 3.
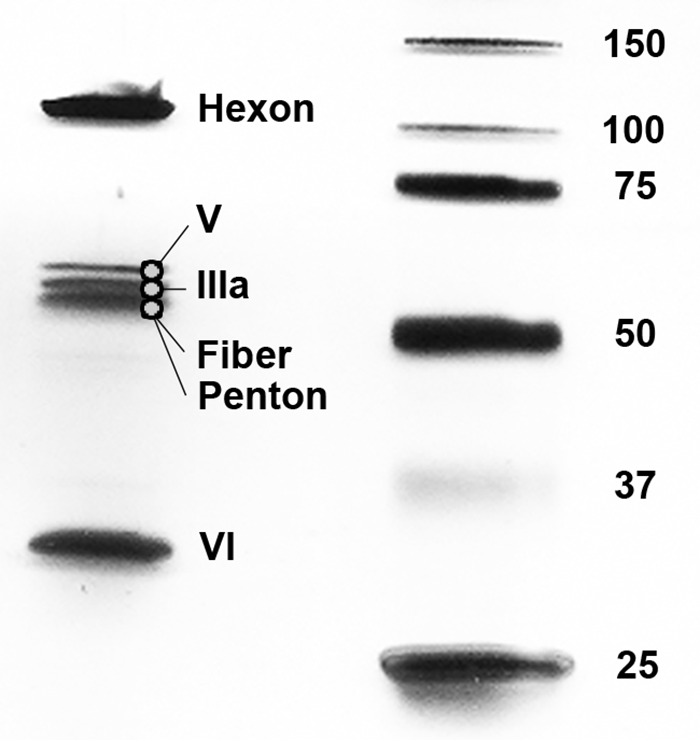
Mass spectrometry of purified virions of BtAdV 250-A. Purified virus was analyzed by SDS-PAGE, and individual protein bands were excised and subjected to nano-HPLC-MS/MS. Peptide sequencing identified the three major capsid proteins (hexon, fiber, and penton), the four minor capsid/cement proteins (IIIa, VI, VIII, and IX), and two of the six core proteins (V and VII). Note that the lower band in the central group of triplet bands was a mixture of both fiber and penton proteins (estimated molecular masses of 56.1 and 54.5 kDa, respectively). Peptides corresponding to proteins VII, VIII, and IX were detected in a band migrating at approximately ≤10-kDa (not visible after separation of the V-IIIa-fiber-penton cluster).
Cryo-EM reconstruction of the BtAdV 250-A capsid.
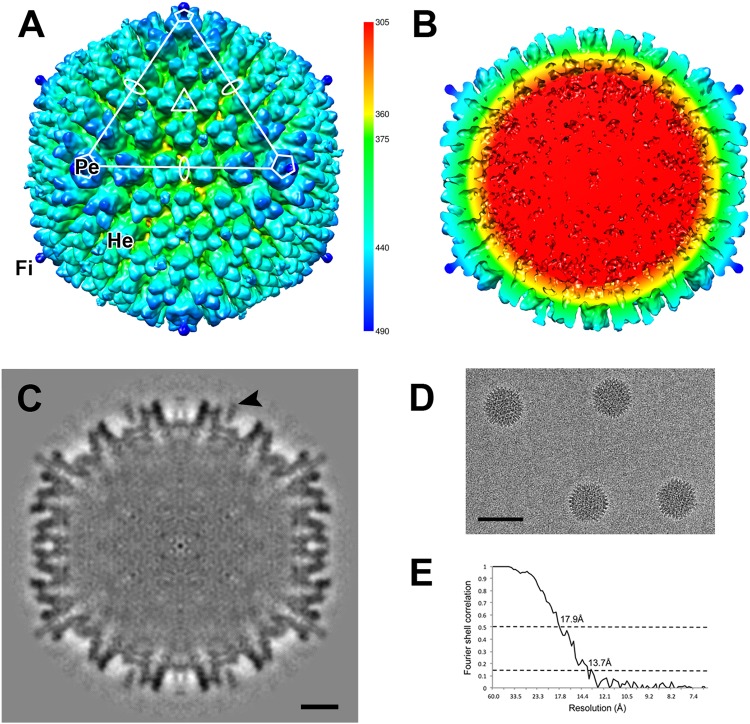
To better understand the structural similarities and/or differences that exist between BtAdVs and other AdVs for which cryo-EM or X-ray crystallographic structures are available (e.g., 36, 44–47), in particular CAdV A, a 3-D reconstruction of BtAdV 250-A capsid was calculated. The surface rendition of the BtAdV capsid showed a prototypical mastadenovirus morphology (Fig. 4A), with a penton protein present at each of the 12 5-fold axes, and the base stalk of the elongated fiber (that extends from the center of the penton) visible. Twelve individual hexon trimers can be viewed within one triangular icosahedral facet, thereby demonstrating that the core capsid structure is composed of 240 hexons. The cross-section of the map revealed the internal densities of these three major capsid proteins (hexon, penton, and fiber), along with the density of the 31.4-kb dsDNA genome which appears amorphous (disorganized) since it lacks the icosahedral symmetry of the capsid, and therefore this density does not represent the true position of the genome within the capsid (Fig. 4B). The density displayed by the structural proteins in the central section confirmed the quality of the map (Fig. 4C) and the Fourier shell correlation curve indicated a resolution of 17.9 Å at a Fourier shell correlation of 0.5 (Fig. 4E).
FIG 4.
Three-dimensional reconstruction of the BtAdV 250-A capsid. (A) The surface-rendered BtAdV map is colored according to radius (key) to illustrate the topology of the capsid. An asymmetric unit is highlighted (white) with the axes of icosahedral symmetry denoted by an oval (2-fold), triangle (3-fold), and a pentagon (5-fold). Only a short portion of the fiber (Fi) protein, shown extending from the penton (Pe) base at the 5-fold axis, could be reconstructed due to its flexible nature (see full-length protein in Fig. 1D). Hexon (He) proteins are also denoted. (B) A cross section of the BtAdV particle, color-coded by radius, reveals a disordered genome (red) and the internal structures of the hexons and pentons that comprise the capsid shell. (C) The central cross-section of BtAdV shows external capsid densities are distinct, but less well defined internally. Weak density connects protein IX (arrowhead) to the capsid. Scale bar, 10 nm. (D) Image of BtAdV capsids, embedded in vitreous ice, that were used for the reconstruction. Scale bar, 100 nm. (E) The Fourier shell correlation is shown as a function of resolution indicating 17.9 and 13.7 Å, where the curve crosses 0.5 and 0.143, respectively.
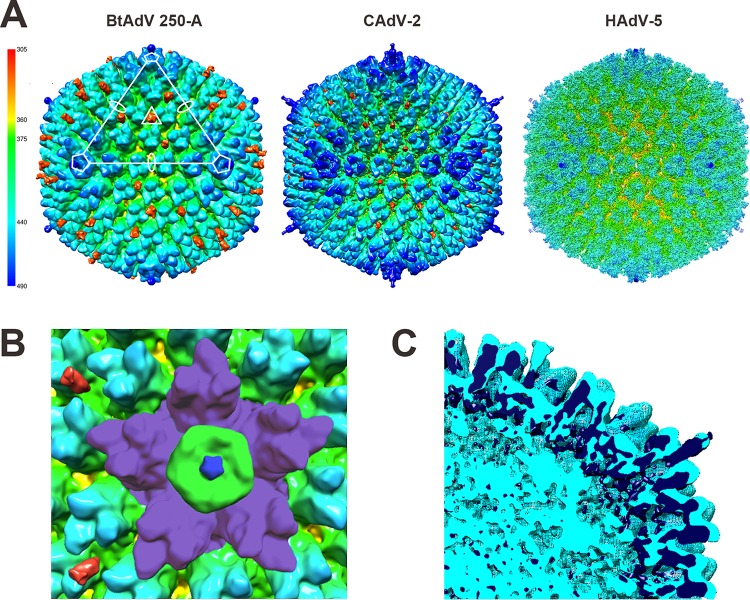
To compare the BtAdV 250-A capsid to other mastadenoviruses, the 12-Å resolution map of CAdV A (CAdV-2) (36) and the 3.6-Å resolution map (45) of Human mastadenovirus C (HAdV C) serotype 5 (HAdV-5) were rendered. The three maps revealed similar morphologies of prototypical adenovirus particles (Fig. 5A), including the five hexons adjacent to the pentons, known as the peripentonal hexons, being more elevated than hexons in other locations of the capsid (Fig. 5B). In addition, protein IX in BtAdV (protein designation based on the CAdV-2 structure [36]), which is a flexible cement protein found on the outer capsid (47–49), was noticeably prominent. Protein IX extends outward from the capsid similar to CAdV-2 (36), although more pronounced and is also reminiscent of the extensive protrusion of LH3 observed in the Ovine atadenovirus D (OAdV D) serotype 7 capsid (44). However, protein IX appears detached from the capsid in the BtAdV central section and surface-rendered map due to weak connecting density (Fig. 4C, arrowhead; Fig. 5A and B). Other minor capsid proteins that lie either at the external base of the hexons and pentons in HAdVs, such as protein IIIa, or that form an internal complex underneath the peripentonal hexons at each 5-fold axis, such as proteins V, VI, or VIII (47), were not resolved. To directly compare the BtAdV 250-A structure to that of CAdV-2, the surface maps of the two viruses were superimposed (Fig. 5C). As anticipated, the densities of both viruses aligned well. Specifically, the internal densities of the hexons, pentons, and N-terminal regions of the fiber overlap.
FIG 5.
BtAdV 250-A has a capsid morphology similar to CAdV-2 and a prominent protein IX extending from its surface. (A) The surface-rendered maps of BtAdV 250-A, CAdV A (serotype CAdV-2) (EMDB 1462), and Human mastadenovirus C (HAdV C; serotype HAdV-5) (EMDB 5172) are colored according to radius (key) to illustrate similar topologies. In addition to the radial coloring, protein IX was highlighted in crimson in BtAdV and CAdV to demonstrate its location and prominence in the capsid. Note that protein IX in BtAdV protrudes well beyond the hexon proteins. (B) Magnification of the icosahedral 5-fold vertex of BtAdV. The five peripentonal hexons (purple), which surround a single tulip-shaped penton base (green) with its central projecting fiber stalk (royal blue), are visible. The C-terminal surface exposed spikes of two protein IX triskelions (crimson) are shown on the left. (C) The central section of the superimposed structures of BtAdV 250-A (light blue) and CAdV-2 (dark blue) reveal overlapping internal structural similarities between the viruses.
Phylogenetic analysis.
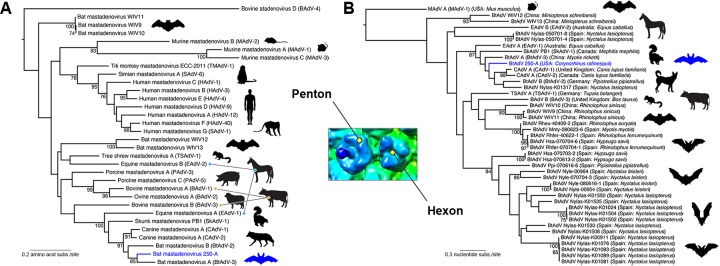
A phylogenetic analysis of BtAdV 250-A was first undertaken using a divergent set of penton amino acid sequences of viruses within the genus Mastadenovirus. This phylogeny indicated a close relationship between BtAdV 250-A and CAdV A, since they formed a monophyletic group with strong bootstrap support (91%) (Fig. 6A). Although BtAdV A and B also clustered in this group, recently identified BtAdVs from China (32) were much more divergent, falling in additional clades throughout the phylogeny. Sequences of the Pol gene are also available for analysis, including BtAdVs from micro- and megabats from India, Japan, and Hungary; however, the sequence alignment is so short that it contains relatively little phylogenetic resolution and thus was excluded from the analysis. A similar relationship to the penton phylogeny was found when analyzing a set of BtAdVs from Asia (China) and Europe (Germany, Spain) using the hexon gene (Fig. 6B). The most striking feature of the hexon tree was that BtAdV 250-A forms a monophyletic group with CAdVs, along with BtAdV A, BtAdV B, and a greater noctule bat (Nyctalus lasiopterus) isolate from Spain (87% bootstrap support). Hence, these results may suggest that BtAdV 250-A is more closely related to CAdV than most other BtAdVs that have been identified to date. Although skunk and equine AdVs also fell within this same clade (Fig. 6B), tentatively suggesting that BtAdVs may have served as a common ancestor for other adenoviruses (which may in part be a function of the wide geographical distribution of bats), the partial sequences of the hexon gene and its variability within groups may also influence this phylogeny (50). Clearly, the future analysis of additional complete hexon sequences may assist in resolving evolutionary relationships. Finally, viruses from two of the Rhinolopus species (horseshoe bats, family Rhinolophidae), which, together with the pteropodid megabats and two other microbat families constitute the suborder Megachiroptera, clustered with viruses recovered from microbats from the suborder Microchiroptera (genera Myotis and Hypsugo) in the hexon tree, potentially suggesting mixing of viruses between the two groups.
FIG 6.
Maximum-likelihood phylogenetic analysis of BtAdV 250-A with other mastadenoviruses. For both the penton and the hexon trees, bootstrap values (>70%) are shown for key nodes. The structural location of the penton and hexon proteins in the BtAdV capsid are shown in the central inset. BtAdV 250-A is highlighted by a blue branch and bat in each tree for reference. (A) Phylogenetic tree of the amino acid sequences of the penton protein for member species of the genus Mastadenovirus (i.e., from a diversity of mammalian taxa). The tree is midpoint rooted for clarity only, and horizontal branch lengths are scaled according to the number of amino acid substitutions per site. GenBank accession numbers for the sequences used in the penton tree were as follows: BAdV-1 (YP_094036), BAdV-2 (NP_597922), BAdV-3 (AEW91335), BAdV-4 (NP_077393), BtAdV-2 (YP_004782104), BtAdV-3 (YP_005271186), BtAdV 250-A (KX871230), BtAdV WIV9 (KT698853), BtAdV WIV10 (KT698854), BtAdV WIV11 (KT698855), BtAdV WIV12 (KT698856), BtAdV WIV13 (KT698852), CAdV-1 (NP_044193), CAdV-2 (AP_000617), EAdV-1 (AEP16413), EAdV-2 (YP_009162351), HAdV-1 (AP_000507), HAdV-3 (YP_002213774), HAdV-4 (AAW33302), HAdV-9 (YP_001974428), HAdV-12 (NP_040920), HAdV-40 (NP_040857), MAdV-1 (NP_015541), MAdV-2 (YP_004123742), MAdV-3 (YP_002822211), PAdV-3 (YP_009205), PAdV-5 (NP_108662), SAdV-1 (YP_213970), SAdV-6 (AFG19591), SkAdV-1 (YP_009162592), TMAdV (YP_007518317), and TSAdV-1 (YP_068064). (B) Evolutionary relationships between BtAdV 250-A and BtAdVs from Europe and Asia were assessed using nucleotide sequences of the hexon gene. Closely related viruses (CAdV-1, CAdV-2, SkAdV-1, EAdV-1, EAdV-2, TSAdV-1, and BAdV-3) as determined from the penton phylogeny in panel A, were also included in the analysis. The tree is rooted using the more divergent sequence of Murine mastadenovirus A (MAdV A), and horizontal branch lengths are scaled according to the number of nucleotide substitutions per site. GenBank accession numbers for the sequences used in the hexon tree were as follows: BAdV-3 (AF030154), BtAdV-2 (JN252129), BtAdV-3 (NC_016895), BtAdV 250-A (KX871230), BtAdV WIV9 (KT698853), BtAdV WIV10 (KT698854), BtAdV WIV11 (KT698855), BtAdV WIV12 (KT698856), BtAdV WIV13 (KT698852), CAdV-1 (AC_000003), CAdV-2 (AC_000020), EAdV-1 (JN418926), EAdV-2 (KT160425), MAdV-1 (M81889), SkAdV-1 (KP238322), TSAdV-1 (AF258784), and BtAdV population set HM856327 to HM856353 (courtesy of I. Casas, S. Vazquez, J. Juste, A. Falcon, C. Aznar, C. Ibanez, F. Pozo, G. Ruiz, J. M. Berciano, I. Garin, J. Aihartza, P. Perez-Brena, and J. E. Echevarria, National Centre of Microbiology, Madrid, Spain).
In vitro host range.
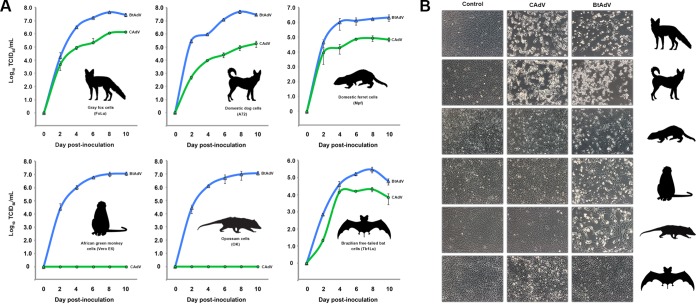
Comparative growth curve analysis between BtAdV 250-A and CAdV-1 demonstrated that there were marked differences in their ability to replicate in different host cells. Although most mastadenoviruses tend to be restricted to one or a few closely related hosts, BtAdV 250-A replicated efficiently in fox fibroblast (FoLu), dog fibroblast (A72), ferret fibroblast (Mpf), opossum epithelial (OK), monkey epithelial (Vero E6), and bat epithelial (Tb1Lu) cells, demonstrating a wide in vitro host range (Fig. 7). In particular, titers over 107.0 TCID50/ml were observed in primate (monkey), carnivore (dog, fox), and marsupial (opossum) cells over the 10-day time course. In contrast, CAdV only replicated efficiently (>105.0 TCID50/ml) in the carnivore cell lines tested (dog, fox, and ferret cells), reaching the highest titer in gray fox cells (106.2 TCID50/ml), the species from which the virus was isolated. Interestingly, although no detectable replication of CAdV was observed in noncarnivoran cells such as opossum or monkey cells, CAdV did grow in bat cells (Tb1Lu) (Fig. 7). Although the CAdV titers in bat cells were comparatively low versus the three carnivore cell lines, most viruses do not propagate very efficiently in this particular cell line (A. B. Allison, personal observation), including BtAdV 250-A. Nevertheless, the recognition that both BtAdV and CAdV replicated in bat cells further reiterates their likely close phylogenetic relationship. Similar to what has been observed with other BtAdVs (21), BtAdV 250-A also replicated in human cells lines such as A549 and HeLa cells (data not shown).
FIG 7.
Comparative in vitro host ranges of BtAdV and CAdV. (A) Gray fox (FoLu), domestic dog (A72), domestic ferret (Mpf), African green monkey (Vero E6), Virginia opossum (OK), and Brazilian free-tailed bat (Tb1Lu) cells were infected with either BtAdV 250-A or CAdV-1 (isolate 24-05), and viral titers (log10 TCID50/ml) were measured at 2-day intervals over a 10-day period. BtAdV 250-A growth curves are shown in blue, whereas CAdV 24-05 growth curves are shown in green. Note that although CAdV was host restricted to cells derived from members of the order Carnivora (fox, dog, and ferret) and bats, BtAdV 250-A replicated efficiently in cells derived from a phylogenetically diverse group of mammals, including primates, as well as a marsupial species (opossum). The data shown are from multistep single growth curve experiments performed in triplicate, with error bars indicating the standard deviations. (B) Cell morphology of the cell lines used in panel A at day 6 postinfection with either BtAdV 250-A or CAdV 24-05. Note the induction of cytopathic effects by BtAdV in all of the cell lines tested beside bat (Tb1Lu) cells.
DISCUSSION
In this report, we present structural, biological, and evolutionary analyses of a novel adenovirus (BtAdV 250-A) isolated from a Rafinesque's big-eared bat. Notably, BtAdV 250-A represents the first isolation of a BtAdV in North America and further suggests that BtAdVs are likely cosmopolitan in nature, despite their first discovery only a decade ago (17). However, if BtAdV 250-A is host specific to Rafinesque's big-eared bats, this would suggest that its distribution in nature would be relatively restricted, since this species is nonabundant and confined primarily to the southeastern United States (51, 52). Currently, there are two BtAdVs formally recognized as species by the International Committee on Taxonomy of Viruses (ICTV): Bat mastadenovirus A (BtAdV-3) from China and Bat mastadenovirus B (BtAdV-2) from Germany (31). In accordance with these designations, we propose that BtAdV 250-A be recognized as a new species, similar to the recently identified BtAdVs from China (32).
Although adenovirus evolution has long been thought to result from virus-host codivergence, on occasion viral transfer to a new host is followed by successful adaptation and sustained spread that may lead to the evolution of a novel virus (3). One possible example of such a transfer appears to be the jump of BtAdVs into terrestrial carnivores, resulting in the emergence of CAdV A (23). Our phylogenetic analysis of the penton protein (Fig. 6A) further supported this notion, since in addition to the close evolutionary relationship between BtAdV 250-A and CAdV A, various equine and bovine mastadenoviruses fell in different clades across the phylogeny, suggesting cross-species transmission events have shaped the evolutionary history of the genus. Notably, new BtAdVs from China (32) also fell in multiple clades, again suggestive of cross-species transfers, although recent surveillance studies also suggest that the unsampled diversity of AdVs is likely to be extensive (30). In contrast to recent host-switching events, other parts of the penton phylogeny are suggestive of long-term virus-host codivergence, such as the strong monophyletic clustering of primate and rodent viruses. Interestingly, Chiroptera (bats) and Carnivora (e.g., domestic dogs and foxes) are closely related in the mammalian phylogeny (53), tentatively suggesting that they might be more conducive to cross-species transfers than more phylogenetically distant orders. As a consequence, the emergence of CAdV may be an instance of cross-species transmission within a background of codivergence.
Although our phylogenetic results are clearly compatible with the cross-species transfer of an adenovirus from bats to carnivores, the large number of bat sequences in the data set and the absence of those from other species may indicate a potential ascertainment bias. For example, it is also possible that there are viruses that are intermediate between BtAdVs and CAdVs presently circulating in other species that have yet to be identified. The recent identification of an adenovirus closely related to BtAdVs from a striped skunk (Mephitis mephitis) in Canada (37) supports this hypothesis. It is of interest, however, that recent surveillance studies in China and Hungary have identified viruses in bats that appear to be more similar to CAdV than to other BtAdVs (i.e., GenBank accession numbers AFG28283 and AIL52733) (26, 30); unfortunately, due to the short length of these sequences (<100 amino acids of the Pol protein), it is presently unclear whether they represent true intermediates between currently well-characterized BtAdVs and CAdVs or possibly a chance spillover event of CAdV from terrestrial carnivores into bats, suggesting that reciprocal transfers could be occurring. To further clarify the evolutionary relationships between such viruses, a more thorough genetic characterization of the wide diversity of BtAdVs that exist is required, in addition to a more extensive sampling of adenoviruses from additional mammalian hosts.
Most adenoviruses that infect mammals (i.e., mastadenoviruses and atadenoviruses) cause relatively benign or subclinical infections, with severe clinical disease usually only associated with predisposing factors such as immunosuppression or concurrent infections (54). Indeed, most adenovirus infections in bats appear to be mild or inapparent (17, 23, 28). However, one notable exception among the mastadenoviruses is CAdV A, which may induce severe clinical manifestations such as encephalitis, paralysis, hepatitis, ocular lesions, and respiratory disease in its normal carnivore hosts (e.g., dogs, wolves, foxes, skunks, bears, and otters) and can be rapidly fatal (55–60). There are two serotypes of CAdV A: CAdV-1, which is the etiological agent of infectious canine hepatitis (61), and CAdV-2, which causes infectious tracheobronchitis (62). CAdV-1 was first described as the etiological agent of encephalitis in foxes in the 1920s, with the virus being first reported in domestic dogs in the 1930s and 1940s (63, 64). Whether the virus was present in domestic dog populations prior to this time period is unknown. Although there are a number of documented cases of dramatic increases in virulence after the cross-species transmission of persistent DNA viruses from their normal, reservoir hosts to new alternate hosts (65), which would be consistent with the differential pathogenesis demonstrated between BtAdVs and CAdV A, the possible increase in virulence of CAdV A could also be an adaptive trait that perhaps evolved to increase transmissibility among carnivore hosts due to alternate routes of infection by the virus (i.e., respiratory spread).
Previously, the cryo-EM structure of the CAdV A (CAdV-2) capsid was solved at a resolution of 12 Å (36). Although a slightly lower resolution of the BtAdV 250-A capsid was achieved here (17.9 Å; 0.5 Fourier shell correction), the reconstruction allowed for a direct comparison between the BtAdV 250-A and CAdV-2 capsids to better understand the overall structural changes imparted during their evolution and to compare their structures to those reported for other adenoviruses (44–47). One striking feature of BtAdV 250-A virions was the prominent extensions or spikes of protein IX (tentative identity based on previous cryo-EM structures [36, 47, 66]) from the capsid surface. It is of interest that a similar (albeit more pronounced) topology is observed in LH3 of OAdV-7, the atadenovirus equivalent of protein IX, where LH3 forms prominent knobs on the surface of the virus believed to be important in maintaining capsid stability (44). Although the N-terminal portion of protein IX in BtAdV 250-A is well conserved relative to CAdV-2 (87.5% amino acid identity in residues 1 to 40), which is likely dependent upon the protein-protein interactions that occur between protein IX and the hexons (47), the C terminus is considerably more variable (46.7% identity; residues 41 to 101). Since this C-terminal region protrudes well beyond the distal tip of the hexons in BtAdV 250-A (Fig. 5A) and would form 80 trimers (240 molecules) on the surface of the virion (47), it is possible that protein IX may play some, yet-to-be-identified, ancillary role in host cell recognition and infection. Interestingly, virions lacking protein IX have been shown to incorporate genomes into their capsids but are incapable of establishing productive infections (67).
Unlike the more conserved hexon, penton, and N-terminal tail (amino acid residues 1 to 40) of the fiber protein, which had overlapping densities when BtAdV 250-A and CAdV-2 internal capsid structures were superimposed (Fig. 5C), the C-terminal regions of their fibers showed more limited conservation (58.6% amino acid identity; residues 41 to 533). The elongated fiber in CAdV-2 (estimated to be 33 nm in length) has been suggested to contain 18.5 repeats (15 to 21 aa per repeat), with the longest repeats (hinge regions) likely imparting a greater degree of flexibility that may aid in subsequent integrin binding by the penton and increase infectivity (36, 68). Although the full-length BtAdV 250-A fiber protein on the virion could not be reconstructed by cryo-EM due to it being disordered as a consequence of its flexible nature, it likely contains a similar topology to CAdV-2 based on transmission electron micrographs of negatively stained virions (Fig. 1D) and the overall conservation of the two hinge regions identified in the CAdV-2 stalk that would impart increased flexibility (36). In addition, BtAdV 250-A shared a higher identity to CAdV-2 than to other BtAdVs (i.e., BtAdV A and B) when comparing the level of amino acid conservation in their fiber proteins.
In vitro host range comparisons demonstrated that CAdV A (CAdV-1) was generally restricted in its ability to replicate in various hosts outside of the order Carnivora, whereas no such host restriction was observed with BtAdV 250-A, since it replicated to high titers in cell lines from multiple divergent mammalian species (Fig. 7). This finding would tentatively be compatible with the potential for successful cross-species transmission of BtAdVs, in addition to their wide geographical distribution. Although CAdV-1 did not replicate in cells derived from noncarnivoran hosts such as primate or marsupial cells as did BtAdV, it did grow in bat cells, further demonstrating the close relationship between bat and carnivore viruses. Whether the fiber proteins of BtAdVs use the CAR to infect cells, as does CAdV-2 (69), is currently unknown. Although a RGD αVβ3 and αVβ5 integrin-binding motif is absent in the BtAdV 250-A penton (as it is in CAdV-2) (36), a Leu-Asp-Val (LDV) motif—used by other AdVs to bind to alternate integrins such as α4β1 (70)—was present in protein IIIa, a minor cement protein that was abundant in BtAdV 250-A capsids (Fig. 3) and is found on the surface exterior near the penton base in HAdVs (47). Although no role of protein IIIa has been ascribed in integrin binding, we found the LDV motif conserved in BtAdV 250-A, BtAdV-2, BtAdV-3, CAdV-1, and CAdV-2. Mutagenesis and subnanometer or near-atomic cryo-EM reconstructions of BtAdV 250-A or other BtAdVs in the future will help to resolve any potential roles such minor capsid proteins (e.g., proteins IIIa and IX) may play in the viral life cycle and also confirm their positions in the capsid. As exemplified by TMAdV (5), a better understanding of the natural host ranges and mechanisms of infection of adenoviruses may be insightful in determining their potential propensity to cross species barriers.
As more research is oriented toward determining the diversity of viruses in insectivorous and fruit bat species around the world, the number of BtAdVs detected will likely continue to increase, as reiterated by recent surveillance studies (30, 32). Such findings will shed more light on the evolutionary history between BtAdVs and other mastadenoviruses and may also aid in the discovery of novel unorthodox adenovirus vectors for use in human therapeutics, as exemplified by CAdV-2 (71). Although bats are now being recognized as reservoir hosts for both RNA and DNA viruses, including some that may pose a risk to human health, it is important to emphasize the key biological role they play in many different ecosystems through pollination, seed dispersal, and pest control (72, 73). Therefore, future research by disease ecologists and virologists into the natural history and epizootiology of bat viruses (such as BtAdVs) should be coupled with the aims of bat conservationists in order to maintain and protect their populations from negative stigmas that may be associated with being potential reservoirs of viral zoonoses.
MATERIALS AND METHODS
Sampling of bats and gross and microscopic examination.
On 21 July 2009, three Rafinesque's big-eared bats were found dead at a known maternity roost in an abandoned building in Mammoth Cave National Park, Edmonson County, Kentucky, and were submitted to the Southeastern Cooperative Wildlife Disease Study for diagnostic evaluation. Routine postmortem gross and microscopic examination was conducted on the three bat carcasses (designated as clinical cases 09-250-A, -B, and -C). Samples of major organs were collected and tested for virus isolation (below) or fixed in 10% buffered formalin for microscopic analysis. Fixed tissues were routinely processed for histology, embedded in paraffin, sectioned at 3 to 5 μm, and stained with hematoxylin and eosin. Special stains and/or fungal cultures were performed as indicated.
Virus isolation and preliminary identification.
Samples of brain, heart, trachea, and gastrointestinal tract from the bats were taken for attempts at virus isolation. Tissue samples (∼0.5 cm3) were mechanically homogenized in 650 μl of minimum essential medium (MEM) supplemented with 5% fetal bovine serum and 400 U/ml penicillin, 400 μg/ml streptomycin, and 1 μg/ml amphotericin B (Sigma, St. Louis, MO). Homogenized tissues were centrifuged for 6,700 × g for 10 min, and clarified supernatant (100 μl) was used to infect 2-day-old 3.8-cm2 confluent monolayers of Vero E6 cells in duplicate (see cell line information below). Supernatant (100 μl) from inoculated cultures was then transferred at weekly intervals and cultures were monitored daily for evidence of cytopathic effects (CPE). Wells exhibiting CPE were harvested and passaged once to generate stock virus. DNA was extracted using a QIAamp DNA minikit (Qiagen, Valencia, CA) and tested by PCR using degenerate adenovirus primers targeting a conserved stretch of the DNA Pol gene (5′-CYTTGTCTGCYTTTTCYTTSGC-3′ and 5′-TATGACATMTGYGGMATGTAYGC-3′) and a GoTaq Flexi DNA polymerase kit (Promega, Madison, WI). Amplicons were cloned using a PCR cloning kit (Qiagen) and sequenced using plasmid-specific primers.
Virus purification.
Confluent Vero E6 cell cultures were grown in 175-cm2 flasks infected with stock virus (250-A) at a multiplicity of infection (MOI) of 0.5 to 1 PFU/cell. Supernatant was harvested at day 4 postinfection and clarified by low-speed centrifugation at 4,400 × g for 30 min at 4°C. Virus was then concentrated using an Amicon Ultra-15 100K centrifugal filter unit (Millipore, Billerica, MA) and layered on a cesium chloride (CsCl) step gradient (1.5, 1.35, and 1.25 g cm−3) (74). The gradient was centrifuged at 169,000 × g for 4 h at 4°C using a Beckman SW 32 Ti rotor in an Optima L-100K ultracentrifuge (Beckman Coulter, Brea, CA). The two virus bands (upper “light” noninfectious particles in the 1.25-g cm−3 phase and lower “heavy” DNA-containing infectious particles in the 1.35-g cm−3 phase) were individually recovered by pipetting. The heavy capsids were then gently mixed with a 1.35-g cm−3 solution of CsCl and centrifuged at 169,000 × g for 18 h at 4°C. The band was recovered as above, dialyzed overnight in purification buffer (10 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2 [pH 7.5]), and concentrated to ≤200 μl using an Amicon Ultra-15 100K centrifugal filter, and then aliquots were frozen at −80°C until further use.
Genomic sequencing and assembly.
Since preliminary genetic evidence suggested that the virus (250-A) isolated from the Rafinesque's big-eared bat was a novel adenovirus, purified virus was subjected to next-generation sequencing to obtain the full genome. Extracted viral DNA was barcoded and sequenced on an Illumina HiSeq2000 platform using 100-bp paired-end chemistry. Reads were demultiplexed using custom in-house software and quality trimmed using the open source trim.pl script (http://wiki.bioinformatics.ucdavis.edu/index.php/Trim.pl). Multiple genome assemblies were generated with the Velvet genome assembler (75), using k-mer sizes ranging from 33 to 65, and an estimated coverage of 250×. Assemblies for each k-mer were BLASTx-searched against a local version of the nr database to predict scaffolds containing viral proteins. The most complete assembly (k-mer 53) constructed an adenovirus into a single segment of 30,386 nt. The ends of the genome were then filled in using primers based on the newly assembled 250-A genome and the conserved termini of BtAdV PPV-1 (BtAdV B) and CAdVs.
Bioinformatic analysis.
The molecular weights of BtAdV 250-A proteins were predicted using the ProtParam tool on the ExPASy server (http://web.expasy.org/protparam/). Potential conserved motifs and protein topologies in putative new genes were assessed by using Quick2D in the Max Planck Institute Bioinformatics Toolkit (http://toolkit.tuebingen.mpg.de/), WU-BLAST on the EMBL-EBI server (http://www.ebi.ac.uk/Tools/sss/wublast/), and the Protein Homology/analogY Recognition Engine V 2.0 server (Phyre2; http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) (76). Nucleotide and protein identities of BtAdV 250-A to other BtAdVs, along with CAdV A serotypes, were determined using the percent identity matrix in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Mass spectrometry.
An aliquot of CsCl-purified BtAdV 250-A virions was subjected to SDS-PAGE to visualize the electrophoretic profiles of the structural proteins and to determine their identities by mass spectrometry. Briefly, 5× Laemmli sample buffer (250 mM Tris-HCl [pH 6.8], 25% β-mercaptoethanol, 10% SDS, 50% glycerol, 0.05% bromophenol blue) was added to CsCl-purified virus, followed by incubation at 100°C for 5 min. The mixture was then electrophoresed by SDS-PAGE, and the molecular masses of the denatured proteins were estimated using a Precision Plus Protein Dual Color Standards ladder (Bio-Rad, Hercules, CA). Proteins were then stained with a SYPRO Ruby protein gel stain (Molecular Probes/Invitrogen, Carlsbad, CA), excised from the gel, digested with trypsin, and analyzed by nano-HPLC-MS/MS using a LTQ-Orbitrap Elite mass spectrometer (Thermo-Fisher Scientific, San Jose, CA) as described previously (77). Peptides were identified using the MascotDaemon search engine (version 2.3.02; Matrix Science, Boston, MA) against a database of BtAdV 250-A proteins deduced from the genomic sequence and Chlorocebus aethiops (Vero cell host) proteins.
Cryo-EM.
Purified BtAdV 250-A virions were vitrified using a Gatan Cryoplunge 3 (Pleasanton, CA). A 3.5-μl aliquot of virus was placed on a glow-discharged continuous carbon supported Quantifoil grid (Jena, Germany) and plunge-frozen in liquid ethane. Imaging was done under cryo conditions with a Gatan 626 cryo holder in a 200kV JEOL 2100 LaB6 transmission electron microscope (TEM) (Peabody, MA). Data were collected under low-dose imaging conditions on a Gatan Ultrascan US4000 CCD, calibrated with a MAG*I*CAL (Ted Pella, Inc.) calibration standard for a pixel size of 2.33 Å and a real magnification of 64,472×. Negative-stain images of BtAdV 250-A virions were prepared using 0.75% uranyl formate and imaged on continuous carbon grids in the 2100 LaB6 TEM.
For the cryo-EM single particle reconstruction, 1,592 particles were extracted from 383 micrographs using a box size of 450 in EMAN/1.9 (78). Using 1,109 particles, a structure with a resolution of 17.9 Å was produced after 20 iterations in auto3dem (79). Continuing the reconstruction or using a more stringent particle selection option (85% of the selected particles) did not further improve the reconstruction (data not shown). These experiments confirmed that the maximum quality of the map with the given number of particles was reached at the 20th iteration. The structure was interpreted in Chimera and the central section was created in Robem (79).
Phylogenetic analysis.
Two adenovirus data sets were compiled for phylogenetic analysis: (i) 32-aa sequences of the full-length penton protein (which is 486 aa in length in BtAdV 250-A) and (ii) 43-nt sequences of the partial hexon gene (spanning the region of nt 1165 to 1824 in BtAdV 250-A). The penton data set contained sequences of all species of the genus Mastadenovirus approved by the ICTV (note that full penton sequences are not currently available for Ovine mastadenovirus B or Porcine mastadenovirus B) (1). Eight additional proposed mastadenoviruses—BtAdV 250-A, TMAdV ECC-2011, a skunk adenovirus (SkAdV-1, isolate PB1) (37), and five new BtAdVs (isolates WIV9 to WIV13) recovered from Rhinolophus sinicus and Miniopterus schreibersii in China (32)—were also included in the analysis. Bovine atadenovirus D (BAdV-4) was used as an outgroup to root the penton tree. The hexon gene data set comprised 35 BtAdVs, along with viruses that were identified to be most closely related to BtAdVs from the penton phylogeny: CAdV A, SkAdV-1, Bovine mastadenovirus B (BAdV-3), Tree shrew mastadenovirus A (TSAdV-1), Equine mastadenovirus A (EAdV-1), and EAdV B (EAdV-2) (38). The homologous sequence from Murine mastadenovirus A (MAdV A) was used as an outgroup to root the hexon tree. See Fig. 6 for the GenBank accession numbers of all sequences used in the phylogenetic analysis.
All sequences were aligned using MUSCLE (80) employing 16 iterations. Because of the highly divergent nature of some parts of the sequence alignment, all gap positions and ambiguous parts of the alignment were removed prior to subsequent analysis using Gblocks (81). This resulted in final data sets of 408 aa residues for the penton protein and 559 nt for the hexon gene. Phylogenetic trees of both data sets were then estimated using the maximum-likelihood method available within the PhyML program (82). The LH+Γ model of amino acid substitution was used in the case of the penton data set, while the GTR+I+Γ nucleotide substitution model was used for the hexon sequences. In both cases, subtree pruning and regrafting branch-swapping was used to find the tree of highest likelihood, with nearest neighbor interchange branch-swapping used in a subsequent bootstrap analysis (1,000 replications).
In vitro host range studies.
The in vitro host ranges of BtAdVs and CAdVs were examined using two viruses: (i) BtAdV 250-A (Vero E6 passage 3) and (ii) CAdV 24-05 (MDCK passage 3). The CAdV 24-05 isolate (serotype 1) was recovered from the liver of a gray fox (Urocyon cinereoargenteus) with infectious canine hepatitis sampled in Georgia in 2005 (83). For comparative replication studies between the two viruses, we used the following bat and carnivore cell lines: (i) Tb1Lu (Mexican free-tailed bat [Tadarida brasiliensis] lung), (ii) FoLu (gray fox lung), (iii) A72 (domestic dog [Canis lupus familiaris] tumor), and (iv) Mpf (domestic ferret [Mustela putorius furo] brain). To determine whether the two viruses could replicate in cells derived from species outside of the orders Chiroptera and Carnivora, we tested them in Vero E6 (African green monkey [Chlorocebus aethiops] kidney) and OK (Virginia opossum [Didelphis virginianus] kidney) cells. Finally, the ability of BtAdV 250-A to replicate in human A549 (carcinomic alveolar basal epithelial) and HeLa (adenocarcinomic cervical) cells was assessed. All cells were obtained from the American Type Culture Collection (Manassas, VA) and grown in MEM with 5% fetal bovine serum (growth media) in a 5% CO2 atmosphere. For growth curve analysis of BtAdV 250-A and CAdV 24-05 in each cell line, cells were seeded at a density of ∼2 × 105 cells/ml in a 3.8-cm2-well format and, once the monolayers were confluent, each well was infected with an MOI of 0.01 tissue culture infection dose 50 (TCID50)/cell. Virus was allowed to absorb to the cells for 2 h; the inocula were then removed, the cells were washed three times in PBS, and growth medium was added. Wells were harvested at 2-day intervals for 10 days and frozen at −80°C until further processing. Titers were assessed by 10-fold titrations in A72 cells in a 96-well plate format and expressed as the log10 TCID50/ml.
Accession number(s).
The complete genome of BtAdV 250-A has been deposited in NCBI GenBank under accession number KX871230. The cryo-EM map of BtAdV 250-A has been deposited in the Electron Microscopy Data Bank (www.emdatabank.org) under accession number EMD-8371.
ACKNOWLEDGMENTS
We thank Steven C. Thomas of the National Park Service at Mammoth Cave National Park (MCNP), Kentucky, for submission of the Rafinesque's big-eared bats and for images of the bats from MCNP; Tom Uhlman for additional bat images from MCNP; Sheng Zhang of the Cornell University Institute of Biotechnology for expertise and support on gel-based protein identifications; and Mark Ruder (University of Georgia), Adam Fitch (University of Pittsburgh), Kristin Shingler (Penn State), Dana Fader (Cornell University), and Alicia Ortega (Cornell University) for their technical and laboratory expertise. Finally, we thank Colin Parrish (Cornell University) for his continuing support.
Funding for this research was provided by start-up funds to A.B.A. from the Department of Biomedical Sciences and Pathobiology, Virginia Tech; by the wildlife management agencies of the SCWDS member states through the Federal Aid to Wildlife Restoration Act (50 Stat. 917); by Pennsylvania Department of Health CURE funds, and by the Office of the Director, National Institutes of Health (NIH), under award number S10OD011986 to S.L.H. E.C.H. is supported by a National Health and Medical Research Council Australia Fellowship (grant AF30). A.B.A. was additionally supported by a Ruth L. Kirschstein National Research Service Award fellowship (F32AI100545) from the National Institute of Allergy and Infectious Diseases, NIH.
REFERENCES
- 1.Harrach B, Benkö M, Both GW, Brown M, Davison AJ, Echavarría M, Hess M, Jones MS, Kajon A, Lehmkuhl HD, Mautner V, Mittal SK, Wadell G. 2012. Family Adenoviridae, p 125–141. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 2.Davison AJ, Benko M, Harrach B. 2003. Genetic content and evolution of adenoviruses. J Gen Virol 84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 3.Benkö M, Harrach B. 2003. Molecular evolution of adenoviruses. Curr Top Microbiol Immunol 272:3–35. [DOI] [PubMed] [Google Scholar]
- 4.Harrach B. 2000. Reptile adenoviruses in cattle? Acta Vet Hung 48:485–490. doi: 10.1556/004.48.2000.4.11. [DOI] [PubMed] [Google Scholar]
- 5.Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. 2011. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog 7:e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupprecht CE, Turmelle A, Kuzmin IV. 2011. A perspective on lyssavirus emergence and perpetuation. Curr Opin Virol 1:662–670. doi: 10.1016/j.coviro.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB. 2010. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J Virol 84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y, Shi Z. 2012. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol 86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 12.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A 102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–557. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 14.Guo WP, Lin XD, Wang W, Tian JH, Cong ML, Zhang HL, Wang MR, Zhou RH, Wang JB, Li MH, Xu J, Holmes EC, Zhang YZ. 2013. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog 9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Rahman SA, Hughes T, Smith C, Field HE, Daszak P, Henipavirus Ecology Research Group . 2011. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg 85:946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith I, Wang LF. 2013. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol 3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda K, Hondo E, Terakawa J, Kiso Y, Nakaichi N, Endoh D, Sakai K, Morikawa S, Mizutani T. 2008. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae). Emerg Infect Dis 14:347–349. doi: 10.3201/eid1402.070932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutcheon JM, Kirsh JW. 2004. Camping in a different tree: results of molecular systematics studies of bats using DNA-DNA hybridization. J Mammal Evol 11:17–47. doi: 10.1023/B:JOMM.0000029144.80747.d2. [DOI] [Google Scholar]
- 19.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 20.Sonntag M, Mühldorfer K, Speck S, Wibbelt G, Kurth A. 2009. New adenovirus in bats, Germany. Emerg Infect Dis 15:2052–2055. doi: 10.3201/eid1512.090646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Ge X, Zhang H, Zhou P, Zhu Y, Zhang Y, Yuan J, Wang LF, Shi Z. 2010. Host range, prevalence, and genetic diversity of adenoviruses in bats. J Virol 84:3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jánoska M, Vidovszky M, Molnár V, Liptovszky M, Harrach B, Benko M. 2011. Novel adenoviruses and herpesviruses detected in bats. Vet J 189:118–121. doi: 10.1016/j.tvjl.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Kohl C, Vidovszky MZ, Mühldorfer K, Dabrowski PW, Radonić A, Nitsche A, Wibbelt G, Kurth A, Harrach B. 2012. Genome analysis of bat adenovirus 2: indications of interspecies transmission. J Virol 86:1888–1892. doi: 10.1128/JVI.05974-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q. 2012. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol 86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raut CG, Yadav PD, Towner JS, Amman BR, Erickson BR, Cannon DL, Sivaram A, Basu A, Nichol ST, Mishra AC, Mourya DT. 2012. Isolation of a novel adenovirus from Rousettus leschenaultii bats from India. Intervirology 55:488–490. doi: 10.1159/000337026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen LH, Wu ZQ, Hu YF, Yang F, Yang J, Jin Q. 2012. Genetic diversity of adenoviruses in bats of China. Bing Du Xue Bao 28:403–408. [PubMed] [Google Scholar]
- 27.Baker KS, Leggett RM, Bexfield NH, Alston M, Daly G, Todd S, Tachedjian M, Holmes CE, Crameri S, Wang LF, Heeney JL, Suu-Ire R, Kellam P, Cunningham AA, Wood JL, Caccamo M, Murcia PR. 2013. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 441:95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lima FE, Cibulski SP, Elesbao F, Carnieli Junior P, Batista HB, Roehe PM, Franco AC. 2013. First detection of adenovirus in the vampire bat (Desmodus rotundus) in Brazil. Virus Genes 47:378–381. doi: 10.1007/s11262-013-0947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conrardy C, Tao Y, Kuzmin IV, Niezgoda M, Agwanda B, Breiman RF, Anderson LJ, Rupprecht CE, Tong S. 2014. Molecular detection of adenoviruses, rhabdoviruses, and paramyxoviruses in bats from Kenya. Am J Trop Med Hyg 91:258–266. doi: 10.4269/ajtmh.13-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidovszky M, Kohl C, Boldogh S, Görföl T, Wibbelt G, Kurth A, Harrach B. 2015. Random sampling of the Central European bat fauna reveals the existence of numerous hitherto unknown adenoviruses. Acta Vet Hung 63:508–525. doi: 10.1556/004.2015.047. [DOI] [PubMed] [Google Scholar]
- 31.Adams MJ, Lefkowitz EJ, King AMQ, Carstens EB. 2014. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol 159:2831–2841. doi: 10.1007/s00705-014-2114-3. [DOI] [PubMed] [Google Scholar]
- 32.Tan B, Yang XL, Ge XY, Peng C, Zhang YZ, Zhang LB, Shi ZL. 2016. Novel bat adenoviruses with an extremely large E3 gene. J Gen Virol 97:1625–1635. doi: 10.1099/jgv.0.000470. [DOI] [PubMed] [Google Scholar]
- 33.Wu L, Zhou P, Ge X, Wang LF, Baker ML, Shi Z. 2013. Deep RNA sequencing reveals complex transcriptional landscape of a bat adenovirus. J Virol 87:503–511. doi: 10.1128/JVI.02332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JS, Lacki MJ, Thomas SC, Grider JF. 2012. Frequent arousals from winter torpor in Rafinesque's big-eared bat (Corynorhinus rafinesquii). PLoS One 7:e49754. doi: 10.1371/journal.pone.0049754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berk AJ. 2013. Adenoviruses, p 1704–1731. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 36.Schoehn G, El Bakkouri M, Fabry CM, Billet O, Estrozi LF, Le L, Curiel DT, Kajava AV, Ruigrok RW, Kremer EJ. 2008. Three-dimensional structure of canine adenovirus serotype 2 capsid. J Virol 82:3192–3203. doi: 10.1128/JVI.02393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak RA, Ackford JG, Slaine P, Li A, Carman S, Campbell D, Welch MK, Kropinski AM, Nagy É. 2015. Characterization of a novel adenovirus isolated from a skunk. Virology 485:16–24. doi: 10.1016/j.virol.2015.06.026. [DOI] [PubMed] [Google Scholar]
- 38.Cavanagh HM, Mahony TJ, Vanniasinkam T. 2012. Genetic characterization of equine adenovirus type 1. Vet Microbiol 155:33–37. doi: 10.1016/j.vetmic.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH. 2012. The major facilitator superfamily (MFS) revisited. FEBS J 279:2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker PJ, Dietzgen RG, Joubert DA, Blasdell KR. 2011. Rhabdovirus accessory genes. Virus Res 162:110–125. doi: 10.1016/j.virusres.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell WC. 2009. Adenoviruses: update on structure and function. J Gen Virol 90:1–20. doi: 10.1099/vir.0.003087-0. [DOI] [PubMed] [Google Scholar]
- 42.Benevento M, Di Palma S, Snijder J, Moyer CL, Reddy VS, Nemerow GR, Heck AJ. 2014. Adenovirus composition, proteolysis, and disassembly studied by in-depth qualitative and quantitative proteomics. J Biol Chem 289:11421–11430. doi: 10.1074/jbc.M113.537498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webster A, Russell S, Talbot P, Russell WC, Kemp GD. 1989. Characterization of the adenovirus proteinase: substrate specificity. J Gen Virol 70:3225–3234. doi: 10.1099/0022-1317-70-12-3225. [DOI] [PubMed] [Google Scholar]
- 44.Pantelic RS, Lockett LJ, Rothnagel R, Hankamer B, Both GW. 2008. Cryoelectron microscopy map of atadenovirus reveals cross-genus structural differences from human adenovirus. J Virol 82:7346–7356. doi: 10.1128/JVI.00764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. 2010. Crystal structure of human adenovirus at 3.5 Å resolution. Science 329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy VS, Nemerow GR. 2014. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Nat Acad Sci 111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furcinitti PS, van Oostrum J, Burnett RM. 1989. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J 8:3563–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vellinga J, van den Wollenberg DJ, van der Heijdt S, Rabelink MJ, Hoeben RC. 2005. The coiled-coil domain of the adenovirus type 5 protein IX is dispensable for capsid incorporation and thermostability. J Virol 79:3206–3210. doi: 10.1128/JVI.79.5.3206-3210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebner K, Pinsker W, Lion T. 2005. Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: phylogenetic, taxonomic, and clinical implications. J Virol 79:12635–12642. doi: 10.1128/JVI.79.20.12635-12642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones C. 1977. Pleotus rafinesquii. Mammal Spec 69:1–4. [Google Scholar]
- 52.Arroyo-Cabrales J, Álvarez-Castañeda ST. 2008. Corynorhinus rafinesquii The International Union for the Conservation of Nature (IUCN) red list of threatened species, version 2014.3. http://www.iucnredlist.org/details/17600/0. [Google Scholar]
- 53.Tobe SS, Kitchener AC, Linacre AMT. 2010. Reconstructing mammalian phylogenies: a detailed comparison of the cytochrome B and cytochrome oxidase subunit I mitochondrial genes. PLoS One 5:e14156. doi: 10.1371/journal.pone.0014156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kojaoghlanian T, Flomenberg P, Horwitz MS. 2003. The impact of adenovirus infection on the immunocompromised host. Rev Med Virol 13:155–171. doi: 10.1002/rmv.386. [DOI] [PubMed] [Google Scholar]
- 55.Karstad L, Ramsden R, Berry TJ, Binn LN. 1975. Hepatitis in skunks caused by the virus of infectious canine hepatitis. J Wildl Dis 11:494–496. doi: 10.7589/0090-3558-11.4.494. [DOI] [PubMed] [Google Scholar]
- 56.Whetstone CA, Draayer H, Collins JE. 1988. Characterization of canine adenovirus type 1 isolated from American black bears. Am J Vet Res 49:778–780. [PubMed] [Google Scholar]
- 57.Park NY, Lee MC, Kurkure NV, Cho HS. 2007. Canine adenovirus type 1 infection of a Eurasian river otter (Lutra lutra). Vet Pathol 44:536–539. doi: 10.1354/vp.44-4-536. [DOI] [PubMed] [Google Scholar]
- 58.Thompson H, O'Keeffe AM, Lewis JC, Stocker LR, Laurenson MK, Philbey AW. 2010. Infectious canine hepatitis in red foxes (Vulpes vulpes) in the United Kingdom. Vet Rec 166:111–114. doi: 10.1136/vr.b4763. [DOI] [PubMed] [Google Scholar]
- 59.Swenson J, Orr K, Bradley GA. 2012. Hemorrhagic and necrotizing hepatitis associated with administration of a modified live canine adenovirus-2 vaccine in a maned wolf (Chrysocyon brachyurus). J Zoo Wildl Med 43:375–383. doi: 10.1638/2011-0192.1. [DOI] [PubMed] [Google Scholar]
- 60.Choi JW, Lee HK, Kim SH, Kim YH, Lee KK, Lee MH, Oem JK. 2014. Canine adenovirus type 1 in a fennec fox (Vulpes zerda). J Zoo Wildl Med 45:947–950. doi: 10.1638/2013-0286.1. [DOI] [PubMed] [Google Scholar]
- 61.Caudell D, Confer AW, Fulton RW, Berry A, Saliki JT, Fent GM, Ritchey JW. 2005. Diagnosis of infectious canine hepatitis virus (CAV-1) infection in puppies with encephalopathy. J Vet Diagn Invest 17:58–61. doi: 10.1177/104063870501700111. [DOI] [PubMed] [Google Scholar]
- 62.Almes KM, Janardhan KS, Anderson J, Hesse RA, Patton KM. 2010. Fatal canine adenoviral pneumonia in two litters of bulldogs. J Vet Diagn Invest 22:780–784. doi: 10.1177/104063871002200524. [DOI] [PubMed] [Google Scholar]
- 63.Green RG, Ziegler NR, Green BB, Dewey ET. 1930. Epizootic fox encephalitis. I General description. Am J Hyg 12:109–129. [Google Scholar]
- 64.Rubarth S. 1947. An acute virus disease with liver lesions in dogs (hepatitis contagiosa canis): a pathologico-anatomical and etiological investigation. Acta Pathol Microbiol Scand Suppl 69:1–222. [Google Scholar]
- 65.Weiss RA. 2002. Virulence and pathogenesis. Trends Microbiol 10:314–317. doi: 10.1016/S0966-842X(02)02391-0. [DOI] [PubMed] [Google Scholar]
- 66.Marsh MP, Campos SK, Baker ML, Chen CY, Chiu W, Barry MA. 2006. Cryoelectron microscopy of protein IX-modified adenoviruses suggests a new position for the C terminus of protein IX. J Virol 80:11881–11886. doi: 10.1128/JVI.01471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sargent KL, Ng P, Evelegh C, Graham FL, Parks RJ. 2004. Development of a size-restricted pIX-deleted helper virus for amplification of helper-dependent adenovirus vectors. Gene Ther 11:504–511. doi: 10.1038/sj.gt.3302107. [DOI] [PubMed] [Google Scholar]
- 68.Wu E, Pache L, Von Seggern DJ, Mullen TM, Mikyas Y, Stewart PL, Nemerow GR. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J Virol 77:7225–7235. doi: 10.1128/JVI.77.13.7225-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soudais C, Boutin S, Hong SS, Chillon M, Danos O, Bergelson JM, Boulanger P, Kremer EJ. 2000. Canine adenovirus type 2 attachment and internalization: coxsackievirus-adenovirus receptor, alternative receptors, and an RGD-independent pathway. J Virol 74:10639–10649. doi: 10.1128/JVI.74.22.10639-10649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wickham TJ, Carrion ME, Kovesdi I. 1995. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther 2:750–756. [PubMed] [Google Scholar]
- 71.Kremer EJ, Nemerow GR. 2015 Adenovirus tales: from the cell surface to the nuclear pore complex. PLoS Pathog 11:e1004821. doi: 10.1371/journal.ppat.1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shilton LA, Altringham JD, Compton SG, Whittaker RJ. 1999. Old World fruit bats can be long-distance seed dispersers through extended retention of viable seeds in the gut. Proc Biol Sci 266:219. doi: 10.1098/rspb.1999.0625. [DOI] [Google Scholar]
- 73.Boyles JG, Cryan PM, McCracken GF, Kunz TH. 2011. Economic importance of bats in agriculture. Science 332:41–42. doi: 10.1126/science.1201366. [DOI] [PubMed] [Google Scholar]
- 74.Jager L, Hausl MA, Rauschhuber C, Wolf NM, Kay MA, Ehrhardt A. 2009. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat Protoc 4:547–564. doi: 10.1038/nprot.2009.4. [DOI] [PubMed] [Google Scholar]
- 75.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Allison AB, Ballard JR, Tesh RB, Brown JD, Ruder MG, Keel MK, Munk BA, Mickley RM, Gibbs SE, Travassos da Rosa AP, Ellis JC, Ip HS, Shearn-Bochsler VI, Rogers MB, Ghedin E, Holmes EC, Parrish CR, Dwyer C. 2015. Cyclic avian mass mortality in the northeastern United States is associated with a novel orthomyxovirus. J Virol 89:1389–1403. doi: 10.1128/JVI.02019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 79.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM: an automated and high-throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 82.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 83.Gerhold RW, Allison AB, Temple DL, Chamberlain MJ, Strait KR, Keel MK. 2007. Infectious canine hepatitis in a gray fox (Urocyon cinereoargenteus). J Wildl Dis 43:734–736. doi: 10.7589/0090-3558-43.4.734. [DOI] [PubMed] [Google Scholar]