ABSTRACT
The retinoic acid-inducible gene 1 (RIG-I) signaling pathway is essential for the recognition of viruses and the initiation of host interferon (IFN)-mediated antiviral responses. Once activated, RIG-I interacts with polyubiquitin chains generated by TRIM25 and binds mitochondrial antiviral signaling protein (MAVS), leading to the production of type I IFN. We now show specific interactions among these key partners in the RLR pathway through the use of bimolecular fluorescence complementation (BiFC) and super-resolution microscopy. Dimers of RIG-I, TRIM25, and MAVS localize into different compartments. Upon activation, we show that TRIM25 is redistributed into cytoplasmic dots associated with stress granules, while RIG-I associates with TRIM25/stress granules and with mitochondrial MAVS. In addition, MAVS competes with TRIM25 for RIG-I binding, and this suggests that upon TRIM25-mediated activation of RIG-I, RIG-I moves away from TRIM25 to interact with MAVS at the mitochondria. For the first time, the distribution of these three proteins was analyzed at the same time in virus-infected cells. We also investigated how specific viral proteins modify some of the protein complexes in the pathway. The protease NS3/4A from hepatitis C virus redistributes the complexes RIG-I/MAVS and MAVS/MAVS but not RIG-I/TRIM25. In contrast, the influenza A virus NS1 protein interacts with RIG-I and TRIM25 in specific areas in the cell cytoplasm and inhibits the formation of TRIM25 homocomplexes but not the formation of RIG-I/TRIM25 heterocomplexes, preventing the formation of RIG-I/MAVS complexes. Thus, we have localized spatially in the cell different complexes formed between RIG-I, TRIM25, and MAVS, in the presence or absence of two viral IFN antagonistic proteins.
IMPORTANCE The first line of defense against viral infections is the innate immune response. Viruses are recognized by pathogen recognition receptors, such as the RIG-I like receptor family, that activate a signaling cascade that induces IFN production. In the present study, we visualized, for the first time in cells, both in overexpression and endogenous levels, complexes formed among key proteins involved in this innate immune signaling pathway. Through different techniques we were able to analyze how these proteins are distributed and reorganized spatially within the cell in order to transmit the signal, leading to an efficient antiviral state. In addition, this work presents a new means by how, when, and where viral proteins can target these pathways and act against the host immune system in order to counteract the activation of the immune response.
KEYWORDS: influenza, innate immunity, microscopy, pathogen recognition receptors, RIG-I, virus
INTRODUCTION
In an infected cell, viruses can be recognized by cellular pathogen recognition receptors (PRRs). These PRRs detect viral genomes and/or viral RNA and trigger the production of type I and III interferon (IFN), mediating the activation of the innate immune response. This innate immune response provides immediate defense against viral infection, leading also to the stimulation of adaptive immune responses (1, 2). Within the cell, there are several families of PRRs that differ in cellular localization and in the activation of specific signaling molecules. The RIG-I-like receptor (RLR) family is composed of cytoplasmic sensors and, to date, three members have been described: RIG-I (retinoic acid-inducible gene 1), MDA5 (melanoma differentiation associated factor 5), and LGP2 (laboratory of genetics and physiology 2). The first RLR identified and the best characterized is RIG-I (3). It contains a DExD/H-box helicase-like domain containing ATPase and translocase activities, a repressor regulatory domain (RD) at the C terminus, and two caspase-associated recruitment domains (CARDs) at the N terminus. RIG-I distinguishes and recognizes the presence of a 5′-triphosphate-containing double-stranded RNA in the cytoplasm of the cell (2, 4–6). In an uninfected cell, RIG-I is in an inactive state where a helicase intermediate domain interacts with the CARDs (7). Upon recognition of the 5′-ppp RNA by the RD, RIG-I hydrolyzes ATP and changes its conformation to an active state (8, 9). The CARDs are then exposed and become K63-linked polyubiquitinated by E3-ligases at different sites of the protein: at lysine 172 (Lys172) by TRIM25 (10) and at Lys849 and Lys851 by RIPLET (11). In addition, RIG-I CARDs bind to free K63 polyubiquitin generated by TRIM25. Ubiquitin-associated RIG-I forms higher-order oligomers (12–14), and it interacts with its downstream adaptor molecule, MAVS, via the CARDs, resulting in MAVS multimerization and activation of IRF-3 and NF-κB, leading to transcriptional activation of IFN and antiviral genes (15, 16).
Since the RLR pathway was discovered, many advances have been made in the understanding of the mechanisms of its activation, but most of the spatial-temporal events and the subcellular localization where the key proteins interact to trigger the signal are still under investigation (17, 18). The analysis of the subcellular localization of the proteins is essential to further understand protein functions. For example, it is known that MAVS protein contains a C-terminal transmembrane domain (TM) that anchors the protein into the mitochondrial membrane, where it forms aggregates once activated (16, 19). The localization of MAVS has been described as essential for the induction of IFN production (20), since alterations in mitochondria or the lack of the TM domain of MAVS provokes the inhibition of the MAVS-mediated antiviral response (15, 20–22). In addition to MAVS, our group and others have shown that TRIM25, the E3 ligase responsible of ubiquitinate RIG-I (10), changes its distribution upon stimulation (10, 23). RIG-I is the main sensor for the recognition of many single-stranded RNA viruses, such as paramyxoviruses, flaviviruses, rhabdoviruses, and orthomyxoviruses (24–26). Most of these viruses have developed strategies to counteract the activation of the RLR pathway. One of the first viral proteins characterized was the protease NS3/4A from hepatitis C virus (HCV) (27, 28). NS3/4A cleaves MAVS in its TM domain, inhibiting type I IFN production, suggesting a link between aberrant localization of MAVS and the evasion of RIG-I activation (28–31).
In IAV, the multifunctional NS1 protein is the major antagonist of host type I IFN system (32). NS1 inhibits the activation of the innate immune response through diverse mechanisms (32, 33). In immunoprecipitation assays, NS1 interacts with RIG-I (34) and binds TRIM25, blocking its multimerization and its capacity to ubiquitinate RIG-I (35).
The bimolecular fluorescence complementation (BiFC) technique has provided us with a unique tool to address the relevance of the spatial distribution and formation of the different interactions among RIG-I, TRIM25, and MAVS to achieve an antiviral state in the cell. These proteins form complexes in very specific and unique subcellular compartments within the cytosol of the cell. Analysis of the expression of these proteins in virus-infected cells, through the use of super-resolution microscopy, has demonstrated the different localizations and interactions among them. Interestingly, the particular granule-like localization of TRIM25 was also detected during activation of the pathway in the absence of its overexpression, colocalizing with stress granule (SG) markers. The viral IFN antagonistic proteins NS3/4A and NS1 inhibit the induction of the antiviral state by dramatically altering the localization and/or formation of specific complexes MAVS/MAVS and RIG-I/MAVS, whereas the distribution of the complex RIG-I/TRIM25 was not affected.
RESULTS
Analysis of RIG-I, TRIM25, and MAVS oligomerization by BiFC.
In order to study the interaction of RIG-I, TRIM25, and MAVS in RLR signaling, a BiFC assay was developed as described in Materials and Methods and as represented in Fig. 1A (36–38). RIG-I, TRIM25, and MAVS are believed to form homo-oligomers in order to be activated and to trigger the signaling cascade (19, 35, 39). To visualize these oligomers in the cell, we fused the YN or YC half-terminal ends of the yellow fluorescent protein (YFP) to the N- or C-terminal sequences of RIG-I, TRIM25, and MAVS (Fig. 1B). Since overexpression of these three proteins is known to result in IFN induction, we confirmed the functional expression of RIG-I, MAVS, and TRIM25 fusion proteins by measuring the activation of the IFN-β promoter after transfection of the plasmids (Fig. 1C). We first investigated the localization of RIG-I oligomers. Cotransfection in HeLa cells of the RIG-I full-length fusion plasmids with the YN and YC halves of YFP led to the recovery of YFP immunofluorescence, indicating oligomer formation, as shown in Fig. 2A (row I). The expression of the proteins was also confirmed by indirect immunofluorescence with an anti-RIG-I antibody, as observed in red in Fig. 2A, and by Western blotting (see Materials and Methods [also data not shown]). The same results were obtained when cells were cotransfected with YN-RIG-I and YC fused to only the CARD of RIG-I (named yc-CARD, Fig. 2A, row II). However, as shown in row III, a BiFC signal was not detected in cells cotransfected with the YN/YC fusions plasmids carrying only the CARD, even though the proteins were detected and functional in activating IFN. Although we cannot exclude that a CARD-CARD dimer is not compatible with the BiFC complex, these results suggest that at least one full-length unit of RIG-I is needed to form a RIG-I dimer/oligomer.
FIG 1.

Representation of the BiFC approach to protein-protein interaction analysis. (A) Half cylinders represent the two fragments of YFP (YN and YC); rectangles represent the protein of interest (PoI) fused to the YFP fragment. The interaction of the proteins brings the two nonfluorescent fragments of YFP together, recovering the autofluorescence (36, 38). (B) Four different combinations tested with the proteins of interest (PoI) based on the fusion of the YN half or the YC half of the YFP to the amino or carboxy terminus of the PoI. (C) IFN-β induction by BiFC fusion constructs. Reporter assays were performed in HEK 293T cells transfected with BiFC fusion plasmids. As a control, a plasmid expressing GFP was used. Experiments were performed in triplicates.
FIG 2.
Oligomerization of RIG-I, MAVS, and TRIM25 detected by BiFC. HeLa cells were transfected with the different fused YN- or YC-tagged plasmids for full-length RIG-I (RIG-I), the CARD of RIG-I (CARD), MAVS, or TRIM25. Schematic representations of the plasmids forming the BiFC are shown on the left sides of the images. At 24 h posttransfection, the cells were fixed and processed for immunofluorescence and analysis by confocal microscopy and super-resolution microscopy. (A) Transfection of the fused YN and YC versions of full-length RIG-I (I), RIG-I/CARD (II), and CARD/CARD (III). (B) Transfection of both fused YN and YC versions of TRIM25 and MAVS. For row I, the cells were stained with an anti-TRIM25 antibody (red color), and images were obtained using confocal microscopy. For row II, the cells were stained with TOM20 (mitochondrial marker, red color), and images were obtained using STED microscopy. The image shows the mean intensity projection of a Z-stack experiment. On the right is a detailed 3D reconstruction of the Z-stacks and a histogram of the intensity of fluorescence profile measured along the dashed line in the zoom. (C) YFP fluorescence is indicated in yellow. RIG-I was stained with polyclonal serum 1C3 (4). MAVS was visualized using a rabbit polyclonal antibody. In Fig. 2C, row II, as in Fig. 2B, row II, the mitochondrial membrane was stained with TOM20, and images were obtained using STED microscopy. The image shows the mean intensity projection of a Z-stack experiment. On the right is a detailed 3D reconstruction of the Z-stacks and a histogram of the intensity of fluorescence profile measured along the dashed line in the zoom. Anti-mouse and/or anti-rabbit Alexa Fluor 647-labeled secondary antibodies were used (red). Nuclei were stained with DAPI (blue). Scale bars, 10 μm.
As shown in Fig. 2, oligomers of TRIM25 (Fig. 2B) and MAVS (Fig. 2C) were also detected by the BiFC assay. Again, the expression of the proteins was confirmed with an anti-TRIM25 and anti-MAVS antibodies (shown in red), respectively. The cellular localization of the three different dimers is different: we found RIG-I complexes homogeneously spread in the cytoplasm of the cell, whereas TRIM25 and MAVS complexes showed a dot-like distribution. Interestingly, the staining with a mitochondrial membrane marker (TOM20) and the use of super-resolution microscopy (row II in Fig. 2B and C) revealed that MAVS complexes were located at the mitochondria (Fig. 2C, row II), as expected. In contrast, TRIM25 oligomers were detected adjunct but dislodged from mitochondria (Fig. 2B, row II). For additional detail, see the enlarged images in row II. The intensity profiles show large areas of overlap in the MAVS/MAVS complex with mitochondria, whereas the profile in the TRIM25/TRIM25 complex suggests only restricted areas of overlapping with the mitochondrial marker.
RIG-I interacts with TRIM25 and MAVS in different cellular compartments.
During RIG-I signaling, RIG-I is known to interact with TRIM25 to acquire K63 polyubiquitin, and this promotes its interactions with MAVS (39). However, it is unclear whether RIG-I, TRIM25, and MAVS form a ternary complex and also where all these interactions take place within the cell. To address this issue, we investigated the cellular localization of RIG-I/TRIM25 and RIG-I/MAVS complexes using BiFC. For this analysis, HeLa cells were transfected with ycRIG-I/ynMAVS (Fig. 3A) and with ycRIG-I/ynTRIM25 (Fig. 3B). RIG-I interacted with MAVS (Fig. 3A) and TRIM25 (Fig. 3B) in the cytoplasm of the cell. The expression of the individual proteins was monitored by indirect immunofluorescence with anti-RIG-I, anti-TRIM25, and anti-MAVS antibodies. TRIM25 and MAVS again show a granular distribution in the cytosol of the transfected cell (red indicates RIG-I detection and green indicates TRIM25 or MAVS, row I). Both of the complexes, RIG-I/MAVS and RIG-I/TRIM25, seemed to be distributed very similarly in the cytoplasm of the cells. However, the stain with TOM20 (red) of the cells transfected with the BiFC showed a different cellular distribution for each protein complex. Stimulated emission depletion (STED) microscopy showed us the detailed distribution of the protein complexes. Images are shown in row II, respectively (three-dimensional [3D] reconstruction was performed in the zoomed areas). The histograms of intensity of fluorescence indicate that the RIG-I/MAVS complex fluorescence overlaps the mitochondrial marker, TOM20 (Fig. 3B, row II), whereas the RIG-I/TRIM25 signal diverges from TOM20 fluorescence (Fig. 3A, row II). We next investigated whether the RIG-I/TRIM25 complexes change after stimulation of the pathway. For this purpose, we transfected SeV RNA in cells expressing the RIG-I/TRIM25 BiFC complex. Although the overexpression of RIG-I/TRIM25 complexes results in activation of the pathway, we expected higher levels of activation after the transfection of RIG-I immunostimulatory RNA, i.e., Sendai virus (SeV) RNA. As shown in Fig. 4, there is an increase in the fluorescence signal 2 h after the transfection of SeV RNA, a finding indicative of an increase in the number of RIG-I/TRIM25 BiFC complexes. In addition, the RIG-I/TRIM25 BiFC complexes appear to be redistributed closer to the mitochondria (stained with TOM20) compared to the mock-treated cells.
FIG 3.
RIG-I interacts with MAVS and TRIM25 in different cellular compartments. HeLa cells were transfected and processed for immunofluorescence. (A) Row I shows images taken by confocal microscopy of the BiFC fusion plasmids ycRIG-I and ynMAVS. In row II the mitochondrial membrane was stained with TOM20, and images were obtained using STED microscopy. The image shows the mean intensity projection of a Z-stack experiment. On the right is a detailed 3D reconstruction of the Z-stacks and a histogram of the intensity of fluorescence profile measured along the line in the zoom. (B) Confocal (upper panel) and STED microscopy images of BiFC fusion plasmids ycRIG-I and ynTRIM25. As in panel A, panel I shows images obtained by confocal microscopy. In row II the mitochondrial membrane was stained with TOM20, and images were obtained using STED microscopy. The image shows the mean intensity projection of a Z-stack experiment. On the right is a detailed 3D reconstruction of the Z-stacks and a histogram of the intensity of fluorescence profile measured along the dashed line in the zoom. The BiFC signal is indicated in yellow. RIG-I was stained using the polyclonal antibody 1C3 (red). MAVS was visualized using a rabbit polyclonal antibody (green); TRIM25 was visualized using a monoclonal antibody (Abcam). Alexa Fluor 488- or Alexa Fluor 647-labeled anti-mouse and anti-rabbit antibodies were used as secondary antibodies, and DAPI was used for the nuclear staining (blue). Scale bars, 10 μm.
FIG 4.
Distribution of the BiFC RIG-I/TRIM25 complex after stimulation of the pathway. HeLa cells were mock transfected or transfected with SeV RNA. After 2 h posttransfection, the cells were fixed and processed for immunofluorescence. The mitochondria were stained with TOM20. Analysis of the intensity of immunofluorescence for BiFC (red line) and TOM20 (yellow line) signals are shown at the bottom of the figure. Alexa Fluor 647-conjugated antibodies were used as secondary antibodies. Nuclei were stained with DAPI. Scale bars, 10 μm.
MAVS does not bind to TRIM25 and competes with TRIM25 in interacting with RIG-I.
Since we observed different localizations of RIG-I/TRIM25 and RIG-I/MAVS complexes, we next looked for a possible TRIM25/MAVS interaction by transfecting HeLa cells with the fused BiFC plasmids ycTRIM25 and ynMAVS. As shown in Fig. 5A, a YFP signal was not detected, although both proteins were expressed and localized in the same areas of the cell (see the zoom images for details).
FIG 5.
MAVS competes with TRIM25 in the interaction with RIG-I. The plasmids ycMAVS and ynTRIM25 were cotransfected in HeLa cells alone (A) or in the presence of HA-tagged MAVS expression plasmid (B), ycRGI-I, ynTRIM25 with HA-MAVS (C), or the three BiFC fusion versions ynRIG-I, ycMAVS, and ycTRIM25 (D). After 24 h, the cells were fixed and processed for immunofluorescence. Mouse monoclonal antibody (MAb) anti-TRIM25 (red), rabbit polyclonal antibody (PAb) anti-MAVS (green), and goat PAb anti-RIG-I were used (light blue). Alexa Fluor 647-, Alexa Fluor 405-, and Alexa Fluor 568-conjugated antibodies, respectively, were used as secondary antibodies. Scale bars, 10 μm.
TRIM25 does not interact with MAVS, but both proteins interact with RIG-I, as we showed in Fig. 3. To further analyze the interactions among the three proteins, HeLa cells were cotransfected in different assays (Fig. 5). Interestingly, in Fig. 5B, the YFP fluorescence signal of the complex RIG-I/TRIM25 was abolished in cells that were also overexpressing MAVS. We show in the zoom image details of the colocalization of the proteins in this assay, where RIG-I (blue) colocalizes with MAVS (green), and TRIM25 (red) is found expressed in close proximity, but at a different location than for the RIG-I/MAVS complexes. However, we observed a YFP fluorescence signal when we cotransfected cells with the BiFC complex ycRIG-I/ynMAVS and HA-TRIM25, indicating the formation of RIG-I/MAVS complexes in the presence of overexpressed TRIM25 (Fig. 5C). These results suggest that, once the pathway is activated, the RIG-I/MAVS complexes might be more stable than the RIG-I/TRIM25 complexes and thus that the overexpression of TRIM25 does not result in preventing RIG-I/MAVS complexes. To confirm these results, we cotransfected cells with the three BiFC fused plasmids (ycRIG-I, ynTRIM25, and ynMAVS). This resulted in the formation of YFP complexes, where TRIM25 was located close to, but not overlapping with, the YFP signal. Staining with anti-RIG-I and MAVS antibodies showed that they localize in the same specific areas of the cell (Fig. 5D, see zoom details), indicating that the YFP complexes are formed by RIG-I and MAVS (BiFC in yellow color) and that TRIM25 is not part of the RIG-I/MAVS complex (stain in red color, 5D). This observation, combined with the different cytoplasmic localizations of the RIG-I/TRIM25 and RIG-I/MAVS complexes, suggests that, during signaling, RIG-I that is found in the cytosol in nonstimulated cells first interacts with TRIM25 to become bound to K63 polyubiquitin and that this promotes release from TRIM25, relocalization, and binding to MAVS.
Analysis of endogenous RIG-I, TRIM25, and MAVS localization in infected cells.
The BiFC technique showed us the spatial location of different interacting complexes within a cell. To analyze the expression of these proteins in the context of a viral infection, HeLa cells were infected with SeV, activating the RIG-I signaling pathway. The cells were fixed and stained with antibodies against RIG-I, TRIM25, and MAVS (see Materials and Methods for details). In noninfected cells, MAVS shows a typical net-like distribution in the cytoplasm that is consistent with its mitochondrial localization (15) (Fig. 6A, row I, left image). Due to the low levels of expression under homeostatic conditions, RIG-I and TRIM25 were not detectable in our immunofluorescence assays, and their expression became evident at later times postinfection (Fig. 6A, row I, right image). Upon SeV infection, MAVS net-like structures appear to concentrate in cytoplasmic perinuclear areas, whereas TRIM25 localizes in aggregates in the cytosol (green and red, respectively). We next performed STED super-resolution microscopy assays in SeV-infected cells. As shown in Fig. 6A, row II, RIG-I at 20 h postinfection (see the 3D rendering images in zooms and arrows for details) is located in specific areas within the cytoplasm of the SeV-infected cell, where it accumulates and interacts in some areas with TRIM25 and in other areas with MAVS (see the gray and black arrows in the histogram profiles, respectively). We barely observed TRIM25/MAVS colocalizations. TRIM25 was previously shown to localize with stress granules (SGs) during Newcastle disease virus (NDV) infection (40). To further characterize the localization of TRIM25 after SeV infection, we performed immunofluorescence assays in mock- and SeV-infected cells with the SG markers TIAR and GB3P. TRIM25 and SG dots became apparent after SeV infection, where they colocalized (Fig. 6B), indicating that TRIM25 associates with SGs. As described previously, a fraction of RIG-I was also found in the virus-induced SGs. (Fig. 6B, lower panel) (41). It was difficult to evaluate the endogenous localization of RIG-I and TRIM25 in mock-infected cells due to their low levels of expression in the absence of stimulation (see, for example, Fig. 6C, upper panels). On the other hand, transfection-based plasmid overexpression of these factors results in some levels of activation. In order to visualize endogenous unstimulated RIG-I, TRIM25, and MAVS, we pretreated cells with IFN. IFN is known to induce the expression of RIG-I and TRIM25 in the absence of their stimulation. IFN-treated cells revealed a clear signal for endogenous RIG-I and TRIM25, which were localized evenly in the cytoplasm and not associated in specific dots or with MAVS (Fig. 6C, lower panels). This observation is in contrast to the localizations seen for these factors in SeV-infected cells (Fig. 6A). In total, our results show that TRIM25 forms cytoplasmic dots associated with SGs but not with MAVS upon SeV infection, whereas activated RIG-I colocalizes either with TRIM25/SGs or with MAVS.
FIG 6.
Analysis of endogenous RIG-I, TRIM25, and MAVS localization in infected cells. HeLa cells were infected with SeV. (A) In row I, noninfected and infected cells (left and right images, respectively) were fixed and processed for indirect immunofluorescence as indicated in Materials and Methods. RIG-I, TRIM25, and MAVS staining is indicated in blue, red, and green, respectively. In row II, an STED super-resolution image was processed. White squares show the 3D reconstruction of the image on the right and left sides. MAVS filaments, TRIM25 dots, and RIG-I distribution are indicated by arrows. Fluorescence intensity histograms measured along the white lines are shown. In the left histogram, gray arrows indicate the overlapping of fluorescence for RIG-I/TRIM25 interactions; in the right histogram, black arrows indicate the overlapping of fluorescence for RIG-I/MAVS interactions. The right and left images show zoom details. Images were processed using ImageJ to show the mean intensity projection of Z-stack images. (B) TRIM25 colocalizes with stress granule (SG) markers in infected cells. HeLa cells were mock treated or infected with SeV. At 20 h postinfection, the cells were fixed and processed for indirect immunofluorescence. The cells were stained for TRIM25 (red) and the SG markers GB3P (purple) and TIAR (green). (C) HeLa cells were either mock infected, IFN treated (1,000 U/ml), or SeV infected. As in panel A, the cells were stained for RIG-I, TRIM25, and MAVS. Images were obtained using confocal microscopy. Nuclei were stained with DAPI. Scale bars, 10 μm.
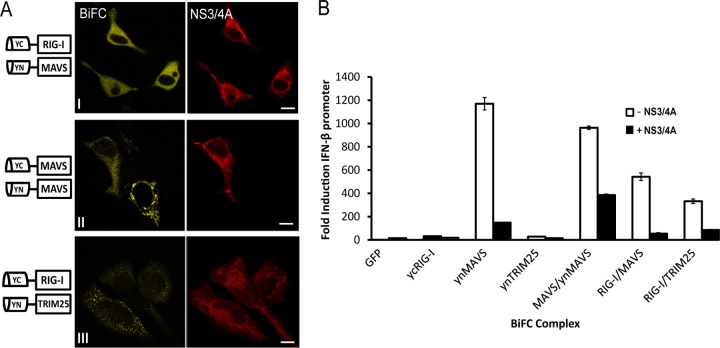
NS3/4A of HCV redistributes MAVS/MAVS and RIG-I/MAVS complexes.
It is well known that the NS3/4A protease cleaves MAVS at Cys-508 in its TM domain, preventing the induction of IFN by the RLR pathways (20, 42). However, whether NS3/4A can also mediate this cleavage and the redistribution of MAVS when in an oligomeric complex with itself or with RIG-I is unclear. In order to investigate this, we cotransfected the BiFC-fused plasmids (interactions RIG/MAVS, MAVS/MAVS, and RIG/TRIM25) with the HCV protease (Fig. 7). Interestingly, NS3/4A redistributed the RIG-I/MAVS complex in the cytoplasm of the cell (Fig. 7A, row I). This complex did not localize any longer at mitochondria, as we previously showed in Fig. 3B, but instead presented a diffuse localization in the cytoplasm. The same results were observed when NS3/4A was cotransfected with the dimer MAVS/MAVS (Fig. 7A, row II; see also Fig. 2C for the MAVS-MAVS distribution in the absence of NS3/4A); the protease did not have any effect in the RIG-I/TRIM25 complex (Fig. 7A, row III). Our results indicate that NS3/4A is not only able to cleave the mitochondrial association domain of MAVS, as previously known, but also preformed MAVS/MAVS and MAVS/RIG-I complexes. As shown in Fig. 7B, NS3/4A blocks the activation of the IFN-β promoter in all the cases.
FIG 7.
HCV NS3/4A redistributes RIG-I/MAVS and MAVS/MAVS. HeLa cells were cotransfected with the BiFC N or C fusion plasmids ycRIG-I/ynMAVS, ycMAVS/ynMAVS, or ycRIG-I/ynTRIM25, together with the HA-NS3/4A (HCV) expression plasmid. (A) At 24 h posttransfection, the cells were fixed, and NS3/4A expression was detected using an anti-HA antibody. Alexa Fluor 633-conjugated anti-mouse antibody (red) was used as the secondary antibody. DAPI nuclear staining is also indicated (blue). Scale bars, 10 μm. (B) A dual-luciferase reporter assay was performed in transfected cells as described in Materials and Methods. The y axis represents the mean nfold relative units versus the average for mock-treated cells. The x axis represents the plasmids transfected in each column in the presence or absence of HA-NS3/4A. The experiments were performed in triplicate.
IAV NS1 inhibits the formation of TRIM25/TRIM25 and RIG-I/MAVS complexes.
Previously, immunoprecipitation assays showed that influenza A virus (IAV) NS1 interacts with RIG-I and TRIM25, preventing RIG-I signaling (34, 35). However, it is still unclear whether NS1 can also target other proteins in the RLR pathway. To test the functional effect of NS1 in the different complexes formed along the activation of the RIG-I pathway, NS1 was coexpressed with the BiFC complexes described previously, as shown in Fig. 8A, panel I. The expression of NS1 was confirmed by indirect immunofluorescence (red color). Quantification of YFP-positive cells by microscopy showed a decrease in yellow fluorescence in cells cotransfected with NS1 and the complexes TRIM25/TRIM25 and RIG-I/MAVS but not the complex RIG-I/TRIM25 (Fig. 8A, panel II).
FIG 8.
IAV NS1 inhibits the formation of the complexes TRIM25/TRIM25 and RIG-I/MAVS. (A) HeLa cells were transfected with theplasmids ycRIG-I/ynMAVS, ycRIG-I/ynTRIM25, or ycMAVS/ynMAVS in the presence or absence of V5-NS1 (PR8) (upper panel). The cells were processed for immunofluorescence, and NS1 was stained with an anti-NS1 antibody, rabbit PAb (red). In the lower panel, the y axis represents the percentage of cells expressing YFP relative to the cells expressing V5-NS1 or V5-empty plasmid. A total of 200 cells per condition were counted and analyzed by microscopy. (B) Whole-cell lysates (WCL) of HEK 293T cells transfected with the plasmids indicated above were immunoprecipitated with an anti-GFP MAb, followed by immunoblot (IB) analysis with rabbit anti-GFP, anti-NS1, or anti-actin PAb (as a loading control; see Materials and Methods for details). (C) HeLa cells were transfected with fusion BiFC plasmids ycRIG-I/ynNS1, ycMAVS/ynNS1, ycTRIM25/ynNS1, and ycTRIM25/ynNS1ΔCCD, as indicated in the schematic drawings on the left. Representative images of confocal microscopy are shown. At 24 h posttransfection, the cells were fixed and immunostained with a rabbit anti-NS1 PAb (red). The autofluorescence of YFP is indicated in yellow. Right, zoom details of the BiFC complexes stained with antibodies anti-RIG-I (blue), anti-TRIM25 (red), and anti-MAVS (green). White arrows indicate areas of colocalization between RIG-I and TRIM25. Nuclei were stained with DAPI (blue). Scale bars, 10 μm.
To confirm the microscopy results, the BiFC complexes were pulled down with an anti-GFP antibody that specifically immunoprecipitates the reconstituted BiFC complexes (Fig. 8B). As shown in Fig. 8, the levels of the RIG-I/MAVS complex decreased in the presence of NS1. In contrast, the levels of the RIG-I/TRIM25 complex remained unaltered.
We have demonstrated the interaction of NS1 with the complex RIG-I/TRIM25 and the inhibition of RIG-I/MAVS complexes by NS1. We were wondering if NS1 could interact with each of the RLR proteins analyzed. To address this, we fused the viral protein with the BiFC plasmids (YN and YC halves), and its interaction was monitored by microscopy as before. NS1 interacts by BiFC with RIG-I and TRIM25 (confocal images are shown in Fig. 8C, rows I and III, respectively) but not with MAVS (see row II). As expected, the deletion of the CCD in TRIM25 inhibited the interaction with NS1 (Fig. 8C, row IV) (35).
Our microscopy analysis allowed us to identify a very specific pattern of distribution when NS1 binds RIG-I or TRIM25, forming speckled spots within the cytoplasm of the cell (see the images in Fig. 8A and C). In order to investigate the composition of these speckles, the cells were costained with antibodies against RIG-I, TRIM25, and MAVS (Fig. 8C, rows I and III [see images on the right]). In line with the results showed in this section, the speckles observed contain RIG-I (blue) and TRIM25 (red) but not MAVS (green).
Overall, these results suggest that NS1 recruits both RIG-I and TRIM25 in the same complex, but not MAVS. The interaction of NS1 with theses complexes may be required for the inhibition of the interaction of RIG-I with MAVS and the decrease in the induction of IFN production.
DISCUSSION
Information on protein subcellular localization is essential to understand the functions of proteins in a cell. The appropriate subcellular localization of proteins is crucial because it provides the physiological context for their function. In signaling proteins, the location is key to interact with the downstream targets involved in pathway activation. At the same time, mislocalization of proteins in a signaling pathway contributes to a loss of function. Since the identification of RIG-I as an important cytoplasmic viral sensor, many efforts have been made to further understand the mechanisms that lead to the activation of the pathway and the downstream propagation of signaling. Nevertheless, there is scarce knowledge on the spatial-temporal location of RIG-I and the proteins downstream of RIG-I activation, TRIM25 and MAVS (18, 43). It is known that RIG-I is found in the cytosol but can relocalize in the cytoplasm in a stimulated cell (12, 39, 41, 44). The downstream adaptor, MAVS, is an organelle-bound protein anchored in the outer membrane of the mitochondria and has recently also been found in the peroxisomes (where it could play a role in the earlier immune response against viral infection) and MAM (mitochondrion-associated membrane), a new signaling site that could serve as a scaffold that coordinates MAVS-dependent signaling between mitochondria and peroxisomes (45). Thus, the need and importance of the redistribution of these components in order to activate the signaling cascade is obvious (14). In the present study, the specific interactions among several members of the RIG-I pathway have been tracked and visualized.
The BiFC technique is a tool widely used in biology to detect protein-protein interactions in situ (38, 46). Through the use of BiFC and super-resolution microscopy, we detected RIG-I, MAVS, and TRIM25 oligomers and analyzed their distribution in the cell. The CARD of RIG-I cannot interact in the absence of other RIG-I domains, suggesting that at least one full-length unit of the protein must dimerize (Fig. 2A). Meanwhile, RIG-I oligomers are cytoplasmic, TRIM25 dimers are found in punctuate dots that are not localized with MAVS dimers (Fig. 2B and C, respectively), and MAVS dimers colocalized with mitochondrial markers, as expected (see the zoom images for details).
RIG-I can form two different and independent complexes with the proteins MAVS and TRIM25. Despite the fact that they are very close each other, the subcellular localization of RIG-I/TRIM25 and RIG-I/MAVS complexes is different. Although RIG-I/MAVS colocalizes with the mitochondrial membrane, as dimers of MAVS (Fig. 3A, row II), RIG-I/TRIM25 does not (Fig. 3B, row II). Consistent with the different localizations, TRIM25 and MAVS do not interact (Fig. 4A). In addition, MAVS inhibits the YFP fluorescence and therefore the formation of the RIG-I/TRIM25 complex when cotransfected with the BiFC plasmids ycRIG-I/ynTRIM25 (Fig. 4B). Meanwhile, we did observe BiFC formation of the complementary complex, ycRIG-I/ynMAVS, together with TRIM25 (Fig. 4C).
The BiFC technique allowed us to capture in space the different interactions through the proteins in the RLR pathway. To investigate these events in the context of a viral infection, we performed super-resolution microscopy and analyzed the endogenous localization of RIG-I, TRIM25, and MAVS after SeV infection. We observed that, compared to noninfected cells, MAVS forms filaments with a perinuclear distribution (in close contact to the mitochondria membrane) and TRIM25 creates aggregates (Fig. 6A, row I). The analysis of super-resolution images showed us that at this time point RIG-I accumulates closer to MAVS in the perinuclear region (Fig. 6A, row II) or close to TRIM25 in another areas. In contrast, we observed negligible colocalization between TRIM25 and MAVS.
Our results suggest that MAVS competes with TRIM25 in the interaction with RIG-I and that the RIG-I/MAVS complex is more stable than the RIG-I/TRIM25 complex. We propose that RIG-I first interacts with TRIM25 to become ubiquitinated. This would lead to the release of TRIM25 from the complex and the subsequent interaction of RIG-I with MAVS close to the mitochondria.
Experiments are ongoing to further characterize the interesting nature and composition of the granules formed by TRIM25. Interestingly, many proteins belonging to the family of TRIM E3 ligases concentrate in dots associated with ubiquitin when active (TRIM ubiquitin-rich bodies [TUBs]) (23). In the case of TRIM25, we show that TRIM25 granules are formed upon SeV stimulation and that they colocalized with the SG markers TIAR and GB3P. This finding, also demonstrated by Yoo et al. (40) in NDV-infected cells, supports a role for SGs in the antiviral innate immune response (40, 41, 47).
To further understand how RLR complexes can be targeted in order to inhibit the IFN response, we combined the BiFC assays with two viral proteins known to antagonize RIG-I signaling (HCV NS3/4A protease and IAV NS1). These viral proteins exhibit very different strategies for manipulating the subcellular interactions of the RIG-I, TRIM25, and MAVS proteins.
Since the overexpression of the RLR pathway proteins to visualize BiFC complexes induces an antiviral response that prevents further infection with viruses, we used plasmid-based expression of the viral proteins to investigate their effects. The HCV protease cleaves MAVS at its TM domain (48). We show that NS3/4A redistributes MAVS oligomers and the complex RIG-I/MAVS in the cytoplasm of the cell but does not affect RIG-I/TRIM25 complexes (Fig. 7A). This result supports our previous findings of two different and separate RIG-I/MAVS and RIG-I/TRIM25 complexes.
An interaction of NS1 with RIG-I and TRIM25 was reported by our lab and others (10, 34, 35). Interestingly, NS1 binds RIG-I and TRIM25 only in specific punctuate dots along the cytoplasm of the transfected cells (Fig. 8A and C). In all of the assays (see Fig. 8C, images on the right), we found that MAVS was excluded from the speckled dots formed by RIG-I, TRIM25, and NS1.
Moreover, in immunofluorescence or immunoprecipitation analyses of the specific BiFC complexes, NS1 inhibited only the formation of the RIG-I/MAVS complex and not that of the RIG-I/TRIM25 complex (Fig. 8A and B). These results and the lack of NS1 interaction with MAVS suggest an early binding of NS1 with RIG-I and TRIM25, either individually or when they are interacting: this appears to be due to the inhibition of the TRIM25 multimers required for its E3 ligase activity, a finding consistent with previous results (35, 49), and the lack of RIG-I ubiquitination may lead to the inhibition of the RIG-I/MAVS interaction.
The BiFC technique provided us with a tool that allowed us to define “space-wise” the different interactions among RIG-I, TRIM25, and MAVS in the RLR pathway. We were also able to show this distribution at a certain time point in the context of viral infection, showing again a striking formation of complexes displayed in different locations within the cell. These results show us that more detailed investigation is needed to address the “five W's”: when, where, and why the RLR proteins are rearranged, what triggers the subcellular locations, and who are the key players in the composition of the specific speckles formed along the pathway.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
A549, HeLa, and HEK 293T cells were purchased from the American Type Culture Collection and maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 100 U of penicillin-streptomycin/ml.
Sequences encoding YFP amino acids 1 to 154 (yn) and 155 to the end (yc) (50) were subcloned from pEYN-NXF1 and pEYC-Y14 (kindly provided by Peter Lichter, German Cancer Research Centre, Heidelberg, Germany [51]) between the ClaI and NheI sites of the mammalian expression vector pCAGGS (52). A short linker (SGLRSRAQASIS), including a unique XhoI site, was added to either the N or the C terminus of yn and yc. The open reading frames of NS1, RIG-I, CARD, TRIM25, and MAVS (10, 34) were amplified from template DNA by PCR and subcloned into the pCAGGS vectors between either ClaI and XhoI (X-yn and X-yc constructs) or XhoI and NheI (yn-X and yc-X constructs). The eight possible combinations of yn and yc constructs for every protein-protein interaction of interest were assayed for BiFC complementation, and combinations showing the strongest BiFC signal were selected for further studies. HA-NS3/4A was kindly provided by Luis Martinez-Sobrido and V5-NS1 was described earlier (53). SeV stocks (924 hemagglutination units/ml) were diluted 1/500 in complete growth medium. Viral infections were performed as described previously (23), and the cells were fixed at 20 h postinfection. For type I IFN, cells were seeded in coverslips, and type I IFN universal was added (1/1,000 IU/ml [PBL]), followed by incubation for 20 h.
Antibodies.
The following antibodies were used in immunofluorescence or Western blot analyses: anti-HA (Sigma), anti-NS1 (155 [53]), anti-RIG-I mouse (1C3 [4]), anti-RIG-I goat (MyBioSource), anti-MAVS (Cell Signaling), anti-TOM20 (Santa Cruz Biotechnology), anti-TRIM25 (Abcam), anti-GFP rabbit (Santa Cruz Biotechnology) and mouse (Abcam), anti-actin (Sigma), anti-GB3P mouse, and anti-TIAR goat (Santa Cruz Biotechnology).
Transfections and reporter assay.
Transfection and reporter assays were performed as described previously (23). Briefly, HEK 293T cells were seeded in a 24-well format plates and, 16 h later, the cells were transfected with 0.6 μl of LT1 (Roche), the indicated plasmids, 50 ng of firefly luciferase reporter plasmid, and 25 ng of Renilla luciferase reporter plasmid. All samples were transfected in triplicate.
At 24 h posttransfection, the cells were lysed in passive lysis buffer (Promega), and the dual-luciferase activity was measured using a dual-luciferase reporter assay system (Promega) in a Biotek Synergy 4 plate reader. Firefly luciferase values were normalized to Renilla values. The fold increase in the level of luciferase reporter was calculated relative to the green fluorescent protein (GFP) plasmid (pcAGGS-GFP) baseline control in each experiment.
Pulldowns.
HEK 293T cells were transfected with the indicated plasmids DNA, and pulldown assays were processed as described before (54). Briefly, the cells were lysed with nondenaturing lysis buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 5 mM EDTA, 0.02% sodium azide, 1% Triton X-100, and complete protease inhibitor cocktail [Roche, Basel, Switzerland]). Cell lysates were sonicated three times at output level 3.0 for 5 s, centrifuged at 13,000 rpm for 15 min at 4°C, and precleared with protein G-agarose beads (Roche) for 4 h at 4°C. Immunoprecipitation was carried out with anti-GFP monoclonal antibody (Abcam), and the samples were analyzed by Western blotting.
Western blotting.
Cells were transfected and collected at 24 h posttransfection in lysis buffer (6 M urea, 2 M β-mercaptoethanol, and 4% sodium dodecyl sulfate), and SDS-PAGE and Western blot analyses were performed as described previously (55). Proteins were resolved on 4 to 20% gradient Ready-Gel precast gels (Bio-Rad) and transferred onto polyvinylidene difluoride membranes. After membrane blocking, proteins were detected by incubation with primary antibodies and horseradish peroxidase-coupled anti-mouse or anti-rabbit antibodies using a chemiluminescence kit (Perkin-Elmer).
Immunofluorescence.
HeLa or A549 cells were cultured and transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol in glass-bottom 12-well plates (MatTek) or 1.5 coverslips (MatTek). In the BiFC experiments, at 24 h posttransfection the plates were incubated for 3 h to 30°C, fixed in ice-cold absolute methanol for 30 min, and blocked with 1% bovine serum albumin in phosphate-buffered saline (PBS). Samples were incubated with the primary antibodies for 2 h, washed three times with PBS, and incubated an additional 1 h with secondary antibodies. Where indicated, nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen) at 1 μg/ml.
Microscope image acquisition.
Confocal laser scanning was performed using a Zeiss LSM 880 Meta (Carl Zeiss Microimaging, Thornwood, NY) fitted with a Plan Apochromatic ×63/1.4 or ×40/1.4 oil objective lens. Z-stack images were taken in a range from 0.1 to 0.5 μm for the images indicated. In all cases, the Z-stacks were deconvolved with AutoQuant-X2. Images were collected at 16 bits and at a resolution of 1024 by 1024 pixels. Super-resolution images were taken with STED SP8 microscope, and the Z-stacks were processed to obtain maximum intensity projection. In the cases indicated, Amira 3D software (FEI Visualization Sciences Group) was used to obtain 3D rendering, and image processing and analysis were carried out using Fiji/ImageJ software.
ACKNOWLEDGMENTS
We thank Peter Lichter (Heidelberg, Germany) for the BiFC plasmids and Luis Martinez-Sobrido for the HA-NS3/4A plasmids. We also acknowledge the help of the Microscopy Shared Resource Facility at the Icahn School of Medicine at Mount Sinai, supported with funding from an NIH Shared Instrumentation Grant (1S10RR024745-01A1).
This study was partly supported by the Center for Research on Influenza Pathogenesis, the National Institute of Allergy and Infectious Disease (NIAID)-funded Center of Excellence for Influenza Research and Surveillance (contract HHSN272201400008C), and by NIAID grant U19AI117873 (to A.G.-S.).
REFERENCES
- 1.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Versteeg GA, Garcia-Sastre A. 2010. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol 13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 4.Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A 107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Kolakofsky D, Kowalinski E, Cusack S. 2012. A structure-based model of RIG-I activation. RNA 18:2118–2127. doi: 10.1261/rna.035949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. 2013. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep 14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrage F, Dutta K, Nistal-Villan E, Patel JR, Sanchez-Aparicio MT, De Ioannes P, Buku A, Aseguinolaza GG, Garcia-Sastre A, Aggarwal AK. 2012. Structure and dynamics of the second CARD of human RIG-I provide mechanistic insights into regulation of RIG-I activation. Structure 20:2048–2061. doi: 10.1016/j.str.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 11.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Jiang QX, Chen ZJ. 2011. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO Rep 13:7–8. doi: 10.1038/embor.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. 2011. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 14.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 17.Goubau D, Deddouche S, Reis ESC. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagan JC. 2012. Signaling organelles of the innate immune system. Cell 151:1168–1178. doi: 10.1016/j.cell.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshiba T, Bashiruddin N, Kawabata S. 2011. Mitochondria and antiviral innate immunity. Int J Biochem Mol Biol 2:257–262. [PMC free article] [PubMed] [Google Scholar]
- 22.West AP, Shadel GS, Ghosh S. 2011. Mitochondria in innate immune responses. Nat Rev Immunol 11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev 227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 26.Yoneyama M, Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev 18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKε molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol 80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 29.Foy E, Li K, Wang C, Sumpter R Jr, Ikeda M, Lemon SM, Gale M Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 30.Foy E, Li K, Sumpter R Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A 102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 33.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 34.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M Jr, Garcia-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerppola TK. 2006. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc 1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerppola TK. 2008. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol 85:431–470. doi: 10.1016/S0091-679X(08)85019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerppola TK. 2009. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev 38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo JS, Takahasi K, Ng CS, Ouda R, Onomoto K, Yoneyama M, Lai JC, Lattmann S, Nagamine Y, Matsui T, Iwabuchi K, Kato H, Fujita T. 2014. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog 10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, Namiki H, Yoneyama M, Fujita T. 2012. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gale M Jr, Foy EM. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 43.Kagan JC. 2012. Defining the subcellular sites of innate immune signal transduction. Trends Immunol 33:442–448. doi: 10.1016/j.it.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang ED, Wang CY. 2009. MAVS self-association mediates antiviral innate immune signaling. J Virol 83:3420–3428. doi: 10.1128/JVI.02623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horner SM, Liu HM, Park HS, Briley J, Gale M Jr. 2011. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A 108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerppola TK. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys 37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujita T, Onoguchi K, Onomoto K, Hirai R, Yoneyama M. 2007. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie 89:754–760. doi: 10.1016/j.biochi.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Baril M, Racine ME, Penin F, Lamarre D. 2009. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol 83:1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A 105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9:789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt U, Richter K, Berger AB, Lichter P. 2006. In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains. J Cell Biol 172:373–381. doi: 10.1083/jcb.200503061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 53.Kerry PS, Ayllon J, Taylor MA, Hass C, Lewis A, Garcia-Sastre A, Randall RE, Hale BG, Russell RJ. 2011. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One 6:e17946. doi: 10.1371/journal.pone.0017946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varga ZT, Grant A, Manicassamy B, Palese P. 2012. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol 86:8359–8366. doi: 10.1128/JVI.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, Garcia-Sastre A. 2010. Inefficient control of host gene expression by the pandemic H1N1 influenza A virus NS1 protein. J Virol 84:6909–6922. doi: 10.1128/JVI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]