ABSTRACT
Human astroviruses (HAstVs) are a leading cause of viral diarrhea in young children, the immunocompromised, and the elderly. There are no vaccines or antiviral therapies against HAstV disease. Several lines of evidence point to the presence of protective antibodies in healthy adults as a mechanism governing protection against reinfection by HAstV. However, development of anti-HAstV therapies is hampered by the gap in knowledge of protective antibody epitopes on the HAstV capsid surface. Here, we report the structure of the HAstV capsid spike domain bound to the neutralizing monoclonal antibody PL-2. The antibody uses all six complementarity-determining regions to bind to a quaternary epitope on each side of the dimeric capsid spike. We provide evidence that the HAstV capsid spike is a receptor-binding domain and that the antibody neutralizes HAstV by blocking virus attachment to cells. We identify patches of conserved amino acids that overlap the antibody epitope and may comprise a receptor-binding site. Our studies provide a foundation for the development of therapies to prevent and treat HAstV diarrheal disease.
IMPORTANCE Human astroviruses (HAstVs) infect nearly every person in the world during childhood and cause diarrhea, vomiting, and fever. Despite the prevalence of this virus, little is known about how antibodies in healthy adults protect them against reinfection. Here, we determined the crystal structure of a complex of the HAstV capsid protein and a virus-neutralizing antibody. We show that the antibody binds to the outermost spike domain of the capsid, and we provide evidence that the antibody blocks virus attachment to human cells. Importantly, our findings suggest that a subunit-based vaccine focusing the immune system on the HAstV capsid spike domain could be effective in protecting children against HAstV disease.
KEYWORDS: antibody, astrovirus, capsid, receptor binding, structure, vaccines, virus neutralization
INTRODUCTION
The Astroviridae family is comprised of two genera, Mamastrovirus and Avastrovirus, which infect mammalian and avian species, respectively (1). Members of the Avastrovirus genus cause a variety of disease manifestations, growth defects, and mortality in poultry (2). Members of the Mamastrovirus genus cause infections in humans and a wide range of mammals (3). Human astroviruses (HAstVs) are classified into eight canonical serotypes (HAstV-1 to HAstV-8 [HAstV-1-8]) within the Mamastrovirus genogroup 1 (4), where HAstV-1 is the predominant serotype worldwide (5, 6). HAstV is a leading cause of viral diarrhea in children, immunocompromised individuals, and the elderly (7). There are approximately 3.9 million cases of viral diarrhea due to HAstV in the United States every year (8). In addition, highly divergent strains of HAstV have recently been attributed to encephalitis in immunocompromised individuals (9–11).
There are no vaccines or antiviral therapeutics for HAstV disease. Several studies provide evidence that antibodies developed by the adaptive immune response during childhood HAstV infection provide protection against subsequent HAstV infection in adulthood. Approximately 75% of children in the United States have acquired antibodies against HAstV by age 10 (12). Clinical studies with healthy adult volunteers found that those with more-severe HAstV diarrheal disease had no detectable anti-HAstV antibodies (13, 14). Finally, immunoglobulin therapy was associated with recovery of an immunocompromised patient with severe and persistent HAstV infection (15). Together, these data suggest that anti-HAstV antibodies acquired by active or passive immunity can provide protection against HAstV infection and disease. However, the locations of neutralizing antibody epitopes are unknown, and this information is critical for rational design of vaccine immunogens.
Astroviruses are nonenveloped icosahedral viruses with an ∼7-kb positive-sense, single-stranded RNA genome. The astrovirus genome has three open reading frames (ORFs); ORF1a and ORF1b encode nonstructural polyproteins, and ORF2 encodes the capsid protein that encapsulates the viral genome (16, 17). The multidomain capsid protein (CP) contains a highly basic N-terminal region, a core domain, a spike domain, and a C-terminal acidic region (18). Newly synthesized CPs spontaneously assemble into viruslike particles inside infected cells (19, 20) and undergo caspase proteolytic removal of the C-terminal acidic domain (21, 22). The immature T=3 HAstV particle is released from cells and becomes further processed by host extracellular proteases to produce the mature HAstV particle. In cell culture, trypsin has been used to produce mature HAstV, which is 105-fold more infectious than immature HAstV (21, 23, 24). Electron cryomicroscopy studies of immature and mature HAstV particles reveal ∼44-nm particles comprised of a T=3 icosahedral shell and globular spikes (25). While the immature T=3 HAstV particle contains 90 dimeric spikes, trypsin proteolysis leaves the mature T=3 HAstV particle with only 30 dimeric spikes located on the 2-fold icosahedral axes. Our lab and others have determined the structures of the dimeric CP spikes from HAstV-1, HAstV-8, and turkey astrovirus 2 (26–28). We also reported the structure of the HAstV CP core domain that forms the icosahedral shell (26). The CP core domain contains two subdomains: a typical jelly roll β-barrel found in many RNA viruses and a squashed β-barrel (26). While it has been shown that mature HAstV gains entry into human cells via clathrin-mediated endocytosis (29), the identity of the host cell receptor(s) and the location of receptor-binding site(s) on the surface of the HAstV particle are unknown.
Here, we determined the structure of the HAstV-2 CP spike bound to a HAstV-neutralizing antibody to 3.2-Å resolution. This structure reveals a quaternary epitope that overlaps several patches of conserved amino acids. We further provide evidence that the spike is a receptor-binding domain and that the antibody blocks HAstV-2 attachment to cells. Our studies provide a roadmap to design vaccine immunogens that induce neutralizing antibodies and also provide a foundation to understand HAstV attachment and entry into human cells.
RESULTS
Specificity and affinity of MAb PL-2 binding to HAstV-2 CP spike.
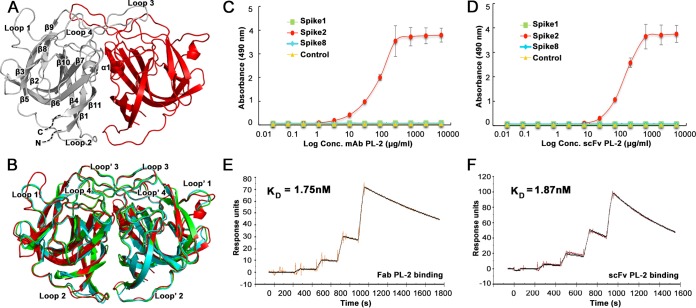
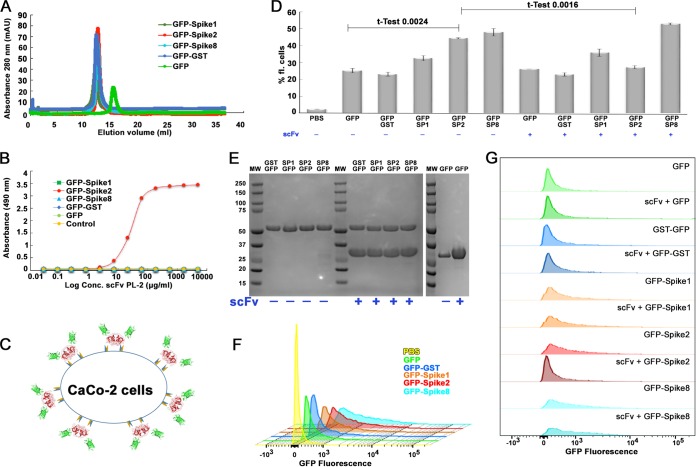
Monoclonal antibody PL-2 (MAb PL-2) was previously generated from mice immunized with purified HAstV-2 virus and was reported to neutralize HAstV-2 virus infectivity in cell culture (30). To determine if MAb PL-2 binds to the HAstV-2 CP spike (Spike2), the outermost domain on the virus surface, we first produced and purified recombinant Spike2 and observed that it elutes as a dimer by size exclusion chromatography, consistent with previous observations for Spike1 and Spike8 (26, 27). To further show that Spike2 is folded correctly, we determined its structure to 1.87 Å (Fig. 1A and Table 1). Spike2 has structural homology to HAstV-1 and HAstV-8 CP spikes (Spike1 and Spike8), with root mean square deviations (RMSDs) of 0.49 Å and 0.57 Å, respectively (Fig. 1B) (26, 27). Unique features of Spike2 include an extended loop 1 and a short alpha helix (α1) within the interface of the dimeric structure (Fig. 1A). To assess binding of MAb PL-2 to Spike2, we performed an enzyme-linked immunosorbent assay (ELISA) (Fig. 1C). We find that MAb PL-2 binds to Spike2 in a dose-dependent manner but does not bind Spike1 or Spike8, consistent with the previously reported specificity of MAb PL-2 for serotype HAstV-2 (30). We then tested PL-2 single-chain variable fragment (scFv PL-2), whose sequence and recombinant production we reported previously (31). We find that scFv PL-2 also binds to Spike2 specifically and in a dose-dependent manner (Fig. 1D). Surface plasmon resonance (SPR) studies were then performed to quantify the binding affinities of MAb PL-2 and scFv PL-2 to Spike2 (Fig. 1E and F). Spike2 containing a 10-histidine tag was captured by anti-His antibody immobilized on the sensor chip. Single-cycle kinetic characterizations were performed by sequentially injecting increasing concentrations of purified MAb PL-2 Fab fragments (Fab PL-2) or scFv PL-2. Fab PL-2 was used for these kinetic studies to eliminate potential complications of avidity with divalent antibody. We found that the dissociation constants (KD) of Fab PL-2 and scFv PL-2 binding to Spike2 were nearly identical at KD of ∼1.8 nM. Taken together, these studies show that MAb PL-2 and scFv PL-2 bind to Spike2 similarly with high affinity and specificity.
FIG 1.
Antibody PL-2 binds with high specificity and affinity to Spike2. (A) Crystal structure of Spike2, with one half of the dimer in gray and the other half in red. The gray side has labeled β-sheets and α-helices. (B) Structural comparison of Spike1 (green), Spike2 (red), and Spike8 (cyan). (C) ELISA results showing that MAb PL-2 binds to Spike2 but not Spike1 or Spike8. (D) ELISA results showing that scFv PL-2 binds to Spike2 but not Spike1 or Spike8. (E) Background-subtracted SPR traces (orange) and curve fit with a 1:1 binding model (black). Sequential injections of increasing concentrations of Fab PL-2 were tested with immobilized Spike2. kon = 3.45 × 105 M−1 s−1, koff = 6.02 × 10−4 s−1, and Chi2 (RU2) = 1.36. (F) Background-subtracted SPR traces (brown) and curve fit (black). Sequential injections of increasing concentrations of scFv PL-2 were tested with immobilized Spike2. kon = 6.55 × 105 M−1 s−1, koff = 1.23 × 10−3 s−1, Chi2 (RU2) = 2.06.
TABLE 1.
Data collection and refinement statisticsa
| Statistic (unit) | Spike2 | scFv PL-2/Spike2 complex |
|---|---|---|
| Data collection | ||
| Space group | P 1 | P 41 |
| Cell dimensions | ||
| a, b, c (Å) | 44.07, 67.72, 90.53 | 200.00, 200.00, 157.52 |
| α, β, γ (°) | 76.53, 80.52, 86.29 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 47.58–1.87 (1.97–1.87) | 50.00–3.20 (3.31–3.20) |
| Rsym or Rmerge | 0.073 (0.445) | 0.152 (0.555) |
| I/σI | 7.3 (2.1) | 13.3 (1.8) |
| Completeness (%) | 96.3 (93.6) | 89.2 (79.0) |
| Redundancy | 2.0 (2.0) | 3.3 (2.5) |
| Refinement | ||
| Resolution (Å) | 87.0–1.87 | 49.31–3.24 |
| No. of reflections | 80,278 | 86,380 |
| Rwork/Rfree | 0.193/0.215 | 0.241/0.288 |
| No. of atoms | 7,459 | 25,962 |
| Protein | 7,124 | 25,887 |
| Ligands | 0 | 56 |
| Water | 335 | 0 |
| B factors | ||
| Protein | 27.69 | 105.64 |
| Ligands | 0 | 159.64 |
| Water | 25.12 | 0 |
| RMS deviations | ||
| Bond lengths (Å) | 0.004 | 0.004 |
| Bond angles (°) | 0.706 | 0.786 |
| No. of TLS groups | 16 | 64 |
| Ramachandran statistics | ||
| Favored (%) | 96.9 | 95.14 |
| Allowed (%) | 3.1 | 4.86 |
| Outliers (%) | 0.0 | 0.0 |
Data from one crystal were used for each structure. Values in parentheses are for the highest-resolution shell. TLS, translation-libration-screw.
Structure of scFv PL-2 bound to HAstV-2 CP spike.
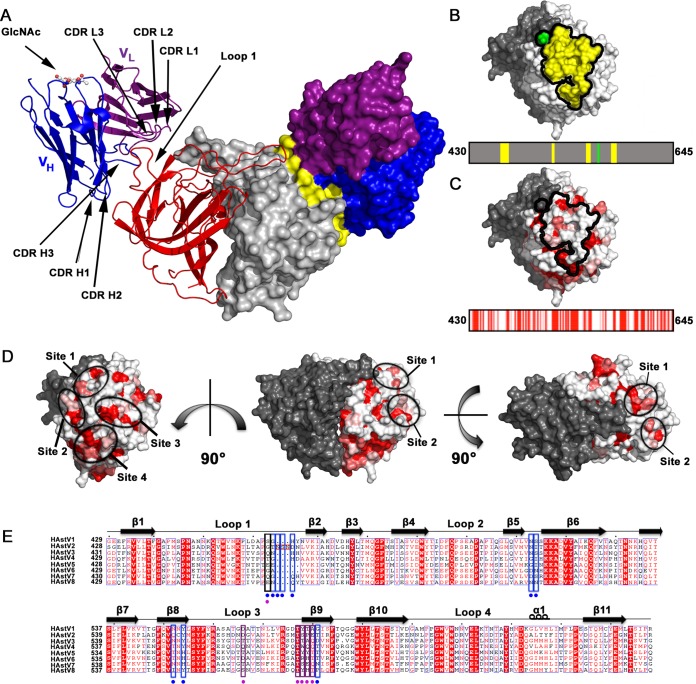
To characterize the MAb PL-2 binding site on Spike2, we cocrystallized and solved the structure of the scFv PL-2/Spike2 complex to 3.2-Å resolution (Fig. 2A and Table 1). At this resolution, electron density maps enabled confident fitting of amino acid side chains and characterization of atomic interactions at the interface between scFv PL-2 and Spike2 (Fig. 3A). The structure confirms size exclusion chromatography data indicating that the scFv binds to Spike2 in a 2:2 ratio, whereby two scFv molecules bind two molecules in the Spike2 dimer (Fig. 2A). scFv PL-2 and Spike2 are intimately associated with each other, and ∼890 Å2 of surface area is buried at each scFv PL-2/Spike2 interface. Each scFv PL-2 binding site is comprised of a conformation-dependent quaternary epitope in which five patches of amino acids from two linear Spike2 peptides come together in the three-dimensional structure (Fig. 2B). Many of the amino acids in the scFv PL-2 epitope are unique to serotype HAstV-2, consistent with ELISA and virus neutralization data showing that the antibody PL-2 is serotype specific (30) (Fig. 2C). However, the scFv PL-2 epitope overlaps several patches of amino acids that are highly conserved between serotypes HAstV-1-8, suggesting that scFv PL-2 may be blocking a conserved functional site on the virus surface (Fig. 2C, D, and E).
FIG 2.
Structure of the scFv PL-2/Spike2 complex and conservation within the epitope. (A) Structure of the scFv PL-2/Spike2 complex in cartoon view (left half) and surface representation (right half). The Spike2 dimer is colored in red and gray. The scFv PL-2 light chains (VL) are colored purple, and the scFv PL-2 heavy chains (VH) are colored blue. On the surface representation, the PL-2 epitope amino acids are colored yellow. The glycosylation site on the scFv PL-2 heavy chain Asn 88 is labeled (GlcNAc). (B) Surface representation of Spike2 showing that the epitope is comprised of four discontinuous amino acid segments of one protomer (in yellow) and amino acid D566 from the other protomer (in green). The PL-2 epitope footprint is shown on top, and the linear schematic of the spike domain is shown on the bottom. (C) Surface representation of Spike2 (top) and linear schematic of spike domain (bottom) with amino acids colored by conservation between serotypes HAstV-1-8 from red (strictly conserved) to white (not conserved). (D) Spike2 structure shown from various sides. One half of the dimer is gray, and the other half is colored by amino acid conservation, whereby conserved, strongly similar, weakly similar, and nonconserved amino acids are colored red, dark pink, light pink, and white, respectively. Patches (sites) of conserved residues that overlap the MAb PL-2 epitope are circled. (E) Sequence alignment of HAstV-1-8 CP spike domains. Extra amino acids in in HAstV-2 CP spike loop 1 are boxed in red. Conserved, strongly similar, weakly similar, and nonconserved amino acids are colored red, dark pink, light pink, and white, respectively. Alignments and mapping of conservation onto the structure were performed with the online ENDscript server. Blue boxes highlight epitope amino acids in Spike2 that interact with the antibody heavy chain. Purple boxes highlight epitope amino acids in Spike2 that interact with the antibody light chain. Black boxes highlight amino acids in Spike2 that interact with both the heavy and light chains. At the bottom of each box is a colored circle to further highlight its interaction with the antibody heavy chain, light chain, or both.
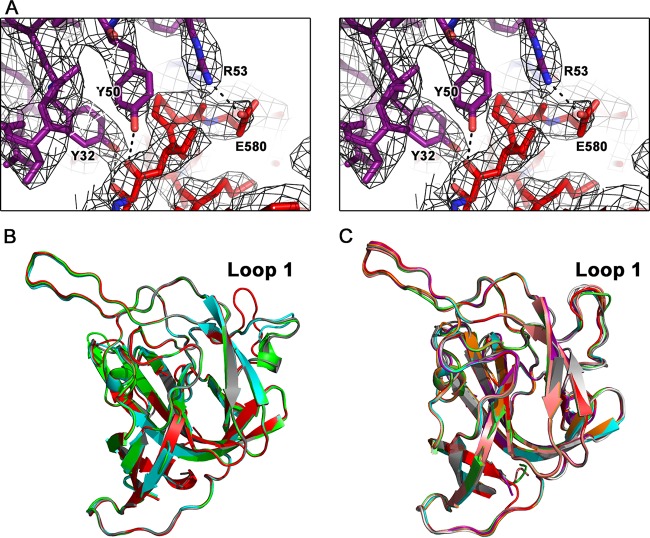
FIG 3.
Electron density maps and flexibility of loop 1. (A) Wall-eyed stereo view of the electron density maps surrounding one of the binding regions between scFv PL-2 and Spike2. Electron density map (black) is contoured at 1.8 σ. (B) Structural comparison of all four chains of Spike2 in the asymmetric unit of the Spike2 crystal. Loop 1 is labeled. (C) Structural comparison of all eight chains of Spike2 in the asymmetric unit of the scFv PL-2/Spike2 complex crystal. Loop 1 is labeled.
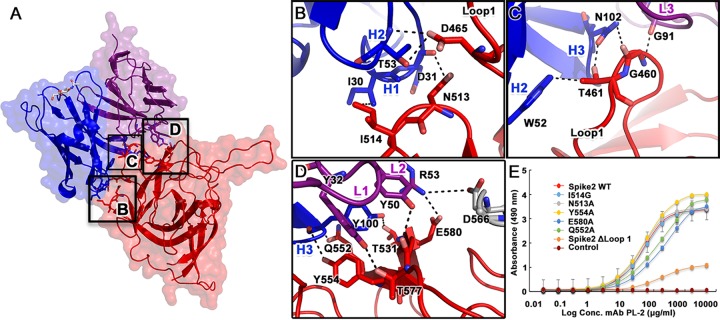
All six complementarity-determining regions (CDRs) from the heavy and light chains of scFv PL-2 interact with Spike2 (Fig. 4A). The interface is characterized by three primary interactions: (i) scFv PL-2 CDR H2 contacts Spike2 loop 1 amino acid D465, and CDR H1 contacts N513 and I514 in the β5-β6 hairpin (Fig. 4B); (ii) scFv PL-2 CDR H2 contacts Spike2 loop 1 amino acid T461, CDR H3 contacts T461 and G460, and CDR L3 contacts G460 (Fig. 4C); and (iii) scFv PL-2 CDR H3 contacts Spike2 amino acids Q552, Y554, and T531, CDR L1 contacts T557, and CDR L2 contacts T577 and E580 in Spike2 strands β8 and β9 and also contacts D566 (Fig. 4D). Notably, scFv PL-2 CDR L2 amino acid R53 forms an electrostatic interaction with D566 in loop L3 of the neighboring protomer of the Spike2 dimer, revealing a quaternary interaction (Fig. 4D). Nearby amino acids D564 and N565 in loop L3 are only 4 to 6 Å away from CDR L1 and may also interact with scFv PL-2 through water molecules. Overall, scFv PL-2 uses an extensive network of hydrogen bonds, electrostatic bonds, and hydrophobic interactions to bind to a quaternary epitope on the surface of the Spike2 dimer.
FIG 4.
Intermolecular interactions at the scFv PL-2/Spike2 interface. (A) Semitransparent surface representation of one half of the scFv PL-2/Spike2 complex. Spike2 is colored in red, scFv PL-2 light chain is colored purple, and scFv PL-2 heavy chain is colored blue. Primary regions of interaction are highlighted in black boxes with labels that correspond to the zoomed-in views in panels B and C. (B) Interactions between antibody CDR loops H1 and H2 with the Spike2 β5-β6 hairpin and the end of loop 1. (C) Interactions between antibody CDR loops H2, H3, and L3 and the Spike2 loop 1. (D) Interactions between antibody CDR loops H3, L1, and L2 and Spike2 strands β8 and β9 (in red) and loop 3 of the neighboring protomer (gray). (E) ELISA results showing binding of MAb PL-2 to Spike2 mutants.
Mutagenesis of HAstV-2 CP spike and MAb PL-2 binding.
To investigate the role of specific amino acids in the MAb PL-2/Spike2 binding interaction, we constructed mutations in Spike2 of amino acids that were shown to make important contacts in the complex structure, and we analyzed MAb PL-2 binding of the mutants by ELISA. All Spike2 mutants eluted as dimers by size exclusion chromatography and were purified to homogeneity (data not shown). However, all of the single-point Spike2 mutants, N513A, I514G, Q552A, Y554A, and E580A, showed binding curves and maxima for MAb PL-2 that were similar to those of wild-type Spike2. We then constructed mutant Spike2 ΔLoop1, in which amino acids G460, T461, and N462 from loop 1 were deleted. This mutant had drastically reduced MAb PL-2 binding compared to wild-type Spike2, highlighting the key role of loop 1 for MAb PL-2 binding (Fig. 4E). We note that loop 1 is quite flexible in the structure of Spike2 alone, and all four Spike2 molecules in the crystallographic asymmetric unit have high B factors and adopt different conformations (Fig. 3B). However, when bound to scFv PL-2, the Spike2 loop 1 adopts the same orientation in all eight molecules in the asymmetric unit (Fig. 3C).
Model of MAb PL-2 bound to the HAstV-2 virion.
We previously reported a model of the HAstV virion using the structures of the HAstV-1 capsid core and spike domains fitted into the 25-Å resolution electron cryomicroscopy density map of the mature T = 3 icosahedral HAstV particle (25, 26). To assess whether MAb PL-2 binds monovalently or divalently to the HAstV-2 virion, we aligned our previously reported Fab PL-2 structure (31) onto each scFv in the scFv PL-2/Spike2 complex, and we then aligned this complex onto each of the 30 spike dimers on the mature T=3 HAstV model. By measuring the respective distances between any two C-terminal ends of the heavy chains of neighboring Fab PL-2 fragments, we are able to show that the angles and distances (∼30 to 35 Å) between certain Fab PL-2 fragments are consistent with those found in an intact MAb (Fig. 5). Thus, our model suggests that MAb PL-2 likely binds divalently to the mature HAstV-2 virion, which would increase the overall avidity of antibody binding.
FIG 5.
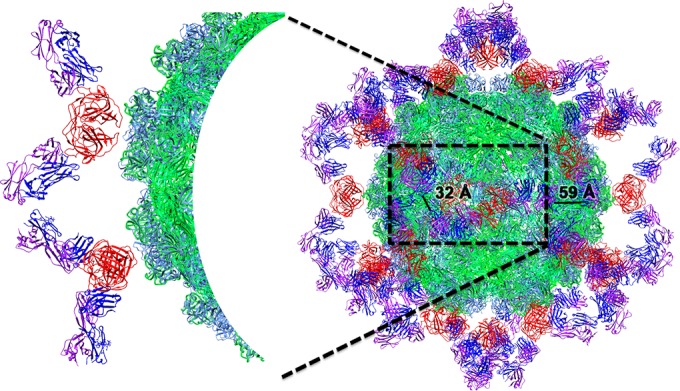
Model of mature HAstV-2 virion bound to Fab PL-2. Cartoon view of mature T=3 HAstV-2 virion bound to Fab PL-2 colored by structural domains, with inner core domain (green), outer core domain (light blue), spike domain (red), Fab PL-2 light chain (purple), and Fab PL-2 heavy chain (blue). Black lines show the distances between C-terminal ends of the heavy chains of neighboring Fab PL-2 fragments. Two Fab fragments with ∼32-Å distance are consistent with those found in an intact MAb. On the left is a zoomed-in view of two antibody PL-2 Fab fragments with a distance of ∼32 Å. The figure was made with Chimera (42).
HAstV-2 CP spike binds to cells and is blocked by scFv PL-2.
To characterize the mechanism of MAb PL-2 neutralization, we sought to test the hypothesis that the HAstV CP spike is a receptor-binding domain and that MAb PL-2 blocks virus binding to human cells. To test our hypothesis, we developed a cell binding assay using fluorescently labeled HAstV CP spike. First, we produced and purified a recombinant fusion protein comprised of green fluorescent protein (GFP) fused at the N terminus of HAstV-2 CP spike (GFP-Spike2). For controls, we fused GFP to the N terminus of serotype HAstV-1 and −8 CP spikes. For additional controls, we produced GFP alone and GFP fused to the N terminus of glutathione S-transferase (GST), which is similar in size to CP Spike and also forms dimers. To determine if GFP fusion affects folding, we first analyzed all samples by size exclusion chromatography (Fig. 6A). We find that GFP-Spike1, GFP-Spike2, GFP-Spike8, and GFP-GST elute at the expected retention volume for a dimer (∼110 kDa) compared to gel filtration standards (Fig. 6A). No aggregates were observed in the void volume. GFP elutes as a monomer (Fig. 6A). Next, to determine if GFP-Spike2 retains its conformation-dependent quaternary epitope for antibody PL-2, we performed an ELISA (Fig. 6B). We found that scFv PL-2 specifically binds GFP-Spike2 and does not bind GFP or any of the other GFP fusion proteins. Taken together, our data show that GFP-Spike2 forms a dimer and retains its conformation-dependent epitope for scFv PL-2.
FIG 6.
scFv PL-2 blocks GFP-Spike2 binding to Caco-2 cells. (A) Superdex 200 10/300 size exclusion chromatography traces of indicated GFP and GFP fusion proteins. (B) ELISA results showing binding of scFv PL-2 to GFP-Spike2 but not GFP, GFP-GST, GFP-Spike1, or GFP-Spike8. (C) Schematic of the Caco-2 cell binding assay showing GFP-Spike dimer attachment to the cell surface. (D) Cell binding assay showing the percentage of fluorescent Caco-2 cells after incubation with indicated proteins. Note abbreviations for GFP-Spike 1 (GFP-SP1), GFP-Spike2 (GFP-SP2), and GFP-Spike8 (GFP-SP8). Samples containing scFv PL-2 are indicated with a plus sign (+) below protein names. Experiments were performed in biological duplicates. Unpaired t test for GFP with GFP-Spike2 and for GFP-Spike2 with GFP-Spike2 + scFv confirms significant differences. (E) Coomassie blue-stained SDS-PAGE of protein samples used for the cell binding assay. Samples containing a 4 M excess scFv PL-2 (band at ∼30 kDa) are indicated. (F) Overlay of histogram plots of Caco-2 cells incubated with PBS, GFP, GFP-GST, GFP-Spike1, GFP-Spike2, and GFP-Spike8. (G) Stacked histogram plots of Caco-2 cells incubated with GFP-fused proteins alone or with scFv PL-2. All histogram plots show fluorescence intensity versus relative cell count.
We then developed a cell binding assay using fluorescence-activated cell sorting (FACS) to quantitatively measure GFP-Spike binding to human colon adenocarcinoma cells (Caco-2 cells), the gold standard cell line for HAstV propagation (Fig. 6C). Purified GFP, GFP-GST, GFP-Spike1, GFP-Spike2, and GFP-Spike8 were incubated overnight with Caco-2 cells, and FACS was used to measure the percentage of fluorescent cells (Fig. 6D to G). We find that Caco-2 cells incubated with GFP-Spike1, GFP-Spike2, and GFP-Spike8 have markedly higher fluorescence than GFP alone or GFP-GST, supporting our hypothesis that HAstV CP spike comprises a receptor-binding domain. As a control for specificity of cell binding, we find that the presence of scFv PL-2 reduces the GFP-Spike2 signal to background fluorescence levels comparable to that of GFP alone. Importantly, scFv PL-2 did not reduce the fluorescence of any of the other GFP fusion proteins or GFP alone (Fig. 6D and G). Together, these data support our hypothesis that the HAstV CP spike is a receptor-binding domain and that MAb PL-2 blocks virus infectivity by blocking virus binding to cells.
DISCUSSION
Here, we present the crystal structure of the HAstV-2 capsid spike domain bound to the neutralizing antibody PL-2. Antibody PL-2 binds to an ∼890-Å2 conformation-dependent quaternary epitope on each side of the dimeric spike. The antibody uses all six CDR loops to form intimate interactions with its epitope, resulting in a binding KD of ∼1.8 nM. Our model of antibody PL-2 bound to the T=3 icosahedral HAstV particle reveals that MAb PL-2 may bind divalently and with high avidity to the HAstV-2 virion.
We provide evidence that the HAstV CP spike is a receptor-binding domain. The CP spike is the outermost domain on the HAstV surface, and the spikes of many other nonenveloped viruses are receptor-binding domains (32–34). Here, we show that GFP-Spike1, GFP-Spike2, and GFP-Spike8 bind to Caco-2 cells, supporting that all canonical HAstV serotypes may use a common mechanism through the CP spike domain to bind to human cells. The sequence identity between CP spike domains from canonical serotypes HAstV-1-8 is 40 to 60%, and we have identified surface-exposed patches of conserved amino acids that overlap the neutralizing antibody PL-2 epitope (Fig. 2C to E). Thus, our studies provide new avenues to investigate both the receptor-binding site(s) on the HAstV surface and the identity of the host cell receptor(s).
We also present evidence for the hypothesis that MAb PL-2 neutralizes HAstV by blocking HAstV receptor binding. Antibodies could potentially block HAstV infection at a number of stages, including (i) extracellular protease cleavage and maturation of the HAstV particle, (ii) HAstV receptor binding, (iii) HAstV endocytosis, and (iv) HAstV capsid uncoating and genome release. We ruled out the possibility that MAb PL-2 blocks protease maturation, as this MAb was previously shown to not be able to bind to immature HAstV CP particles (19). We also ruled out the possibility that MAb PL-2 blocks capsid uncoating, since we find that it binds to the spike and not the core domain that forms the T=3 icosahedral shell of the virus. It is possible that spike dimers also need to disassociate during virus uncoating, although our structural studies suggest that MAb PL-2 binds only slightly to the dimer interface and would not prevent disassociation. Instead, we hypothesize that MAb PL-2 blocks HAstV receptor binding. Here, we show that MAb PL-2 specifically blocks binding of GFP-Spike2, but not GFP or GFP-Spike8, to Caco-2 cells. Thus, our data support our hypothesis that MAb PL-2 neutralizes HAstV-2 infection by blocking virus attachment to cells. Definitive cell binding inhibition studies with radiolabeled HAstV-2 virus can now be pursued.
Finally, our studies lay the foundation to develop novel therapies to prevent and treat HAstV diarrheal disease. We provide direct evidence that the HAstV CP spike is the target of a neutralizing antibody, suggesting that a subunit vaccine immunogen comprised of recombinant HAstV CP spike may elicit a protective neutralizing-antibody response. However, polyclonal antibodies raised against HAstV tend to be very serotype specific (35), suggesting that a vaccine comprised of CP spikes from several serotypes may be required to elicit a broadly protective polyclonal antibody response. Our studies also highlight the potential to develop therapeutic anti-HAstV antibodies. One study describes the rapid recovery of an immunocompromised patient with severe and persistent HAstV disease following high-dose immunoglobulin therapy (15). We predict that a therapeutic anti-HAstV antibody could be useful not only for the treatment of individuals with severe or chronic HAstV infection but also as a prophylaxis against HAstV infection in high-risk and immunocompromised individuals.
MATERIALS AND METHODS
MAb, Fab, and scFv PL-2 production.
MAb PL-2 was purified from mouse ascites fluid as described previously (31). Fab PL-2 was generated by incubation of MAb PL-2 with immobilized papain, followed by Fab fragment purification, as described previously (31). Production of recombinant scFv PL-2 was described previously (31). Briefly, scFv PL-2 containing an N-terminal BiP signal sequence and a C-terminal thrombin cleavage site followed by a Twin Strep-tag was expressed in stably transfected Schneider 2 insect cells. Secreted scFv PL-2 was affinity purified on a StrepTrap column followed by size exclusion chromatography using a Superdex 200 column in phosphate-buffered saline (PBS).
Expression and purification of Spike constructs.
Synthetic genes codon optimized for Escherichia coli encoding HAstV-2 CP spike amino acids 429 to 644 (UniProtKB entry Q82446), HAstV-1 CP spike amino acids 429 to 645 (UniProtKB entry O12792), or HAstV-8 CP spike amino acids 429 to 647 (UniProtKB entry Q9IFX1) were purchased. To make Spike expression plasmids, genes were cloned into pET52b (Addgene) in frame with a C-terminal thrombin cleavage site and a 10-histidine purification tag. To make Spike2 mutant expression plasmids, the Phusion Site-Direct Mutagenesis kit (Thermo Scientific) was used with phosphorylated mutagenesis primers. To make GFP-Spike expression plasmids, genes were cloned into a pET-GFP plasmid (Addgene catalog number 29716) in frame with an N-terminal 6-histidine purification tag and enhanced green fluorescent protein. All plasmids were verified by DNA sequencing. Plasmids were transformed into E. coli strain BL21(DE3), and protein production was induced with 1 mM isopropyl-β-d-thiogalactopyranoside at 18°C for 16 h. E. coli cells were lysed by ultrasonication in 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 20 mM imidazole (buffer A) containing 2 μM MgCl2, 1,250 U Benzonase (Millipore), and 1× protease inhibitor cocktail Set V EDTA-Free (Millipore). Proteins were batch purified from soluble lysates by Talon metal affinity chromatography and eluted with buffer A containing 500 mM imidazole. Proteins were dialyzed into 20 mM Tris-HCl (pH 8.0) and 25 mM NaCl and purified by anion-exchange chromatography on a HiTrap Q FF column with a gradient elution to 20 mM Tris-HCl (pH 8.0) and 1 M NaCl. Proteins were buffer exchanged into PBS and further purified by size exclusion chromatography on a Superdex 200 column in PBS.
ELISA.
Purified Spike or GFP-Spike proteins at a concentration of 5 μg/ml in PBS (total volume, 150 μl) were incubated overnight at room temperature in 96-well ELISA microtiter plates. Plates were then washed three times with PBS containing 0.05% Tween 20 (PBST). Wells were blocked by addition of 150 μl of 5% bovine serum albumin (BSA) in PBS and incubation at room temperature for 1 h followed by three PBST washes. Antibody sample MAb PL-2 or scFv PL-2 was diluted to 5 μg/ml with 1% BSA in PBS and serially diluted 1:3 with 1% BSA in PBS. Wells were incubated with 150 μl antibody for 1 h at room temperature, and the plates were washed three times with PBST. For ELISAs in which the primary antibody was MAb PL-2, plates were then incubated for 1 h at room temperature with 150 μl horseradish peroxidase (HRP)-conjugated, secondary goat anti-mouse IgG antibody diluted 1:5,000 in 1% BSA in PBS. For ELISAs in which the primary antibody was scFv PL-2, plates were then incubated for 1 h at room temperature with 150 μl HRP-conjugated Strep-Tactin protein, diluted 1:5,000 in 1% BSA in PBS. Plates were washed three times with PBST and developed by adding peroxidase substrate o-phenylenediamine dihydrochloride (OPD) in 0.05 M phosphate-citrate buffer (pH 5.0) and 1.5% hydrogen peroxide for 10 min at room temperature. The reactions were stopped by incubation with 2 N sulfuric acid for 10 min at room temperature, and the absorbance was measured at 490 nm.
Surface plasmon resonance.
Surface plasmon resonance data were collected on a Biacore X100 instrument at 25°C. A histidine capture kit (GE Healthcare) was employed to covalently immobilize anti-His antibody onto a CM5 sensor chip surface using amine coupling at 12,000 response units (RU), allowing for subsequent immobilization of the His-tagged Spike2 ligand. The reference surface was prepared in the same way without the antibody. The running buffer for experiments consisted of 10 mM HEPES (pH 7.4), 150 mM NaCl, 3.4 mM EDTA, and 0.005% (vol/vol) Surfactant P20. Spike2 containing a C-terminal 10×His tag was diluted to 0.3 μg/ml after scouting, such that 120 to 150 RU of Spike2 ligand was bound to the surface following a 60-s injection at 10 μl/min. A single-cycle kinetics approach was employed because the extended dissociation time of 2,400 s made a multicycle kinetics approach impractical. The analyte scFv PL-2 or Fab PL-2 was flowed over the sensor surface at 0 nM, 1.12 nM, 3.74 nM, 11.23 nM, and 37.44 nM. Each injection had a contact time of 60 s at 30 μl/min. Sensor surfaces were regenerated with glycine buffer (pH 1.5) using a 30-s injection at 30 μl/min. Biacore X100 Evaluation software (v2.x) was used to determine unique kinetic constants using the 1:1 fitting algorithm. Start-up cycle and blank injection runs prior to and after triplicate sample runs were used to correct for background, as established with the single-cycle kinetics analysis. For single-cycle kinetics, the analyte is injected with increasing concentrations in a single cycle without the need of regeneration of the chip between runs.
Spike2 structure determination.
Purified Spike2 was dialyzed into 10 mM Tris-HCl (pH 8.0) and 150 mM NaCl and concentrated to 11.5 mg/ml. Crystals were grown by hanging drop vapor diffusion at 22°C with a well solution of 12% polyethylene glycol (PEG) 3350, 0.1 M ammonium sulfate, and 0.1 M sodium citrate (pH 7.0). Crystals were transferred into a cryoprotectant solution of 20% PEG 3350, 0.1 M ammonium sulfate, 0.1 M sodium citrate (pH 7.0), and 25% glycerol and flash frozen in liquid nitrogen. Diffraction data were collected at cryogenic temperature at the Advanced Photon Source on beamline 23-ID-B using a wavelength of 1.033 Å. Diffraction data from a single crystal were processed with iMosflm (36) and Scala (37) (Table 1). The Spike2 structure was solved by molecular replacement, using the HAstV-8 CP spike (PDB ID 3QSQ) (27) and the program Phaser (38). The Spike2 structure was refined and manually rebuilt using Phenix (39) and Coot (40), respectively. The final Spike2 structure (PDB 5KOU) had two dimers in the asymmetric unit of the crystal.
Formation and structure determination of the scFv PL-2/Spike2 complex.
Complex formation was performed by incubation of 1.9 mg of Spike2 with 7.6 mg of scFv PL-2 (∼3.5 molar excess of scFv PL-2) overnight at 4°C. Purification tags from both Spike2 and the scFv PL-2 were removed by digestion of the complex with thrombin protease overnight at 4°C. The scFv PL-2/Spike2 complex was purified by size exclusion chromatography on a Superdex 200 column in PBS. The complex coeluted at an apparent molecular mass of ∼110 kDa compared to gel filtration standards, consistent with a 2:2 (Spike2:scFv) complex in solution (data not shown). The purified complex was dialyzed into 10 mM Tris-HCl (pH 8.0) and 150 mM NaCl and concentrated to 12.3 mg/ml. Crystals were grown by hanging drop vapor diffusion at 22°C with a well solution of 20% PEG 3350 and 0.2 M sodium phosphate monobasic. Crystals were transferred into a cryoprotectant solution of 24% PEG 3350, 0.2 M sodium phosphate monobasic, and 25% glycerol and flash frozen in liquid nitrogen. Diffraction data from a single crystal were collected at cryogenic temperature at the Advanced Photon Source on beamline 23-ID-B using a wavelength of 1.033 Å. Diffraction data were processed with HKL2000 (41). The structure was solved by molecular replacement using the Spike2 structure (PDB ID 5KOU) and the variable domain of Fab PL-2 (heavy-chain amino acids 1 to 117 and light chain amino acids 1 to 108) (PDB ID 5I30) (31) and the program Phaser (38). To obtain the initial model, both structures were inserted in the search in a 2:2 ratio. The results showed most complexes having many clashes; one spike monomer, however, showed good electron density and no clashes with an scFv molecule. A PDB was built with this half of the complex and a symmetry mate was built to form the full dimer. The new complex was used in a new molecular replacement search, which gave the final results. The scFv PL-2/Spike2 complex structure was refined and manually rebuilt using Phenix (39) and Coot (40), respectively. The final scFv PL-2/Spike2 complex structure (PDB 5KOV) had a total of four scFv/Spike2 complexes in the asymmetric unit of the crystal. The high Rmerge is due to the crystal being anisotropic, likely due to the packing of the scFv PL-2/Spike2 complex in the crystal. Two poorly ordered scFv molecules (chains Q and W) contribute to high B factors. These scFv molecules had weak electron density and high B factors due to the lack of molecular contacts with neighboring molecules. However, the good redundancy of the data and signal-to-noise ratio in the highest-resolution shell gives us confidence in our final model.
Caco-2 cell binding assays.
The human colon adenocarcinoma cell line Caco-2 was obtained from ATCC (HTB-37). Cells were propagated in prewarmed Dulbecco's modified Eagle medium (DMEM) containing 15% fetal bovine serum and 100 U/ml penicillin-streptomycin. Caco-2 cells were subcultured every 3 to 4 days at 37°C in a 5% CO2 atmosphere. For cell binding assays, 3 ml of 1 × 105 cells/ml Caco-2 cells were plated into wells of a 6-well tissue culture treated plate and incubated overnight. This resulted in 90 to 100% confluence, which matches conditions for HAstV propagation in Caco-2 cells (29). On day 2, medium was aspirated, followed by the addition of 1,125 μl prewarmed medium premixed with 375 μl protein sample in PBS. The final concentration of GFP and GFP-Spike samples was 5 μM. For samples containing scFv PL-2, the final concentration of scFv PL-2 was 20 μM. Caco-2 cells were incubated overnight. On day 3, medium and protein were aspirated, and cell monolayers were washed three times with 3 ml cold PBS. To detach cells, 500 μl prewarmed trypsin-EDTA (0.25%) was added to each well and cells were incubated for 10 min at 37°C in a 5% CO2 atmosphere. Cells were resuspended in 9.5 ml growth medium and pelleted by centrifugation for 10 min at 200 × g. Cell pellets were washed by resuspension in 10 ml PBS and pelleted again by centrifugation for 10 min at 200 × g. Cell pellets (∼0.5 × 106 cells) were resuspended in 500 μl PBS and kept on ice. Cells were analyzed by fluorescence-activated cell sorting on an LSR II flow cytometer (BD Biosciences) with a 488-nm excitation wavelength, a 505 long-pass (LP) mirror, and a 525/50 band pass (BP) filter. Scatter gating was used to exclude small debris and large clumps of cells. Ten thousand events were measured. The percentage of fluorescent cells was then determined by gating at a FITC-A signal of 800, where the percentage of fluorescent cells incubated with PBS-only samples is ∼1%. FlowJo Software was used for data analysis and display.
Accession number(s).
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5KOU and 5KOV.
ACKNOWLEDGMENTS
We thank Phillip Berman for the use of his BIAcore SPR instrument, Alicia Sanchez-Fauquier for donation of MAb PL-2 in ascites fluid, Bari Holm Nazario for assistance with flow cytometry data collection and analysis, and Kevin Johnson and Lena Meyer for assistance with DNA cloning and protein production. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases grant AI095369 (to R.M.D.) and by the Hellman Fellows Fund (to R.M.D.). The National Institutes of Health “Initiative for Maximizing Student Development” provided fellowship support (to W.A.B.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Mendez E, Arias CF. 2013. Astroviruses, p 609–628. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Koci MD, Schultz-Cherry S. 2002. Avian astroviruses. Avian Pathol 31:213–227. doi: 10.1080/03079450220136521. [DOI] [PubMed] [Google Scholar]
- 3.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. 2011. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol 11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King AMQ, Lefkowitz E, Adams MJ, Carstens EB (ed). 2011. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Amsterdam, Netherlands. [Google Scholar]
- 5.Koopmans MP, Bijen MH, Monroe SS, Vinje J. 1998. Age-stratified seroprevalence of neutralizing antibodies to astrovirus types 1 to 7 in humans in The Netherlands. Clin Diagn Lab Immunol 5:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtz JB, Lee TW. 1984. Human astrovirus serotypes. Lancet ii:1405. [DOI] [PubMed] [Google Scholar]
- 7.Bosch A, Pinto RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg Infect Dis 5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, Brooks T, Paine SM, Anderson G, Virasami A, Tong CY, Clark DA, Plagnol V, Jacques TS, Qasim W, Hubank M, Breuer J. 2015. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis 60:881–888. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, Grant P, Samayoa E, Federman S, Miller S, Lunn MP, Gant V, Chiu CY. 2015. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurtz J, Lee T. 1978. Astrovirus gastroenteritis age distribution of antibody. Med Microbiol Immunol 166:227–230. doi: 10.1007/BF02121154. [DOI] [PubMed] [Google Scholar]
- 13.Kurtz JB, Lee TW, Craig JW, Reed SE. 1979. Astrovirus infection in volunteers. J Med Virol 3:221–230. doi: 10.1002/jmv.1890030308. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell DK. 2002. Astrovirus gastroenteritis. Pediatr Infect Dis J 21:1067–1069. doi: 10.1097/00006454-200211000-00018. [DOI] [PubMed] [Google Scholar]
- 15.Bjorkholm M, Celsing F, Runarsson G, Waldenstrom J. 1995. Successful intravenous immunoglobulin therapy for severe and persistent astrovirus gastroenteritis after fludarabine treatment in a patient with Waldenstrom's macroglobulinemia. Int J Hematol 62:117–120. doi: 10.1016/0925-5710(95)00396-A. [DOI] [PubMed] [Google Scholar]
- 16.Lewis TL, Greenberg HB, Herrmann JE, Smith LS, Matsui SM. 1994. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J Virol 68:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang B, Monroe SS, Koonin EV, Stine SE, Glass RI. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc Natl Acad Sci U S A 90:10539–10543. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishna NK. 2005. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol 18:17–26. doi: 10.1089/vim.2005.18.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Risco C, Carrascosa JL, Pedregosa AM, Humphrey CD, Sanchez-Fauquier A. 1995. Ultrastructure of human astrovirus serotype 2. J Gen Virol 76(Part 8):2075–2080. [DOI] [PubMed] [Google Scholar]
- 20.Mendez E, Aguirre-Crespo G, Zavala G, Arias CF. 2007. Association of the astrovirus structural protein VP90 with membranes plays a role in virus morphogenesis. J Virol 81:10649–10658. doi: 10.1128/JVI.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendez E, Fernandez-Luna T, Lopez S, Mendez-Toss M, Arias CF. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J Virol 76:7996–8002. doi: 10.1128/JVI.76.16.7996-8002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendez E, Salas-Ocampo E, Arias CF. 2004. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J Virol 78:8601–8608. doi: 10.1128/JVI.78.16.8601-8608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee TW, Kurtz JB. 1981. Serial propagation of astrovirus in tissue culture with the aid of trypsin. J Gen Virol 57:421–424. doi: 10.1099/0022-1317-57-2-421. [DOI] [PubMed] [Google Scholar]
- 24.Bass DM, Qiu S. 2000. Proteolytic processing of the astrovirus capsid. J Virol 74:1810–1814. doi: 10.1128/JVI.74.4.1810-1814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dryden KA, Tihova M, Nowotny N, Matsui SM, Mendez E, Yeager M. 2012. Immature and mature human astrovirus: structure, conformational changes, and similarities to hepatitis E virus. J Mol Biol 422:650–658. doi: 10.1016/j.jmb.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.York RL, Yousefi PA, Bogdanoff W, Haile S, Tripathi S, DuBois RM. 2015. Structural, mechanistic, and antigenic characterization of the human astrovirus capsid. J Virol 90:2254–2263. doi: 10.1128/JVI.02666-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong J, Dong L, Mendez E, Tao Y. 2011. Crystal structure of the human astrovirus capsid spike. Proc Natl Acad Sci U S A 108:12681–12686. doi: 10.1073/pnas.1104834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DuBois RM, Freiden P, Marvin S, Reddivari M, Heath RJ, White SW, Schultz-Cherry S. 2013. Crystal structure of the avian astrovirus capsid spike. J Virol 87:7853–7863. doi: 10.1128/JVI.03139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez E, Munoz-Yanez C, Sanchez-San Martin C, Aguirre-Crespo G, Banos-Lara Mdel R, Gutierrez M, Espinosa R, Acevedo Y, Arias CF, Lopez S. 2014. Characterization of human astrovirus cell entry. J Virol 88:2452–2460. doi: 10.1128/JVI.02908-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Fauquier A, Carrascosa AL, Carrascosa JL, Otero A, Glass RI, Lopez JA, San Martin C, Melero JA. 1994. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology 201:312–320. doi: 10.1006/viro.1994.1296. [DOI] [PubMed] [Google Scholar]
- 31.Bogdanoff WA, Morgenstern D, Bern M, Ueberheide BM, Sanchez-Fauquier A, DuBois RM. 2016. De novo sequencing and resurrection of a human astrovirus-neutralizing antibody. ACS Infect Dis 2:313–321. doi: 10.1021/acsinfecdis.6b00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludert JE, Feng N, Yu JH, Broome RL, Hoshino Y, Greenberg HB. 1996. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J Virol 70:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. 2013. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan M, Hegde RS, Jiang X. 2004. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J Virol 78:6233–6242. doi: 10.1128/JVI.78.12.6233-6242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrmann JE, Hudson RW, Perron-Henry DM, Kurtz JB, Blacklow NR. 1988. Antigenic characterization of cell-cultivated astrovirus serotypes and development of astrovirus-specific monoclonal antibodies. J Infect Dis 158:182–185. doi: 10.1093/infdis/158.1.182. [DOI] [PubMed] [Google Scholar]
- 36.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]