Abstract
Mitochondria are double‐membrane‐bound organelles that are present in all nucleated eukaryotic cells and are responsible for the production of cellular energy in the form of ATP. Mitochondrial function is under dual genetic control – the 16.6‐kb mitochondrial genome, with only 37 genes, and the nuclear genome, which encodes the remaining ∼1300 proteins of the mitoproteome. Mitochondrial dysfunction can arise because of defects in either mitochondrial DNA or nuclear mitochondrial genes, and can present in childhood or adulthood in association with vast clinical heterogeneity, with symptoms affecting a single organ or tissue, or multisystem involvement. There is no cure for mitochondrial disease for the vast majority of mitochondrial disease patients, and a genetic diagnosis is therefore crucial for genetic counselling and recurrence risk calculation, and can impact on the clinical management of affected patients. Next‐generation sequencing strategies are proving pivotal in the discovery of new disease genes and the diagnosis of clinically affected patients; mutations in >250 genes have now been shown to cause mitochondrial disease, and the biochemical, histochemical, immunocytochemical and neuropathological characterization of these patients has led to improved diagnostic testing strategies and novel diagnostic techniques. This review focuses on the current genetic landscape associated with mitochondrial disease, before focusing on advances in studying associated mitochondrial pathology in two, clinically relevant organs – skeletal muscle and brain. © 2016 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: mitochondria, mitochondrial disease, mtDNA, respiratory chain deficiency, genetic diagnosis, muscle pathology, immunohistochemistry, neuropathology
Introduction
Mitochondria are double‐membrane‐bound organelles present in all nucleated eukaryotic cells, and are responsible for numerous cellular processes, including calcium homeostasis, iron–sulphur cluster biogenesis, apoptosis, and the production of cellular energy (ATP) by oxidative phosphorylation (OXPHOS) 1, 2. With bacterial origins, a historical symbiotic relationship evolved during which mitochondria became normal constituents of eukaryotic cells 3. Their ancestry remains apparent from their own multicopy genetic material [mitochondrial DNA (mtDNA)], with copy number varying greatly between individuals and across different tissues from the same individual. The 16.6‐kb circular mtDNA molecule encodes 13 subunits of the OXPHOS components, 22 mitochondrial tRNAs, and two subunits of the mitoribosomes 4. Additionally, the mitoproteome requires a further ∼1300 nuclear‐encoded proteins for producing, assembling or supporting the five multimeric OXPHOS complexes (I–V) and ancillary mitochondrial processes 5. It stands to reason that mitochondrial dysfunction can result from either mtDNA or nuclear gene defects, and can occur as a primary, congenital condition or a secondary, age‐associated effect attributable to somatic mutation 6.
The umbrella term ‘mitochondrial disease’ refers to a clinically heterogeneous group of primary mitochondrial disorders in which the tissues and organs that are most often affected are those with the highest energy demands. Clinical symptoms can arise in childhood or later in life, and can affect one organ in isolation or be multisystemic 7; the minimum disease prevalence in adults is ∼12.5 per 100 000 8, and ∼4.7 per 100 000 in children 9. There is a general lack of genotype–phenotype correlations in many mitochondrial disorders, which means that establishing a genetic diagnosis can be a complicated process, and remains elusive for many patients. This review provides a concise update on three areas where there have been major advances in our understanding in recent years 10, i.e. the molecular genetics, muscle pathology and neuropathology associated with mitochondrial disease, highlighting the range of new techniques that are improving the diagnosis of patients with suspected mitochondrial disease, with the aim of providing options to families at risk of an otherwise incurable condition.
The genetics of mitochondrial disease
Mitochondrial disease caused by mtDNA
Unlike nuclear DNA, which is diploid and follows Mendelian laws of inheritance, mtDNA is exclusively maternally inherited 11. The multicopy nature of mtDNA gives rise to heteroplasmy, a unique aspect of mtDNA‐associated genetics that occurs when there is coexistence of a mix of mutant and wild‐type mtDNA molecules (heteroplasmy). In contrast, homoplasmy occurs when all of the mtDNA molecules have the same genotype. Heteroplasmic mutations often have a variable threshold, i.e. a level to which the cell can tolerate defective mtDNA molecules 12. When the mutation load exceeds this threshold, metabolic dysfunction and associated clinical symptoms occur. Point mutations and large‐scale mtDNA deletions represent the two most common causes of primary mtDNA disease, the former usually being maternally inherited, and the latter typically arising de novo during embryonic development.
mtDNA point mutations
mtDNA point mutations (including small indel mutations) constitute a significant cause of human disease, with an estimated population prevalence of one in 200 13. Mutations have been reported in every mtDNA gene, and have been associated with clinical symptoms ranging from non‐syndromic sensorineural deafness to MELAS, a devastating syndromic neurological condition whose predominant features, i.e. mitochondrial encephalopathy, lactic acidosis, and stroke‐like episodes, give rise to the acronym. Clinical symptoms can present in child or adulthood, and mutations can be inherited (∼75% cases) or occur de novo (∼25% cases) 14. Maternally transmitted mtDNA defects may involve a clinically unaffected mother who harbours the familial mtDNA mutation below the threshold required for cellular dysfunction, although her oocytes harbour varying mutation loads, owing to the selection pressures of the mitochondrial bottleneck 15. It is therefore almost impossible to predict the recurrence risk for subsequent pregnancies, although prenatal testing of embryonic tissues by the use of chorionic villus biopsy or amniocentesis can provide an accurate measure of mtDNA heteroplasmy in the fetus, which can inform reproductive choices 16. The recurrence risk of de novo mtDNA point mutations is very low, except for the risk of germline mosaicism in maternal oocytes 14.
Single, large‐scale mtDNA deletions
Single, large‐scale mtDNA deletions have a population frequency of 1.5/100 000 8, with three main associated phenotypes: chronic progressive external ophthalmoplegia (PEO) (∼65% of cases), Kearns–Sayre syndrome (KSS) (∼30% of cases), and Pearson syndrome (<5% of cases) 17. Pearson syndrome is the most severe presentation associated with single, large‐scale mtDNA deletions; patients present early in life with sideroblastic anaemia and pancreatic dysfunction, and the condition is often fatal in infancy 18. KSS patients present before the of age 20 years with ptosis and/or PEO and pigmentary retinopathy, and may have multisystem involvement, including myopathy, ataxia, or cardiac conduction defects 17. PEO is the more benign presentation attributable to single mtDNA deletions, and is associated with ophthalmoplegia, ptosis, and myopathy 19. Unlike nuclear gene rearrangements, single, large‐scale mtDNA deletions often arise sporadically during embryonic development and have a low recurrence risk 20. Clinically affected women who harbour a large‐scale mtDNA deletion have a low (<10%) risk of transmission 20, and prenatal testing is informative for at‐risk pregnancies 16.
Secondary mtDNA mutations
Large‐scale mtDNA deletions and point mutations represent primary mtDNA defects, but secondary defects are other common causes of mitochondrial disease. Defective mtDNA maintenance, transcription, or protein translation, or a defective ancillary process such as mitochondrial import, can cause either quantitative (depletion of mtDNA copy number) or qualitative (affecting mtDNA genome integrity, resulting in multiple large mtDNA deletions) effects. These result from mutations affecting nuclear genes, and inheritance occurs in a Mendelian (or de novo) fashion.
Mitochondrial disease caused by nuclear mitochondrial genes
The majority of the genes encoding the mitoproteome are in the nuclear genome 5 and follow Mendelian inheritance patterns. De novo, X‐linked, dominant and recessive inheritance cases have been reported in the literature 21, 22, 23, 24. The first nuclear mitochondrial gene mutation was identified in 1995 in SDHA, encoding a structural subunit of complex II 25, and there has been monumental progress in the discovery of mitochondrial disease candidate genes since then. New proteomic and transcriptomic approaches are being applied to models of human disease to uncover new candidates 26, 27, and patient analyses are validating their involvement in human pathology 28. The traditional approach of linkage analysis by the use of multiple affected family members has given way to massively parallel sequencing strategies, including whole exome sequencing (WES), either of affected singletons or of proband–parent trios, and new disease genes are still emerging over 20 years later. Of the ∼1300 proteins in the mitoproteome, mutations have been reported in >250 genes 29, and both new genes and new mechanisms involving genes already implicated in human disease through alternative pathways are being reported 30. It is apparent that more severe clinical phenotypes are often associated with recessive defects, presumably because of varying heteroplasmy levels in clinically affected tissues and the dichotomous effect of recessive mutations; therefore, mtDNA mutations are more common in adults, whereas nuclear gene defects are overrepresented in paediatric cases 31.
In this review, we delineate the nuclear mitochondrial disease genes into those that cause isolated and those that cause multiple respiratory chain complex deficiencies, for simplicity and brevity.
Mitochondrial disease caused by nuclear mitochondrial genes: isolated respiratory chain complex deficiencies
Histochemical and biochemical evidence of an isolated respiratory chain complex deficiency can be suggestive of a mutation affecting either a structural subunit or an assembly/ancillary factor of one of the five OXPHOS complexes. Our current knowledge of the structural subunits and ancillary factors for each complex is summarized in Figure 1.
Figure 1.
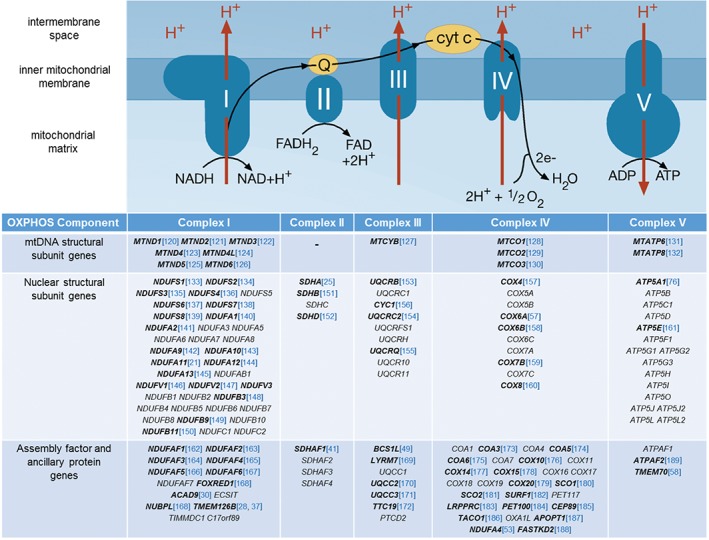
Schematic of the OXPHOS complexes, their component subunits, and associated ancillary factors. Multimeric protein complexes I–IV shuttle electrons along the respiratory chain, facilitated by the reduction of the cofactors coenzyme Q10 (Q) and cytochrome c (cyt c). Electron transfer is coupled to the transfer of protons (H+) across the inner mitochondrial membrane to generate a proton motive force, which is used by complex V (ATP synthase) to synthesize ATP. Characterization of OXPHOS complexes has identified the constitutive subunits that are either mtDNA‐encoded or nuclear‐encoded, and many of the nuclear‐encoded proteins involved in complex assembly, biogenesis, or ancillary function; genes in which mutations have been identified are shown in bold, and the first report of disease‐causing mutations is shown in blue.
Isolated complex I deficiency
Complex I (NADH dehydrogenase) is composed of 44 structural subunits (seven of which are mtDNA‐encoded) with at least 14 ancillary/assembly factors 32, 33. Isolated complex I deficiency represents the biochemical phenotype for ∼30% of paediatric patients 34, of whom 70–80% have a nuclear gene defect 35. The clinical symptoms associated with complex I deficiency are heterogeneous, although the prognosis is typically poor, with rapid progression. Lactic acidosis is a common feature, although it is often present with other symptoms, such as cardiomyopathy or leukodystrophy. Mutations have been identified in 19 of the 37 structural subunits, and in 10 of 14 identified assembly factors. Although there are a few exceptions, such as the p.Trp22Arg NDUFB3 36 and p.Gly212Val TMEM126B European founder mutations 28, 37, and the p.Cys115Tyr NDUFS6 Caucasus Jewish founder mutation 38, studies have revealed the majority of complex I deficiency mutations to be private and non‐recurrent 39. NDUFS2 and ACAD9 mutations account for a significant proportion of diagnoses, although it is likely that clearer genetic diagnostic trends will emerge from large diagnostic next‐generation sequencing (NGS) datasets 40.
Isolated complex II deficiency
Succinate dehydrogenase (SDH), unlike any of the other complexes of the mitochondrial OXPHOS system, is entirely nuclear‐encoded, and is involved in both the tricarboxylic acid cycle (where it metabolizes succinate to fumarate) and the respiratory chain (transferring electrons from FADH2 to reduce ubiquinone to ubiquinol). Complex II deficiency is rare (2–8% of mitochondrial disease cases 41, 42), with <50 patients having been reported. Biallelic mutations have been associated with congenital metabolic presentations, predominantly affecting either the central nervous system (CNS) or heart (hypertrophic cardiomyopathy, leukodystrophy, Leigh syndrome, and encephalopathy) 43, whereas heterozygous mutations are associated with cancer susceptibility, particularly pheochromocytoma and paraganglioma 44. Although SDH was initially believed to have distinct genotype–phenotype relationships (SDHA and SDHAF1 being linked to mitochondrial disease, and SDHB/SDHC/SDHD/SDHAF2 being linked with cancer susceptibility), it is emerging that there is phenotypic overlap, prompting tumour surveillance of unaffected relatives heterozygous for SDHx mutations 45, 46.
Isolated complex III deficiency
Ubiquinol–cytochrome c oxidoreductase, complex III of the respiratory chain, functions as a homodimer to transfer electrons from ubiquinol to cytochrome b, and then to cytochrome c. Complex III is composed of 11 structural subunits plus two heme groups and the Rieske iron–sulphur protein. Exercise intolerance is the clinical phenotype reported for >50% of patients with mutations in the mtDNA MTCYB gene, but cardiomyopathy and encephalomyopathy have also been noted 47. Pathogenic mutations have been reported in four of the nuclear‐encoded structural subunits plus five assembly/ancillary factors 48, with presentations including developmental delay, encephalopathy, lactic acidosis, liver dysfunction, renal tubulopathy, and muscle weakness 48, 49.
Isolated complex IV deficiency
Cytochrome c oxidase (COX), complex IV of the respiratory chain, is embedded in the inner mitochondrial membrane, and functions as a dimer, with two copper‐binding sites, two heme groups, one magnesium ion, and one zinc ion 50. Complex IV pumps protons across the inner mitochondrial membrane, contributing to the proton motive force for ATP synthase exploitation, and donates electrons to oxygen at the respiratory chain termini to form water. Complex IV has 13 structural subunits, and at least 26 additional proteins involved in assembly and biogenesis 51. NDUFA4 was originally described as a complex I subunit gene, but has since been reassigned to complex IV, following functional studies 52 supported by the presence of NDUFA4 defects in a patient with severe COX deficiency 53. Mutations have been reported in structural COX subunits, but most defects affect biogenesis/assembly proteins. Some proteins are linked tightly with specific aspects of COX biogenesis (e.g. COA6, involved in copper‐dependent COX2 biogenesis 54), and others have more diverse roles 55. Clinically, presentations are often early onset and devastating, predominantly affecting the heart and CNS (e.g. SURF1, in which >80 different mutations have been reported to cause Leigh syndrome 56), although a milder Charcot–Marie–Tooth phenotype has been associated with biallelic COX6A1 variants 57.
Isolated complex V deficiency
ATP synthase, complex V, is the multimeric molecular motor that drives ATP production through phosphorylation of ADP. Utilizing the proton motive force generated by electron transport and proton pumping by the respiratory chain, the 600‐kDa complex consists of 13 different subunits (some of which have different isoforms; for example, ATP5G1, ATP5G2 and ATP5G3 encode subunit c isoforms), and involves at least three ancillary factors. Defects have been reported in only four nuclear complex V genes to date, with varied clinical phenotypes. The most common defects involve TMEM70, including a Roma TMEM70 founder mutation causing lactic acidosis and cardiomyopathy 58, although encephalopathy and cataracts have been reported in other populations 59.
Mitochondrial disease caused by nuclear mitochondrial genes: multiple respiratory chain defects
Mitochondrial function is regulated and maintained by ∼1300 nuclear genes; these nuclear genes are translated by cytosolic translational machinery, and the 5′ mitochondrial targeting sequence directs transport of the translated proteins into the mitochondrion, where they are required for diverse functions. These include the transcription of mitochondrial mRNA (e.g. POLRMT 60), mitochondrial DNA maintenance (e.g. POLG 61), regulation of mitochondrial dNTP pools (e.g. RRM2B 62), cellular signalling (e.g. SIRT1 63), and the translation of mtDNA‐derived proteins. Numerous subgroups of proteins are involved in mitochondrial gene translation: mitochondrial aminoacyl tRNA synthetases, which are responsible for charging each mitochondrial tRNA molecule with the appropriate amino acid (e.g. AARS2 64), proteins involved in RNA processing (e.g. MTPAP 65), mitoribosomal proteins (e.g. MRPL44 66), and proteins involved in mitochondrial tRNA modification (e.g. TRMU 67). Defects in ≥250 nuclear mitochondrial genes have now been reported in association with multiple respiratory chain defects and clinical mitochondrial disease 29. The genetic diagnostic pathway for these disorders is complex, and WES is often the most successful strategy 68.
Non‐OXPHOS mitochondrial disease
Not all mitochondrial disease patients have evidence of respiratory chain enzyme dysfunction, but have other evidence of mitochondrial disease, such as elevated lactate levels, suggestive magnetic resonance imgaing brain changes, and multisystem involvement. Genetic causes include defective enzymes of the Krebs cycle (e.g. aconitase/ACO2 69) or cofactor transport (e.g. thiamine transporter/SLC19A3 70).
Molecular genetic analysis of mitochondrial disease
In the absence of effective treatments, provision of a firm genetic diagnosis facilitates genetic counselling and access to reproductive options for patients and their families. Given the small size of the mtDNA genome, this is often sequenced in suspected mitochondrial disease patients to exclude a primary mtDNA defect before nuclear genes are scrutinized. NGS‐based testing is becoming more prevalent 71, and also provides an accurate measure of mtDNA heteroplasmy. NGS technologies are revolutionizing the genetic testing pipeline in the diagnostic genetic laboratory, with Sanger sequencing of candidate genes on a sequential basis being replaced with powerful, high‐throughput analysis. A variety of options are currently being implemented – targeted panels of candidate genes 36, unbiased WES 72, and whole genome sequencing (WGS) 73 (Figure 2). Custom, panel‐based NGS strategies can be very successful in providing a rapid genetic diagnosis in the clinical setting, but this success depends on the degree of characterization to ensure that the appropriate candidate genes are targeted. Stratification according to respiratory chain defect can be appropriate for many patients in whom muscle biopsy is available, but even then it may be misleading – a number of patients with an isolated complex I deficiency have, in fact, a defect of mitochondrial translation 40; moreover, this strategy can be ineffective for genes that show inconsistent biochemical profiles 74. Stratification according to clinical phenotype is similarly complicated by genetic heterogeneity 75.
Figure 2.
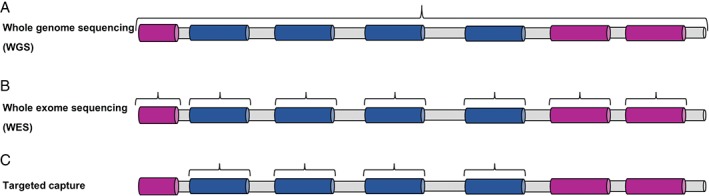
NGS strategies employed in the genetic diagnosis of mitochondrial disease. (A) WGS analyses all coding and non‐coding regions of the genome. (B) WES targets only the coding exons plus immediate intron–exon boundaries. (C) Target capture facilitates sequencing of a predetermined genomic region or list of candidate disease genes. Non‐coding/intronic regions are shaded grey, exons of candidate genes are shaded blue, and exons of non‐candidate genes are shaded pink.
Despite a proven track record in a research setting and the increasing availability of affordable NGS options to diagnostic laboratories, the case has yet to be made regarding the clinical validity of unrestricted WES within a diagnostic setting. One solution to the stratification dilemma, and one that has been successfully implemented for the analysis of other heterogeneous Mendelian disorders, is a combination of unbiased WES with targeted analysis of ‘virtual’ gene panels 76, 77; this allows informative reporting of negative results, and removes the possibility of incidental findings. Further analysis of the WES data for patients lacking a diagnosis following virtual panel analysis could be subsequently undertaken in a research setting. Indeed, most of the candidate genes included in diagnostic virtual panels have their origins in research. WES has been incredibly fruitful in elucidating genes involved in human pathology, including heterogeneous mitochondrial clinical phenotypes such as cardiomyopathy, with mutations identified in AARS2 78, MRPL3 79, MTO1 80, and ACAD9 72. New candidate genes continue to be discovered in a research setting, and are then included in diagnostic screening; one success is exemplified by the report of patients harbouring mutations in TMEM126B, a candidate gene identified by research‐based complexome profiling 27, 28, 37. Similarly, characterization of predicted mitochondrial proteins of unknown function is another critical strategy for identifying novel disease candidate genes 26.
Investigating muscle pathology associated with mitochondrial disease
As discussed above, the laboratory investigation of suspected mitochondrial disease is complex, and algorithms employ a multidisciplinary approach using clinical and functional studies to guide genetic analysis 81. Although mitochondrial disorders are characterized by a wide spectrum of clinical presentations, owing to the high metabolic requirements, muscle is frequently affected – either exclusively (e.g. myopathy and chronic progressive external ophthalmoplegia) or as a predominant feature in multisystem phenotypes 81, 82. In both scenarios, muscle involvement can arise from mutations in nuclear or mtDNA genes, and the association with distinctive histopathological hallmarks makes muscle an excellent postmitotic surrogate for the study of many multisystem mitochondrial disorders. Diagnostic centres specializing in mitochondrial disorders employ numerous techniques to assess mitochondrial function, including the assessment of individual mitochondrial OXPHOS activities in vitro 83. Although useful for identifying widespread mitochondrial defects, this technique has some limitations; it requires large quantities of muscle (typically 50–100 mg of tissue) and may fail to detect subtle OXPHOS deficiencies, especially when only a few muscle fibres are affected (e.g. mild mosaic deficiencies). Furthermore, only complexes I–IV can be reliably assessed in frozen muscle.
The histological and histochemical examination of serially‐sectioned muscle can provide crucial evidence of mitochondrial pathology. Haematoxylin and eosin (H&E) and modified Gomori trichrome stains assess basic muscle morphology, providing information on fibre size and the presence of any abnormal inclusions or central nuclei which are indicative of muscle denervation (Figure 3). The modified Gomori trichrome stain 84, 85 specifically highlights connective tissue (light blue), muscle fibres (blue), and mitochondria (red), and allows the detection of ragged‐red fibres (RRFs) 86. RRFs are characterized by a ‘fibre cracking’ appearance and abnormal subsarcolemmal proliferation of mitochondria, resulting from a compensatory response to a respiratory chain biochemical defect 87. RRFs can show either normal oxidative enzyme activities (often reported in association with the m.3243A>G mutation or some sporadic MTCYB mutations) or COX deficiency associated with a wide range of mtDNA‐related genetic disorders 88. They represent a characteristic histopathological feature of mitochondrial disorders, however, they are not entirely diagnostic, as they are also seen with normal ageing 6 and other muscle conditions 89, 90.
Figure 3.
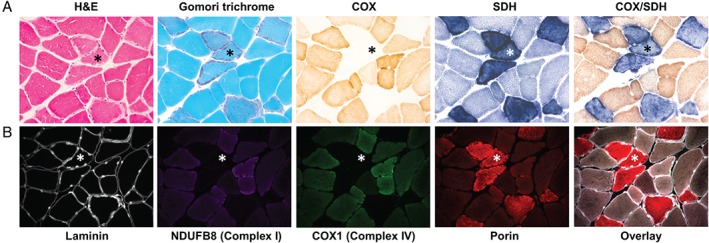
Histological, histochemical and immunohistochemical hallmarks of mitochondrial pathology in primary mtDNA‐related disease. (A) Serial skeletal muscle (vastus lateralis) sections from a patient with a single, large‐scale mtDNA deletion were stained with H&E and modified Gomori trichrome to assess basic muscle morphology and the presence of RRFs, respectively. The individual COX, SDH and sequential COX/SDH histochemical reactions show fibres manifesting mitochondrial accumulation and focal COX deficiency. (B) The lack of histochemical assays to assess other OXPHOS complex activities prompted the development of a quadruple immunofluorescence assay that can quantify the levels of complex I (NDUFB8 subunit), complex IV (COX1 subunit), laminin, and a mitochondrial mass marker (porin), all within a single 10‐µm section. A highlighted COX‐deficient fibre (*) shows focal accumulation of sarcolemmal mitochondria around the periphery of the fibre, and downregulated expression of both complex I and IV proteins. (All images taken at ×20 objective magnification.)
Sequential COX/SDH histochemistry is the standard method used to assess mitochondrial respiratory chain function in muscle cryosections 91, 92, the activities of the partially mtDNA‐encoded complex IV (COX), and the fully nuclear‐encoded complex II (SDH). By combining both reactions in a single slide (Figure 3A, COX/SDH panel), fibres or cells with mitochondrial dysfunction are easily identifiable, and are seen as a mosaic reduction or loss of COX activity with preserved SDH activity (blue fibres), indicative of an underlying mtDNA‐related abnormality 93, 94. The absence of routine histochemical assays to evaluate other OXPHOS complexes, such as complex I, which is the largest and most commonly affected OXPHOS complex in mitochondrial disorders 95, has prompted the recent development of a novel high‐throughput immunofluorescence assay to fill the gap in the diagnostic repertoire 96. This technique enables accurate quantification of the two most commonly affected OXPHOS components, namely complexes I and IV 97, together with a mitochondrial mass marker (porin) in individual muscle fibres on a single 10‐µm tissue section (Figure 3B). The semi‐automatic quantification of a large number of muscle fibres is facilitated by labelling laminin to define fibre boundaries. Image analysis is exclusively based on intensity measurements, increasing its accuracy and reliability, and is automated (http://iah‐rdevext.ncl.ac.uk/immuno/). We are currently optimizing the immunodetection of antibodies to assess complex III and complex V, in order to better quantify the full extent of mitochondrial respiratory deficiency in patient muscle sections, but the opportunity to assess this at a single‐fibre level shows great potential for both diagnostic and research applications (Figure 4).
Figure 4.
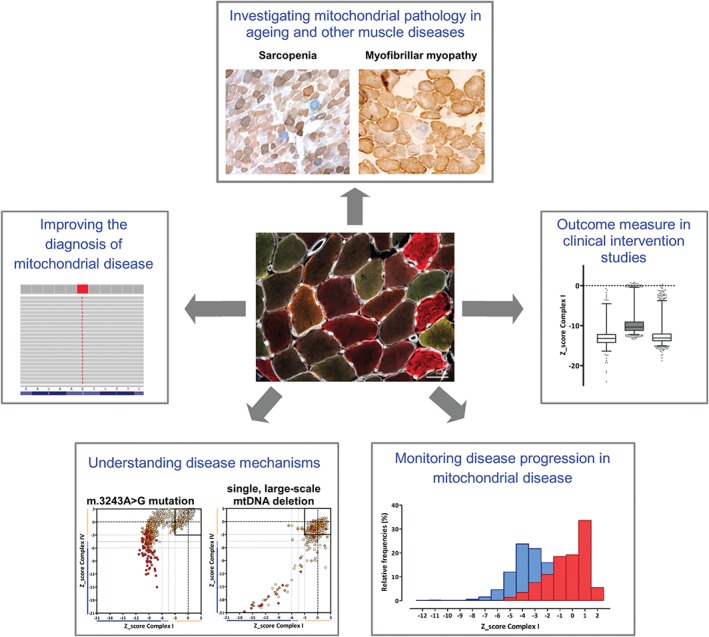
Current and future applications of a quantitative, quadruple OXPHOS immunofluorescence assay. Given its capacity to interrogate levels of both complex I and IV – and additional OXPHOS components – at a single muscle fibre level, we believe that the quadruple immunofluorescence assay can be applied to several areas of diagnostic and research activity in the laboratory to help investigate the role of mitochondrial biochemical defects 96. We are already implementing this methodology in a diagnostic setting, validating the assay with biopsies from patients showing a range of mtDNA‐related and nuclear genetic diagnoses of mitochondrial disease. The assay also shows promise as a powerful tool with which to investigate the mitochondrial pathological changes observed in ageing and other myopathies (e.g. myofibrillar myopathies 90), to investigate molecular disease mechanisms and mitochondrial disease progression, as well as providing an extremely sensitive outcome measure in clinical therapeutic intervention studies (e.g. pharmacological agents or exercise) aimed at improving muscle oxidative capacity in patients with mitochondrial disease.
Neuropathology associated with mitochondrial disease
Neurological symptoms are particularly common, and may be devastating in patients with mitochondrial disease, including sensorineural deafness, cerebellar ataxia, peripheral neuropathy, dementia, and epilepsy 81. In recent years, a number of neuropathological studies have documented the characteristic features of neurodegeneration in patients with mitochondrial disease, and these have spurred the development of novel tools with which to understand the mechanisms underlying neural dysfunction and cell death.
New insights into mechanisms of neurodegeneration
Upon neuropathological investigation, the brains from patients with mitochondrial disease often show atrophy, cortical lesions, evidence of neuronal cell loss, and mitochondrial OXPHOS abnormalities in the remaining cells. Patients with the heteroplasmic m.3243A>G mutation and a MELAS phenotype often develop foci of cortical necrosis on the surface of the brain (Figure 5A). These are often referred to as ischaemic‐like lesions, as they resemble stroke penumbra but do not conform to a particular vascular territory. It is proposed that these lesions evolve during stroke‐like episodes, and may be initiated by mitochondrial respiratory abnormalities in neurons that act to alter the balance of excitation and inhibition in neural networks, promoting neuronal hyperexcitability 98. This is important, as seizures are frequently detected by electroencephalography in patients who have had a stroke‐like episode 99. Although focal necrotic changes associated with the m.3243A>G mutation have been commonly documented, it is important to note that patients harbouring other genetic defects (e.g. the m.8344A>G mutation 100 and autosomal recessive POLG mutations 101, 102) also develop cortical lesions, suggesting shared mechanisms underpinning their formation. These lesions typically affect posterior brain regions, including the occipital, parietal and temporal lobes, and feature microvacuolation and neuronal cell dropout (Figure 5B, C), neuronal eosinophilia, astrogliosis, and secondary myelin loss. Recent studies have proposed that vulnerability of GABAergic interneurons could underpin neuronal hyperexcitability, as dramatic downregulation of OXPHOS subunits constituting complexes I and IV has been observed within interneurons (Figure 5D) 103; other theories suggest that aggregation of abnormally enlarged mitochondria and the presence of mitochondrial respiratory chain abnormalities in the cerebral microvasculature may contribute to impaired cerebral perfusion 104, 105. Although the precise mechanisms are not known, the emergence of lesions in the brain reflect an acute process leading to rapid neuronal loss that can occur on the background of more chronic and protracted cell loss throughout the brain.
Figure 5.
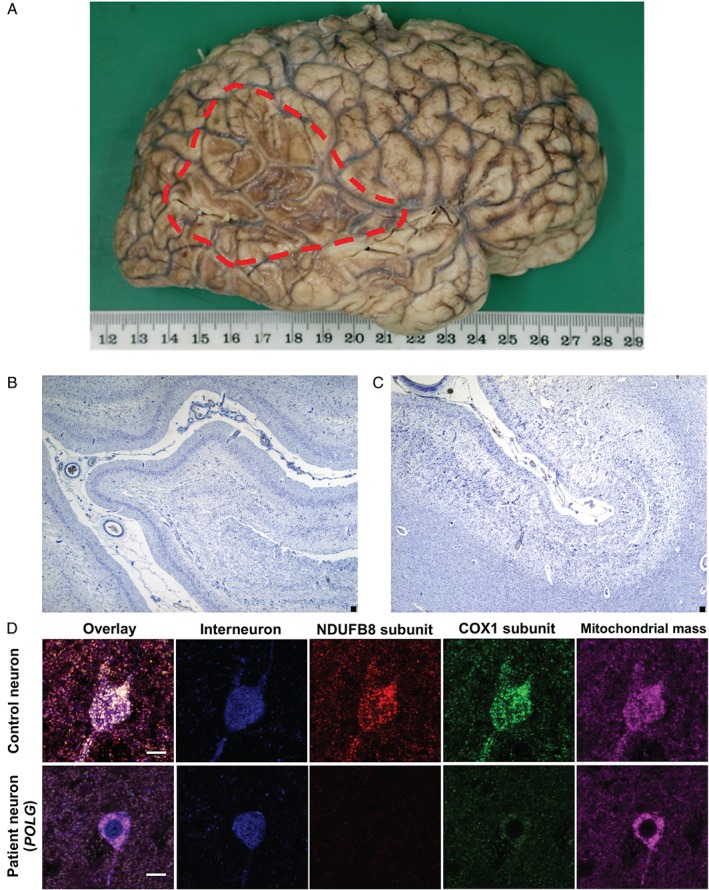
Neuropathological changes associated with stroke‐like episodes in patients with mitochondrial disease. (A) Extensive cortical necrosis affecting the occipital, temporal and parietal lobes in a brain from a patient harbouring the m.3243A>G mutation. (B, C) Microscopic analysis reveals atrophy, microvacuolation and severe neuronal loss in the frontal cortex of a patient with the m.3243A>G mutation [(B) Cresyl fast violet staining] and in the temporal cortex of a patient with the m.8344A>G mutation [(C) Cresyl fast violet staining]. (D) Respiratory chain abnormalities include downregulation of subunits constituting complex I (red; NDUFB8 subunit) and complex IV (green; COXI) relative to intact mitochondrial mass (magenta; porin) in inhibitory interneurons (blue; GAD 65–67) in a patient harbouring autosomal recessive POLG mutations. Scale bar: 10 µm.
The cerebellum is frequently involved in mitochondrial disease, with many patients developing cerebellar ataxia. Neuropathologically, the cerebellum reveals signs of lesions (Figure 6A) similar to those observed in the cortex, global Purkinje cell dropout (Figure 6B), and loss of dentate nucleus neurons 106. Recent work has shown downregulation of protein subunits constituting complex I in remaining Purkinje cells, their GABAergic synapses, and dentate nucleus neurons (Figure 6C). In conjunction, there is evidence of neuronal network remodelling with thickened dendritic arborizations, axonal torpedoes, and altered synaptic density 107, 108, 109. There is a distinct lack of correlation between the severity of cell loss and the heteroplasmy level of mutated mtDNA in surviving neurons, suggesting that other factors must be important in determining cell loss 110.
Figure 6.
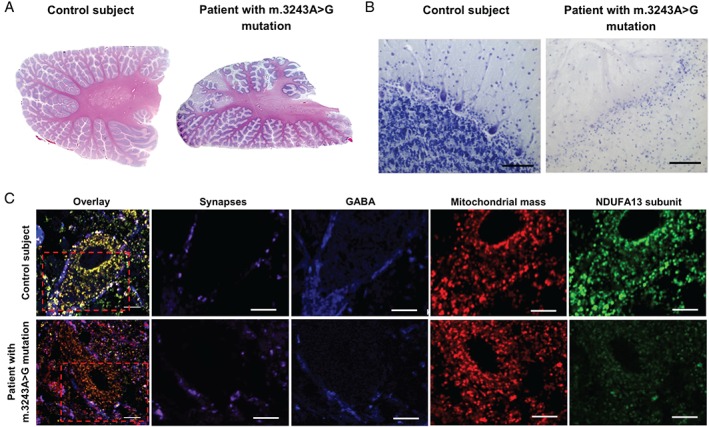
Cerebellar pathology in patients with the m.3243A>G mutation. (A) Numerous areas of necrosis are evident throughout the cerebellar cortex of a patient in comparison with control cerebellum (H&E staining). (B) Extreme neuronal loss is seen microscopically, affecting Purkinje cells and granule cells in the cortex (Cresyl fast violet staining). Scale bar: 100 µm). (C) In dentate nucleus neurons and in GABAergic (blue; GAD 65–67) synapses (magenta; synaptophysin) from Purkinje cells, there is downregulation of complex I (green; NDUFA13) relative to mitochondrial mass (red; COX4I2). Scale bar: 10 µm).
Patients harbouring a single large‐scale mtDNA deletion may develop KSS, which is associated with severe demyelination and spongiosis of the white matter tracts of the brain, including the cerebrum, cerebellum, spinal cord, and brainstem 111. The loss of myelin is proposed to be attributable to specific vulnerability of mature oligodendrocytes, the myelin‐producing glia, where a loss of respiratory chain activity resulting from the mtDNA deletion causes a distal oligodendrogliopathy and subsequent loss of myelin products 112. It is not known why the mtDNA deletion preferentially affects oligodendrocytes.
In summary, neuropathological studies have shown that neuronal cell loss can occur via two different processes: an acute event, such as in stroke‐like lesions, or a global, protracted loss of cells. There is no evidence of protein accumulation within neurons, surviving neurons frequently show respiratory chain deficiency, including downregulation of complex I subunits, and there is a lack of correlation of cell loss and mtDNA heteroplasmy in remaining neurons.
Tools to aid the study of mitochondrial neuropathology
Recently, a number of novel methods have been developed to provide further insights into potential mechanisms of neurodegeneration, particularly for understanding the early events leading to irreversible neuronal cell loss. Clear lipid‐exchanged acrylamide‐hybridized rigid imaging/immunostaining/in situ‐hybridization‐compatible tissue hydrogel has paved the way for large volumes of archived, postmortem material to be investigated with three‐dimensional analysis of the neuronal networks 113. This will enable a greater understanding of neuronal vulnerability in mitochondrial disease 114. The recent development of induced pluripotent stem cell technology allows the cellular transfection of human patient fibroblasts with four key transcription factors to confer pluripotency. These pluripotent cells can subsequently be differentiated into neurons and glial cells, and the effects of both the nuclear genome and mitochondrial genome can be investigated to determine disease mechanisms, efficacy of drug treatment, and cell replacement therapies 115, 116. Additionally, a number of transgenic mouse models utilizing Cre/Lox technology to selectively knock out nuclear mitochondrial genes within specific populations of neurons and glial cells are promising for the understanding of specific disease mechanisms 117, 118, 119.
Challenges for the future
Developing an effective treatment for mitochondrial disease is an enormous challenge that is dependent on the integration of clinical understanding of disease progression, molecular genetic mechanisms, and neuropathological features in mitochondrial disease. Patient‐based clinical, molecular genetic and histopathology studies can then inform the development of appropriate disease model systems to determine mechanisms and treatment to ultimately improve the lives of patients with mitochondrial disease.
Author contributions statement
All authors contributed to the drafting of the manuscript and its critical revision for important intellectual content.
Acknowledgements
Work in our laboratories is supported by a Wellcome Trust Strategic Award (096919/Z/11/Z), the MRC Centre for Neuromuscular Diseases (G0601943), Newcastle University Centre for Ageing and Vitality [supported by the Biotechnology and Biological Sciences Research Council and Medical Research Council (G016354/1)], the UK NIHR Biomedical Research Centre in Age and Age Related Diseases award to the Newcastle upon Tyne Hospitals NHS Foundation, the MRC/ESPRC Newcastle Molecular Pathology Node, the UK National Health Service Highly Specialised ‘Rare Mitochondrial Disorders of Adults and Children’ service, and the Lily Foundation. CLA is in receipt of a National Institute for Health Research (NIHR) doctoral fellowship (NIHR‐HCS‐D12‐03‐04). The views expressed are those of the authors and not necessarily of the NHS, NIHR, or the Department of Health. The authors would like to thank Alexia Chrysostomou, Hannah Rosa and Amy Vincent for contributing images shown in the figures.
No conflicts of interest were declared.
References
- 1. Duchen MR. Mitochondria and calcium: from cell signalling to cell death. J Physiol 2000; 529: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stehling O, Lill R. The role of mitochondria in cellular iron–sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harbor Perspect Biol 2013; 5: a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science (New York, NY) 2001; 292: 1096–1099. [DOI] [PubMed] [Google Scholar]
- 4. Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature 1981; 290: 457–465. [DOI] [PubMed] [Google Scholar]
- 5. Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 2016; 44: D1251–D1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greaves LC, Nooteboom M, Elson JL, et al. Clonal expansion of early to mid‐life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet 2014; 10: e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lightowlers RN, Taylor RW, Turnbull DM. Mutations causing mitochondrial disease: what is new and what challenges remain? Science (New York, NY) 2015; 349: 1494–1499. [DOI] [PubMed] [Google Scholar]
- 8. Gorman GS, Schaefer AM, Ng Y, et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol 2015; 77: 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skladal D, Halliday J, Thorburn DR. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain 2003; 126: 1905–1912. [DOI] [PubMed] [Google Scholar]
- 10. Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. J Pathol 2012; 226: 274–286. [DOI] [PubMed] [Google Scholar]
- 11. Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA 1980; 77: 6715–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet 2015; 16: 530–542. [DOI] [PubMed] [Google Scholar]
- 13. Chinnery PF, Elliott HR, Hudson G, Samuels DC, Relton CL. Epigenetics, epidemiology and mitochondrial DNA diseases. Int J Epidemol 2012; 41: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sallevelt SC, de Die‐Smulders CE, Hendrickx AT, et al. De novo mtDNA point mutations are common and have a low recurrence risk. J Med Genet 2016; DOI: 10.1136/jmedgenet-2016-103876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson IJ, Carling PJ, Alston CL, et al. Mitochondrial DNA sequence characteristics modulate the size of the genetic bottleneck. Hum Mol Genet 2016; 25: 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nesbitt V, Alston CL, Blakely EL, et al. A national perspective on prenatal testing for mitochondrial disease. Eur J Hum Genet 2014; 22: 1255–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mancuso M, Orsucci D, Angelini C, et al. Redefining phenotypes associated with mitochondrial DNA single deletion. J Neurol 2015; 262: 1301–1309. [DOI] [PubMed] [Google Scholar]
- 18. Rotig A, Cormier V, Blanche S, et al. Pearson's marrow‐pancreas syndrome. A multisystem mitochondrial disorder in infancy. J Clin Invest 1990; 86: 1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pitceathly RD, Rahman S, Hanna MG. Single deletions in mitochondrial DNA – molecular mechanisms and disease phenotypes in clinical practice. Neuromusc Disord 2012; 22: 577–586. [DOI] [PubMed] [Google Scholar]
- 20. Chinnery PF, DiMauro S, Shanske S, et al. Risk of developing a mitochondrial DNA deletion disorder. Lancet 2004; 364: 592–596. [DOI] [PubMed] [Google Scholar]
- 21. Berger I, Hershkovitz E, Shaag A, Edvardson S, Saada A, Elpeleg O. Mitochondrial complex I deficiency caused by a deleterious NDUFA11 mutation. Ann Neurol 2008; 63: 405–408. [DOI] [PubMed] [Google Scholar]
- 22. Sperl W, Fleuren L, Freisinger P, et al. The spectrum of pyruvate oxidation defects in the diagnosis of mitochondrial disorders. J Inherit Metab Dis 2015; 38: 391–403. [DOI] [PubMed] [Google Scholar]
- 23. Tang S, Wang J, Lee NC, et al. Mitochondrial DNA polymerase gamma mutations: an ever expanding molecular and clinical spectrum. J Med Genet 2011; 48: 669–681. [DOI] [PubMed] [Google Scholar]
- 24. Thompson K, Majd H, Dallabona C, et al. Recurrent de novo dominant mutations in SLC25A4 cause severe early‐onset mitochondrial disease and loss of mitochondrial DNA copy number. Am J Hum Genet 2016; 99: 860–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bourgeron T, Rustin P, Chretien D, et al. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet 1995; 11: 144–149. [DOI] [PubMed] [Google Scholar]
- 26. Floyd BJ, Wilkerson EM, Veling MT, et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell 2016; 63: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heide H, Bleier L, Steger M, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab 2012; 16: 538–549. [DOI] [PubMed] [Google Scholar]
- 28. Alston CL, Compton AG, Formosa LE, et al. Biallelic mutations in TMEM126B cause severe complex i deficiency with a variable clinical phenotype. Am J Hum Genet 2016; 99: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mayr JA, Haack TB, Freisinger P, et al. Spectrum of combined respiratory chain defects. J Inherit Metab Dis 2015; 38: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nouws J, Nijtmans L, Houten SM, et al. Acyl‐CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab 2010; 12: 283–294. [DOI] [PubMed] [Google Scholar]
- 31. DiMauro S, Schon EA. Mitochondrial respiratory‐chain diseases. N Engl J Med 2003; 348: 2656–2668. [DOI] [PubMed] [Google Scholar]
- 32. Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature 2013; 494: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirst J. Mitochondrial complex I. Annu Rev Biochem 2013; 82: 551–575. [DOI] [PubMed] [Google Scholar]
- 34. Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology 1999; 52: 1255–1264. [DOI] [PubMed] [Google Scholar]
- 35. Swalwell H, Kirby DM, Blakely EL, et al. Respiratory chain complex I deficiency caused by mitochondrial DNA mutations. Eur J Hum Genet 2011; 19: 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alston CL, Howard C, Olahova M, et al. A recurrent mitochondrial p.Trp22Arg NDUFB3 variant causes a distinctive facial appearance, short stature and a mild biochemical and clinical phenotype. J Med Genet 2016; 53: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanchez‐Caballero L, Ruzzenente B, Bianchi L, et al. Mutations in complex I assembly factor TMEM126B result in muscle weakness and isolated complex i deficiency. Am J Hum Genet 2016; 99: 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spiegel R, Shaag A, Mandel H, et al. Mutated NDUFS6 is the cause of fatal neonatal lactic acidemia in Caucasus Jews. Eur J Hum Genet 2009; 17: 1200–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pagniez‐Mammeri H, Loublier S, Legrand A, Benit P, Rustin P, Slama A. Mitochondrial complex I deficiency of nuclear origin I. Structural genes. Mol Genet Metab 2012; 105: 163–172. [DOI] [PubMed] [Google Scholar]
- 40. Haack TB, Haberberger B, Frisch EM, et al. Molecular diagnosis in mitochondrial complex I deficiency using exome sequencing. J Med Genet 2012; 49: 277–283. [DOI] [PubMed] [Google Scholar]
- 41. Ghezzi D, Goffrini P, Uziel G, et al. SDHAF1, encoding a LYR complex‐II specific assembly factor, is mutated in SDH‐defective infantile leukoencephalopathy. Nat Genet 2009; 41: 654–656. [DOI] [PubMed] [Google Scholar]
- 42. Parfait B, Chretien D, Rotig A, Marsac C, Munnich A, Rustin P. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum Genet 2000; 106: 236–243. [DOI] [PubMed] [Google Scholar]
- 43. Jain‐Ghai S, Cameron JM, Al Maawali A, et al. Complex II deficiency – a case report and review of the literature. Am J Hum Genet 2013; 161a: 285–294. [DOI] [PubMed] [Google Scholar]
- 44. Timmers HJ, Gimenez‐Roqueplo AP, Mannelli M, Pacak K. Clinical aspects of SDHx‐related pheochromocytoma and paraganglioma. Endocr Relat Cancer 2009; 16: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hensen EF, Siemers MD, Jansen JC, et al. Mutations in SDHD are the major determinants of the clinical characteristics of Dutch head and neck paraganglioma patients. Clin Endocrinol 2011; 75: 650–655. [DOI] [PubMed] [Google Scholar]
- 46. Alston CL, Ceccatelli Berti C, Blakely EL, et al. A recessive homozygous p.Asp92Gly SDHD mutation causes prenatal cardiomyopathy and a severe mitochondrial complex II deficiency. Human Genet 2015; 134: 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lott MT, Leipzig JN, Derbeneva O, et al. mtDNA variation and analysis using Mitomap and Mitomaster. Curr Protoc Bioinformatics 2013; 44: 1.23.21–1.23.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mordaunt DA, Jolley A, Balasubramaniam S, et al. Phenotypic variation of TTC19‐deficient mitochondrial complex III deficiency: a case report and literature review. Am J Hum Genet A 2015; 167: 1330–1336. [DOI] [PubMed] [Google Scholar]
- 49. de Lonlay P, Valnot I, Barrientos A, et al. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat Genet 2001; 29: 57–60. [DOI] [PubMed] [Google Scholar]
- 50. Shoubridge EA. Cytochrome c oxidase deficiency. Am J Med Genet 2001; 106: 46–52. [DOI] [PubMed] [Google Scholar]
- 51. Rak M, Benit P, Chretien D, et al. Mitochondrial cytochrome c oxidase deficiency. Clin Sci 2016; 130: 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Balsa E, Marco R, Perales‐Clemente E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab 2012; 16: 378–386. [DOI] [PubMed] [Google Scholar]
- 53. Pitceathly RD, Rahman S, Wedatilake Y, et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep 2013; 3: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stroud DA, Maher MJ, Lindau C, et al. COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA‐encoded COX2 . Hum Mol Genet 2015; 24: 5404–5415. [DOI] [PubMed] [Google Scholar]
- 55. Mourier A, Ruzzenente B, Brandt T, Kuhlbrandt W, Larsson NG. Loss of LRPPRC causes ATP synthase deficiency. Hum Mol Genet 2014; 23: 2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wedatilake Y, Brown RM, McFarland R, et al. SURF1 deficiency: a multi‐centre natural history study. Orphanet J Rare Dis 2013; 8: 96 DOI: 10.1186/1750-1172-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tamiya G, Makino S, Hayashi M, et al. A mutation of COX6A1 causes a recessive axonal or mixed form of Charcot–Marie–Tooth disease. Am J Hum Genet 2014; 95: 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cizkova A, Stranecky V, Mayr JA, et al. TMEM70 mutations cause isolated ATP synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nat Genet 2008; 40: 1288–1290. [DOI] [PubMed] [Google Scholar]
- 59. Spiegel R, Khayat M, Shalev SA, et al. TMEM70 mutations are a common cause of nuclear encoded ATP synthase assembly defect: further delineation of a new syndrome. J Med Genet 2011; 48: 177–182. [DOI] [PubMed] [Google Scholar]
- 60. Bestwick ML, Shadel GS. Accessorizing the human mitochondrial transcription machinery. Trends Biochem Sci 2013; 38: 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Agostino A, Valletta L, Chinnery PF, et al. Mutations of ANT1, Twinkle, and POLG1 in sporadic progressive external ophthalmoplegia (PEO). Neurology 2003; 60: 1354–1356. [DOI] [PubMed] [Google Scholar]
- 62. Bourdon A, Minai L, Serre V, et al. Mutation of RRM2B, encoding p53‐controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet 2007; 39: 776–780. [DOI] [PubMed] [Google Scholar]
- 63. Marcu R, Wiczer BM, Neeley CK, Hawkins BJ. Mitochondrial matrix Ca(2)(+) accumulation regulates cytosolic NAD(+)/NADH metabolism, protein acetylation, and sirtuin expression. Mol Cell Biol 2014; 34: 2890–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Diodato D, Ghezzi D, Tiranti V. The mitochondrial aminoacyl tRNA synthetases: genes and syndromes. Int J Cell Biol 2014; 2014: 787956 DOI: 10.1155/2014/787956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wolf AR, Mootha VK. Functional genomic analysis of human mitochondrial RNA processing. Cell Rep 2014; 7: 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carroll CJ, Isohanni P, Poyhonen R, et al. Whole‐exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J Med Genet 2013; 50: 151–159. [DOI] [PubMed] [Google Scholar]
- 67. Umeda N, Suzuki T, Yukawa M, et al. Mitochondria‐specific RNA‐modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J Biol Chem 2005; 280: 1613–1624. [DOI] [PubMed] [Google Scholar]
- 68. Taylor RW, Pyle A, Griffin H, et al. Use of whole‐exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA 2014; 312: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Metodiev MD, Gerber S, Hubert L, et al. Mutations in the tricarboxylic acid cycle enzyme, aconitase 2, cause either isolated or syndromic optic neuropathy with encephalopathy and cerebellar atrophy. J Med Genet 2014; 51: 834–838. [DOI] [PubMed] [Google Scholar]
- 70. Gerards M, Kamps R, van Oevelen J, et al. Exome sequencing reveals a novel Moroccan founder mutation in SLC19A3 as a new cause of early‐childhood fatal Leigh syndrome. Brain 2013; 136: 882–890. [DOI] [PubMed] [Google Scholar]
- 71. Tang S, Wang J, Zhang VW, et al. Transition to next generation analysis of the whole mitochondrial genome: a summary of molecular defects. Hum Mut 2013; 34: 882–893. [DOI] [PubMed] [Google Scholar]
- 72. Haack TB, Danhauser K, Haberberger B, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet 2010; 42: 1131–1134. [DOI] [PubMed] [Google Scholar]
- 73. Hartmannova H, Piherova L, Tauchmannova K, et al. Acadian variant of Fanconi syndrome is caused by mitochondrial respiratory chain complex I deficiency due to a non‐coding mutation in complex I assembly factor NDUFAF6 . Hum Mol Genet 2016; DOI: 10.1093/hmg/ddw245. [DOI] [PubMed] [Google Scholar]
- 74. Tetreault M, Fahiminiya S, Antonicka H, et al. Whole‐exome sequencing identifies novel ECHS1 mutations in Leigh syndrome. Hum Genet 2015; 134: 981–991. [DOI] [PubMed] [Google Scholar]
- 75. Blok MJ, van den Bosch BJ, Jongen E, et al. The unfolding clinical spectrum of POLG mutations. J Med Genet 2009; 46: 776–785. [DOI] [PubMed] [Google Scholar]
- 76. Lieber DS, Calvo SE, Shanahan K, et al. Targeted exome sequencing of suspected mitochondrial disorders. Neurology 2013; 80: 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wortmann SB, Koolen DA, Smeitink JA, van den Heuvel L, Rodenburg RJ. Whole exome sequencing of suspected mitochondrial patients in clinical practice. J Inherit Metab Dis 2015; 38: 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gotz A, Tyynismaa H, Euro L, et al. Exome sequencing identifies mitochondrial alanyl‐tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am J Hum Genet 2011; 88: 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Galmiche L, Serre V, Beinat M, et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum Mut 2011; 32: 1225–1231. [DOI] [PubMed] [Google Scholar]
- 80. Ghezzi D, Baruffini E, Haack TB, et al. Mutations of the mitochondrial‐tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am J Hum Genet 2012; 90: 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McFarland R, Taylor RW, Turnbull DM. A neurological perspective on mitochondrial disease. Lancet Neurol 2010; 9: 829–840. [DOI] [PubMed] [Google Scholar]
- 82. Taylor RW, Schaefer AM, Barron MJ, McFarland R, Turnbull DM. The diagnosis of mitochondrial muscle disease. Neuromusc Disord 2004; 14: 237–245. [DOI] [PubMed] [Google Scholar]
- 83. Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biol 2007; 80: 93–119. [DOI] [PubMed] [Google Scholar]
- 84. Gomori G. A rapid one‐step trichrome stain. Am J Clin Pathol 1950; 20: 661–664. [DOI] [PubMed] [Google Scholar]
- 85. Engel WK, Cunningham GG. Rapid examination of muscle tissue. an improved trichrome method for fresh‐frozen biopsy sections. Neurology 1963; 13: 919–923. [DOI] [PubMed] [Google Scholar]
- 86. Egger J, Lake BD, Wilson J. Mitochondrial cytopathy. A multisystem disorder with ragged red fibres on muscle biopsy. Arch Dis Childhood 1981; 56: 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moraes CT, Ricci E, Bonilla E, DiMauro S, Schon EA. The mitochondrial tRNA(Leu(UUR)) mutation in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am J Hum Genet 1992; 50: 934–949. [PMC free article] [PubMed] [Google Scholar]
- 88. Petruzzella V, Moraes CT, Sano MC, Bonilla E, DiMauro S, Schon EA. Extremely high levels of mutant mtDNAs co‐localize with cytochrome c oxidase‐negative ragged‐red fibers in patients harboring a point mutation at nt 3243. Hum Mol Genet 1994; 3: 449–454. [DOI] [PubMed] [Google Scholar]
- 89. Rygiel KA, Tuppen HA, Grady JP, et al. Complex mitochondrial DNA rearrangements in individual cells from patients with sporadic inclusion body myositis. Nucleic Acids Res 2016; 44: 5313–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Vincent AE, Grady JP, Rocha MC, et al. Mitochondrial dysfunction in myofibrillar myopathy. Neuromusc Disord 2016; 26: 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Old SL, Johnson MA. Methods of microphotometric assay of succinate dehydrogenase and cytochrome c oxidase activities for use on human skeletal muscle. Histochem J 1989; 21: 545–555. [DOI] [PubMed] [Google Scholar]
- 92. Sciacco M, Bonilla E. Cytochemistry and immunocytochemistry of mitochondria in tissue sections. Methods Enzymol 1996; 264: 509–521. [DOI] [PubMed] [Google Scholar]
- 93. Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Distribution of wild‐type and common deletion forms of mtDNA in normal and respiration‐deficient muscle fibers from patients with mitochondrial myopathy. Hum Mol Genet 1994; 3: 13–19. [DOI] [PubMed] [Google Scholar]
- 94. Johnson MA, Turnbull DM, Dick DJ, Sherratt HS. A partial deficiency of cytochrome c oxidase in chronic progressive external ophthalmoplegia. J Neurol Sci 1983; 60: 31–53. [DOI] [PubMed] [Google Scholar]
- 95. Loeffen JL, Smeitink JA, Trijbels JM, et al. Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum Mut 2000; 15: 123–134. [DOI] [PubMed] [Google Scholar]
- 96. Rocha MC, Grady JP, Grunewald A, et al. A novel immunofluorescent assay to investigate oxidative phosphorylation deficiency in mitochondrial myopathy: understanding mechanisms and improving diagnosis. Sci Rep 2015; 5: 15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jackson MJ, Schaefer JA, Johnson MA, Morris AA, Turnbull DM, Bindoff LA. Presentation and clinical investigation of mitochondrial respiratory chain disease. A study of 51 patients. Brain 1995; 118(Pt 2): 339–357. [DOI] [PubMed] [Google Scholar]
- 98. Iizuka T, Sakai F, Suzuki N, et al. Neuronal hyperexcitability in stroke‐like episodes of MELAS syndrome. Neurology 2002; 59: 816–824. [DOI] [PubMed] [Google Scholar]
- 99. Canafoglia L, Franceschetti S, Antozzi C, et al. Epileptic phenotypes associated with mitochondrial disorders. Neurology 2001; 56: 1340–1346. [DOI] [PubMed] [Google Scholar]
- 100. Tanji K, Gamez J, Cervera C, et al. The A8344G mutation in mitochondrial DNA associated with stroke‐like episodes and gastrointestinal dysfunction. Acta Neuropathol 2003; 105: 69–75. [DOI] [PubMed] [Google Scholar]
- 101. Deschauer M, Tennant S, Rokicka A, et al. MELAS associated with mutations in the POLG1 gene. Neurology 2007; 68: 1741–1742. [DOI] [PubMed] [Google Scholar]
- 102. Tzoulis C, Tran GT, Coxhead J, et al. Molecular pathogenesis of polymerase gamma‐related neurodegeneration. Ann Neurol 2014; 76: 66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lax NZ, Grady J, Laude A, et al. Extensive respiratory chain defects in inhibitory interneurones in patients with mitochondrial disease. Neuropathol Appl Neurobiol 2016; 42: 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Betts J, Jaros E, Perry RH, et al. Molecular neuropathology of MELAS: level of heteroplasmy in individual neurones and evidence of extensive vascular involvement. Neuropathol Appl Neurobiol 2006; 32: 359–373. [DOI] [PubMed] [Google Scholar]
- 105. Koga Y, Akita Y, Junko N, et al. Endothelial dysfunction in MELAS improved by l‐arginine supplementation. Neurology 2006; 66: 1766–1769. [DOI] [PubMed] [Google Scholar]
- 106. Lax NZ, Hepplewhite PD, Reeve AK, et al. Cerebellar ataxia in patients with mitochondrial DNA disease: a molecular clinicopathological study. J Neuropathol Exp Neurol 2012; 71: 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chrysostomou A, Grady JP, Laude A, Taylor RW, Turnbull DM, Lax NZ. Investigating complex I deficiency in Purkinje cells and synapses in patients with mitochondrial disease. Neuropathol Appl Neurobiol 2016; 42: 477–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tanji K, DiMauro S, Bonilla E. Disconnection of cerebellar Purkinje cells in Kearns–Sayre syndrome. J Neurol Sci 1999; 166: 64–70. [DOI] [PubMed] [Google Scholar]
- 109. Mori O, Yamazaki M, Ohaki Y, et al. Mitochondrial encephalomyopathy with lactic acidosis and stroke like episodes (MELAS) with prominent degeneration of the intestinal wall and cactus‐like cerebellar pathology. Acta Neuropathol 2000; 100: 712–717. [DOI] [PubMed] [Google Scholar]
- 110. Fukutani Y, Nakamura I, Matsubara R, Kobayashi K, Isaki K. Pathology of the cerebellar dentate nucleus in sporadic olivopontocerebellar atrophy: a morphometric investigation. J Neurol Sci 1996; 137: 103–108. [DOI] [PubMed] [Google Scholar]
- 111. Oldfors A, Fyhr IM, Holme E, Larsson NG, Tulinius M. Neuropathology in Kearns–Sayre syndrome. Acta Neuropathol 1990; 80: 541–546. [DOI] [PubMed] [Google Scholar]
- 112. Lax NZ, Campbell GR, Reeve AK, et al. Loss of myelin‐associated glycoprotein in Kearns–Sayre syndrome. Arch Neurol 2012; 69: 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods 2013; 10: 508–513. [DOI] [PubMed] [Google Scholar]
- 114. Phillips J, Laude A, Lightowlers R, Morris CM, Turnbull DM, Lax NZ. Development of passive CLARITY and immunofluorescent labelling of multiple proteins in human cerebellum: understanding mechanisms of neurodegeneration in mitochondrial disease. Sci Rep 2016; 6: 26013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hatakeyama H, Goto YI. Heteroplasmic mitochondrial DNA mutations and mitochondrial diseases: toward iPSC‐based disease modeling, drug discovery, and regenerative therapeutics. Stem Cells 2016; 34: 801–808. [DOI] [PubMed] [Google Scholar]
- 116. Prigione A, Lichtner B, Kuhl H, et al. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell‐like metabolic reprogramming. Stem Cells 2011; 29: 1338–1348. [DOI] [PubMed] [Google Scholar]
- 117. Iommarini L, Peralta S, Torraco A, Diaz F. Mitochondrial diseases Part II: Mouse models of OXPHOS deficiencies caused by defects in regulatory factors and other components required for mitochondrial function. Mitochondrion 2015; 22: 96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Peralta S, Torraco A, Iommarini L, Diaz F. Mitochondrial diseases Part III: Therapeutic interventions in mouse models of OXPHOS deficiencies. Mitochondrion 2015; 23: 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Torraco A, Peralta S, Iommarini L, Diaz F. Mitochondrial diseases Part I: mouse models of OXPHOS deficiencies caused by defects in respiratory complex subunits or assembly factors. Mitochondrion 2015; 21: 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kirby DM, McFarland R, Ohtake A, et al. Mutations of the mitochondrial ND1 gene as a cause of MELAS. J Med Genet 2004; 41: 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ugalde C, Hinttala R, Timal S, et al. Mutated ND2 impairs mitochondrial complex I assembly and leads to Leigh syndrome. Mol Genet Metab 2007; 90: 10–14. [DOI] [PubMed] [Google Scholar]
- 122. Crimi M, Papadimitriou A, Galbiati S, et al. A new mitochondrial DNA mutation in ND3 gene causing severe Leigh syndrome with early lethality. Pediatr Res 2004; 55: 842–846. [DOI] [PubMed] [Google Scholar]
- 123. Singh G, Lott MT, Wallace DC. A mitochondrial DNA mutation as a cause of Leber's hereditary optic neuropathy. N Engl J Med 1989; 320: 1300–1305. [DOI] [PubMed] [Google Scholar]
- 124. Brown MD, Torroni A, Reckord CL, Wallace DC. Phylogenetic analysis of Leber's hereditary optic neuropathy mitochondrial DNA's indicates multiple independent occurrences of the common mutations. Hum Mut 1995; 6: 311–325. [DOI] [PubMed] [Google Scholar]
- 125. Santorelli FM, Tanji K, Kulikova R, et al. Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochem Biophysic Res Commun 1997; 238: 326–328. [DOI] [PubMed] [Google Scholar]
- 126. Jun AS, Brown MD, Wallace DC. A mitochondrial DNA mutation at nucleotide pair 14459 of the NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc Natl Acad Sci USA 1994; 91: 6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Dumoulin R, Sagnol I, Ferlin T, Bozon D, Stepien G, Mousson B. A novel gly290asp mitochondrial cytochrome b mutation linked to a complex III deficiency in progressive exercise intolerance. Mol Cell Probes 1996; 10: 389–391. [DOI] [PubMed] [Google Scholar]
- 128. Bruno C, Martinuzzi A, Tang Y, et al. A stop‐codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am J Hum Genet 1999; 65: 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wong LJ, Dai P, Tan D, et al. Severe lactic acidosis caused by a novel frame‐shift mutation in mitochondrial‐encoded cytochrome c oxidase subunit II. Am J Hum Genet 2001; 102: 95–99. [DOI] [PubMed] [Google Scholar]
- 130. Hanna MG, Nelson IP, Rahman S, et al. Cytochrome c oxidase deficiency associated with the first stop‐codon point mutation in human mtDNA. Am J Hum Genet 1998; 63: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Holt IJ, Harding AE, Petty RK, Morgan‐Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet 1990; 46: 428–433. [PMC free article] [PubMed] [Google Scholar]
- 132. Jonckheere AI, Hogeveen M, Nijtmans LG, et al. A novel mitochondrial ATP8 gene mutation in a patient with apical hypertrophic cardiomyopathy and neuropathy. J Hum Genet 2008; 45: 129–133. [DOI] [PubMed] [Google Scholar]
- 133. Benit P, Chretien D, Kadhom N, et al. Large‐scale deletion and point mutations of the nuclear NDUFV1 and NDUFS1 genes in mitochondrial complex I deficiency. Am J Hum Genet 2001; 68: 1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Loeffen J, Elpeleg O, Smeitink J, et al. Mutations in the complex I NDUFS2 gene of patients with cardiomyopathy and encephalomyopathy. Ann Neurol 2001; 49: 195–201. [DOI] [PubMed] [Google Scholar]
- 135. Benit P, Slama A, Cartault F, et al. Mutant NDUFS3 subunit of mitochondrial complex I causes Leigh syndrome. J Hum Genet 2004; 41: 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Budde SM, van den Heuvel LP, Janssen AJ, et al. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem Biophysic Res Commun 2000; 275: 63–68. [DOI] [PubMed] [Google Scholar]
- 137. Kirby DM, Salemi R, Sugiana C, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest 2004; 114: 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Triepels RH, van den Heuvel LP, Loeffen JL, et al. Leigh syndrome associated with a mutation in the NDUFS7 (PSST) nuclear encoded subunit of complex I. Ann Neurol 1999; 45: 787–790. [DOI] [PubMed] [Google Scholar]
- 139. Loeffen J, Smeitink J, Triepels R, et al. The first nuclear‐encoded complex I mutation in a patient with Leigh syndrome. Am J Hum Genet 1998; 63: 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Fernandez‐Moreira D, Ugalde C, Smeets R, et al. X‐linked NDUFA1 gene mutations associated with mitochondrial encephalomyopathy. Ann Neurol 2007; 61: 73–83. [DOI] [PubMed] [Google Scholar]
- 141. Hoefs SJ, Dieteren CE, Distelmaier F, et al. NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet 2008; 82: 1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. van den Bosch BJ, Gerards M, Sluiter W, et al. Defective NDUFA9 as a novel cause of neonatally fatal complex I disease. J Med Genet 2012; 49: 10–15. [DOI] [PubMed] [Google Scholar]
- 143. Hoefs SJ, van Spronsen FJ, Lenssen EW, et al. NDUFA10 mutations cause complex I deficiency in a patient with Leigh disease. Eur J Hum Genet 2011; 19: 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ostergaard E, Rodenburg RJ, van den Brand M, et al. Respiratory chain complex I deficiency due to NDUFA12 mutations as a new cause of Leigh syndrome. J Med Genet 2011; 48: 737–740. [DOI] [PubMed] [Google Scholar]
- 145. Angebault C, Charif M, Guegen N, et al. Mutation in NDUFA13/GRIM19 leads to early onset hypotonia, dyskinesia and sensorial deficiencies, and mitochondrial complex I instability. Hum Mol Genet 2015; 24: 3948–3955. [DOI] [PubMed] [Google Scholar]
- 146. Schuelke M, Smeitink J, Mariman E, et al. Mutant NDUFV1 subunit of mitochondrial complex I causes leukodystrophy and myoclonic epilepsy. Nat Genet 1999; 21: 260–261. [DOI] [PubMed] [Google Scholar]
- 147. Benit P, Beugnot R, Chretien D, et al. Mutant NDUFV2 subunit of mitochondrial complex I causes early onset hypertrophic cardiomyopathy and encephalopathy. Hum Mut 2003; 21: 582–586. [DOI] [PubMed] [Google Scholar]
- 148. Calvo SE, Compton AG, Hershman SG, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next‐generation sequencing. Sci Transl Med 2012; 4: 118ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Haack TB, Madignier F, Herzer M, et al. Mutation screening of 75 candidate genes in 152 complex I deficiency cases identifies pathogenic variants in 16 genes including NDUFB9 . J Med Genet 2012; 49: 83–89. [DOI] [PubMed] [Google Scholar]
- 150. van Rahden VA, Fernandez‐Vizarra E, Alawi M, et al. Mutations in NDUFB11, encoding a complex I component of the mitochondrial respiratory chain, cause microphthalmia with linear skin defects syndrome. Am J Hum Genet 2015; 96: 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Alston CL, Davison JE, Meloni F, et al. Recessive germline SDHA and SDHB mutations causing leukodystrophy and isolated mitochondrial complex II deficiency. J Med Genet 2012; 49: 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Jackson CB, Nuoffer JM, Hahn D, et al. Mutations in SDHD lead to autosomal recessive encephalomyopathy and isolated mitochondrial complex II deficiency. J Med Genet 2014; 51: 170–175. [DOI] [PubMed] [Google Scholar]
- 153. Haut S, Brivet M, Touati G, et al. A deletion in the human QP‐C gene causes a complex III deficiency resulting in hypoglycaemia and lactic acidosis. Hum Genet 2003; 113: 118–122. [DOI] [PubMed] [Google Scholar]
- 154. Miyake N, Yano S, Sakai C, et al. Mitochondrial complex III deficiency caused by a homozygous UQCRC2 mutation presenting with neonatal‐onset recurrent metabolic decompensation. Hum Mut 2013; 34: 446–452. [DOI] [PubMed] [Google Scholar]
- 155. Barel O, Shorer Z, Flusser H, et al. Mitochondrial complex III deficiency associated with a homozygous mutation in UQCRQ . Am J Hum Genet 2008; 82: 1211–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Gaignard P, Menezes M, Schiff M, et al. Mutations in CYC1, encoding cytochrome c1 subunit of respiratory chain complex III, cause insulin‐responsive hyperglycemia. Am J Hum Genet 2013; 93: 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Shteyer E, Saada A, Shaag A, et al. Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am J Hum Genet 2009; 84: 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Massa V, Fernandez‐Vizarra E, Alshahwan S, et al. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus‐encoded subunit of cytochrome c oxidase. Am J Hum Genet 2008; 82: 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Indrieri A, van Rahden VA, Tiranti V, et al. Mutations in COX7B cause microphthalmia with linear skin lesions, an unconventional mitochondrial disease. Am J Hum Genet 2012; 91: 942–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Hallmann K, Kudin AP, Zsurka G, et al. Loss of the smallest subunit of cytochrome c oxidase, COX8A, causes Leigh‐like syndrome and epilepsy. Brain 2016; 139: 338–345. [DOI] [PubMed] [Google Scholar]
- 161. Mayr JA, Havlickova V, Zimmermann F, et al. Mitochondrial ATP synthase deficiency due to a mutation in the ATP5E gene for the F1 epsilon subunit. Hum Mol Genet 2010; 19: 3430–3439. [DOI] [PubMed] [Google Scholar]
- 162. Dunning CJ, McKenzie M, Sugiana C, et al. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J 2007; 26: 3227–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest 2005; 115: 2784–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Saada A, Vogel RO, Hoefs SJ, et al. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)‐interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am J Hum Genet 2009; 84: 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Saada A, Edvardson S, Rapoport M, et al. C6ORF66 is an assembly factor of mitochondrial complex I. Am J Hum Genet 2008; 82: 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sugiana C, Pagliarini DJ, McKenzie M, et al. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am J Hum Genet 2008; 83: 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008; 134: 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Calvo SE, Tucker EJ, Compton AG, et al. High‐throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet 2010; 42: 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Invernizzi F, Tigano M, Dallabona C, et al. A homozygous mutation in LYRM7/MZM1L associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex III activity. Hum Mut 2013; 34: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Tucker EJ, Wanschers BF, Szklarczyk R, et al. Mutations in the UQCC1‐interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet 2013; 9: e1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Wanschers BF, Szklarczyk R, van den Brand MA, et al. A mutation in the human CBP4 ortholog UQCC3 impairs complex III assembly, activity and cytochrome b stability. Hum Mol Genet 2014; 23: 6356–6365. [DOI] [PubMed] [Google Scholar]
- 172. Ghezzi D, Arzuffi P, Zordan M, et al. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat Genet 2011; 43: 259–263. [DOI] [PubMed] [Google Scholar]
- 173. Ostergaard E, Weraarpachai W, Ravn K, et al. Mutations in COA3 cause isolated complex IV deficiency associated with neuropathy, exercise intolerance, obesity, and short stature. J Med Genet 2015; 52: 203–207. [DOI] [PubMed] [Google Scholar]
- 174. Huigsloot M, Nijtmans LG, Szklarczyk R, et al. A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am J Hum Genet 2011; 88: 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Ghosh A, Trivedi PP, Timbalia SA, et al. Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum Mol Genet 2014; 23: 3596–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Valnot I, von Kleist‐Retzow JC, Barrientos A, et al. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum Mol Genet 2000; 9: 1245–1249. [DOI] [PubMed] [Google Scholar]
- 177. Weraarpachai W, Sasarman F, Nishimura T, et al. Mutations in C12orf62, a factor that couples COX I synthesis with cytochrome c oxidase assembly, cause fatal neonatal lactic acidosis. Am J Hum Genet 2012; 90: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Antonicka H, Mattman A, Carlson CG, et al. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early‐onset fatal hypertrophic cardiomyopathy. Am J Hum Genet 2003; 72: 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Szklarczyk R, Wanschers BF, Nijtmans LG, et al. A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum Mol Genet 2013; 22: 656–667. [DOI] [PubMed] [Google Scholar]
- 180. Valnot I, Osmond S, Gigarel N, et al. Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal‐onset hepatic failure and encephalopathy. Am J Hum Genet 2000; 67: 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Papadopoulou LC, Sue CM, Davidson MM, et al. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet 1999; 23: 333–337. [DOI] [PubMed] [Google Scholar]
- 182. Zhu Z, Yao J, Johns T, et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 1998; 20: 337–343. [DOI] [PubMed] [Google Scholar]
- 183. Mootha VK, Lepage P, Miller K, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA 2003; 100: 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Lim SC, Smith KR, Stroud DA, et al. A founder mutation in PET100 causes isolated complex IV deficiency in Lebanese individuals with Leigh syndrome. Am J Hum Genet 2014; 94: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. van Bon BW, Oortveld MA, Nijtmans LG, et al. CEP89 is required for mitochondrial metabolism and neuronal function in man and fly. Hum Mol Genet 2013; 22: 3138–3151. [DOI] [PubMed] [Google Scholar]
- 186. Weraarpachai W, Antonicka H, Sasarman F, et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late‐onset Leigh syndrome. Nat Genet 2009; 41: 833–837. [DOI] [PubMed] [Google Scholar]
- 187. Melchionda L, Haack TB, Hardy S, et al. Mutations in APOPT1, encoding a mitochondrial protein, cause cavitating leukoencephalopathy with cytochrome c oxidase deficiency. Am J Hum Genet 2014; 95: 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Ghezzi D, Saada A, D'Adamo P, et al. FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am J Hum Genet 2008; 83: 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. De Meirleir L, Seneca S, Lissens W, et al. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12 . J Med Genet 2004; 41: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]


