Abstract
Background & Aims
The occurrence of drug‐induced liver injury (DILI) is a major issue in all phases of drug development. To identify novel biomarker candidates associated with DILI, we utilised an affinity proteomics strategy, where antibody suspension bead arrays were applied to profile plasma and serum samples from human DILI cases and controls.
Methods
An initial screening was performed using 4594 randomly selected antibodies, representing 3450 human proteins. Resulting candidate proteins together with proposed DILI biomarker candidates generated a DILI array of 251 proteins for subsequent target analysis and verifications. In total, 1196 samples from 241 individuals across four independent cohorts were profiled: healthy volunteers receiving acetaminophen, patients with human immunodeficiency virus and/or tuberculosis receiving treatment, DILI cases originating from a wide spectrum of drugs, and healthy volunteers receiving heparins.
Results
We observed elevated levels of cadherin 5, type 2 (CDH5) and fatty acid‐binding protein 1 (FABP1) in DILI cases. In the two longitudinal cohorts, CDH5 was elevated already at baseline. FABP1 was elevated after treatment initiation and seemed to respond more rapidly than alanine aminotransferase (ALT). The elevations were verified in the DILI cases treated with various drugs. In the heparin cohort, CDH5 was stable over time whereas FABP1 was elevated.
Conclusions
These results suggest that CDH5 may have value as a susceptibility marker for DILI. FABP1 was identified as a biomarker candidate with superior characteristics regarding tissue distribution and kinetics compared to ALT but likely with limited predictive value for the development of severe DILI. Further studies are needed to determine the clinical utility of the proposed markers.
Keywords: drug‐induced liver injury, biomarker discovery, affinity proteomics, plasma profiling, suspension bead arrays
Abbreviations
- ALP
alkaline phosphatase
- ALT
alanine aminotransferase
- APAP
acetaminophen
- AST
aspartate aminotransferase
- AUC
area under the curve
- CDH5
cadherin 5, type 2
- DILI
drug‐induced liver injury
- FABP1
fatty acid‐binding protein 1
- HIV
human immunodeficiency virus
- HV
healthy volunteers
- MFI
median fluorescence intensity
- TB
tuberculosis
Key points.
An affinity proteomics screening using 4594 antibodies was performed followed by a targeted array for profiling of 241 individuals (1196 samples) from four independent cohorts with drug‐induced liver injuries (DILI).
CDH5 (cadherin 5, type 2) was found to be elevated in DILI cases and already in baseline samples of the longitudinal cohorts.
FABP1 (fatty acid‐binding protein 1) was found to be elevated in DILI cases and showed a treatment‐dependent profile in the longitudinal cohorts.
We propose CDH5 as a potential susceptibility marker and FABP1 as a potential marker with superior tissue distribution and kinetics compared to ALT.
Drug‐induced liver injury (DILI) is the single leading cause for termination of drug development and safety‐related withdrawal of approved drugs from the market 1. In clinical practice, it accounts for more than 50% of liver failure cases and represents a major safety issue for patients 2. In some patients, DILI can cause severe injury leading to acute liver failure that can be life‐threatening and require liver transplantation 3, 4. The estimated incidence of DILI is about 14 cases per 100 000 inhabitants 5. Because of the low incidence of DILI, there are often too few individuals in a clinical trial to detect the adverse effects 2.
The biomarkers commonly used to detect liver injury are serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP) and total bilirubin levels. Serum elevations of ALT and AST indicate hepatocellular damage and increased ALP is related to hepatobiliary injury. Those markers are useful for the diagnosis of severe DILI, but lack the ability to differentiate between severe and mild cases at the onset of DILI and predict if a patient will adapt or develop severe injury of the liver. Mild elevations of liver enzymes are commonly observed and may be because of different factors, such as diet, exercise or fatty liver. Furthermore, mild elevations of ALT can be observed after treatment with drugs that do not have the potential to cause severe DILI and in many cases those elevations are of transient nature and will resolve despite of continued treatment 6. There are no adequate predictive methods or reliable biomarkers for DILI and the diagnosis is a matter of excluding other conditions, such as viral hepatitis, and finding a causative connection between drug exposure and potential drug‐related injury 3, 4. There is a clear need for more specific markers for liver injury compared with current standard tests and markers that predict susceptibility to DILI of individual patients.
In the present study, we aimed at the discovery of novel biomarkers for DILI by the application of affinity proteomics to human DILI cohorts based on a variety of drug treatments. Affinity proteomics uses capture reagents such as antibodies for comparative proteomic characterization of samples and has proven successful in various previous studies within e.g. malaria and multiple sclerosis 7, 8. Antibodies with relevance for liver toxicity were used in exploratory multiplexed assays and biomarker candidates were identified based on DILI cases from a healthy volunteer (HV) cohort treated with acetaminophen (APAP) and a cohort with patients treated for human immunodeficiency virus (HIV) and/or tuberculosis (TB). The results were subsequently verified using DILI cases from a variety of drug treatments collected by the SAFE‐T consortium, a public−private partnership under the framework of the EU Innovative Medicines Initiative 9 that focuses on the clinical qualification of biomarkers for drug‐induced injuries to the liver, kidney and vascular system. Furthermore, the findings were compared with asymptomatic liver enzyme elevations induced by heparins that have not been associated with clinical significant liver injury.
See supplementary information for Materials and methods.
Results
To enable a broad‐scale screening and protein profiling for the potential identification of novel biomarker candidates associated with DILI, we here set out an explorative affinity proteomics strategy. An initial non‐targeted discovery screening that encompassed 4594 antibodies targeting 3450 unique proteins, representing approximately 17% of the human gene products, was combined with targeted profiling using a carefully designed DILI array. The screening and verification strategy was based on the analysis of 1196 plasma and serum samples from 241 individuals divided across four independent cohorts (HV APAP, HIV/TB, SAFE‐T DILI and HV Heparin, see Supplementary demographics tables and Table S2) in combination with an antibody suspension bead array approach (Fig. 1).
Figure 1.
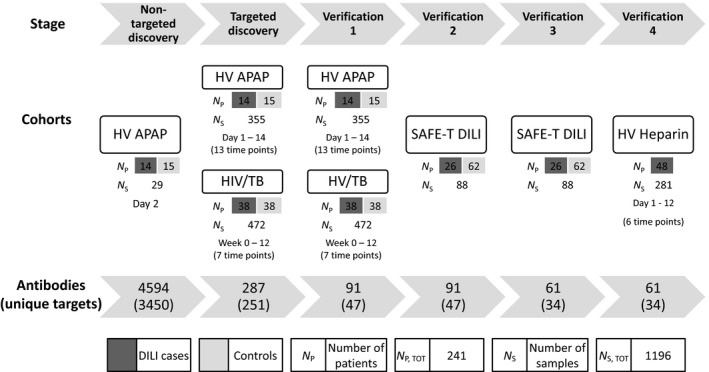
Study design. Overview of the study design with information regarding the various cohorts (number of samples and collection time points) and the antibody arrays (number of antibodies and unique protein targets) used in the different stages.
Design of the DILI antibody array
In the non‐targeted discovery stage, a screening array of 4594 antibodies and a subset of the HV APAP cohort (day 2, baseline) were used. The baseline samples served to identify potential DILI susceptibility markers that could be included in the DILI array. This resulted in the selection of 113 antibodies. These were in combination with a selection of antibodies from previous in‐house protein profiling efforts and thorough literature mining, pathway analysis, gene and protein expression data and information on liver‐enriched expression of proteins from the Human Protein Atlas project 10 used to generate the targeted DILI array of 287 antibodies representing 251 proteins (Table S1).
Experimental profiling setup
In the targeted discovery stage, the DILI array was used to profile two independent cohorts of longitudinal design, the complete HV APAP cohort (355 samples, 14 cases, 15 controls, 13 time points) and the HIV/TB cohort (472 samples, 38 cases and 38 controls, 7 time points). In a first verification, the same cohorts were again profiled but with a smaller set of antibodies defined by the results from the targeted discovery stage and incorporating additional antibodies per protein, resulting in a set of 91 antibodies targeting 47 proteins (Table S3). The SAFE‐T DILI cohort (88 samples, 26 cases, 62 controls) was included in Verification 2, to replicate the initial findings. In Verification 3, an even smaller set of 34 selected targets represented by 61 antibodies and more antibodies per protein target was used. Finally, the HV Heparin cohort (281 samples, 48 cases, 6 time points), was profiled in Verification 4 and repeated.
Elevated levels of CDH5 and FABP1
Two proteins, CDH5 (cadherin 5, type 2) and FABP1 (fatty acid‐binding protein 1), were identified to be elevated in DILI cases. The longitudinal dependence of these markers was first investigated by profiling serum and plasma from the HV APAP and HIV/TB cohort, covering a time frame of 14 days and 12 weeks respectively. In HV APAP, CDH5 showed elevated median levels in DILI cases already in the pre‐dose/baseline samples (day 1–3). Although only significant at day 11, the trend of increased levels was sustained across the studied time frame including time points of dosing as well as during washout with the exception of day 6. The same trend with baseline‐elevated levels was observed in the HIV/TB cohort, with significant differences for week 2 and 4 as well as for week 8 and 12 (Fig. 2b). ALT and FABP1 displayed a treatment‐dependent appearance with increasing levels in DILI cases after initiation of the dosing in both the HV APAP and the HIV/TB cohort (Fig. 2a+c). In HV APAP, a significant elevation in FABP1 was observed one day prior to ALT (day 8 compared to day 9). The elevation for both remained significant during the continued dosing as well as after treatment had stopped. Interestingly, towards the end of the study period, FABP1 levels started to decrease (compare day 9 with day 14) whereas ALT remained unchanged. For the HIV/TB cohort, a significantly higher level of FABP1 was found for week 6 and onwards. The elevated levels were confirmed in the SAFE‐T DILI cohort with significant differences of 10−5 and 10−10 for CDH5 and FABP1 respectively (Fig. 2b,c). Furthermore, profiling of the Heparin cohort showed an elevation in FABP1 levels across the studied time frame whereas CDH5 levels were relatively stable (Fig. 3b,c).
Figure 2.
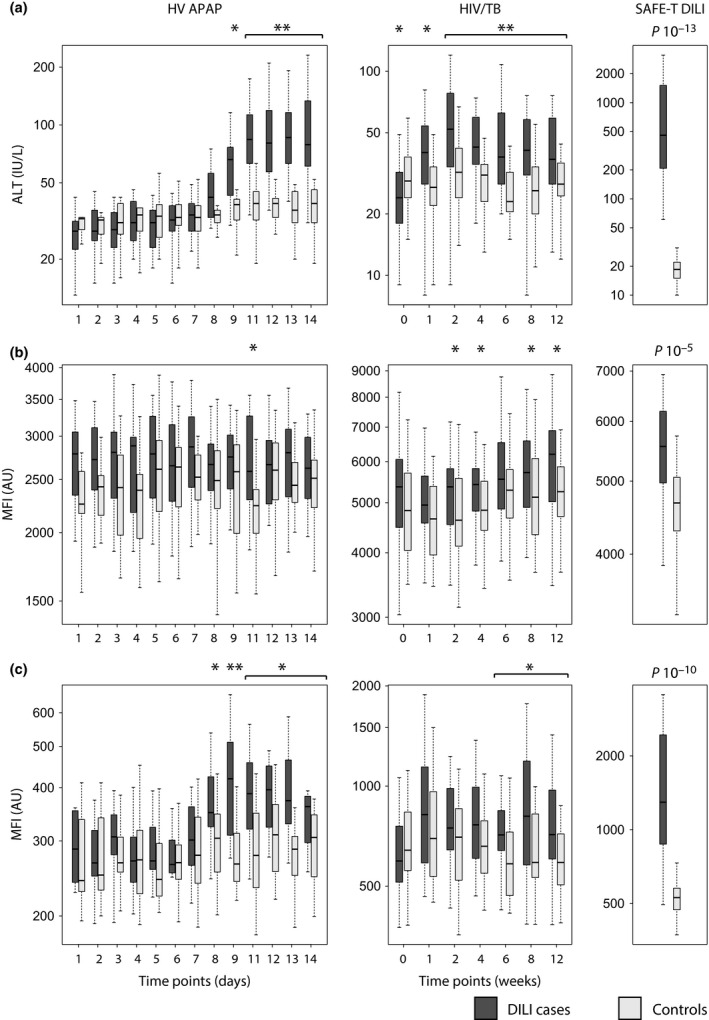
Protein profiles in the HV APAP, HIV/TB and SAFE‐T DILI cohorts. (a) ALT, (b) CDH5 and (c) FABP1 in DILI cases and controls of three independent cohorts measured with a standard clinical test (enzymatic activity assay) or with the antibody suspension bead array assay. In HV APAP, acetaminophen was administered between day 4 and day 11. For patients in HIV/TB, the treatment started at week 1 and continued across the analysed time frame. Antibodies targeting CDH5 and FABP1 detected elevated levels of the proteins in DILI cases in all the three cohorts. * P < 0.05, ** P < 0.01 based on Wilcoxon's rank sum tests.
Figure 3.
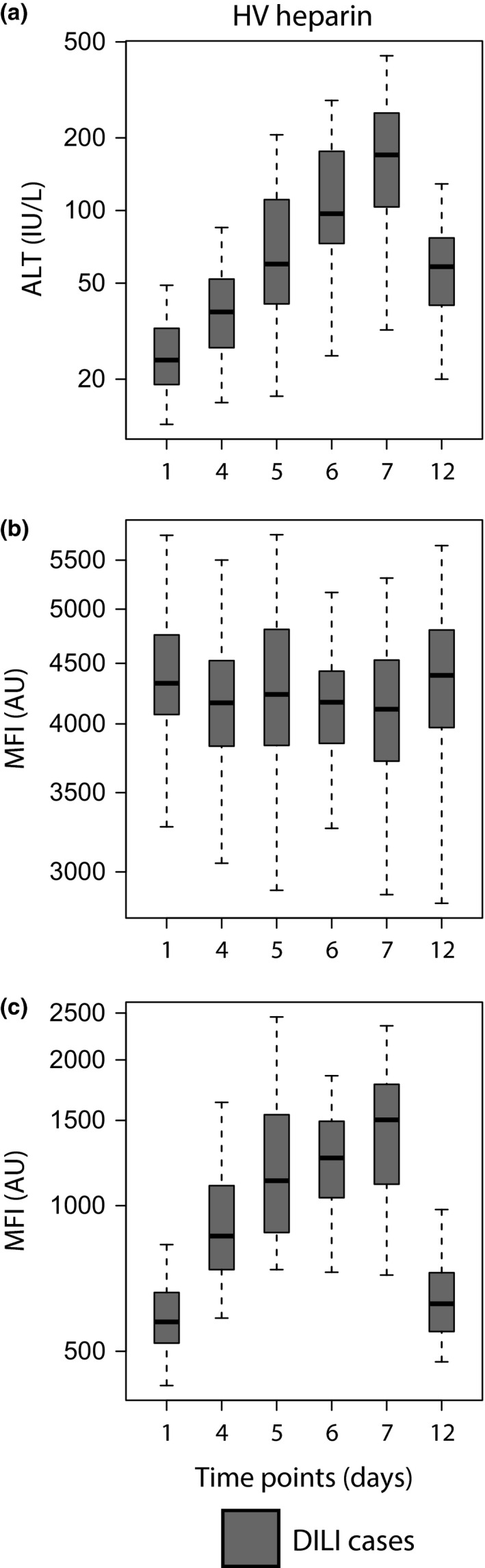
Protein profiles in the HV Heparin cohort. (a) ALT, (b) CDH5 and (c) FABP1in DILI cases of the HV Heparin cohort. The profiles for CDH5 were relatively constant across the studied time points, whereas profiles for FABP1 showed a time‐dependent elevation comparable to that of ALT. Heparin was administered from day 1 (baseline) to day 5 (and serum samples collected before dosing).
As shown in Table S2, the HIV/TB cohort was balanced in age and gender. The HV APAP had too small sample size to perform stratified analysis. When the SAFE‐T cohort was stratified by gender or age, same trends of fold changes between cases and controls were observed for both markers although not significant in all age groups (Fig. S1 and S2).
To assess the diagnostic power of CDH5 and FABP1, receiver operating characteristic analysis was performed on the SAFE‐T DILI cohort, yielding an area under the curve (AUC) of 0.74 and 0.94 respectively. ALT, which was used for classification of DILI, gave an expected AUC of 1 in this sample material. Combining CDH5 and FABP1 in the analysis did not further improve the AUC.
Target verification and distribution
To demonstrate the reproducibility in measuring CDH5 and FABP1, the average Pearson correlation taken over the different cohorts was determined as 0.85 and 0.95 respectively. For correlation plots between repeated experiments, see Figure S3.
Antibody targeting with HPA030562 and HPA028275 for CDH5 and FABP1, respectively, was confirmed with Western blot of serum samples from the SAFE‐T DILI cohort, a recombinant CDH5 standard and FABP1 overexpression lysate. Bands at the predicted molecular mass were detected (Fig. S4). Further evidence of on‐target detection was given by the comparison to bead‐based sandwich immunoassays (Fig. S5).
The presence, distribution and abundance of CDH5 and FABP1 in relation to protein and RNA expression were investigated in healthy human tissue. Immunohistochemical analysis of liver, kidney, heart muscle and skeletal muscle tissue using anti‐FABP1 antibodies showed strong protein expression in the hepatocytes but no staining of the bile duct cells. In kidney, FABP1 expression was present in cells of the tubules but not in cells of the glomeruli. No staining was detected in myocytes of either heart or skeletal muscle tissue (Fig. 4). On a more global level, FABP1 showed protein expression in seven of 83 analysed healthy tissue cell types. In addition to high and medium expression levels in liver and kidney, medium levels were also seen for the digestive tract (duodenum, small intestine, appendix, colon and rectum). Staining using anti‐CDH5 antibodies revealed protein expression in all four tissues (Fig. 4). More globally, protein expression of medium to high levels in 54 of 81 normal tissue cell types was observed. Transcriptome analysis using RNA sequencing showed highest expression of FABP1 in liver tissue followed by duodenum, small intestine and colon. CDH5 was found in all tissues except the bone marrow, with placenta, lung and adipose tissue being the highest ranked (Fig. S6).
Figure 4.
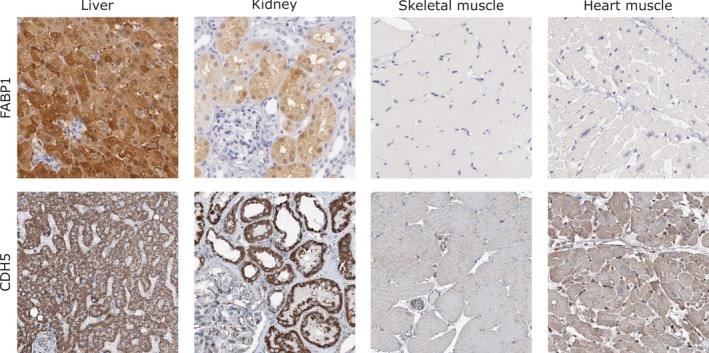
Immunohistochemical staining. Immunohistochemistry analysis using anti‐FABP1 and anti‐CDH5 antibodies showed cytoplasmic expression patterns in liver and kidney. Skeletal and heart muscle showed negative or weak protein expression in myocytes for both candidates. CDH5 expression was observed in blood vessels in all tissues included.
Discussion
In the current study, affinity proteomics was applied to profile plasma and serum samples from human DILI cases and controls. We identified two potential candidate markers for DILI. They each showed different characteristics that may be useful in prediction or diagnosis.
Our screening and verification strategy was based on samples from 241 individuals divided across four independent cohorts, comprising in total 1196 samples. Firstly, a cohort of healthy volunteers receiving acetaminophen (HV APAP cohort) and a cohort of patients with HIV and/or TB receiving treatment were profiled (HIV/TB cohort). Both of these cohorts included repeated measurements of each individual over time covering the time before and after treatment initiation, allowing assessment of the longitudinal dependence of biomarkers and providing potential to find markers that can predict, diagnose and monitor DILI. Secondly, a cohort where DILI was confirmed at the time of sampling (SAFE‐T DILI cohort) was used to assess a first diagnostic power of biomarkers found in the screening. Finally, potential markers were investigated in a cohort of healthy volunteers receiving heparins (HV Heparin cohort) for further evaluation in a setting with asymptomatic and transient ALT elevations. In the end, two proteins were identified to show elevated levels in the DILI cases.
CDH5 encodes a calcium‐dependent cell adhesion protein, cadherin‐5 (also called VE‐cadherin), that is specific to endothelial cells and a major component of endothelial adherens junctions. It plays a key role in endothelial cell biology and is expressed in blood vessels throughout the body. Through both its adhesive and signalling properties, CDH5 maintains the balance between intercellular junction plasticity and integrity, a requirement for endothelial cells to maintain proper barrier function of blood vessels while still being capable of responding to inflammatory and growth factor signalling 11. Endothelial dysfunction is a hallmark of many pathological and disease states, including inflammation, atherosclerosis, hypertension, diabetes and tumour metastasis. Elevated levels of soluble CDH5 have been reported to be associated with diabetic retinopathy and coronary atherosclerosis 12, 13. However, the mechanisms involved in this process were not studied. Furthermore, increased levels of soluble CDH5 were shown to be associated with poor outcome in severe sepsis 14. In rheumatoid arthritis patients, soluble CDH5 correlated with disease activity 15. The study also showed that tumour necrosis factor (TNF) alpha induced time‐dependent shedding of CDH5 into cell media of human umbilical vein endothelial cell cultures. In addition to TNF alpha, numerous other endogenous mediators of inflammation have been shown to increase microvascular permeability by targeting CDH5 16. The cleavage of the CDH5 extracellular domain by proteinases is one of the molecular mechanisms involved in neutrophil progression across the endothelial junctions during inflammatory processes and endothelial apoptosis 17. Based on our finding of elevated levels of soluble CDH5 in blood from DILI cases compared to controls prior to treatment with different drugs, we suggest that the protein may have use as a potential susceptibility marker for DILI that reflects an ongoing inflammation. It remains to be proven that the measurement of CDH5 in parallel with conventional liver injury markers improves the identification of individuals at risk to develop DILI prior to drug treatment.
Fatty acid‐binding proteins belong to a family of intracellular lipid‐binding proteins that are involved in uptake, transport and metabolism of fatty acids. FABP1 encodes a fatty acid‐binding protein found in hepatocytes of the liver, but in small quantities, also in kidney (proximal tubules) and small intestine as confirmed by the immunohistochemistry data using Human Protein Atlas antibodies. Tissue content of FABP1 in liver tissue is 2.7 mg/g and the content in human intestine ranges from 26 to 198 μg/g 18. In previous studies, urinary FABP1 levels were better suited to allow the accurate and earlier detection of both histopathological and functional kidney injuries compared with conventional renal markers 19, 20. It was also proposed as a predictive serum marker for recovery of graft function after kidney transplantations 21. In context of the liver, FABP1 has been proposed as a sensitive serum marker of acute hepatocellular damage in liver transplant recipients 22 and a diagnostic marker to detect liver injury because of chronic Hepatitis C infection 23. In a controlled trial, FABP1 was used in the place of aminotransferases as a more sensitive alternative to detect small differences in hepatocellular injury that can be induced during liver surgery. When hepatocytes are damaged, FABP1 is released to the circulation 24. Some factors that will affect the plasma concentration of a biomarker for liver injury are the tendency for the biomarker to be released during liver injury, its half‐life and stability in plasma and the relative abundance in the liver compared with other organs 25. FABP1 makes up 1–5% of the cytosolic protein content in hepatocytes 26 and with a molecular mass of 14 kDa it has a plasma half‐life of 11 min before cleared by the kidneys 27. This can be compared to ALT, which is a relatively large molecule (96 kDa) with tissue expression in liver, kidney, pancreas, heart and skeletal muscle 28 and a half‐life of 40–60 h 29. Although FABP1 exhibits extrahepatic expression, one major advantage compared to ALT as a biomarker for DILI is the lack of expression in cardiac and skeletal muscle tissue (Fig. 4). FABP1 could therefore serve as a complement to ALT and add additional information when muscle injury cannot be excluded as a cause for increased ALT. The low molecular mass, high relative abundance in the liver and the low serum baseline levels together with a short half‐life in the circulation make FABP1 a promising new biomarker for an ongoing liver injury. However, in the HV Heparin study, FABP1 elevations correlated very well with ALT serum levels. Therefore, a use of FABP1 as a prognostic biomarker that distinguishes between patients with transient ALT elevations that adapt to the treatment and those that progress to severe DILI cannot be proposed.
This study has its main limitation in the difficulty to access DILI samples. DILI is a rare clinical event. A number of international collaborations, such as the Spanish DILI Registry or the Drug‐induced Liver Injury Network, have been created to establish large patient databases as well as a standardized case definition of DILI. Still, the access to well‐characterized clinical samples from DILI cases is the main limiting factor for the discovery and qualification of new DILI biomarkers. Furthermore, the SAFE‐T DILI cohort was based on a large heterogeneity of causative drugs collected during the DILI episode without available baseline samples. Relatively small liver enzyme elevations were observed in the HV APAP cohort. Here, subjects received the therapeutic dose of 4 g APAP per day and responders showed transient elevations lower than in cases of APAP overdose. The dosing was also discontinued if ALT or AST reached >3× the upper limit of normal. The HIV/TB cohort also showed small liver enzyme elevations. Direct comparison of the extent of an increase in ALT between the healthy and patient cohorts may not be applicable. A slight increase in ALT and AST level is more damaging or fatal in sick immunocompromised HIV and TB patients. In most cases, this rise is accompanied by clinical symptoms of liver toxicity and consequently treatment is discontinued before ALT reaches the levels observed in the HV APAP or SAFE‐T cohort. However, for both the HV APAP and HIV/TB cohorts the observed time course of DILI was valuable. Other limitations of the study are an imbalance in age and gender distributions as well as the slightly varying DILI case definition used for the different cohorts, as the standardization in case definition has evolved during the course of the study.
This exploratory study aimed to identify potential protein markers that could have use as DILI biomarkers irrespective of the drug inducing the injury. The antibody suspension bead array assay in the employed format is a method for relative quantification. The format is convenient in the discovery stages of biomarker research, generating multiplex data for hundreds of proteins in hundreds of samples in one single assay. Although it could be of interest to reveal potential drug‐related differences in the proteins across the four cohorts or within cohorts, the division of subjects into subgroups of agents would generate groups too small to be able to draw any conclusion. The underlying disease would also need to be considered leading to further subgroups. In addition, this would demand that the same DILI criteria were used for all cohorts and would require absolute quantitative data. For the same reasons, comparisons between cohorts were not possible.
In summary, this biomarker discovery study has been based on affinity proteomics using human samples from both healthy volunteers and patients with DILI caused by a wide spectrum of therapeutical agents. We decided here to utilise a very stringent approach by only focusing on those proteins that were differentiated in all four cohorts. Two biomarker candidates were thereby identified as significantly elevated across the different DILI studies. CDH5 may potentially have use as a susceptibility marker for DILI. FABP1 showed superior characteristics regarding tissue distribution and kinetics compared to ALT but limited predictive value for the development of severe DILI. We propose CDH5 and FABP1 as complementary markers to ALT. Both biomarker candidates have been taken up in the clinical biomarker qualification performed by the SAFE‐T consortium. Clinically applicable ELISA assays have been validated according to the SAFE‐T biomarker qualification strategy for large‐scale analysis of clinical samples. Further analysis of CDH5 and FABP1 in comparison to and in combination with current standard assays in samples from large cohorts of healthy volunteers, patients with drug‐ and none drug‐induced liver injury and with common disorders is currently ongoing to establish the performance characteristics and potential use for CDH5 and FABP1 as biomarkers. Finally, the safety biomarker community together with regulators will need to determine the utility of these potential novel DILI biomarkers in drug development and clinical practice.
Supporting information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1111/liv.13174/suppinfo
Acknowledgements
Members of the SAFE‐T DILI work package are acknowledged as follows: M Merz, G A Kullak‐Ublick, S Ormarsdottir, Joachim Tillner and Frances Hackman.
Financial support: This work has received support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking (SAFE‐T, grant no 115003), ProNova VINN Excellence Centre for Protein Technology and the KTH Center for Applied Proteomics funded by the Erling‐Persson Family Foundation.
Conflict of interest: Commercial affiliations as following: JR and IF ‐ Momenta Pharmaceuticals, ISK ‐ AstraZeneca.
Liver Int. 2017; 37: 132–140. DOI: 10.1111/liv.13174
Handling Editor: Carmen Berasain
References
- 1. Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov 2005; 4: 489–99. [DOI] [PubMed] [Google Scholar]
- 2. Lee WM. Drug‐induced hepatotoxicity. New Engl J Med 2003; 349: 474–85. [DOI] [PubMed] [Google Scholar]
- 3. Aithal GP, Watkins PB, Andrade RJ, et al Case definition and phenotype standardization in drug‐induced liver injury. Clin Pharmacol Ther 2011; 89: 806–15. [DOI] [PubMed] [Google Scholar]
- 4. Navarro VJ, Senior JR. Drug‐related hepatotoxicity. New Engl J Med 2006; 354: 731–9. [DOI] [PubMed] [Google Scholar]
- 5. Sgro C, Clinard F, Ouazir K, et al Incidence of drug‐induced hepatic injuries: a French population‐based study. Hepatology 2002; 36: 451–5. [DOI] [PubMed] [Google Scholar]
- 6. Watkins PB, Seligman PJ, Pears JS, Avigan MI, Senior JR. Using controlled clinical trials to learn more about acute drug‐induced liver injury. Hepatology 2008; 48: 1680–9. [DOI] [PubMed] [Google Scholar]
- 7. Bachmann J, Burte F, Pramana S, et al Affinity proteomics reveals elevated muscle proteins in plasma of children with cerebral malaria. PLoS Pathog 2014; 10: e1004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bystrom S, Ayoglu B, Haggmark A, et al Affinity proteomic profiling of plasma, cerebrospinal fluid, and brain tissue within multiple sclerosis. J Proteome Res 2014; 13: 4607–19. [DOI] [PubMed] [Google Scholar]
- 9. Matheis K, Laurie D, Andriamandroso C, et al A generic operational strategy to qualify translational safety biomarkers. Drug Discov Today 2011; 16: 600–8. [DOI] [PubMed] [Google Scholar]
- 10. Uhlen M, Fagerberg L, Hallstrom BM, et al Proteomics. Tissue‐based map of the human proteome. Science 2015; 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 11. Harris ES, Nelson WJ. VE‐cadherin: at the front, center, and sides of endothelial cell organization and function. Curr Opin Cell Biol 2010; 22: 651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Navaratna D, Mcguire PG, Menicucci G, Das A. Proteolytic degradation of VE‐cadherin alters the blood‐retinal barrier in diabetes. Diabetes 2007; 56: 2380–7. [DOI] [PubMed] [Google Scholar]
- 13. Soeki T, Tamura Y, Shinohara H, et al Elevated concentration of soluble vascular endothelial cadherin is associated with coronary atherosclerosis. Circ J 2004; 68: 1–5. [DOI] [PubMed] [Google Scholar]
- 14. Zhang RY, Liu YY, Li L, et al Increased levels of soluble vascular endothelial cadherin are associated with poor outcome in severe sepsis. J Int Med Res 2010; 38: 1497–506. [DOI] [PubMed] [Google Scholar]
- 15. Sidibe A, Mannic T, Arboleas M, et al Soluble VE‐cadherin in rheumatoid arthritis patients correlates with disease activity: evidence for tumor necrosis factor alpha‐induced VE‐cadherin cleavage. Arthritis Rheum 2012; 64: 77–87. [DOI] [PubMed] [Google Scholar]
- 16. Wallez Y, Huber P. Endothelial adherens and tight junctions in vascular homeostasis, inflammation and angiogenesis. Biochim Biophys Acta 2008; 1778: 794–809. [DOI] [PubMed] [Google Scholar]
- 17. Herren B, Levkau B, Raines EW, Ross R. Cleavage of beta‐catenin and plakoglobin and shedding of VE‐cadherin during endothelial apoptosis: evidence for a role for caspases and metalloproteinases. Mol Biol Cell 1998; 9: 1589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pelsers MM, Namiot Z, Kisielewski W, et al Intestinal‐type and liver‐type fatty acid‐binding protein in the intestine. Tissue distribution and clinical utility. Clin Biochem 2003; 36: 529–35. [DOI] [PubMed] [Google Scholar]
- 19. Negishi K, Noiri E, Doi K, et al Monitoring of urinary L‐type fatty acid‐binding protein predicts histological severity of acute kidney injury. Am J Pathol 2009; 174: 1154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nielsen SE, Sugaya T, Hovind P, et al Urinary liver‐type fatty acid‐binding protein predicts progression to nephropathy in type 1 diabetic patients. Diabetes Care 2010; 33: 1320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawai A, Kusaka M, Kitagawa F, et al Serum liver‐type fatty acid‐binding protein predicts recovery of graft function after kidney transplantation from donors after cardiac death. Clin Transplant 2014; 28: 749–54. [DOI] [PubMed] [Google Scholar]
- 22. Pelsers MM, Morovat A, Alexander GJ, et al Liver fatty acid‐binding protein as a sensitive serum marker of acute hepatocellular damage in liver transplant recipients. Clin Chem 2002; 48: 2055–7. [PubMed] [Google Scholar]
- 23. Akbal E, Koklu S, Kocak E, et al Liver fatty acid‐binding protein is a diagnostic marker to detect liver injury due to chronic hepatitis C infection. Arch Med Res 2013; 44: 34–8. [DOI] [PubMed] [Google Scholar]
- 24. Van Den Broek MA, Bloemen JG, Dello SA, et al Randomized controlled trial analyzing the effect of 15 or 30 min intermittent Pringle maneuver on hepatocellular damage during liver surgery. J Hepatol 2011; 55: 337–45. [DOI] [PubMed] [Google Scholar]
- 25. Amacher DE. The discovery and development of proteomic safety biomarkers for the detection of drug‐induced liver toxicity. Toxicol Appl Pharm 2010; 245: 134–42. [DOI] [PubMed] [Google Scholar]
- 26. Haunerland NH, Spener F. Fatty acid‐binding proteins–insights from genetic manipulations. Prog Lipid Res 2004; 43: 328–49. [DOI] [PubMed] [Google Scholar]
- 27. Van De Poll MC, Derikx JP, Buurman WA, et al Liver manipulation causes hepatocyte injury and precedes systemic inflammation in patients undergoing liver resection. World J Surg 2007; 31: 2033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rafter I, Graberg T, Kotronen A, et al Isoform‐specific alanine aminotransferase measurement can distinguish hepatic from extrahepatic injury in humans. Int J Mol Med 2012; 30: 1241–9. [DOI] [PubMed] [Google Scholar]
- 29. Ozer J, Ratner M, Shaw M, Bailey W, Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology 2008; 245: 194–205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1111/liv.13174/suppinfo


