Abstract
Background. Long noncoding RNAs (lncRNAs) play key roles in a wide range of biological processes and their deregulation results in human disease, including keloids. Earlobe keloid is a type of pathological skin scar, and the molecular pathogenesis of this disease remains largely unknown. Methods. In this study, microarray analysis was used to determine the expression profiles of lncRNAs and mRNAs between 3 pairs of earlobe keloid and normal specimens. Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to identify the main functions of the differentially expressed genes and earlobe keloid-related pathways. Results. A total of 2068 lncRNAs and 1511 mRNAs were differentially expressed between earlobe keloid and normal tissues. Among them, 1290 lncRNAs and 1092 mRNAs were upregulated, and 778 lncRNAs and 419 mRNAs were downregulated. Pathway analysis revealed that 24 pathways were correlated to the upregulated transcripts, while 11 pathways were associated with the downregulated transcripts. Conclusion. We characterized the expression profiles of lncRNA and mRNA in earlobe keloids and suggest that lncRNAs may serve as diagnostic biomarkers for the therapy of earlobe keloid.
1. Introduction
Keloids are defined as pathologically formed scars that exceed the boundary of the original wound [1]. They are also deemed as benign dermal tumors that are unique to humans. Etiologically, keloids may occur because of minor skin injury, such as body piercing and insect bites. In addition, it is widely agreed that the incidence rate of keloid is significantly higher in populations with darker skin, such as Africans and Asians. The external ear is one of the most common sites for keloid formation [2]. Many different treatment modalities such as surgical excision, intralesional corticosteroids, radiotherapy, and pressure earrings have been used for keloids [3, 4]. Although it has unclear etiology, the development of keloid could be considered as a process of abnormal wound healing, during which redundant extracellular collagen fibers as well as proteoglycans are deposited [5]. It is known that various molecular factors contribute to this process, for example, growth factors [6, 7], cytokines [8], and related gene pathways [9]. Some among them may be the key points that could stop or reverse this pathologic process. For example, transforming growth factor-β (TGF-β) receptor was recently reported to be a potential target in treating keloid [10]. However, deeper understanding of the molecular mechanism of keloid formation is still required for detecting critical biological factors and for the further development of effective therapies.
It is known that 90% of the human genome is transcribed to RNAs that do not code proteins (noncoding RNAs). A lot of evidence suggests that long noncoding RNA (lncRNA; >200 nucleotides) regulates protein-coding genes at the transcriptional and posttranscriptional levels, as well as transcription control [11, 12]. It is known that lncRNAs play important roles in cellular differentiation, development, and disease [11, 12]. However, for earlobe keloids, the expression or function of lncRNAs has not been studied to date.
It the present study, global expression profiles of the lncRNAs and the mRNAs from 3 pairs of earlobe keloid specimens and normal skin tissues were detected using a microarray technique, from which significantly dysregulated lncRNAs and mRNAs were screened. These results indicated that the aberrant expression levels of lncRNAs may have important roles in the development of earlobe keloid and that knowing the differently expressed lncRNAs might provide useful biomarkers for earlobe keloid therapy and diagnosis.
2. Materials and Methods
2.1. Patients and Specimens
The study procedures were approved by the Ethics Review Board of Wuhan General Hospital of Guangzhou Military Command of the People's Liberation Army and it was carried out in accordance with the Declaration of Helsinki (2008) of the World Medical Association. Keloid was diagnosed by the overgrowth of a scar that obviously exceeded the boundary of the original wound. Demographic and clinical characteristics of the patients were extracted from their medical records. Earlobe keloid specimens were obtained from the resected keloid at our outpatient clinic. The normal skin specimens were obtained from the ear of the same patient. All patients were fully informed of the aim and protocol of the study and gave written informed consent to participate in the study.
2.2. RNA Isolation, Quantification, and Quality Control
Total RNA was extracted with the mirVana miRNA Isolation Kit (Applied Biosystems) and then eluted with 100 mL of nuclease-free water. Total RNA was quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific) and the integrity of RNA was determined using an Agilent 2100 bioanalyzer and RNA 6000 Nano Kit (Agilent Technologies).
2.3. RNA Labeling and Array Hybridization
RNA sample preparation and microarray hybridization were performed according to Agilent One-Color Microarray-Based Gene Expression Analysis Protocol (Agilent Technologies, Santa Clara, CA, USA) with minor modifications. RNA was purified from 100 μg total RNA after removal of rRNA using RNeasy Mini Kit (Qiagen). After that, specimens were amplified and transcribed into cRNA, and cyanine-3-CTP was applied to label the cRNA (Quick Amp Labeling Kit: One-Color; Agilent). Labeled cRNA was once again purified with the RNeasy Mini Kit (Qiagen) and quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific).
The cRNA was fragmented and hybridized using an Agilent Gene Expression Hybridization Kit (Agilent): 0.6 μg labeled cRNA was fragmented by adding 5.0 μL 10x blocking agent and 1.0 μL 25x fragmentation buffer, and then the mixture was heated at 60°C for 30 minutes. After that, 25 μL 2x GEx Hybridization Buffer was added to stop the fragmentation reaction. Finally, 50 mL hybridization solution was dispensed into the gasket slide and assembled to the lncRNA expression microarray slide. The slides were incubated for 17 hours at 65°C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed, and scanned with using the Agilent DNA Microarray Scanner (part number, G2505C).
2.4. Data Analysis
Data were extracted with Agilent Feature Extraction software 11.7.1.1. GeneSpring GX 12.5 (Agilent Technologies) was used to normalize the quantiles of the raw data. The lncRNAs are carefully constructed using the quality-controlled, public transcriptome databases (RefSeq, UCSC Known Genes, lncRNAWiki, LNCipedia, NONCODE v4, fRNAdb v3.4, Broad lincRNA, GENCODE, etc.), as well as landmark publications. After that, lncRNAs and mRNAs with significant differential expression between the two groups were identified, and the volcano plot was drawn. Hierarchical clustering was performed using MeV 4.9.0 (http://www.tm4.org/mev.html), and heat maps were obtained by this analysis. Gene Ontology (GO) analysis was performed based on Gene Ontology (www. geneontology.org), which provided three structured networks of defined terms that describe gene product functions. Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) database was used for pathway analysis of the differentially expressed genes.
2.5. Quantitative Real-Time PCR (qRT-PCR)
The total RNA was isolated using mirVana miRNA Isolation Kit (Applied Biosystems) and was then reverse-transcribed using PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time; TaKaRa). The expression of five upregulated lncRNAs and five downregulated lncRNAs was measured by qRT-PCR using SYBR Green assays (TaKaRa), and GAPDH was used as an internal control. The expression level of each lncRNA was represented as a fold change using 2−ΔΔCt methods. The expression levels of lncRNAs differentially expressed between earlobe keloid specimens and normal skin specimens were analyzed using Student's t-test with SPSS version 17.0 [13].
2.6. Statistics
Statistical analysis was performed with SPSS version 19.0. The differences in expression levels of tested lncRNAs and mRNAs between earlobe keloid and normal skin tissues were assessed using Student's t-test, and fold change ≥ 2.0 and P < 0.05 were considered significant. Fisher's exact test was used for GO analysis and KEGG pathway analysis. P < 0.05 was considered significant.
3. Results
3.1. Differentially Expressed lncRNAs
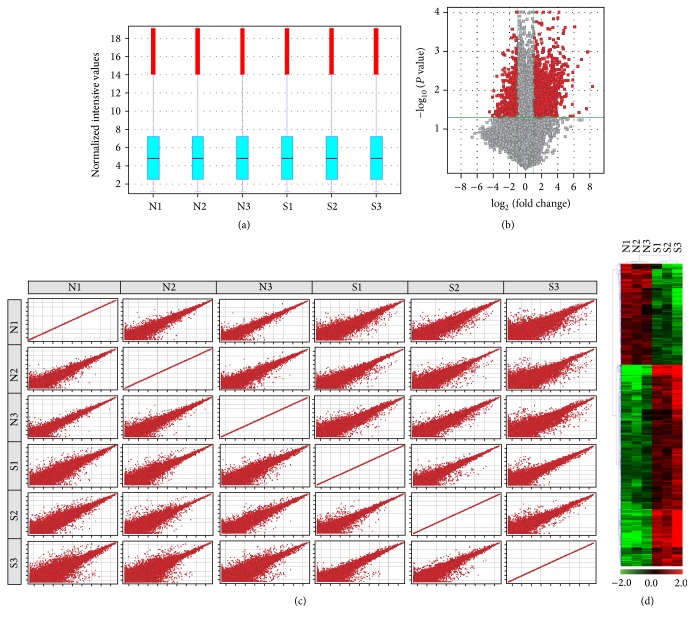
The baseline data for the 3 patients (3 pairs of specimens) included in the study are shown in Table 1. In order to compare the distributions of intensities from all samples, we used a box plot to visualize the distributions of a dataset. Box-whisker plotting suggested similar distribution of the data from six RNA gene chips (Figure 1(a)). The expression profiles of 2068 lncRNAs indicated that they were differentially expressed (fold change ≥ 2.0 and P < 0.05) between earlobe keloid specimens and normal skin specimens (shown in the lncRNA profiling). Variations in lncRNA expression among specimens were shown by volcano plotting and scatter plotting (Figures 1(b) and 1(c)). Among these lncRNAs, 1290 were upregulated more than twofold in the earlobe keloid specimens compared to the normal skin specimens, while 778 lncRNAs were downregulated more than twofold. lncRNA expression data are deposited at Gene Expression Omnibus under accession number GSE83286. The top 20 differentially expressed lncRNAs are listed in Tables 2 and 3. Finally, to infer the relationships among specimens, hierarchical clustering was performed to show distinguishable lncRNA expression patterns among samples (Figure 1(d)).
Table 1.
Baseline data of included patients.
| Case | Age (years) | Gender | Reason of skin injury | Size of keloid (cm × cm × cm) | History of keloid (months) |
|---|---|---|---|---|---|
| 1 | 21 | Female | Earlobe piercing | 2.0 × 1.3 × 0.8 | 10 |
| 2 | 34 | Female | Earlobe piercing | 1.8 × 1.5 × 1.2 | 14 |
| 3 | 24 | Female | Earlobe piercing | 2.7 × 2.0 × 1.5 | 17 |
Figure 1.
Expression profiles of lncRNAs in earlobe keloid and normal skin specimens. (a) Box-whisker plots of lncRNAs showed the distributions of intensities from all samples. (b) Volcano plots showed variation in lncRNA expression. The vertical lines correspond to 2.0-fold upregulation and downregulation and the horizontal line represents a P value of 0.05. (c) Scatter plots show variation in lncRNA expression. (d) Hierarchical clustering shows lncRNA expression profiling. Cluster analysis arranges samples into groups based on their expression levels, which allows us to hypothesize the relationships among samples. “Red” indicates highly relative expression, and “green” indicates lowly relative expression.
Table 2.
The top 20 upregulated lncRNAs.
| Seq. name | Source | Fold change | Chrom. | Strad. | txStrat | txEnd | Associated gene name |
|---|---|---|---|---|---|---|---|
| NONHSAT120157 | NONCODE v4 | 302.566 | chr7 | − | 37946864 | 37949441 | SFRP4 |
| NONHSAT062994 | NONCODE v4 | 198.76 | chr19 | − | 18896928 | 18897844 | COMP |
| ENST00000424523 | Ensembl | 187.8763 | chr7 | + | 92484223 | 92546465 | |
| NONHSAT016934 | NONCODE v4 | 121.1942 | chr10 | − | 127823937 | 127843874 | ADAM12 |
| NONHSAG007229 | NONCODE v4 | 98.20973 | chr10 | − | 134634754 | 134637851 | TTC40 |
| NONHSAT135001 | NONCODE v4 | 85.73119 | chr9 | + | 131745793 | 131747541 | NUP188 |
| NONHSAT016933 | NONCODE v4 | 66.81899 | chr10 | − | 127779305 | 127798357 | ADAM12 |
| NONHSAT033754 | NONCODE v4 | 59.65379 | chr13 | + | 50191636 | 50192101 | |
| NONHSAT102388 | NONCODE v4 | 57.30576 | chr5 | + | 79377827 | 79379011 | THBS4 |
| NONHSAT016928 | NONCODE v4 | 56.89644 | chr10 | − | 127700956 | 127703336 | ADAM12 |
| NONHSAT076769 | NONCODE v4 | 55.34357 | chr2 | + | 216476099 | 216669548 | LINC00607 |
| NONHSAT033252 | NONCODE v4 | 45.90168 | chr13 | − | 38137358 | 38144948 | POSTN |
| NONHSAG013256 | NONCODE v4 | 45.0158 | chr13 | − | 38136835 | 38145672 | POSTN |
| ENST00000557618 | Ensembl | 40.71083 | chr14 | + | 60981837 | 61021634 | |
| ENST00000597626 | Ensembl | 37.49051 | chr21 | + | 35287852 | 35341659 | |
| NONHSAG030448 | NONCODE v4 | 36.29881 | chr2 | − | 216232403 | 216237205 | FN1 |
| NONHSAT056875 | NONCODE v4 | 34.17572 | chr18 | + | 907552 | 909671 | ADCYAP1 |
| NONHSAG052055 | NONCODE v4 | 32.53344 | chr9 | + | 34084331 | 34096676 | DCAF12 |
| NONHSAT100815 | NONCODE v4 | 31.91256 | chr5 | + | 28524293 | 28602803 | |
| NONHSAT077639 | NONCODE v4 | 30.05823 | chr2 | − | 238241611 | 238243429 | COL6A3 |
Table 3.
The top 20 downregulated lncRNAs.
| Seq. name | Source | Fold change | Chrom. | Strad. | txStrat | txEnd | Associated gene name |
|---|---|---|---|---|---|---|---|
| NONHSAT053431 | NONCODE v4 | 22.78803 | chr17 | + | 37395854 | 37400623 | FBXL20 |
| NONHSAT030286 | NONCODE v4 | 17.52123 | chr12 | + | 101988749 | 102021958 | MYBPC1 |
| FR244962 | fRNAdb v3.4 | 16.8388 | chr7 | + | 31551811 | 31552010 | |
| ENST00000601148 | Ensembl | 15.31924 | chr19 | + | 51843949 | 51847370 | |
| TCONS_l2_00026076 | Broad lincRNA | 13.31326 | chr7 | + | 80804833 | 80828289 | |
| FR193036 | fRNAdb v3.4 | 12.53563 | chr19 | − | 56526608 | 56527152 | |
| NONHSAT125631 | NONCODE v4 | 12.31555 | chr8 | + | 25398695 | 25408293 | |
| TCONS_l2_00016248 | Broad lincRNA | 12.02637 | chr20 | + | 37230676 | 37256614 | |
| NONHSAT030224 | NONCODE v4 | 12.02533 | chr12 | − | 100560001 | 100562998 | GOLGA2P5 |
| NONHSAT076673 | NONCODE v4 | 11.6689 | chr2 | − | 211074313 | 211081443 | ACADL |
| NONHSAT137402 | NONCODE v4 | 11.66708 | chrX | + | 69454505 | 69457167 | AWAT1 |
| ENST00000580420 | Ensembl | 11.61363 | chr18 | + | 29522538 | 29524119 | |
| FR174595 | fRNAdb v3.4 | 11.10483 | chr6 | + | 118888744 | 118889041 | |
| NONHSAT077942 | NONCODE v4 | 11.03293 | chr2 | − | 242455829 | 242457154 | |
| NONHSAT060814 | NONCODE v4 | 10.87916 | chr19 | + | 7410710 | 7411049 | |
| NONHSAT097800 | NONCODE v4 | 10.56074 | chr4 | + | 110897243 | 110898692 | EGF |
| NONHSAT070090 | NONCODE v4 | 10.32572 | chr2 | + | 36991437 | 36993016 | VIT |
| TCONS_00022478 | Broad lincRNA | 10.21703 | chr14 | + | 38205181 | 38208450 | |
| ENST00000556024 | Ensembl | 9.802968 | chr14 | − | 38025363 | 38036300 | |
| NONHSAT102735 | NONCODE v4 | 9.773005 | chr5 | + | 90142207 | 90144638 | GPR98 |
3.2. Differentially Expressed mRNAs
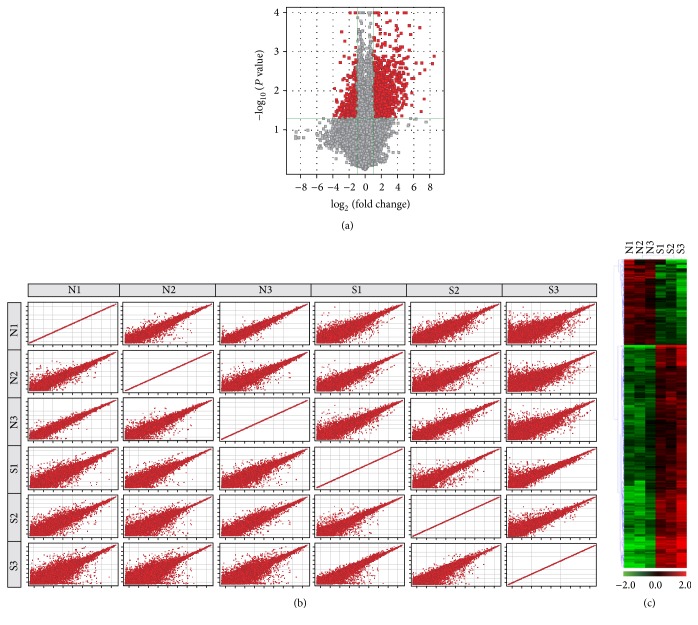
A total of 1511 mRNAs were differentially expressed between the two tissues (fold change ≥ 2.0 and P < 0.05). A total of 1092 of 1511 mRNAs were expressed significantly higher in earlobe keloid specimens and 419 mRNAs were expressed significantly lower compared to normal skin specimens (shown in the mRNA profiling). mRNA expression data are deposited at Gene Expression Omnibus under accession number GSE83286. The top 20 differentially expressed mRNAs are listed in Supplemental Tables 1 and 2 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/5893481. Variations in the mRNA expression among specimens were shown by volcano plotting and scatter plotting (Figures 2(a) and 2(b)). Hierarchical clustering showed that mRNA expression modes among samples were distinguishable (Figure 2(c)).
Figure 2.
Expression profiles of mRNAs in earlobe keloid and normal skin specimens. (a and b) Volcano and scatter plots show differences in expression. The vertical green lines delimit 2.0-fold upregulation and downregulation. Red plots represent mRNAs with >2.0-fold change and corrected P value < 0.05. (c) Hierarchical clustering shows mRNA expression profiling. Cluster analysis arranges samples into groups based on their expression levels, which allows us to hypothesize the relationships among samples. “Red” indicates highly relative expression, and “green” indicates lowly relative expression.
3.3. GO Analysis
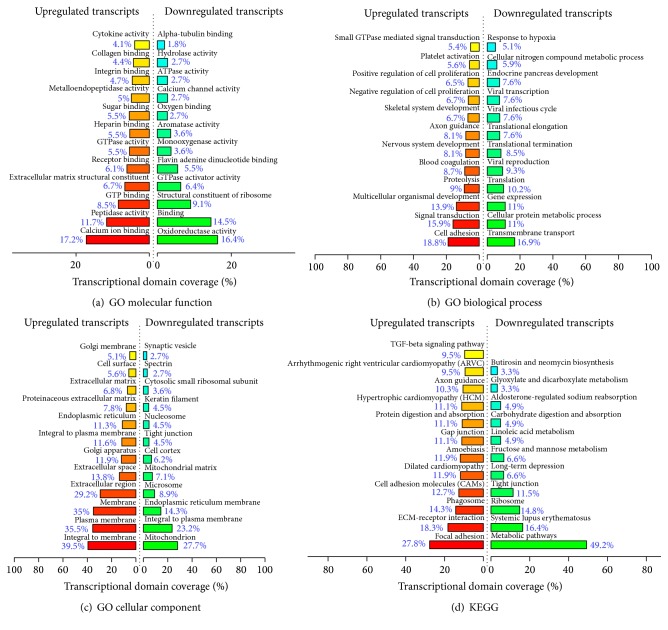
The GO project is a collaborative effort to construct and use ontologies to facilitate the biologically meaningful annotation of genes and their products in a wide variety of organisms [14]. We performed GO analysis for lncRNAs to determine molecular function, biological processes, and cellular components. For molecular function (Figure 3(a)), calcium ion binding (GO:0005509) had the highest transcriptional domain coverage (TDC, 17.2%) in upregulated transcripts, while oxidoreductase activity (GO:0016491; TDC, 16.4%) was highest in downregulated transcripts. In biological processes (Figure 3(b)), it was found that upregulated genes were enriched most in the process of cell adhesion (GO:0007155; TDC, 18.8%). In contrast, downregulated genes were enriched most in the process of transmembrane transport (GO:0007155; TDC, 16.9%). In the cellular components (Figure 3(c)), it was detected that integral to membrane (GO:0016021) had the highest enrichment of upregulated genes (TDC, 39.5%), and mitochondria had the highest enrichment of downregulated genes (GO:0005886; TDC, 27.7%).
Figure 3.
(a) Top 12 enriched GO terms for differentially expressed mRNAs for molecular function. The bar plot shows the transcriptional domain coverage. (b) Top 12 enriched GO terms for differentially expressed mRNAs for biological processes. The bar plot shows the transcriptional domain coverage. (c) Top 12 enriched GO terms for differentially expressed mRNAs for cellular components. The bar plot shows the transcriptional domain coverage. (d) The results of KEGG pathway enrichment analysis. The bar plot shows the transcriptional domain coverage of the enrichment pathway.
3.4. KEGG Analysis
KEGG pathway enrichment analysis was used for differentially expressed genes to identify pathways represented among the lncRNAs identified in the earlobe keloid gene expression signature. KEGG analysis suggested that 24 pathways were significantly correlated with upregulated gene expression. The focal adhesion pathway had the highest enrichment of increased transcription (TDC, 27.8%) and comprised 35 targets genes. Pathway analysis also revealed that 11 pathways corresponded to downregulated transcripts and that the most enriched network was metabolic pathways (TDC, 49.2%), which comprised 30 target genes (Figure 3(d)). Many of these pathways are reported to be associated with keloid, including the gene category focal adhesion pathway [15], TGF-β signaling pathway [16–18], mitogen-activated protein kinase (MAPK) pathway [19], and gap junction pathway [20, 21].
3.5. QRT-PCR Validation
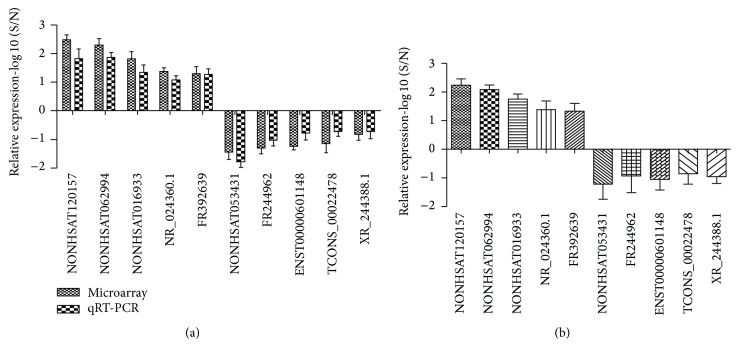
To verify the microarray data, five upregulated lncRNAs (NONHSAT120157, NONHSAT062994, NONHSAT016933, NR_024360.1, and FR39263) and five downregulated lncRNAs (NONHSAT053431, FR244962, ENST00000601148, TCONS_00022478, and XR_244388.1) were randomly selected from the differentially expressed lncRNAs. We detected the expression levels of these lncRNAs in 10 earlobe keloids tissues and normal skin samples (Supplemental Table 3) using qRT-PCR. As shown in Figure 4, the qRT-PCR results and microarray data are consistent.
Figure 4.
Quantitative RT-PCR validation of 10 differentially expressed lncRNAs. (a) Comparison of fold change [log10(S/N)] of lncRNAs between microarray and quantitative RT-PCR results (S: earlobe keloid specimens; N: normal skin specimens). (b) Relative expression levels of lncRNAs in 10 other pairs of earlobe keloid and normal skin specimens (P < 0.05).
4. Discussion
Emerging evidence shows that a set of noncoding RNAs (for example, miRNA) is involved in the mechanism of keloid formation [22–25]. lncRNAs are larger than miRNAs and have more complex structure. Deregulated expression of lncRNA disrupts cellular physiology and then leads to pathology [26–28]. Thus, we suggest that lncRNAs may play crucial roles in many biological processes and are vital to the formation of earlobe keloids. However, the profile and biological function of lncRNAs for earlobe keloid remain largely unknown. Thus, in the present study, we established the expression profile of lncRNAs in human earlobe keloids.
We analyzed lncRNA and mRNA expression profiles in the tissues of earlobe keloid and control tissues to reveal the potential roles of lncRNAs in the pathogenesis of earlobe keloid. High-throughput microarray techniques uncovered differential expression between 3 pairs of earlobe keloid and normal skin specimens. We identified that 1290 lncRNAs and 1092 mRNAs were upregulated and 778 lncRNAs and 419 mRNAs were downregulated in all 3 earlobe keloid and normal tissues (fold change ≥ 2.0, P < 0.05). GO and KEGG pathway analysis were used to explore the possible biological functions and potential mechanisms of lncRNAs and mRNAs in earlobe keloids. In fact, Liang and colleagues have previously identified differential expression of lncRNAs and mRNAs between 3 pairs of keloid and normal skin tissue by microarray (32). Compared with their results, our study has several differences. First, tissues used here were earlobe keloid and normal specimens, and the expression profiles of lncRNAs were significantly different from the previous results. Second, to verify the microarray data, the expression levels of five upregulated lncRNAs and five downregulated lncRNAs were detected in 10 earlobe keloids tissues and normal skin samples using qRT-PCR, and the results were consistent with microarray data.
An integrative method including pathway was developed to identify possible functional relationships between the different RNA molecules. Based on the differentially expressed mRNAs, pathway analysis revealed which biological functions and mechanisms were involved in earlobe keloid formation. Our results suggest that different biological processes, such as cell-cell adhesion, cell migration, cell death, cell junction formation, epithelial to mesenchymal transition (EMT), TGF-β, and MAPK, are among the significantly enriched mRNAs. Most of these pathways are involved in the process of tissue fibrosis. For example, studies in a wide range of experimental models have revealed that TGF-β is a central mediator of keloid fibrogenesis. It is reported that Loureirin B attenuated the contraction of fibroblasts which was induced by TGF-β in hypertrophic scar formation (33). Yan et al. have reported that EMT plays crucial roles in keloid formation [29]. Gobin et al. have shown that emodin-loaded liposomes decrease survival rate of keloids which express high levels of receptor tyrosine kinase (RTK) (included in the focal adhesion pathway) [30]. Among these related pathways, we found that focal adhesion, extracellular matrix receptor interaction, cell adhesion molecules, and gap junction-associated pathway showed significant changes in upregulated and downregulated mRNAs. For example, 35 differentially expressed mRNAs were involved in the focal adhesion pathway and 16 differentially expressed mRNAs were enriched in cell adhesion molecules. Otherwise, 7 of which were downregulated in the tight junction related pathway. These results indicated that cell adhesion and tight associated signaling may play an important role in the mechanism of earlobe keloid formation, which is not identified by other researches.
Our study used microarray data to analyze systematically and comprehensively differentially expressed lncRNAs and mRNAs between normal skin and earlobe keloid tissues. Many differentially expressed lncRNAs could play a vital role in regulating earlobe keloid formation through various pathways. In our present study, we found that TGF-β, MAPK, cell tight, and adhesion related signaling series mRNAs may interact with IncRNAs. Previous reports demonstrated that ADAM proteins are involved in cell adhesion, cell fusion, cell signaling, and proteolysis. ADAM33 is a member of ADAM family that is associated with keloid scars in the northeastern Chinese population (36). ADAM12 are reluctant to adhere to fibronectin, a key ECM protein in keloids (37). Patients suffering from collagen VI related myopathies caused by mutations in COL6A1, COL6A2, and COL6A3 often also display skin abnormalities, like formation of keloids or “cigarette paper” scars, dry skin, striae rubrae, and keratosis pilaris (follicular keratosis) (38). Keloid fibroblasts were propagated in culture and their proliferative behaviour and response to the epidermal growth factor (EGF) were studied (39). Our results found that NONHSAT016934, NONHSAT016933, NONHSAT016928, and NONHSAT077639 expressions were increased, whereas NONHSAT097800 expression was decreased in earlobe keloid and normal tissues. These lncRNAs were associated with the related genes of keloid (ADAM12, COL6A3, and EGF). We will carry out further studies of these differentially expressed lncRNAs to establish their functions in earlobe keloid formation.
In conclusion, we studied the differential expression profile of lncRNAs and mRNAs in earlobe keloid and normal skin tissues. Our microarray analysis indicated that lncRNAs are involved in the pathological process of earlobe keloid formation. Therefore, subgroup analysis of lncRNAs should be performed to explore this relationship in the future. In addition, we will select numbers of samples to deepen the research into the lncRNA molecular mechanism and biochemical function in order to provide a novel accurate method for therapy of earlobe keloid.
Supplementary Material
The top 20 up- and down-regulated mRNAs.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (Grant nos. 81301638 and 81301639) and a grant from the Natural Science Foundation of Hubei Province (Grant no. 2012FFB06808).
Competing Interests
All the authors declare that they have no competing interests to disclose.
Authors' Contributions
Liang Guo and Kai Xu contributed equally to this study.
References
- 1.Pérez-Espadero A., Escudero Nafs F. J. Bilateral keloids secondary to otoplasty treated by excision and keloids flaps. Anales del Sistema Sanitario de Navarra. 2013;36(2):333–338. doi: 10.4321/S1137-66272013000200019. [DOI] [PubMed] [Google Scholar]
- 2.Cha H. W., Jung H. J., Lim H. J., Lee S.-J., Kim D. W., Lee W. J. The efficacy of complete surgical excision of keloid and piercing sinus tract on earlobe keloid. Annals of Dermatology. 2013;25(3):370–373. doi: 10.5021/ad.2013.25.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Leeuwen M. C., van der M. B., Wal A. E., et al. Intralesional cryotherapy for treatment of keloid scars: a prospective study. Plastic and Reconstructive Surgery. 2015;135(2):580–589. doi: 10.1097/PRS.0000000000000911. [DOI] [PubMed] [Google Scholar]
- 4.Erol O. O., Gurlek A., Agaoglu G., Topcuoglu E., Oz H. Treatment of hypertrophic scars and keloids using intense pulsed light (IPL) Aesthetic Plastic Surgery. 2008;32(6):902–909. doi: 10.1007/s00266-008-9161-7. [DOI] [PubMed] [Google Scholar]
- 5.Syed F., Ahmadi E., Iqbal S. A., Singh S., McGrouther D. A., Bayat A. Fibroblasts from the growing margin of keloid scars produce higher levels of collagen I and III compared with intralesional and extralesional sites: clinical implications for lesional site-directed therapy. British Journal of Dermatology. 2011;164(1):83–96. doi: 10.1111/j.1365-2133.2010.10048.x. [DOI] [PubMed] [Google Scholar]
- 6.Ooi B. N. S., Mukhopadhyay A., Masilamani J., et al. Hepatoma-derived growth factor and its role in keloid pathogenesis. Journal of Cellular and Molecular Medicine. 2010;14(6):1328–1337. doi: 10.1111/j.1582-4934.2009.00779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z.-C., Tang B., Guo D., et al. Expression of insulin-like growth factor-1 receptor in keloid and hypertrophic scar. Clinical and Experimental Dermatology. 2014;39(7):822–828. doi: 10.1111/ced.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q., Yamaza T., Kelly A. P., et al. Tumor-like stem cells derived from human keloid are governed by the inflammatory niche driven by IL-17/IL-6 axis. PLoS ONE. 2009;4(11) doi: 10.1371/journal.pone.0007798.e7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan T. T., See P., Tran E., et al. Suppression of insulin-like growth factor signalling pathway and collagen expression in keloid-derived fibroblasts by quercetin: its therapeutic potential use in the treatment and/or prevention of keloids. British Journal of Dermatology. 2003;148(3):544–552. doi: 10.1046/j.1365-2133.2003.05174.x. [DOI] [PubMed] [Google Scholar]
- 10.Bian D., Zhang J., Wu X., et al. Asiatic acid isolated from Centella asiatica inhibits TGF-β1-induced collagen expression in human keloid fibroblasts via PPAR-γ activation. International Journal of Biological Sciences. 2013;9(10):1032–1042. doi: 10.7150/ijbs.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kung J. T. Y., Colognori D., Lee J. T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapusta A., Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends in Genetics. 2014;30(10):439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Wu K., Yang Z., et al. Multidrug-resistance related long non-coding RNA expression profile analysis of gastric cancer. PLoS ONE. 2015;10(8) doi: 10.1371/journal.pone.0135461.e0135461 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Ruan Z., Sun X., Sheng H., Zhu L. Long non-coding RNA expression profile in atrial fibrillation. International Journal of Clinical and Experimental Pathology. 2015;8(7):8402–8410. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Nie F., Chen X., et al. Upregulated periostin promotes angiogenesis in keloids through activation of the ERK 1/2 and focal adhesion kinase pathways, as well as the upregulated expression of VEGF and angiopoietin-1. Molecular Medicine Reports. 2015;11(2):857–864. doi: 10.3892/mmr.2014.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song R., Li G., Li S. Aspidin PB, a novel natural anti-fibrotic compound, inhibited fibrogenesis in TGF-β1-stimulated keloid fibroblasts via PI-3K/Akt and Smad signaling pathways. Chemico-Biological Interactions. 2015;238:66–73. doi: 10.1016/j.cbi.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Dong X., Zhang C., Ma S., Wen H. Mast cell chymase in keloid induces profibrotic response via transforming growth factor-β1/Smad activation in keloid fibroblasts. International Journal of Clinical and Experimental Pathology. 2014;7(7):3596–3607. [PMC free article] [PubMed] [Google Scholar]
- 18.Liang C.-J., Yen Y.-H., Hung L.-Y., et al. Thalidomide inhibits fibronectin production in TGF-β1-treated normal and keloid fibroblasts via inhibition of the p38/Smad3 pathway. Biochemical Pharmacology. 2013;85(11):1594–1602. doi: 10.1016/j.bcp.2013.02.038. [DOI] [PubMed] [Google Scholar]
- 19.Kuo Y.-R., Wu W.-S., Wang F.-S. Flashlamp pulsed-dye laser suppressed TGF-β1 expression and proliferation in cultured keloid fibroblasts is mediated by MAPK pathway. Lasers in Surgery and Medicine. 2007;39(4):358–364. doi: 10.1002/lsm.20489. [DOI] [PubMed] [Google Scholar]
- 20.Pistorio A. L., Ehrlich H. P. Modulatory effects of connexin-43 expression on gap junction intercellular communications with mast cells and fibroblasts. Journal of Cellular Biochemistry. 2011;112(5):1441–1449. doi: 10.1002/jcb.23061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu F., Gao J., Ogawa R., Hyakusoku H. Variations in gap junctional intercellular communication and connexin expression in fibroblasts derived from keloid and hypertrophic scars. Plastic and Reconstructive Surgery. 2007;119(3):844–851. doi: 10.1097/01.prs.0000255539.99698.f4. [DOI] [PubMed] [Google Scholar]
- 22.Makino K., Jinnin M., Hirano A., et al. The downregulation of microRNA let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. The Journal of Immunology. 2013;190(8):3905–3915. doi: 10.4049/jimmunol.1200822. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., Wang X., Yang D., Xiao Z., Chen X. MicroRNA-21 affects proliferation and apoptosis by regulating expression of PTEN in human keloid fibroblasts. Plastic and Reconstructive Surgery. 2014;134(4):561e–573e. doi: 10.1097/prs.0000000000000577. [DOI] [PubMed] [Google Scholar]
- 24.Kashiyama K., Mitsutake N., Matsuse M., et al. MiR-196a downregulation increases the expression of type I and III collagens in keloid fibroblasts. Journal of Investigative Dermatology. 2012;132(6):1597–1604. doi: 10.1038/jid.2012.22. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z.-Y., Lu L., Liang J., Guo X.-R., Zhang P. H., Luo S.-J. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genetics and Molecular Research. 2014;13(2):2727–2738. doi: 10.4238/2014.April.14.2. [DOI] [PubMed] [Google Scholar]
- 26.Ponting C. P., Oliver P. L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Gesualdo F. D., Capaccioli S., Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5(22):10976–10996. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bida O., Gidoni M., Ideses D., Efroni S., Ginsberg D. A novel mitosis-associated lncRNA, MA-linc1, is required for cell cycle progression and sensitizes cancer cells to Paclitaxel. Oncotarget. 2015;6(29):27880–27890. doi: 10.18632/oncotarget.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan L., Cao R., Wang L., et al. Epithelial-mesenchymal transition in keloid tissues and TGF-β1-induced hair follicle outer root sheath keratinocytes. Wound Repair and Regeneration. 2015;23(4):601–610. doi: 10.1111/wrr.12320. [DOI] [PubMed] [Google Scholar]
- 30.Gobin A. S., Rhea R., Newman R. A., Mathur A. B. Silk-fibroin-coated liposomes for long-term and targeted drug delivery. International Journal of Nanomedicine. 2006;1(1):81–87. doi: 10.2147/nano.2006.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The top 20 up- and down-regulated mRNAs.