Abstract
Serial femtosecond crystallography (SFX) employing high-intensity X-ray free-electron laser (XFEL) sources has enabled structural studies on microcrystalline protein samples at non-cryogenic temperatures. However, the identification and optimization of conditions that produce well diffracting microcrystals remains an experimental challenge. Here, we report parallel SFX and transmission electron microscopy (TEM) experiments using fragmented microcrystals of wild type (WT) homoprotocatechuate 2,3-dioxygenase (HPCD) and an active site variant (H200Q). Despite identical crystallization conditions and morphology, as well as similar crystal size and density, the indexing efficiency of the diffraction data collected using the H200Q variant sample was over 7-fold higher compared to the diffraction results obtained using the WT sample. TEM analysis revealed an abundance of protein aggregates, crystal conglomerates and a smaller population of highly ordered lattices in the WT sample as compared to the H200Q variant sample. While not reported herein, the 1.75 Å resolution structure of the H200Q variant was determined from ~16 minutes of beam time, demonstrating the utility of TEM analysis in evaluating sample monodispersity and lattice quality, parameters critical to the efficiency of SFX experiments.
Keywords: Transmission Electron Microscopy, Serial Femtosecond Crystallography, X-ray Free Electron Laser, Liquid-jet Injector
Introduction
The X-ray Free Electron Laser (XFEL) has advanced the field of X-ray crystallography by enabling structure determination using radiation sensitive sub-micrometer sized crystals (Aquila et al., 2012; Hunter and Fromme, 2011; Miller, 2014; Schlichting and Miao, 2012). X-ray damage is overcome through the use of ultrashort X-ray pulses that produce a diffraction pattern before radiation induced structural rearrangements occur within the crystal. These experiments are often conducted in a serial mode using liquid injectors to deliver crystals to the XFEL pulses. Unlike more conventional goniometer-based crystallography experiments, where one or a few crystals at cryogenic temperatures are used to obtain a complete dataset, liquid injector serial femtosecond crystallography (SFX) experiments are contingent upon the collection of several thousand diffraction patterns often using millions of crystals delivered in random orientations (Boutet et al., 2012; Kirian et al., 2010; Redecke et al., 2013).
Such injector systems have been successful for high resolution structure determination at near physiological temperatures (Boutet et al., 2012; Chapman et al., 2011; Ginn et al., 2015; Kang et al., 2015; Liu et al., 2013; Redecke et al., 2013; Weierstall et al., 2014). For these experiments, crystal slurries are focused into a continuous liquid jet that delivers single crystals of a few microns to submicron dimensions to the XFEL pulses. Recently, development of viscous media injectors have lessened sample consumption, a major drawback of liquid injector-based SFX experiments, by significantly reducing the sample flow to better match the XFEL repetition rate (for example, 120Hz at the Linac Coherent Light Source (LCLS) at the Stanford Linear Accelerator (SLAC) (Botha et al., 2015; Conrad et al., 2015; Fromme et al., 2015; Sugahara et al., 2015; Weierstall et al., 2014). While improvements in liquid injectors and the analysis of sparse datasets are active and dynamic research areas involving many groups (Ginn et al., 2015; Hattne et al., 2014; Kabsch, 2014; Qu et al., 2014; Weierstall et al., 2014), less focus has been geared towards understanding the intrinsic characteristics of microcrystalline slurries and their amenability to the injection process.
Properties that could affect injector performance and/or diffraction quality include crystal concentration, size distribution, quality and uniformity of the crystal lattices, or the presence of protein and/or crystal aggregates. Various methods have been used to identify microcrystals with homogenous physical properties. Such methods include dynamic light scattering (Schubert, 2015), second-order nonlinear imaging of chiral crystals (Luft, 2015), as well as microfluidic technologies (Abdallah et al., 2015; Kupitz et al., 2014) and transmission electron microscopy (TEM) (Gomery et al., 2013; Redecke et al., 2013; Stevenson et al., 2014b). In the latter case, we previously showed that not only is TEM capable of identifying conditions amenable to microcrystal formation, but also is capable of characterizing the intrinsic properties of the microcrystals (I.E. size, lattice quality, aggregation, etc.) (Stevenson et al., 2016). In this report, we provide evidence that demonstrates the utility of TEM in the characterization of microcrystalline slurries to maximize the success of SFX studies, through the comparative analysis of homoprotecatechuate 2,3-dioxygenase (HPCD) samples with significantly different indexing efficiencies.
Experimental Procedure
Enzyme purification, crystallization and sample preparation
Recombinant homoprotocatechuate 2,3-dioxygenase from B. fuscum (E.C. 1.13.11.15) was expressed in E.coli and purified using procedures described previously (Groce and Lipscomb, 2005; Mbughuni et al., 2012). The enzyme preparations used in this study included a wild type (WT) enzyme and an active site variant with His200 substituted for Gln (H200Q). Previously published crystallization conditions and procedures that produce diffraction quality macro-crystals of 2,3-HPCD enzymes (Kovaleva and Lipscomb, 2007, 2012; Kovaleva et al., 2015) were modified for injector-based experiments to scale up production of micro-crystal slurries using an over-nucleation approach. Protein solutions (7 – 10 mg/ml) were gently mixed in a 1:1 ratio with crystallization solution consisting of 12 – 14% PEG6000, 0.1 M calcium chloride, and 0.1 M MES pH 5.8, ensuring that no precipitate forms at these concentrations of components. After a few minutes of equilibration, a 20 μl aliquot of the 1:1 stock mixture (protein and crystallization solution) was seeded with a few roughly crushed macro-crystals in an eppendorf tube, and growth of many needle-like crystals was observed after a few hours of incubation at 20 °C. Every 2 – 4 hours 50-200 μL of a fresh mixture of protein and crystallization solution was added into the crystallization tubes to maintain maximal growth of needle-like crystals, followed by a final 12 hour period of growth to achieve saturation in a total of 2 – 3 ml volume. The WT enzyme preparation produced slightly larger population of initial crystals (thin rods), whereas the H200Q variant preparation produced slurries with needle-like crystals.
A mixture of glass beads (0.1 and 0.5 mm) was used to crush the crystals to achieve a smaller and more uniform sample size for injector-based studies. The WT sample, consisting of thin rods, was vortexed with the glass beads mixture several times at medium to high speed for 60 seconds, followed by a 10-minute rest period. Slurries of needle-like crystals of the H200Q variant were concentrated by centrifugation at 2,000 rpm for 5 to 10 minutes prior to the addition of the glass beads mixture and additional centrifugation for 2 minutes. The efficiency of crystal crushing using either the vortexing or microcentrifugation methods were monitored qualitatively by light microscopy, and by visually observing the light scattering of sample medium using a flashlight. Crushed samples were passed thru a 20 μm filter to remove particulates and larger crystalline material prior to loading into the sample reservoir for diffraction studies.
SFX data collection, processing and refinement
Diffraction data were collected at the CXI end station of LCLS using 8.8 keV X-ray pulses with 40 fs duration and 1 μm beam focus at the X-ray interaction point. Sample delivery using gas dynamic virtual nozzle (GDVN) was carried out at a flow rate of 20 μl/sec at 16 °C. Diffraction images were collected at 120 Hz using a CSPAD detector and processed using ccbtx.xfel software package (Hattne et al., 2014; Sauter et al., 2014). The WT enzyme sample resulted in 814 indexable images during 6.7 min of data collection. A total of 15.6 min of data collection using the H200Q variant sample resulted in a dataset based on 14026 merged images. During indexing, secondary lattices indicative of multiple crystals were identified by first indexing, then removing primary lattices and then attempting to index the remaining bright reflections (Hattne et al., 2014; Sauter and Poon, 2010).
Quantification of microcrystal slurries
To quantify the microcrystals, samples were diluted in mother liquor 300 – fold and a 10 μL aliquot was applied to a bright-line hemocytometer (Hausser Scientific). Using a Jan Scientific Jansi UVEX microscope with nominal 5-, 15-, and 40x magnifications the sample chamber grids were visualized using brightfield and UV fluorescence with exposures of 0.1 and 5 seconds, respectively. Images were evaluated using the Jan Scientific CrystalDetect software and nominal 15x magnification images were recorded (Infinity 2-3C Camera, Lumenera Scientific). Manual quantification was achieved using the Vi-CELL XR 2.03 software program (Beckman Coulter). A crystal was defined as having a minimum dimension of 6 pixels (~2.5 μm), with intensity values 3 standard deviations above the background threshold.
Transmission electron microscopy
Microcrystal samples were analyzed by transmission electron microscopy as previously described (Stevenson et al., 2014a). Briefly, 10 μL of sample was applied to glow charged 400 square mesh copper grids with a continuous carbon film (Electron Microscopy Sciences). Following sample incubation on the grid for 30 seconds, excess liquid was removed and the grid stained with 2% (w/v) uranyl acetate. Protein crystallinity of each sample was determined by screening for the presence of lattices. Lattice quality was evaluated by performing a fast Fourier transform of the lattice to obtain the reciprocal lattice reflections or Bragg spots. Bragg spot order of three or above was considered to be of high quality. A single-tile specimen holder was used to deliver samples to a FEI Tecnai T12 electron microscope. The microscope was operated at 120 kV and images were captured with a 2k × 2k Gatan UltraScan 1000 CCD camera.
Results
Preparation and SFX data collection of HPCD microcrystalline samples
In this study, microcrystalline suspensions were prepared using two HPCD stocks, a WT enzyme and an H200Q variant. The single isosteric substitution in the active site (H200Q) has no effect on the crystal morphology (rod-like), spacegroup, unit cell dimensions or diffraction quality as established previously in structural synchrotron-based studies using macrocrystals at cryo-temperatures (Kovaleva et al., 2015). Therefore, these enzyme stocks were used to account for prep-to-prep variability for optimization of the batch crystallization procedures.
Due to rod/needle-like morphology of HPCD crystals and much faster crystal growth in one dimension, fragmentation was required to reduce the length of microcrystals and improve uniformity for injector-based studies. In addition to vortexing, a commonly used fragmentation method, sonication, and centrifugation fragmentation methods were tested (see experimental procedure section). However, because of the limited amount of beam time allocated for this project, diffraction experiments for only two bulk fragmentation methods (centrifugation or vortexing, for H200Q and WT samples respectively), were carried out on the CXI instrument (LCLS) using the gas dynamic virtual nozzle (GDVN) injector (DePonte, 2008).
Diffraction data for both samples were collected using the same experimental settings, as described in the Experimental Procedure section. All still diffraction images acquired during beam time were used for indexing and integration with ccbtx.xfel (Table 1). Therefore, to enable comparison between samples (Figure 1A,B), the overall “indexing efficiency” is defined here as a percentage of the total images, collected at the repetition rate of 120 Hz, which could be indexed successfully.
Table 1.
Comparison of physical properties and processing characteristics for the HPCD samples used for SFX data collection
| (H200Q HPCD) | (WT HPCD) | |
|---|---|---|
| Density (crystals/ml)a | 1.2 × 108 (±0.1) | 1.53 × 108 (±0.07) |
| Size distributionb | ||
| a ~ b (μm) | 2.5 (±0.5) | 2.6 (±0.6) |
| c (μm) | 11.7 (±5.6) | 10.4 (±4.1) |
| Number of Bragg Spotsc | 4th order | 3rd order |
| Indexing efficiency (%)d | 12.2 | 1.7 |
| 2nd lattice hits (%)e | 16.1 | 8.4 |
Values for crystal density quantification using nominal 15x magnification (n=12) as described in Experimental Procedure.
Size distribution for fragmented microcrystal samples was determined manually from UV imaging data at nominal 15x magnification (n=15). Mean values for short (a, b) and long (c) dimensions for rod-like morphology.
Bragg Spot order calculated from FFT transforms of observable lattices (median derived from n=8).
The percentage of collected images that contained indexed patterns (single hits and 2nd lattices).
Percentage of double hits in the total number of indexed patterns.
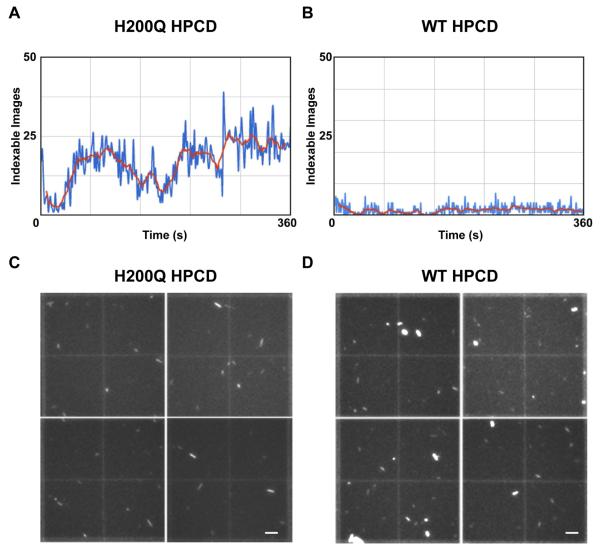
Figure 1. Representative observed indexing rates and crystal density quantification for both H200Q and WT HPCD slurries.
A plot of the number of indexable patterns observed during a representative 6 minutes of data collection using the GDVN injector with the H200Q HPCD microcrystal sample (A) and with the WT HPCD microcrystal sample (B) illustrates the reduced hit rate observed for the WT sample. The number of indexable hits/second from cctbx.xfel processing are plotted, where the blue trace indicates total number of indexable images per second and the red trace is the averaged indexing rate every 11 sec. Representative hemocytometer quadrants, reconstructed from 4 separate images using a 15x objective on a JanScientific Jansi UVEX microscope, illustrate fairly similar crystal densities for H200Q (C) and WT (D) slurries. Crystal quantification was achieved for both slurries using manual counting with ImageJ software (see experimental section, scale bars – C,D: 50 μm) (n=12)
During 6.7 min of data collection using the WT enzyme sample (which showed a high tendency of nozzle clogging), a total of 814 diffraction patterns were indexed, corresponding to 1.7% of the total images collected (Table 1). The observed indexing efficiency of 1.7% for the WT enzyme sample compares well with the typical values from previously published SFX studies, where 0.5 – 4% of the still diffraction images acquired at the same repetition rate (120 Hz) were successfully indexed and merged (Boutet et al., 2012; Chapman et al., 2011; Hattne et al., 2014; Schmidt et al., 2015; Tenboer et al., 2014). In contrast, 12.2% of the total images acquired using the H200Q enzyme sample during 15.6 min of beam time were successfully indexed (Table 1).
Characterizing microcrystalline density and size distribution
We next examined whether differences in the crystal density and size of the microcrystalline slurries could explain the 7.3-fold difference in indexing efficiency of H200Q vs WT, as these are important considerations for sample delivery and sample consumption in liquid injector-based experiments. Previously, crystal densities of ~109 crystals/mL have been proposed as the benchmark to attain high hit rates while reducing the occurrence of double hits during GDVN experiments (Boutet et al., 2012; Kern et al., 2014; Schlichting and Miao, 2012), although sample densities of 1010 to 1011 crystals/mL are frequently used as well (Hattne et al., 2014; Tenboer et al., 2014). Therefore, in order to quantify accurately the number of crystals for both H200Q and the WT slurries, we applied crystal slurries to a hemocytometer and determined the number of crystals/ml from images obtained during UV-microscopy (Figure 1C,D). Both samples showed similar crystal densities of ~108 crystals/ml (Table 1), suggesting that crystal density between slurries was not a contributing factor to the ~7-fold difference in the indexing rate between them. The occurrence of a second lattice was found in 16.1% and 8.4% of indexed images for the H200Q and the WT samples, respectively (Table 1). This data suggests that lower crystal densities may be practical for liquid injector-based experiments.
Next, we examined the crystal size distribution using brightfield and UV microscopy for the WT and the H200Q variant samples (Table 1). In an effort to reduce clogging of the GDVN injector nozzle, samples are typically filtered to limit the maximum crystal size. If the crystal size distribution of the two samples was significantly different, the different indexing rates observed during data collection could potentially be attributed to increased clogging of the GDVN nozzle, which focuses the crystal slurry to a stream of about 5 μm in diameter. However, our analysis revealed no size difference between the WT and the H200Q variant samples (Table 1). Many of the crystals from both samples were long needles with a significant fraction retaining a length of 10 – 20 μm following the fragmentation and filtration procedures.
Analysis of crystal lattice quality and monodispersity with TEM
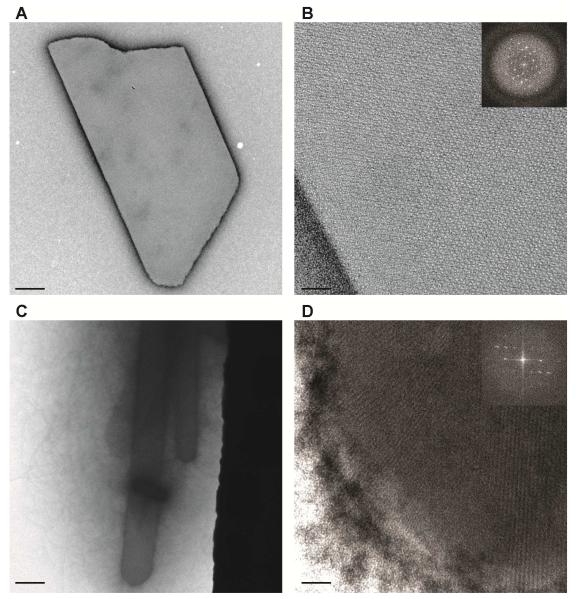
The homogeneity and intrinsic order of microcrystals are critical parameters for SFX studies that also cannot be assessed effectively by bright-field microscopy. Similarities between crystal morphology, size, and density raise the possibility that observed differences in the indexing efficiency may be attributed to the quality of the crystal lattice in the two samples. Negative-stain TEM analysis, conducted on the two microcrystalline samples of HPCD, revealed both major differences in the lattice quality and homogeneity (Figure 2). In addition to higher order Bragg spots (> 4th order) observed in H200Q crystals, the EM landscape indicated a highly monodispersed sample (Figure 2A,B). In contrast, WT microcrystals showed a high degree of sample aggregation, protein conglomerates, and a limited number of crystal lattices with > 3rd order Bragg spots (Figure 2C,D). Moreover, delivery of the WT sample was prone to nozzle clogging, which would be consistent with the high degree of protein aggregation observed by TEM and the low indexing rate for this sample (Figure 1B).
Figure 2. Negative-stain TEM images of HPCD microcrystals.
A,B. Representative TEM sampling of H200Q crystals, illustrates sample monodispersity (A), with high quality crystal lattices (B) as determined from its FFT profile (inset). On average, 4th order Bragg spots were recorded for H200Q crystals.
C,D. Representative TEM sampling of WT crystals, illustrates sample aggregation or crystal conglomerates (C), typically not seen with brightfield or UV microscopy. Lattice quality was of high quality (D), with an average of 3rd order Bragg spots observed for WT samples. (Scale bars – A,C: 0.5 μm; B,D: 50 nm)
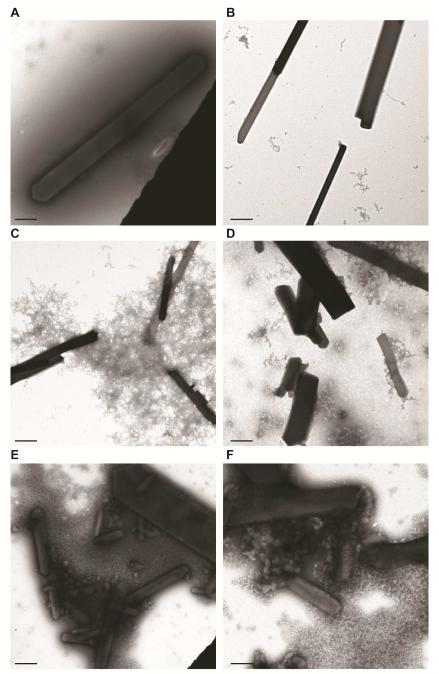
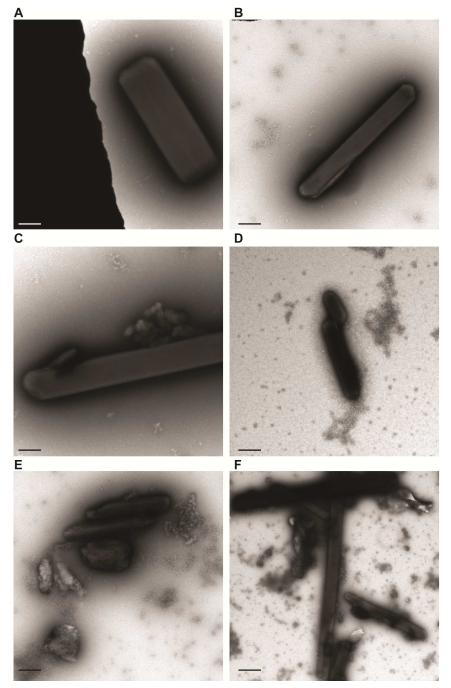
Since bulk fragmentation of the WT and H200Q microcrystals differed, we performed TEM experiments on the fragmentation protocols to determine its effects on sample monodispersity, lattice quality and crystal aggregation. While we observed no major differences in the lattice quality among HPCD samples prepared by different fragmentation methods, we did observe minor differences in the amount of crystal aggregation. TEM analysis revealed fewer aggregates for crystals fragmented via centrifugation or vortexing (with a glass bead cocktail) when compared to crystals prepared by sonication, irrespective of the characteristics of the starting material prior to fragmentation (Figures 3 and 4). Given that cryo-cooled macrocrystals generated for fragmentation of both H200Q and WT show no difference in diffraction or crystal quality (Kovaleva et al., 2015), the observed variance in the indexing rates between the two otherwise similar samples could stem from the differences in monodispersity and degree of protein or crystal aggregation generated during protein sample preparation or crystallization experiments. The extent and influence of these factors are speculative and will require additional testing to demonstrate a clear significant correlation.
Figure 3. Negative-stain TEM analysis of bulk fragmentation methods for rod-like 2,3 – HPCD microcrystals.
A,B. TEM images of H200Q (A) or WT (B) crystals after fractionation with centrifugation.
C,D. TEM images of H200Q (C) or WT (D) crystals after fractionation with vortexing.
E,F. TEM images of H200Q (E) or WT (F) crystals after fractionation with sonication.
Lattice quality was evaluated when single crystals were observed and median values of 3rd order Bragg spots (not shown) for all samples across the varying fragmentation protocols. (Scale bars – A-F: 1 μm)
Figure 4. Negative-stain TEM analysis of bulk fragmentation methods for needle-like 2,3 – HPCD microcrystals.
A,B. TEM images of H200Q (A) or WT (B) crystals after fractionation with centrifugation.
C,D. TEM images of H200Q (C) or WT (D) crystals after fractionation with vortexing.
E,F. TEM images of H200Q (E) or WT (F) crystals after fractionation with sonication.
Lattice quality was evaluated when single crystals were observed and median values of 3rd order Bragg spots (not shown) for all samples across the varying fragmentation protocols. (Scale bars – A-F: 1 μm)
Discussion
Successful liquid injector-based SFX experiments require consistent delivery of well diffracting single crystals into the XFEL beam path. The inherent diffraction quality of the microcrystals, crystalline monodispersity and the compatibility of the sample to the injection process are all important factors. Our parallel SFX and TEM experiments with HPCD microcrystals illustrate that the use of optimized samples with the GDVN injector, such as the ordered monodispersed H200Q variant microcrystals, will result in efficient SFX experiments. While not reported herein, approximately 16 minutes of protein crystal screening time at CXI (LCLS) resulted in an enzyme structure at 1.75 Å resolution, which is one of the most rapidly collected SFX datasets at atomic resolution using a GDVN injector (manuscript in preparation). In contrast, the presence of protein aggregates or crystal conglomerates can greatly decrease the occurrence of useful diffraction images (even if the crystal lattice is of high quality), requiring much longer data collection time and increased sample volumes to acquire enough data for a complete dataset. Therefore, pre-characterization of samples to identify and optimize conditions that promote monodispersity with highly ordered crystal lattices prior to XFEL beam time would lead to increased efficiency and higher-quality SFX data.
The application of TEM is particularly advantageous in this regard, for systems where crystals are too small (< 5 μm) for X-ray diffraction screening measurements at micro-focused synchrotron beamlines. X-ray powder diffraction (XPD) is a commonly used tool to assess the diffraction capability of microcrystalline preparations, but much like TEM, powder patterns will not exhibit the same resolution to which single crystals diffract at XFEL sources (Schlichting, 2015). Thus, while this information would be useful where TEM is not accessible, it would not provide details beyond the crystallinity of the sample. The usefulness in TEM compared to other sample preparation tools (I.E. dynamic light scattering, SONICC, UV-fluorescence, XPD, etc.) is that it not only is capable of identifying crystalline material, but also provides the experimenter with additional information about further optimizing or discarding crystals grown under certain conditions (Stevenson et al., 2016).
In addition to microcrystal quality, our data also raises the possibility that crystal density may play a greater role in data collection efficiency than previously thought. Assessment of our data and previously published results illustrate negative correlation between increasing overall indexing efficiency with decreasing crystal densities (Boutet et al., 2012; Brewster et al., 2015; Chapman et al., 2011; Hattne et al., 2014; Redecke et al., 2013; Schmidt et al., 2015; Tenboer et al., 2014). Furthermore, when considering the monodispersed H200Q sample (as visualized by TEM) which had lower crystal densities than those routinely used for GDVN based experiments, we still observed ~20% double lattice hits for indexed images. This raises the possibility that lower crystal densities may be advantageous and practical for liquid-jet experiments. However, since TEM experiments to analyze monodispersity and overall quality of crystal lattices were not performed for previously published samples, we cannot rule out that these factors played a role in their overall indexing hit rates. Yet taken together, these data suggest that monodisperse samples analyzed by TEM with crystal densities of 108 crystals/mL may improve efficiency and lessen sample consumption, which is applicable to challenging protein targets (I.E. membrane proteins, multi-protein complexes, etc.).
Acknowledgements
This work was supported by SLAC and Laboratory Directed Research Development grant 0006-15 (E.G.K. and A.E.C.). Use of the Linac Coherent Light Source (LCLS), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. We thank Michael M. Mbughuni for protein purification, and staff at LCLS-CXI for assistance with data collection. This work was supported by the National Institute of General Medical Sciences (NIGMS) of the US National Institutes of Health (NIH) under Award numbers GM112686 (G.C), GM112497 (C.O.B), and GM102520 (A. S. B.). H.P.S and G.C acknowledge support from BioXFEL-STC1231306.
Footnotes
Author Contributions:
E.G.K conducted crystallization, sample preparation, and X-ray data collection. A.S.B. and E.L.B performed processing of diffraction image data. D.P.D. provided technical assistance with sample delivery during XFEL data collection. C.O.B., H.P.S., X.F. and G.C. conducted UV imaging and TEM analysis. E.G.K, C.O.B., A.E.C., and G.C wrote the paper with contributions from all authors.
References
- Abdallah BG, Roy-Chowdhury S, Coe J, Fromme P, Ros A. High Throughput Protein Nanocrystal Fractionation in a Microfluidic Sorter. Analytical chemistry. 2015;87:4159–4167. doi: 10.1021/acs.analchem.5b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquila A, Hunter MS, Doak RB, Kirian RA, Fromme P, White TA, Andreasson J, Arnlund D, Bajt S, Barends TR, et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Optics express. 2012;20:2706–2716. doi: 10.1364/OE.20.002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha S, Nass K, Barends TR, Kabsch W, Latz B, Dworkowski F, Foucar L, Panepucci E, Wang M, Shoeman RL, et al. Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams. Acta crystallographica Section D, Biological crystallography. 2015;71:387–397. doi: 10.1107/S1399004714026327. [DOI] [PubMed] [Google Scholar]
- Boutet S, Lomb L, Williams GJ, Barends TR, Aquila A, Doak RB, Weierstall U, DePonte DP, Steinbrener J, Shoeman RL, et al. High-resolution protein structure determination by serial femtosecond crystallography. Science (New York, NY) 2012;337:362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster AS, Sawaya MR, Rodriguez J, Hattne J, Echols N, McFarlane HT, Cascio D, Adams PD, Eisenberg DS, Sauter NK. Indexing amyloid peptide diffraction from serial femtosecond crystallography: new algorithms for sparse patterns. Acta crystallographica Section D, Biological crystallography. 2015;71:357–366. doi: 10.1107/S1399004714026145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman HN, Fromme P, Barty A, White TA, Kirian RA, Aquila A, Hunter MS, Schulz J, DePonte DP, Weierstall U, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470:73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad CE, Basu S, James D, Wang D, Schaffer A, Roy-Chowdhury S, Zatsepin NA, Aquila A, Coe J, Gati C, et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ. 2015;2:421–430. doi: 10.1107/S2052252515009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePonte DP, Weierstall U, Schmidt K, Warner J, Starodub D, Spence JC, Doak RB. Gas dynamic virtual nozzle for generation of microscopic droplet streams. Journal of Physics D: Applied Physics. 2008;41:195505. [Google Scholar]
- Fromme R, Ishchenko A, Metz M, Chowdhury SR, Basu S, Boutet S, Fromme P, White TA, Barty A, Spence JC, et al. Serial femtosecond crystallography of soluble proteins in lipidic cubic phase. IUCrJ. 2015;2:545–551. doi: 10.1107/S2052252515013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn HM, Messerschmidt M, Ji X, Zhang H, Axford D, Gildea RJ, Winter G, Brewster AS, Hattne J, Wagner A, et al. Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data. Nature communications. 2015;6:6435. doi: 10.1038/ncomms7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomery K, Humphrey EC, Herring R. Imaging and diffraction of protein crystallization using TEM. Microscopy (Oxford, England) 2013;62:363–368. doi: 10.1093/jmicro/dfs088. [DOI] [PubMed] [Google Scholar]
- Groce SL, Lipscomb JD. Aromatic ring cleavage by homoprotocatechuate 2,3-dioxygenase: role of His200 in the kinetics of interconversion of reaction cycle intermediates. Biochemistry. 2005;44:7175–7188. doi: 10.1021/bi050180v. [DOI] [PubMed] [Google Scholar]
- Hattne J, Echols N, Tran R, Kern J, Gildea RJ, Brewster AS, Alonso-Mori R, Glockner C, Hellmich J, Laksmono H, et al. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Nature methods. 2014;11:545–548. doi: 10.1038/nmeth.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MS, Fromme P. Toward structure determination using membrane-protein nanocrystals and microcrystals. Methods (San Diego, Calif) 2011;55:387–404. doi: 10.1016/j.ymeth.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Processing of X-ray snapshots from crystals in random orientations. Acta crystallographica Section D, Biological crystallography. 2014;70:2204–2216. doi: 10.1107/S1399004714013534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J, Hattne J, Tran R, Alonso-Mori R, Laksmono H, Gul S, Sierra RG, Rehanek J, Erko A, Mitzner R, et al. Methods development for diffraction and spectroscopy studies of metalloenzymes at X-ray free-electron lasers. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014;369:20130590. doi: 10.1098/rstb.2013.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirian RA, Wang X, Weierstall U, Schmidt KE, Spence JC, Hunter M, Fromme P, White T, Chapman HN, Holton J. Femtosecond protein nanocrystallography-data analysis methods. Optics express. 2010;18:5713–5723. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva EG, Lipscomb JD. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science (New York, NY) 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva EG, Lipscomb JD. Structural basis for the role of tyrosine 257 of homoprotocatechuate 2,3-dioxygenase in substrate and oxygen activation. Biochemistry. 2012;51:8755–8763. doi: 10.1021/bi301115c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva EG, Rogers MS, Lipscomb JD. Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200. Biochemistry. 2015;54:5329–5339. doi: 10.1021/acs.biochem.5b00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupitz C, Grotjohann I, Conrad CE, Roy-Chowdhury S, Fromme R, Fromme P. Microcrystallization techniques for serial femtosecond crystallography using photosystem II from Thermosynechococcus elongatus as a model system. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014;369:20130316. doi: 10.1098/rstb.2013.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wacker D, Gati C, Han GW, James D, Wang D, Nelson G, Weierstall U, Katritch V, Barty A, et al. Serial femtosecond crystallography of G protein-coupled receptors. Science (New York, NY) 2013;342:1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft JR, Wolfley JR, Franks EC, Lauricella AM, Gualtieri EJ, Snell EH, Xiao R, Everett JK, Montelione GT. The detection and subsequent volume optimization of biological nanocrystals. Struct Dyn. 2015;2:041710. doi: 10.1063/1.4921199. PMC4711624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbughuni MM, Meier KK, Munck E, Lipscomb JD. Substrate-mediated oxygen activation by homoprotocatechuate 2,3-dioxygenase: intermediates formed by a tyrosine 257 variant. Biochemistry. 2012;51:8743–8754. doi: 10.1021/bi301114x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RJ. Femtosecond crystallography with ultrabright electrons and x-rays: capturing chemistry in action. Science (New York, NY) 2014;343:1108–1116. doi: 10.1126/science.1248488. [DOI] [PubMed] [Google Scholar]
- Qu K, Zhou L, Dong YH. An improved integration method in serial femtosecond crystallography. Acta crystallographica Section D, Biological crystallography. 2014;70:1202–1211. doi: 10.1107/S1399004714002004. [DOI] [PubMed] [Google Scholar]
- Redecke L, Nass K, DePonte DP, White TA, Rehders D, Barty A, Stellato F, Liang M, Barends TR, Boutet S, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science (New York, NY) 2013;339:227–230. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter NK, Hattne J, Brewster AS, Echols N, Zwart PH, Adams PD. Improved crystal orientation and physical properties from single-shot XFEL stills. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2014;70(12):3299–3309. doi: 10.1107/S1399004714024134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter NK, Poon BK. Autoindexing with outlier rejection and identification of superimposed lattices. J Appl Crystallogr. 2010;43:611–616. doi: 10.1107/S0021889810010782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting I. Serial femtosecond crystallography: the first five years. IUCrJ. 2015;2:246–255. doi: 10.1107/S205225251402702X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting I, Miao J. Emerging opportunities in structural biology with X-ray free-electron lasers. Current opinion in structural biology. 2012;22:613–626. doi: 10.1016/j.sbi.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Pande K, Basu S, Tenboer J. Room temperature structures beyond 1.5 A by serial femtosecond crystallography. Struct Dyn. 2015;2:041708. doi: 10.1063/1.4919903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert R, Meyer A, Dierks K, Kapis S, Reimer R, Einspahr H, Perbandt M, Betzel C. Reliably distinguishing protein nanocrystals from amorphous precipitate by means of depolarized dynamic light scattering. Journal of Applied Crystallography. 2015;48:1476–1484. [Google Scholar]
- Stevenson HP, DePonte DP, Makhov AM, Conway JF, Zeldin OB, Boutet S, Calero G, Cohen AE. Transmission electron microscopy as a tool for nanocrystal characterization pre- and post-injector. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014a;369:20130322. doi: 10.1098/rstb.2013.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson HP, Lin G, Barnes CO, Sutkeviciute I, Krzysiak T, Weiss SC, Reynolds S, Wu Y, Nagarajan V, Makhov AM, et al. Transmission electron microscopy for the evaluation and optimization of crystal growth. Acta crystallographica Section D, Biological crystallography. 2016 doi: 10.1107/S2059798316001546. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson HP, Makhov AM, Calero M, Edwards AL, Zeldin OB, Mathews II, Lin G, Barnes CO, Santamaria H, Ross TM, et al. Use of transmission electron microscopy to identify nanocrystals of challenging protein targets. Proceedings of the National Academy of Sciences of the United States of America. 2014b;111:8470–8475. doi: 10.1073/pnas.1400240111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara M, Mizohata E, Nango E, Suzuki M, Tanaka T, Masuda T, Tanaka R, Shimamura T, Tanaka Y, Suno C, et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nature methods. 2015;12:61–63. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, Frank M, Hunter M, Boutet S, Williams GJ, Koglin JE, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science (New York, NY) 2014;346:1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierstall U, James D, Wang C, White TA, Wang D, Liu W, Spence JC, Bruce Doak R, Nelson G, Fromme P, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nature communications. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]