Abstract
Purpose
Mutations in membrane frizzled-related protein (MFRP) are associated with nanophthalmia, hyperopia, foveoschisis, irregular patches of RPE atrophy, and optic disc drusen in humans. Mouse mfrp mutants show retinal degeneration but no change in eye size or refractive state. The goal of this work was to generate zebrafish mutants to investigate the loss of Mfrp on eye size and refractive state, and to characterize other phenotypes observed.
Methods
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 methods were used to generate multiple frameshift mutations in zebrafish mfrp causing premature translational stops in Mfrp. Spectral-domain optical coherence tomography (SD-OCT) was used to measure eye metrics and refractive state, and immunohistochemistry was used to study adult eyes. Gene expression levels were measured using quantitative PCR.
Results
Zebrafish Mfrp was shown to localize to apical and basal regions of RPE cells, as well as the ciliary marginal zone. Loss of Mfrp in mutant zebrafish was verified histologically. Zebrafish eyes that were mfrp mutant showed reduced axial length causing hyperopia, RPE folding, and macrophages were observed subretinally. Visual acuity was reduced in mfrp mutant animals.
Conclusions
Mutation of zebrafish mfrp results in hyperopia with subretinal macrophage infiltration, phenocopying aspects of human and mouse Mfrp deficiency. These mutant zebrafish will be useful in studying the onset and progression of Mfrp-related nanophthalmia, the cues that initiate the recruitment of macrophages, and the mechanisms of Mfrp function.
Keywords: mfrp, zebrafish, hyperopia, macrophage infiltration
Refractive error and its management place a substantial burden on quality of life, and the global loss of productivity associated with uncorrected refractive error (URE) was estimated to cost hundreds of billions of dollars, with URE-related blindness affecting 39 million people and URE-related visual impairment affecting 285 million people globally according to recent estimates.1,2 Myopia, hyperopia, or astigmatism occur when the refractive apparatus of the eye (cornea and lens) and the axial length of the eye are not correctly coordinated, and incoming light rays are not brought to a sharp point of focus on the retina, causing blurred vision. Hyperopia refers to an eye whose axial length is shorter than the focal length, or whose lens has low convergent power because of weak action of ciliary muscles. Hyperopic patients have higher incidence rates of strabismus and amblyopia,3–5 and loss of vision that cannot be corrected can occur as a result.
Nanophthalmia (also known as posterior/partial microphthalmia) is an extreme form of hyperopia. Nanophthalmic eyes of human patients are small but functional, with refractive errors of +8.00 to +25.00 diopters, and show thickening of both the choroidal vascular bed and sclera.6,7 Although the scleras of nanophthalmic eyes are excessively small, the retinas are normally sized, and the crowding of the retina inside the sclera often leads to separation of the retina and RPE, causing folds to form in the macula with deformation or loss of the fovea, known as foveoschisis.8 Additionally, nanophthalmic eyes are prone to angle-closure glaucoma and pigmentary retinal dystrophy.9,10 Currently, nanophthalmia is linked with four loci, termed nanophthalmos (NNOS) 1 to 4. NNOS 1 and 3 have only been localized to chromosomal regions (11p; 2q), and NNOS 2 and 4 are caused by mutations in membrane frizzled-related protein (MFRP) and TMEM98, respectively.11,12
Membrane frizzled-related protein is a type II transmembrane receptor with an extracellular frizzled-related cysteine-rich domain, two cubulin domains, and two low-density lipoprotein receptor class A (LDLA) repeats13 (Fig. 1A). Membrane frizzled-related protein is expressed in the RPE and ciliary body of the eye.14,15 Mutations in MFRP are associated with NNO2 (OMIM 609549), microphthalmia (OMIM 611040), high hyperopia, retinitis pigmentosa, foveoschisis, irregular patches of RPE atrophy, and optic disc drusen in humans, and retinal degeneration in mice.11,16–20 Membrane frizzled-related protein has been reported to be transcribed dicistronically with a downstream gene located in its 3′ untranslated region: C1q and tumor necrosis factor related protein 5 (C1qTNF5).21 However, it has also been shown that the C1qTNF5 gene has its own independent promoter.22 Membrane frizzled-related protein and C1qTNF5 protein have been shown to co-immunoprecipitate, indicating that the proteins have the potential to interact in vivo.23 Furthermore, although a premature stop codon in mouse Mfrp might be expected to lead to nonsense-mediated decay of its mRNA transcript, this is not the case. Quantitative PCR (qPCR) analyses show that both Mfrp and C1qtnf5 transcripts are significantly upregulated in the rdx/Mfrp174delG mutant mouse. C1qTNF5 is also upregulated at the protein level in Mfrp mutants.22 Although it has been speculated that this could reflect an autoregulatory feedback mechanism controlling expression of both genes, this idea is confounded by the close proximity of the two genes.
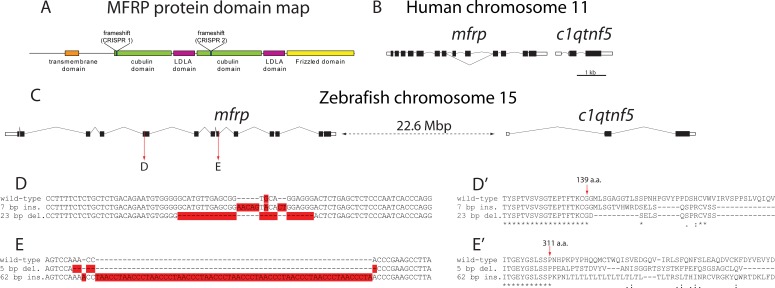
Figure 1.
Schematic of human and zebrafish mfrp loci. (A) Membrane frizzled-related protein contains a transmembrane domain, two cubulin domains, two LDLA domains, and a Frizzled domain. Zebrafish frameshift mutants were made by separately targeting the two cubulin domains using CRISPR guide RNAs. (B, C) Human and zebrafish mfrp chromosomal regions are shown with c1qtnf5. Human mfrp is immediately adjacent to c1qtnf5 but is separated by more than 22 Mbp in zebrafish. Scale bar: 1 kb. (D, D') Alignments show DNA and protein sequences, respectively, of wild-type and mutant alleles of zebrafish mfrp generated using CRISPR 1. (E, E') Alignments show the DNA and protein sequences of the wild-type and mutant alleles generated using CRISPR 2. Red highlighting indicates edited bases.
Previous animal models of mfrp mutation have failed to recapitulate the nanophthalmic phenotype observed in humans. The rd6/Mfrprd6 mouse mutant skips exon 4 of mfrp as a result of a splice donor mutation, removing part of the transmembrane domain,14 and has been used as a model for Stargardt's disease and other flecked retina disorders, as it exhibits white subretinal spots regularly spaced across the eye, and shows slow photoreceptor degeneration throughout the life of the mouse, as well as an increase in the number of apical microvilli of RPE cells.15 Photoreceptors in mfrprd6 mice are disorganized and degenerate postnatally.24 Another mfrp mouse mutant, rdx/Mfrp174delG, is missing a single base pair from exon 3 (giving rise to only the N-terminal cytosolic domain followed by 83 nonsense amino acids), and shows eventual RPE atrophy in addition to white fundus flecks and progressive retinal degeneration.25 The white subretinal spots were found to be macrophages derived directly from circulating monocytes, and the activity of the alternative complement pathway was found to be depressed in rdx/Mfrp174delG mice.25,26 Importantly, however, neither mfrp mutant mouse shows evidence of nanophthalmia or hyperopia. It has been speculated that the species differences between mice and humans may underlie this disparity, although the exact cause or mechanism is unknown.27
To investigate the effects of mfrp loss in another animal model, we chose the zebrafish, Danio rerio, whose cone-rich retina and diurnal lifestyle contrast with the crepuscular mouse. Using a clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 approach, we induced premature stop codons in zebrafish mfrp and show that loss of mfrp causes reduced eye size when normalized to either body axis length, or to the lens of the eye. This reduction in eye size, first seen at 1 month postfertilization (mpf), leads to a significant increase in relative refractive error such that the fish are highly hyperopic. Retinas of mfrp−/− zebrafish had no gross abnormalities and laminated normally. However, the eyes showed frequent folds in the RPE, as well as bulging of the retinal layers, likely secondary complications caused by the normally sized retina detaching from the too-small sclera. In addition, subretinal macrophages were observed in mfrp−/− eyes. Here we demonstrate that inactivation of zebrafish mfrp leads to a nanophthalmic-hyperopic phenotype that recapitulates that seen in human patients deficient in functional MFRP.
Materials and Methods
Zebrafish Husbandry
Zebrafish were housed in recirculating filtered water system using reverse-osmosis purified water built by Aquatic Habitats (Aquatic Habitats, Apopka, FL, USA). Water was supplemented with Instant Ocean salt (60 mg/L) and maintained at 28.5°C with a 14-hour light and 10-hour dark cycle. Before experimental manipulation or tissue fixation, fish were anesthetized in 0.2 mg/mL ethyl 3-aminobenzoate methanesulfonate (tricaine) (Sigma-Aldrich, St. Louis, MO, USA). All experiments were performed in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. The protocol was approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin, protocol number AUA1378.
Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Synthesis and Gene Editing
Clustered regularly interspaced short palindromic repeats were designed against regions of zebrafish mfrp exons using ZiFiT Targeter Version 4.2 (http://zifit.partners.org/ZiFiT, in the public domain).28,29 The targeted sequences were as follows: mfrp CRISPR 1, 5′-GGGGGCATGTTGAGCGGTGC-3′, targeting exon 5; mfrp CRISPR 2 5′- GGGTAAGGCTTCGGGTGGTT-3′, targeting exon 8. Clustered regularly interspaced short palindromic repeats guide RNA templates were generated by cloning annealed oligonucleotides with appropriate overhangs into BsaI-digested pDR274 plasmid.30 Clustered regularly interspaced short palindromic repeats guide RNAs were synthesized using the MEGAshortscript T7 Transcription Kit and purified using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). Zebrafish codon-optimized cas9 was synthesized using the mMESSAGE mMACHINE Kit (Ambion) and polyadenylated using the Poly(A) Tailing Kit (Ambion). Clustered regularly interspaced short palindromic repeats guide RNAs and cas9 mRNA were injected into 1- to 4-cell zebrafish embryos at 12.5 ng/μL and 300 ng/μL, respectively, and surviving embryos raised to adulthood before outcrossing to identify founder fish carrying germline edits in mfrp. Offspring from these fish were raised to adulthood, then finclipped for genotyping. Screening for CRISPR 1 editing used primers CRISPR 1 amplicon-F (5′-ACAGCAGCTGAACGTGGCCCTT-3′) and CRISPR 1 amplicon-R (5′-AGGTCCCTCGATAGCCAGAGAC-3′) and assaying for loss of a BsrBI restriction site (23-bp deletion) or gain of an RsaI restriction site (7-bp insertion). Screening for CRISPR 2 editing used primers CRISPR 1 amplicon-F (5′-TGCAGCCACAAGCACAGAGGT-3′) and CRISPR 1 amplicon-R (5′-TCAGCACAGCCCTGATAGAGCAT-3′) and assaying for gain of an Hpy166II restriction site (5-bp deletion) or appearance of a higher molecular weight amplicon (62-bp insertion).
Histology
Eyes for semithin plastic histology were enucleated and pierced with an 18-gauge needle and immersed in fixative (2% glutaraldehyde, 2% formaldehyde, in 80 mM cacodylate buffer) for 1 hour. Aldehydes were washed away in 80 mM cacodylate buffer and the tissue was postfixed in 1% osmium tetroxide. The samples were then taken through a graded methanol series, washed with acetonitrile, and embedded in resin (Embed 812; Electron Microscopy Systems, Hatfield, PA, USA). The embedded tissue was sectioned axially with a thickness of 0.5 μm and stained with a 1% methylene blue solution before imaging. Eyes for cryosectioning and immunohistochemistry were fixed overnight in 4% formaldehyde, cryoprotected in 20% sucrose, and mounted in HistoPrep frozen tissue embedding medium (Thermo Fisher Scientific, Waltham, MA, USA). Cryosections were cut and thaw-mounted onto Superfrost Plus glass slides (Thermo Fisher Scientific) before rehydration in PBS. Tissues were blocked at room temperature using donkey serum in PBS with 1% Tween-20 and 1% Triton X-100 before applying antibodies at 4°C overnight. Antibodies used were goat anti-Mfrp at 1:200 dilution (R&D Systems, Minneapolis, MN, USA); mouse 4C4 at 1:200 dilution (kind gift from Peter Hitchcock, University of Michigan Kellogg Eye Center); mouse antiproliferating cell nucleic antigen (PCNA) at 1:200 (clone PC10; Cell Signaling Technology, Danvers, MA, USA); and rabbit anti-phospho histone H3 at 1:200 (Sigma-Aldrich). After incubation with primary antibodies, cryosections were washed with PBS before incubation with species-appropriate fluorescently conjugated secondary antibodies for 2 hours at room temperature. Cryosections were counterstained with TO-PRO-3 (Thermo Fisher Scientific) at 1 μM to label nuclei. Confocal microscopy was performed using a Nikon Eclipse E600FN microscope equipped with 488-nm and 635-nm excitation lasers, and appropriate filter sets. Images were generated using the Nikon EZ-C1 viewer (Nikon Instruments, Melville, NY, USA) and assembled and annotated using Adobe Illustrator (Adobe Systems, Inc., San Jose, CA, USA) software.
In Vivo Eye Measurement and Relative Refractive Error Calculation
Zebrafish were anesthetized and placed on the imaging stage. Axial length, body length, and retinal radius were measured as previously described.31 Relative refractive error was calculated as 1 − (retinal radius/F), where F, an idealized retinal radius = lens radius × 2.324.
Optokinetic Response (OKR) Assay for Visual Acuity in Adult Zebrafish
Zebrafish were placed in a plastic spectrophotometer cuvette and positioned at the center of a testing arena composed of four computer monitors that served as the chamber walls. The monitors are capable of presenting a moving grating, creating the effect of a virtual rotating cylinder with black and white stripes. These stimuli were generated by the Moving Grating v.1.3 software program (http://michaelbach.de/stim, in the public domain) with spatial frequency and contrast settings varied. Stimuli were presented to fish for 1 minute with clockwise rotation, then 1 minute of counterclockwise rotation, and saccades counted for both sessions. Saccades were counted as smooth eye pursuits in the direction of grating motion followed by a rapid reset eye movement in the opposite direction. Spatial frequencies were converted to cycles per degree (cpd) as previously described.32 Contrast testing was carried out at a constant spatial frequency of 7.7 cpd, with settings ranging from 0 (no contrast, gray screen) to 5 (full contrast, black and white stripes).
Quantitative PCR
Zebrafish eyes were disrupted with a motorized pestle and the RNA was purified using RNeasy kit (Qiagen, Germantown, MD, USA). cDNA was synthesized (iScript kit; Bio-Rad, Hercules, CA, USA), and quantitative PCR was performed (iCycler; Bio-Rad) using iQ-Sybr Green (Bio-Rad). Quantitative real-time PCR was performed using the following primers: zfMFRP-qPCR-F1 (5′-CCGATGAAGACAATGAGGAA-3′), zfMFRP-qPCR-R1 (5′-ACCAACACTGCTCTGTGT-3′), zfMFRP-qPCR-F2 (5′-GGTCAGAGAGGAGACAGAG-3′), zfMFRP-qPCR-R2 (5′-AGTGCCACCAATGCTTC-3′), zfC1QTNF5-qPCR-F1 (5′-GCAGGGATGGACGAG-3′), zfC1QTNF5-qPCR-R1 (5′-CCTCGGTCTCCTGTGA-3′), zfC1QTNF5-qPCR-F2 (5′-AGAAAGAGGGGAGAGCG-3′), zfC1QTNF5-qPCR-R2 (5′-GGCGCTAAAGGCTGATT-3′), ef1α F (5′-TCTCTCAATCTTGAAACTTATCAATCA-3′), ef1α R (5′-AACACCCAGGCGTACTTGAA-3′).
Statistical Analysis
At least six individual eyes were used for each experiment involving ocular metrics, except in Supplementary Figure S1, where only four eyes were available for mfrp−/−. For analysis of eye metrics and refractive state, sample sizes were as follows: 7 days postfertilization (dpf), n = 10 fish (20 eyes); 15 dpf, n = 10 fish (20 eyes); 31 dpf, n = 10 fish (20 eyes); 84 dpf, n = 10 fish (20 eyes); 149 dpf, n = 10 fish (20 eyes). Ten fish per genotype were assessed for adult optokinetic response. One-way ANOVA analyses were performed for all experiments, except quantitative PCR, when t-tests were used to compare expression levels of target genes in mfrp−/− and mfrp+/+ eyes. All statistical calculations were performed using GraphPad Prism (GraphPad, La Jolla, CA, USA).
Results
Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Inactivation of Zebrafish mfrp Results in Loss of Mfrp Protein From the RPE
Zebrafish Mfrp, a type II membrane protein with extracellular cubulin, LDLA, and frizzled domains, has 49% identity with human MFRP, and 64% conservation of amino acids (Fig. 1A). Unlike in human and mouse, the genes for zebrafish mfrp and c1qtnf5 are not located adjacent to one another, and instead are separated by more than 22 Mbp on chromosome 15 (Figs. 1B, 1C). This means that zebrafish mfrp is unlikely to be transcriptionally regulated with c1qtnf5, and mutations in mfrp should not have a direct effect on c1qtnf5 expression; however, it is still possible that each protein may affect the transcription of the other in trans. We designed CRISPRs to target exons 5 or 8 of zebrafish mfrp, toward the start of each cubulin domain. Injection of these CRISPRs along with cas9 mRNA into developing embryos generated four germline mfrp-disrupting alleles: a 7-bp insertion (mw77) and a 23-bp deletion (mw76) in exon 5 (CRISPR 1), and a 5-bp deletion (mw78) and a 62-bp insertion (mw79) in exon 8 (CRISPR 2) (Fig. 1). Each allele is predicted to change the reading frame of the Mfrp protein and cause premature truncation with some missense amino acid incorporation (Figs. 1D', 1E').
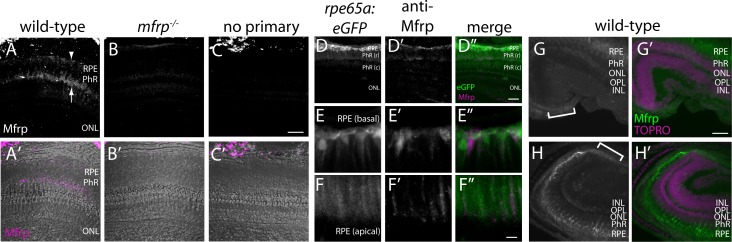
Using an MFRP-specific antibody, cryosections of wild-type zebrafish showed that Mfrp protein is localized in the RPE, and can be seen on both the apical and basal sides of the cells (Figs. 2A, 2A'). No staining was seen in cryosections incubated without anti-MFRP antibody. When cryosections of mfrpmw78/mw78 (referred to as mfrp−/−) siblings of wild-types were examined, no Mfrp immunostaining was visible, indicating that the epitope recognized by the antibody is no longer expressed, likely rendering the protein nonfunctional (Fig. 2B). When cryosections from transgenic rpe65a:eGFP (R. Collery, unpublished data, 2016) zebrafish eyes with cytoplasmic enhanced green fluorescent protein (eGFP) delineating RPE cells were immunostained with anti-MFRP antibody, staining of the RPE cells was seen in both the basal part of the cell (Figs. 2 D, 2E), as well as the fine apical microvillar processes (Fig. 2F). Mfrp was also detected in the ciliary epithelium contiguous with the RPE (Figs. 2G, 2H). Despite repeated attempts, the anti-Mfrp antibody was unable to detect zebrafish protein on Western blots, and so could not be used to further demonstrate loss of Mfrp protein in mutants.
Figure 2.
Zebrafish Mfrp is localized to the apical and basal RPE and ciliary region. (A, B, C) An anti-Mfrp antibody raised in mouse detected Mfrp in zebrafish apical and basal RPE (arrow, arrowhead, respectively). (A', B', C') Immunofluorescent signal in (A–C) is pseudocolored magenta to contrast with superimposed phase images of the cryosections. Note that rod outer segments (PhR) are found in the interval between basal and apical RPE. Cone inner and outer segments (not labeled) are in the interval between PhR and the ONL. No staining was seen when control cryosections were processed without anti-Mfrp primary antibody. Mfrp protein was seen in wild-type zebrafish RPE, but not in age-matched mutant (mfrp−/−) zebrafish with homozygous 5 bp deletions in mfrp (B, B'). Bright staining behind the eye is nonspecific staining by the secondary antibody as seen in (C). (D) Low-power image similar to (A) that includes RPE along with higher power images of basal RPE (E) and apical RPE (F) from transgenic fish expressing eGFP under control of the rpe65a promoter to delineate the RPE. Mfrp staining (D', E', F') in the RPE labels the same cells as those filled with cytoplasmic eGFP (D, E, F). Mfrp staining was seen on both apical (E') and basal (F') RPE. (D”, E”, F'') are merged images of eGFP (green) and MFRP (magenta). Note that because of the very high eGFP signal compared with that for Mfrp, (E) and (F) are single Z-slices of eGFP, whereas (E') and (F') are Z-projections through the same cells of Mfrp staining. (G, H) Mfrp staining was also seen in both the dorsal (G) and ventral (H) ciliary epithelium (brackets), contiguous with the RPE. In (G') and (H'), the Mfrp immunofluorescence is shown in green as an overlay with nuclei labeled with TO-PRO-3 (magenta). Scale bars: (A–D) 100 μm; (E–F) 20 μm; (G–H) 100 μm. PhR, photoreceptors (r, rods; c, cones); ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer.
Inactivation of mfrp Leads to Nanophthalmos With Reduced Size in the Posterior Segment of the Eye
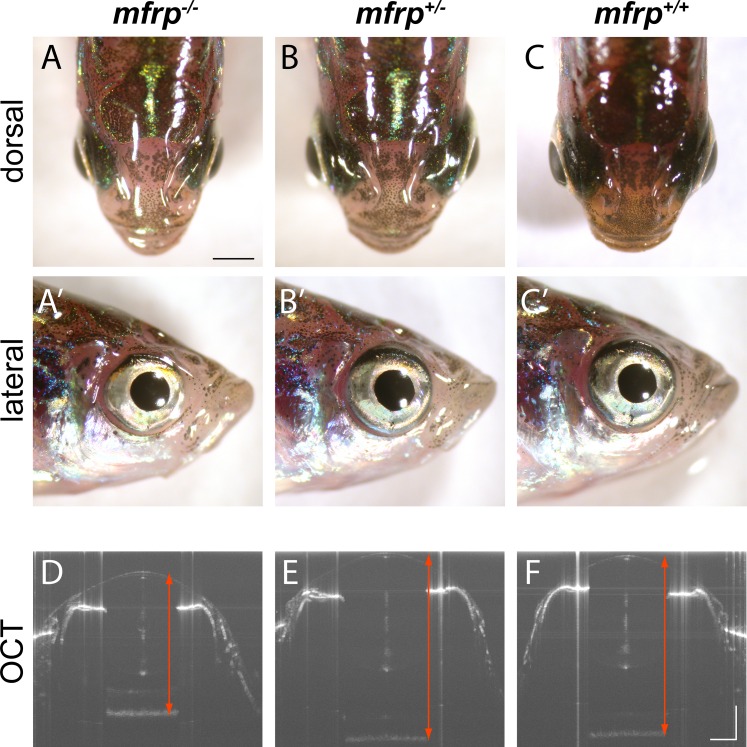
Confirmed heterozygous mfrp5bp/+ zebrafish (referred to as mfrp+/−) were incrossed to give offspring that were wild-type, heterozygous, and homozygous with respect to mfrp mutation. Adult offspring were genotyped and gross images taken dorsally and laterally at 5 mpf. Eyes of mfrp−/− mutants protrude less from the head than eyes of mfrp+/− or mfrp+/+ fish, suggesting that they are smaller in size (Fig. 3). Lateral views of the head show that eyes of mfrp−/− mutants are slightly smaller in dorsal-ventral and nasal-caudal aspects. However, it is the anterior-posterior or axial dimension that is most significantly altered. Spectral-domain optical coherence tomography (SD-OCT) confirmed that the axial length (cornea to back of RPE, axial length being the eye metric that is the largest contributor to refractive error leading to myopia33) of mfrp−/− mutants was significantly shorter than heterozygotes or wild-types in vivo (Figs. 3D–F). Interocular distances were measured and found normal in mfrp mutants, indicating that hypotelorism was not responsible for the apparent reduced eye size (Supplementary Fig. S1).
Figure 3.
Zebrafish mfrp mutants show reduced eye size consistent with nanophthalmia. (A, B, C) Dorsal views of mfrp mutant zebrafish and sibling heterozygous and wild-type controls show mfrp homozygotes have smaller eye globes that do not project from the head as much as in their control siblings. (A', B', C') Reduced eye size is also apparent in lateral views. (D, E, F) In vivo SD-OCT imaging shows the reduced axial length (front of cornea to back of RPE; red arrows) associated with homozygous mfrp mutants. Scale bars: (A–C), 1 mm; (D–F), 300 μm.
Retinas with RPEs attached were dissected from fixed mfrp−/−, mfrp+/−, and mfrp+/+ fish and imaged anteriorly and posteriorly (Figs. 4A–C). Retinal cups of mfrp−/− are smaller than controls, and both the RPE and neural retina show folds characteristic of excess mechanical constraint of sclera too small for the retina. Lamination of mfrp−/− retinas appeared normal, and no loss of rods or cones was observed.
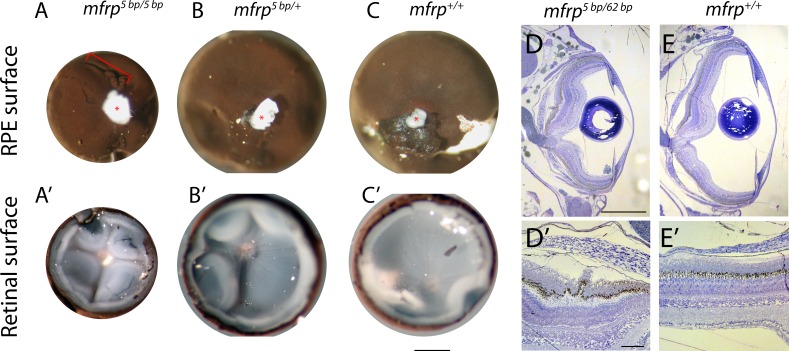
Figure 4.
Zebrafish mfrp mutant eyes show RPE folds and compressed retinas. (A, B, C) Dissected eyecups (RPE + retina, dissected after whole-eye fixation) from mfrp mutants are smaller than sibling controls. Mutants of mfrp show folds in the RPE (red bracket), and retinas show more involutions due to compression within a smaller sclera. Optic nerves are highlighted with red asterisks. (D, E) Semithin plastic histologic sections show that mfrp mutant eyes are smaller than control siblings. RPE folds can be seen clearly at higher magnifications. Scale bars: (A–C) 1 mm; (D–E) 500 μm; (D', E') 20 μm.
Eyes from adult mfrp−/− mutants along with wild-type sibling zebrafish were fixed for semithin plastic histology at 3 mpf and 5 mpf. At 3 mpf, mfrp−/− eyes were noticeably smaller than controls (Figs. 4D, 4E). Posterior segment size of mfrp−/− eyes was most severely affected, whereas anterior segments appeared to be similar in size to controls. At higher magnification, RPE and retinal folds could be clearly seen in mfrp−/− eyes but not in controls.
To address whether the retina may be hyperproliferative and contribute to the folding phenotype, cryosections were immunostained for PCNA. This mitotic cell marker, however, did not show differences in the number of proliferating cells between mfrp retinas relative to controls (Supplementary Fig. S2). These data suggest that RPE-retinal folding is due to the reduced scleral dimensions and not a result of retinal overgrowth.
Eyes That Are mfrp−/− Show Subretinal Microglia/Macrophages
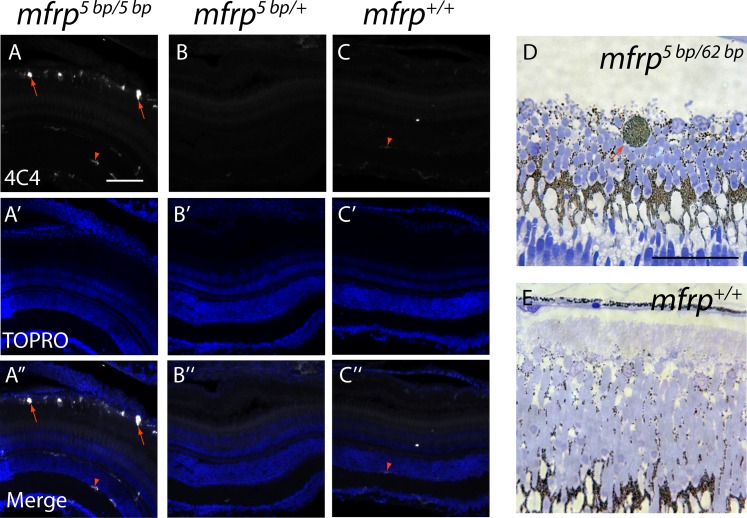
Eyes that are mfrp−/−, mfrp+/−, and mfrp+/+ were fixed for cryosectioning before staining with 4C4 antibody, which recognizes activated microglia/macrophages in zebrafish, as shown by morphologic analysis following nerve lesion.34 Subretinal cells in the posterior part of mfrp−/− eyes stained positively for microglia/macrophages and showed an amoeboid appearance, contrasting with resident macrophage/microglia that show a slender morphology (Fig. 5).35 Plastic sections of mfrp−/− eyes showed round pigmented cells located subretinally. Based on their similarity to pigmented microglia/macrophages in mice,25 it is likely that these represent the 4C4-positive cells in Figure 5A that were identified as macrophages. No subretinal pigmented cells were seen in control eyes.
Figure 5.
Zebrafish mfrp mutants show subretinal microglia/macrophages. (A, B, C) Cells in the subretinal space of mfrp mutant zebrafish stain positively with a microglia/macrophage marker (antibody 4C4; red arrows), whereas control heterozygous and wild-type sibling zebrafish lack such staining in the subretinal space. Resident microglia in the inner retina also stain with this marker (red arrowheads). (D, E) A pigmented subretinal cell, likely a macrophage, is visible among the photoreceptor outer segments in an mfrp mutant plastic section (red arrow). These pigmented cells are not seen in controls. Scale bars: (A–F) 100 μm; (G–H) 20 μm.
Inactivation of mfrp Results in Hyperopia
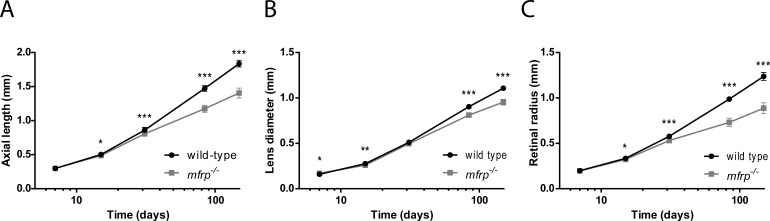
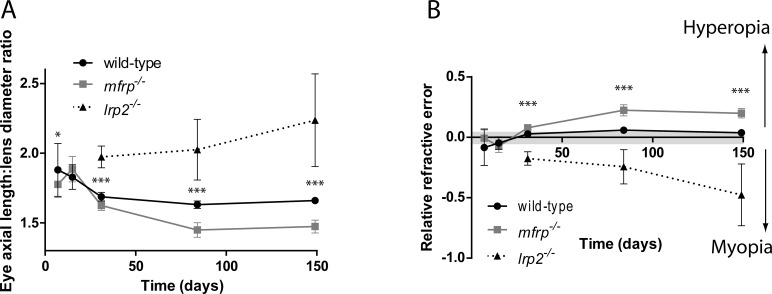
Eye sizes of mfrp−/− mutants at various ages along with wild-type sibling zebrafish were measured using SD-OCT. Axial length, retinal diameter, and retinal radius (center of lens to back of RPE) were measured, along with body axis length, and used to calculate eye size as a ratio of independent anatomical metrics to normalize between individual animals whose size varies naturally within a given population (Fig. 6). Relative refractive error, a measure of the degree of deviation from emmetropia, where zero represents an emmetropic eye, negative numbers indicate myopia, and positive numbers indicate hyperopia, was also calculated.31 At 7 dpf, eye sizes and refractive state of mfrp−/− mutants cannot be distinguished from wild-type controls (Figs. 6, 7). However, at 15 dpf, axial length, lens diameter, and retinal radius are significantly reduced in mfrp−/− mutants, and by 1 mpf, these fish show significant hyperopia when relative refractive error is calculated. At 13 weeks and 26 weeks, mfrp−/− eyes were smaller than both mfrp+/− heterozygotes and wild-type when normalized with respect to either body axis length (global independent metric) or lens diameter (independent ocular metric). Eyes that were mfrp−/− were also significantly more hyperopic than heterozygotes or wild-types, and these changes in eye size and refractive state were even more pronounced at 3 months. In Figures 6 and 7, mfrp−/− refers to the mfrp5bp/5bp allele (mw78). Eye axial length: lens diameter ratios and relative refractive errors were also measured for mfrp7bp/23bp homozygotes, heterozygotes, and wild-types at 2 mpf, and these homozygotes were also found to have decreased eye axial lengths and increased hyperopia with respect to controls (Supplementary Fig. S3).
Figure 6.
Zebrafish mfrp mutants show smaller eye metrics than wild-types. (A) Eye axial length is significantly shorter in mfrp mutants than wild-type controls at 15 dpf, and this difference becomes more pronounced as the fish age. (B) Lens diameter is significantly smaller in mfrp mutants than wild-type controls at 7 dpf, and this difference becomes more pronounced as the fish age. (C) Retinal radius, a proxy for focal length of the eye, is significantly shorter in mfrp mutants than wild-type controls at 15 dpf, and this difference becomes more pronounced as the fish age. ***P < 0.0001; **P < 0.01; *P < 0.05. Sample sizes are indicated in the Statistical Analysis section of Materials and Methods.
Figure 7.
Zebrafish mfrp mutants exhibit hyperopia. (A, B) Relative refractive error measurements of mfrp5bp/5bp mutant zebrafish show that they are significantly hyperopic compared with wild-type controls at 30 dpf, and continue to be hyperopic as they age. A previously published zebrafish model of myopia, lrp2−/−,40 is also graphed to show refractive error in the opposite direction. The gray bar in B shows the normal range of relative refractive error observed in wild-type zebrafish. ***P < 0.0001; *P < 0.05. Significance asterisks indicate comparisons between wild-type and mfrp mutants, although lrp2 mutants are also significantly different from both. Sample sizes are indicated in the Statistical Analysis section of Materials and Methods.
Zebrafish Mutants for mfrp Show Reduced Optokinetic Response
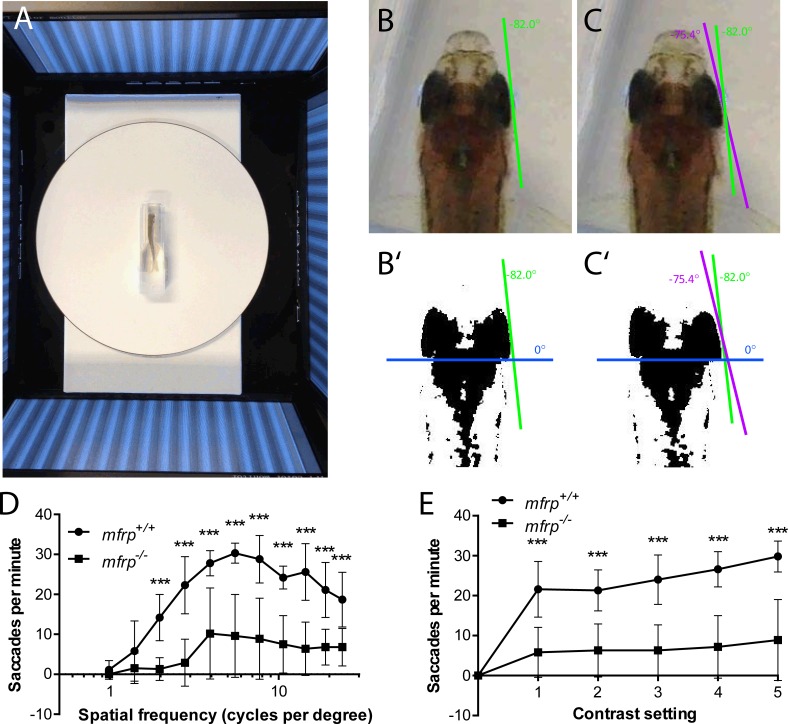
Adult zebrafish that were mfrp+/+ and mfrp−/− were tested for optokinetic response at 12 mpf. Using our system, mfrp+/+ fish demonstrated a visual acuity (VA) threshold of 1.395 cpd, with a mean number of 5.8 saccades per minute, exceeding the VA cutoff of 3 saccades as described by Tappeiner and colleagues36 (Figs. 8D, 8E). However, mfrp−/− fish did not reach the VA cutoff until a spatial frequency of 3.922 cpd, with an average of 10.2 saccades. At lower spatial frequencies, mfrp−/− fish showed fewer saccades per minute, indicating that loss of Mfrp leads to reduced VA.
Figure 8.
Zebrafish mutant for mfrp show reduced optokinetic response. (A) Zebrafish undergoing OKR assay are placed in a plastic cuvette at the center of a testing arena made of four computer monitors that simulate a rotating drum. (B, B', C, C') Images of zebrafish eye positions were taken immediately before (B) and after (C) a rapid eye reset following a smooth pursuit, or saccade. Images were thresholded in ImageJ (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA) to highlight the highly melanized posterior part of the eye used to define eye position. (D) Fish that were mfrp−/− showed lower visual acuity than mfrp+/+ siblings at 12 mpf by OKR assay. (E) Fish that were mfrp−/− showed fewer saccades per minute than mfrp+/+ siblings by OKR assay when contrast settings were reduced; however, this appeared to be due to reduced visual acuity rather than lack of contrast sensitivity. Graphs show mean saccades per minute ± SD.
Maintaining the spatial frequency at 7.7 cpd, a setting that pilot studies showed to be close to the optimum frequency for wild-type visual response, mfrp+/+ and mfrp−/− fish OKRs were assessed at sequentially lower contrast settings. Although mfrp−/− fish showed fewer saccades than mfrp+/+ fish at each contrast setting, this appeared to be constant rather than changing based on contrast level. This reduction is likely due to lower VA and not a diminished response to contrast levels.
Levels of c1qtnf5 Are Not Altered in mfrp−/− Mutant Eyes
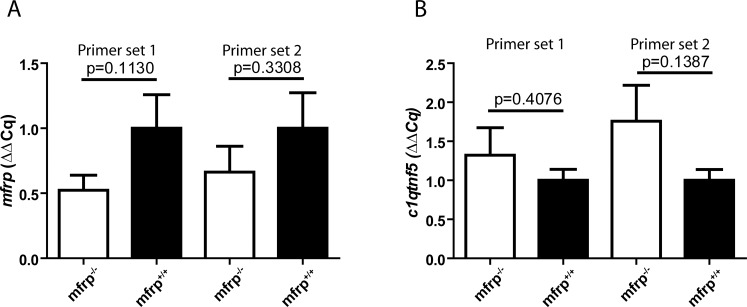
In mice, mutations in Mfrp result in upregulation of C1qtn5, although this could reflect the tight linkage of the two coding frames rather than a regulatory relationship. In a preliminary investigation of whether loss of Mfrp affected levels of c1qtnf5 in mutant eyes, RNA was extracted from mfrp−/− and wild-type adult eyes and used to synthesize cDNA. Two independent primer sets designed against c1qtnf5 were used for qPCR, using ef1α to normalize between samples. For both primer sets, no significant change in c1qtnf5 mRNA levels was seen, suggesting that presence or absence of Mfrp may have little effect on transcription of c1qtnf5 (Fig. 9).
Figure 9.
Zebrafish mfrp mutants do not show significant changes in mfrp or c1qtnf5 mRNA levels. (A) Quantitative RT-PCR on cDNA synthesized from eyes of mfrp mutants showed no significant change in levels of mfrp mRNA compared with wild-type controls assessed by t-test. (B) Similarly, no significant change in levels of c1qtnf5 mRNA compared with wild-type controls. In both experiments, two primer sets were independently used.
Similarly, levels of mfrp mRNA were measured in mfrp−/− and wild-type adult eyes using two independent primer sets. No significant change was seen in mfrp levels for either genotype, suggesting that nonsense-mediated decay is insufficient to significantly reduce mfrp mRNA levels. Because the RNA used for qPCR analysis was extracted from whole eyes, and not exclusively from tissues expressing Mfrp, we cannot rule out the possibility that mfrp and c1qtnf5 may regulate one another at a transcriptional level. Future experiments on purified RPE samples may help to answer this question.
Discussion
Mutations in MFRP have been associated with extreme hyperopia and retinal flecking in humans, but an animal model that recapitulates these phenotypes together has not previously been developed. Although the flecked retinal phenotype and photoreceptor degeneration has been characterized in rd6 and rdx mice, and has provided insight into the macrophage-based nature of the fundus flecks, neither nanophthalmia nor hyperopia were observed in these animals.14,25,26 It has been speculated that species differences, genetic differences, or perhaps nocturnal lifestyle associated with the mouse eye may underlie the difference in eye size response to Mfrp deficiency.27 In this work, we disrupted translation of full-length zebrafish Mfrp protein at multiple locations in the coding region and found that these mutant alleles uniformly led to nanophthalmia, hyperopia, and increases in subretinal microglia/macrophages. Like mice, the zebrafish eye is much smaller than that of humans, reaching only 1 to 2 mm in adulthood. However, like humans, zebrafish have a diurnal lifestyle and an abundance of cone photoreceptors. As circadian-based vision is known to affect eye growth, perhaps these shared features explain the similar phenotypes between zebrafish and humans in response to loss of Mfrp. For these reasons, zebrafish represent an emerging model for studying the genetic and cellular mechanisms underlying refractive errors in humans.
Human patients with nanophthalmia and hyperopia caused by MFRP mutations suffer from serious visual problems, and accompanying pathologies threaten to cause complete blindness. Hyperopic patients are prone to retinitis pigmentosa, optic disc drusen, and reduced electroretinographic responses from rod and cone photoreceptors. Although mutations in MFRP are responsible for a reduction in eye size as well as the retinal pathology, the link between the two sets of symptoms is unclear. In addition, it is not known whether reduced eye size, with its accompanying mechanical stress, is the cause of subsequent inflammatory responses in the MFRP retina, or if both processes take place independently. Mice with mfrp mutations have been shown to undergo retinal degeneration, with RPE atrophy and reduced ERGs.24,25 Furthermore, mfrp mutant mice show the appearance of white retinal flecks in fundus images, which in the case of rdx mice were shown to be due to monocyte-derived subretinal macrophages.26 In this work, microglia/macrophages were seen subretinally in mfrp−/− zebrafish adults. However, no retinal degeneration or photoreceptor loss was seen in histologic sections of any of the zebrafish mfrp mutants, the oldest examined being 5 months of age. It is interesting to note that accumulation of subretinal microglia/macrophages is seen in both mouse and zebrafish models of Mfrp deficiency, which may reflect a change in the RPE barrier due to loss of MFRP. Although there is much interest in the role of microglia/macrophages in retinal pathology,37–40 in zebrafish such cells are present in the subretinal space in the absence of any evidence of photoreceptor loss. Although this could reflect the inherent regenerative capacity of zebrafish, it should be noted that in mice, recent unpublished work designed to deplete subretinal microglia/macrophage suggests that those cells do not overtly cause photoreceptor loss (Besharse et al., in preparation).
In contrast to Mfrp mice, which do not exhibit hyperopia or nanophthalmia, mfrp mutant zebrafish show reduced eye size relative to controls, with concomitant hyperopic refractive error. A reduction in functional vision is also seen, as evidenced by OKR, which has been proven to be a useful measure of visual acuity in zebrafish.32,41 Using our OKR testing system, wild-type fish show a VA threshold of 1.395 cpd at 12 mpf, whereas mfrp−/− fish show a VA threshold of 3.922 cpd, a clear reduction in visual acuity likely caused by refractive error. The axial length of the mfrp eye is decreased when considered as a single metric, and also when normalized to an independent metric, such as the lens or the overall body length. The reduction in eye size is particularly apparent in the posterior segment, recapitulating nanophthalmia/posterior microphthalmia seen in clinical human studies of MFRP mutation. Shortening of the posterior segment of the eye with normally sized anterior segment and lens, accompanied with foveoschisis-like retinal folds detached from the sclera, reinforces the value of mfrp mutant zebrafish in modeling these aspects of mfrp-related eye disease. Interestingly, Mfrp mutations have been associated with altered levels of Prss56, a serine protease associated with angle-closure glaucoma, posterior microphthalmia, and myopia in humans.41–44
Unlike Mfrp mice, however, mice with Prss56 mutations show reduced eye size and hyperopia.45 It may be that the mechanism that prevents mouse Mfrp mutations from recapitulating human hyperopia does not affect eye size regulation at the same level as Prss56, because Prss56 mutants phenocopy human refractive error.
The RPE is vital to transmitting emmetropization signals from the light-sensing retina to the choroid and sclera, which ultimately regulate the overall size of the eye globe. In this work, we focus on Mfrp, which is expressed on the apical and basal surfaces of the RPE, as well as the ciliary marginal zone (CMZ), and causes hyperopia when mutated. We have previously published on another protein found on the RPE, Lrp2, which causes myopia when mutated.46 That mutating or ablating a single membrane-bound factor from the RPE cell surface can lead to dramatic changes in refractive error, in both hyperopic and myopic directions, demonstrates the importance for proper RPE activity and function for emmetropization. Zebrafish mutants that exhibit extreme hyperopia and myopia, respectively, form an important toolkit to carry out further study on the nature of eye size regulation and its modulators.
The zebrafish mfrp mutant presented here will provide valuable insight into the pre- and postsymptomatic hyperopic eye, and may be used to better understand the nature of the altered cell signaling resulting from the loss of Mfrp from the RPE. Furthermore, this animal model can be used to investigate the cues that initiate macrophage recruitment in the mfrp eye. Finally, the mechanisms that may protect the zebrafish photoreceptors from undergoing degeneration in mfrp mutants may be useful in identifying potential targets or therapeutics that may aid in the treatment of patients suffering from photoreceptor degeneration and other retinal pathologies.
Supplementary Material
Acknowledgments
The authors thank Michael Cliff and William Hudzinski for excellent zebrafish husbandry. Supported by National Institutes of Health/National Eye Institute R01 research grants EY03222 (JCB) and EY016060 (BAL) as well as a Core Grant for Vision Research (P30 EY001931).
Disclosure: R.F. Collery, None; P.J. Volberding, None; J.R. Bostrom, None; B.A. Link, None; J.C. Besharse, None
References
- 1. Smith TST,, Frick KD,, Holden BA,, Fricke TR,, Naidoo KS. Potential lost productivity resulting from the global burden of uncorrected refractive error. Bull World Health Organ. 2009; 87: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pascolini D,, Mariotti SP. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012; 96: 614–618. [DOI] [PubMed] [Google Scholar]
- 3. Atkinson J,, Braddick O,, Robier B,, et al. Two infant vision screening programmes: prediction and prevention of strabismus and amblyopia from photo- and videorefractive screening. Eye (Lond). 1996; 10: 189–198. [DOI] [PubMed] [Google Scholar]
- 4. Ingram RM,, Walker C,, Wilson JM,, Arnold PE,, Lucas J,, Dally S. A first attempt to prevent amblyopia and squint by spectacle correction of abnormal refractions from age 1 year. Br J Ophthalmol. 1985; 69: 851–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anker S,, Atkinson J,, Braddick O,, Nardini M,, Ehrlich D. Non-cycloplegic refractive screening can identify infants whose visual outcome at 4 years is improved by spectacle correction. Strabismus. 2004; 12: 227–245. [DOI] [PubMed] [Google Scholar]
- 6. Vingolo EM,, Steindl K,, Forte R,, et al. Autosomal dominant simple microphthalmos. J Med Genet. 1994; 31: 721–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yue BY,, Kurosawa A,, Duvall J,, Goldberg MF,, Tso MO,, Sugar J. Nanophthalmic sclera. Fibronectin studies. Ophthalmology. 1988; 95: 56–60. [DOI] [PubMed] [Google Scholar]
- 8. Serrano JC,, Hodgkins PR,, Taylor DS,, Gole GA,, Kriss A. The nanophthalmic macula. Br J Ophthalmol. 1998; 82: 276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgoyne C,, Tello C,, Katz LJ. Nanophthalmia and chronic angle-closure glaucoma. J Glaucoma. 2002; 11: 525–528. [DOI] [PubMed] [Google Scholar]
- 10. Khairallah M,, Messaoud R,, Zaouali S,, Ben Yahia S,, Ladjimi A,, Jenzri S. Posterior segment changes associated with posterior microphthalmos. Ophthalmology. 2002; 109: 569–574. [DOI] [PubMed] [Google Scholar]
- 11. Sundin OH,, Leppert GS,, Silva ED,, et al. Extreme hyperopia is the result of null mutations in MFRP, which encodes a Frizzled-related protein. Proc Natl Acad Sci U S A. 2005; 102: 9553–9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Awadalla MS,, Burdon KP,, Souzeau E,, et al. Mutation in TMEM98 in a large white kindred with autosomal dominant nanophthalmos linked to 17p12-q12. JAMA Ophthalmol. 2014; 132: 970–977. [DOI] [PubMed] [Google Scholar]
- 13. Katoh M. Molecular cloning and characterization of MFRP, a novel gene encoding a membrane-type Frizzled-related protein. Biochem Biophys Res Commun. 2001; 282: 116–123. [DOI] [PubMed] [Google Scholar]
- 14. Kameya S,, Hawes NL,, Chang B,, Heckenlively JR,, Naggert JK,, Nishina PM. Mfrp, a gene encoding a frizzled related protein, is mutated in the mouse retinal degeneration 6. Hum Mol Genet. 2002; 11: 1879–1886. [DOI] [PubMed] [Google Scholar]
- 15. Hawes NL,, Chang B,, Hageman GS,, et al. Retinal degeneration 6 (rd6): a new mouse model for human retinitis punctata albescens. Invest Ophthalmol Vis Sci. 2000; 41: 3149–3157. [PubMed] [Google Scholar]
- 16. Ayala-Ramirez R,, Graue-Wiechers F,, Robredo V,, Amato-Almanza M,, Horta-Diez I,, Zenteno JC. A new autosomal recessive syndrome consisting of posterior microphthalmos, retinitis pigmentosa, foveoschisis, and optic disc drusen is caused by a MFRP gene mutation. Mol Vis. 2006; 12: 1483–1489. [PubMed] [Google Scholar]
- 17. Zenteno JC,, Buentello-Volante B,, Quiroz-González MA,, Quiroz-Reyes MA. Compound heterozygosity for a novel and a recurrent MFRP gene mutation in a family with the nanophthalmos-retinitis pigmentosa complex. Mol Vis. 2009; 15: 1794–1798. [PMC free article] [PubMed] [Google Scholar]
- 18. Kannabiran C,, Singh H,, Sahini N,, Jalali S,, Mohan G. Mutations in TULP1, NR2E3, and MFRP genes in Indian families with autosomal recessive retinitis pigmentosa. Mol Vis. 2012; 18: 1165–1174. [PMC free article] [PubMed] [Google Scholar]
- 19. Crespí J,, Buil JA,, Bassaganyas F,, et al. A novel mutation confirms MFRP as the gene causing the syndrome of nanophthalmos-retinitis pigmentosa-foveoschisis-optic disk drusen. Am J Ophthalmol. 2008; 146: 323–328. [DOI] [PubMed] [Google Scholar]
- 20. Wasmann RA,, Wassink-Ruiter JSK,, Sundin OH,, Morales E,, Verheij JBGM,, Pott JWR. Novel membrane frizzled-related protein gene mutation as cause of posterior microphthalmia resulting in high hyperopia with macular folds. Acta Ophthalmol (Copenh). 2014; 92: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayward C,, Shu X,, Cideciyan AV,, et al. Mutation in a short-chain collagen gene, CTRP5, results in extracellular deposit formation in late-onset retinal degeneration: a genetic model for age-related macular degeneration. Hum Mol Genet. 2003; 12: 2657–2667. [DOI] [PubMed] [Google Scholar]
- 22. Chavali VRM,, Sommer JR,, Petters RM,, Ayyagari R. Identification of a promoter for the human C1Q-tumor necrosis factor-related protein-5 gene associated with late-onset retinal degeneration. Invest Ophthalmol Vis Sci. 2010; 51: 5499–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mandal MNA,, Vasireddy V,, Jablonski MM,, et al. Spatial and temporal expression of MFRP and its interaction with CTRP5. Invest Ophthalmol Vis Sci. 2006; 47: 5514–5521. [DOI] [PubMed] [Google Scholar]
- 24. Won J,, Smith RS,, Peachey NS,, et al. Membrane frizzled-related protein is necessary for the normal development and maintenance of photoreceptor outer segments. Vis Neurosci. 2008; 25: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fogerty J,, Besharse JC. 174delG mutation in mouse MFRP causes photoreceptor degeneration and RPE atrophy. Invest Ophthalmol Vis Sci. 2011; 52: 7256–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fogerty J,, Besharse JC. Subretinal infiltration of monocyte derived cells and complement misregulation in mice with AMD-like pathology. Adv Exp Med Biol. 2014; 801: 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sundin OH. The mouse's eye and Mfrp: not quite human. Ophthalmic Genet. 2005; 26: 153–155. [DOI] [PubMed] [Google Scholar]
- 28. Sander JD,, Zaback P,, Joung JK,, Voytas DF,, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007; 35: W599–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sander JD,, Maeder ML,, Reyon D,, Voytas DF,, Joung JK,, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010; 38: W462–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hwang WY,, Fu Y,, Reyon D,, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013; 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collery RF,, Veth KN,, Dubis AM,, Carroll J,, Link BA. Rapid, accurate, and non-invasive measurement of zebrafish axial length and other eye dimensions using SD-OCT allows longitudinal analysis of myopia and emmetropization. PLoS One. 2014; 9: e110699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cameron DJ,, Rassamdana F,, Tam P,, et al. The optokinetic response as a quantitative measure of visual acuity in zebrafish. J Vis Exp. 2013; 80: e50832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Atchison DA,, Jones CE,, Schmid KL,, et al. Eye shape in emmetropia and myopia. Invest Ophthalmol Vis Sci. 2004; 45: 3380–3386. [DOI] [PubMed] [Google Scholar]
- 34. Becker T,, Becker CG. Regenerating descending axons preferentially reroute to the gray matter in the presence of a general macrophage/microglial reaction caudal to a spinal transection in adult zebrafish. J Comp Neurol. 2001; 433: 131–147. [DOI] [PubMed] [Google Scholar]
- 35. Davis EJ,, Foster TD,, Thomas WE. Cellular forms and functions of brain microglia. Brain Res Bull. 1994; 34: 73–78. [DOI] [PubMed] [Google Scholar]
- 36. Tappeiner C,, Gerber S,, Enzmann V,, Balmer J,, Jazwinska A,, Tschopp M. Visual acuity and contrast sensitivity of adult zebrafish. Front Zool. 2012; 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Combadière C,, Feumi C,, Raoul W,, et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007; 117: 2920–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ambati J,, Anand A,, Fernandez S,, et al. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat Med. 2003; 9: 1390–1397. [DOI] [PubMed] [Google Scholar]
- 39. Fischer AJ,, Zelinka C,, Milani-Nejad N. Reactive retinal microglia, neuronal survival, and the formation of retinal folds and detachments. Glia. 2015; 63: 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kohno H,, Maeda T,, Perusek L,, Pearlman E,, Maeda A. CCL3 production by microglial cells modulates disease severity in murine models of retinal degeneration. J Immunol. 2014; 192: 3816–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mueller KP,, Neuhauss SCF. Quantitative measurements of the optokinetic response in adult fish. J Neurosci Methods. 2010; 186: 29–34. [DOI] [PubMed] [Google Scholar]
- 42. Soundararajan R,, Won J,, Stearns TM,, et al. Gene profiling of postnatal Mfrprd6 mutant eyes reveals differential accumulation of Prss56, visual cycle and phototransduction mRNAs. PLoS One. 2014; 9: e110299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gal A,, Rau I,, El Matri L,, et al. Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease. Am J Hum Genet. 2011; 88: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Orr A,, Dubé M-P,, Zenteno JC,, et al. Mutations in a novel serine protease PRSS56 in families with nanophthalmos. Mol Vis. 2011; 17: 1850–1861. [PMC free article] [PubMed] [Google Scholar]
- 45. Nair KS,, Hmani-Aifa M,, Ali Z,, et al. Alteration of the serine protease PRSS56 causes angle-closure glaucoma in mice and posterior microphthalmia in humans and mice. Nat Genet. 2011; 43: 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Veth KN,, Willer JR,, Collery RF,, et al. Mutations in zebrafish lrp2 result in adult-onset ocular pathogenesis that models myopia and other risk factors for glaucoma. PLoS Genet. 2011; 7: e1001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.