Abstract
Post‐transcriptional control of gene expression is aberrant in cancer cells. Sustained stabilization and enhanced translation of specific mRNAs are features of tumor cells. AU‐rich elements (AREs), cis‐acting mRNA decay determinants, play a major role in the posttranscriptional regulation of many genes involved in cancer processes. This review discusses the role of aberrant ARE‐mediated posttranscriptional processes in each of the hallmarks of cancer, including sustained cellular growth, resistance to apoptosis, angiogenesis, invasion, and metastasis. WIREs RNA 2017, 8:e1368. doi: 10.1002/wrna.1368
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Posttranscriptional Control and AU‐Rich Elements
Normal cells possess well‐regulated and complex posttranscriptional mechanisms of gene and protein expression to ensure cellular homeostasis. These mechanisms can occur in different multiple and interacting stages of gene expression, including 5′ capping, 3′ polyadenylation, cleavage, pre‐mRNA splicing, export, mRNA decay, and translation. In tumor cells, aberrations in any of these processes are observed and can participate in the maintenance and progression of cancer. Both cis‐acting sequences and transacting factors comprise the regulatory features of mRNA decay, a focus of this review. The 3′‐untranslated region (3′UTR) at the 3′ end of the mRNA transcript is the main hub for the most important sequences that influence mRNA decay. Among the most studied sequence in the 3′UTR is AU‐rich elements (AREs), which are characterized not only by an enrichment in A and U but, importantly, by their patterns, repeats, and context as well (Box 1). The minimum ARE is not simply AUUUA but rather is the pentamer in AU context; an example is UAUUUAU or UUAUUUAU or two or more of overlapping repeats of the pentamer. AREs can be classified in two ways, namely, class clustering (classes: I, II, and III) and pentamer cluster‐based classification (five clusters based on the number of repeats). Currently, more than 4000 ARE‐containing transcripts are found in the human genome, but only a fraction has been experimentally validated. Other cis‐acting regulatory sequences, such as GU‐rich elements (GREs)1, 2 regulate mRNA decay, but their roles in cancer are not well defined.
BOX 1. ARE SNAPSHOT.
ARE Classes
Class classification:
Class I: Dispersed AUUUA in U‐rich context
Class II: Overlapping (AUUUA)n
Class II: U‐rich stretch
Cluster classification (Revised):
Cluster 1: WWWUAUUUAUWW
Cluster 2: WWAUUUAUUUAWW
Cluster 3: AUUUAUUUAUUUA
Cluster 4: WAUUUAUUUAUUUAUUUAW
Cluster 5: WAUUUAUUUAUUUAUUUAW
Signaling regulating ARE–mRNA decay and translation
Mitogen‐activated protein kinase pathway
Phosphatidylinositol 3 (PI3)‐kinase/AKT pathway
Wnt signaling pathway
RNA‐binding proteins impacting ARE (examples)
mRNA‐stability promoting proteins
ELAVL1 (HuR) RRM domain
AUF1 RRM domain
mRNA‐decay promoting proteins
ZFP36 (TTP) CCCH domain
ZFP36L1 CCCH domain
ZFP36L2 CCCH domain
KSRP K homology domain
AUF1 RRM domain
Animal models or RNA‐binding proteins
See Box 3
Resources and databases
AU‐rich element‐containing mRNA database (ARED): http://brp.kfshrc.edu.sa/ARED/
AREsite: http://nibiru.tbi.univie.ac.at/AREsite2/welcome
AREscore: http://arescore.dkfz.de/arescore.pl
Atlas for 3′UTR regulatory activity (AURA): http://aura.science.unitn.it/cite/
Scan for Motifs: http://bioanalysis.otago.ac.nz/sfm/sfm_main.pl
CLIPdb: http://lulab.life.tsinghua.edu.cn/clipdb/
UTRsite and UTRdb: http://utrsite.ba.itb.cnr.it/ http://utrdb.ba.itb.cnr.it/
RNA‐binding proteins database: http://rbpdb.ccbr.utoronto.ca/
In general, cis‐acting sequences are specifically recognized and regulated by trans‐acting factors, which can be either proteins (RNA‐binding proteins, RBPs) or noncoding RNAs such as the microRNAs (miRNAs). The latter are a conserved class of small noncoding RNAs (approximately 22 nucleotides) that imperfectly base pair with their targets and cause mRNA degradation or translational repression. miRNAs themselves can cooperate with AREs in mRNA decay and can also affect RBPs and thus indirectly control ARE‐mediated mRNA decay and translation.3 A recent and thorough account of the interactions of miRNAs and RBPs is available.4
RBPS: EXPRESSION AND ACTIVITY IN NORMAL AND CANCER CELLS
Estimates indicate that RBPs can be in thousands, and a manually curated list of 1542 RBPs has been created.5 Several distinct domains distinguish RBPs; the most characterized ones are the RNA‐recognition motif (RRM), K homology domain, and the Cys–Cys–Cys–His zinc finger domain. Several ARE–RBPs have been explored in detail. Among the RRM family is the embryonic lethal abnormal vision‐like protein group that comprises four members (HuB/Hel‐N, HuC, HuD, and HuR). HuR is the most ubiquitously expressed and largely studied member and has been a focus for many years, particularly in cancer research. It is more of a universal RNA stability factor that binds loosely defined AU/U‐rich sequences, both in the 3′UTR and in introns, and in numerous transcripts.6, 7 HuR has been thought to bind first to its pre‐mRNA, and both are transported to the cytoplasm, where HuR‐bound and stabilized mRNA can be efficiently translated.8 The CCCH ZFP36 family consists of the following members: ZFP36 (TTP, TIS11, GOS24, and Nup475), ZFP36L1 (BRF‐1, Tis11b, Berg36, and ERF‐1), and ZFP36L2 (BRF2, Tis11d, and ERF‐2). ZFP36/TTP is the most widely studied ARE–mRNA decay‐promoting RBP, particularly in the context of its strong anti‐inflammatory role; in recent years, it has also been studied for a tumor suppressor role. The AU‐rich‐binding factor 1 (AUF1), also called heterogeneous nuclear ribonucleoprotein D (HNRNPD), has two quasi RRM domains and comprises the following four isoforms: p37, p40, p42, and p45.9 These isoforms differ in their ability to modulate mRNA decay or stability on the basis of the specific isoform, cell type, and, most probably, interactions with other RBPs, namely, HuR and TTP.10, 11 T‐cell intracellular antigen‐1 (TIA‐1) and TIA‐related (TIAR) proteins recognize AREs and cause translational inhibition. These and other RBPs have been covered by several reviews and can be found elsewhere.
The specificity of RBPs toward AREs and other RNA decay sequence determinants depends on different factors, including type of ARE, its length and context, secondary structure microenvironment, posttranscriptional modifications of the RBPs themselves, and interactions with other RBPs. RBP–RBP interactions are complex and can involve either competitive roles toward AREs, such as in the case of TTP and HuR or cooperative modes, e.g., TTP and KSRP affecting the affinity and stereochemistry to AREs. An important interaction observed in healthy cells as opposed to cancer cells and is frequently mentioned in this review is the TTP–HuR axis and balance. In this process, TTP binds and controls HuR mRNA through competition for the binding of HuR itself (auto‐regulation) to its 3′UTR.12, 13 ARE–RBPs can be subjected to a number of modifications that affect their function, such as phosphorylation in response to signals or cellular changes. Several signaling pathways, such as mitogen‐activated protein kinase and PI3 kinase pathways, control ARE and RBP interactions (Box 1).
RBPs can exist either in the nucleus, cytoplasm, or both as shuttling proteins; when in the nucleus, they exist as heterogeneous ribonucleoprotein particles (hnRNPs) as a complex with the pre‐mRNA. This compartment localization is subject to tight regulation in normal cells and can be affected by different stimuli. For example, under unstimulated conditions in normal cells, HuR largely resides within the nucleus, whereas in cancer cells or cells under stress, its preferred localization is in the cytoplasm. The nucleocytoplasmic translocation of HuR appears to be controlled by several posttranslational modifications, such as HuR phosphorylation, methylation, ubiquitination, isomerization, and cleavage. In case of phosphorylation, several kinases have been shown to mediate this process, including the protein kinase C, AMP‐activated protein kinase, the mitogen‐activated protein kinase p38 MAPK, cyclin‐dependent kinases 1 and 5 (CDK1 and CDK5), checkpoint kinase 2, Janus kinase 3, SRC, and Abl‐1 tyrosine kinases.14, 15, 16, 17, 18 Cytoplasmic translocation appears to be necessary for the ability of HuR and other RBPs to regulate mRNA stability or translation through binding to their 3′UTR. HuR may modulate translation of several ARE–mRNAs by a process that is independent of ARE or 3′UTR, which involves binding of HuR to IRES in the 5′UTR.19, 20, 21
AREs mediate their effects on mRNA decay by first enhancing deadenylation (polyA shortening), a first step in mRNA decay facilitated by CCR4/NOT and PAN2/PAN complexes. Through their cognate RBPs, AREs recruit the degradation machinery, the exosome, which is a complex of exonucleases that degrade the mRNA at 3′ to 5′. Also, the removal of 5′ cap decapping enzymes, followed by the 5′–3′ decay‐mediated process, is another mRNA decay pathway. Many of the ARE–mRNAs are localized to processing (P)‐bodies which are cytoplasmic RNA granules and they are the sites in which 5′ ARE–mRNA degradation and translational repression occur. P‐bodies can also occur as part of the stress granules that contain additional proteins and translationally inactive mRNAs. Several ARE–RBPs are associated with either P‐bodies or stress granules. For example, TIA‐1 and TIAR are a part of stress granules, whereas HuR and TTP can be recruited to both. A different player in cellular ARE–mRNA decay, ribonuclease L (RNase L) has been recently identified; it is an endoriboculease that usually targets UU/UA ribonucleotides in viral mRNAs. RNase L promotes the decay of HuR and other cell cycle mRNAs, probably through interactions with TTP; cellular growth is suppressed as a result.22, 23, 24 The cellular mRNA substrates for RNase L appear to be also specified by cooperation with specific miRNA signatures, specifically miR‐17/miR‐29/miR‐200.25
Several players and mechanisms, such as ZFP36 family members and KSRP, participate in the translational repression of ARE–mRNA by RBPs. In normal cells, particularly when not undergoing exposure to stress or external stimuli, AREs ensures that the mRNA is translationally silent. Deadenylation, which causes mRNA decay, can also attenuate ARE–mRNA translation by reducing the occupancy of the poly‐A tail‐binding protein (along its interacting partners); this process can be either promoted or attenuated by TTP or HuR, respectively.26 Furthermore, TTP promotes 5′‐decapping, which can also affect translation. TTP promotes the binding of eukaryotic initiation factor 4E2 (eIF4E2), a translational inhibitor during normal conditions, to ARE–mRNAs to block its translation.27 Another mechanism of TTP‐mediated protein repression is interacting with translational repressor RCK to impart ARE–mRNA translational inhibition.28 Another interacting co‐factor in TTP‐mediated translational repression is the cap‐binding translation repression 4EHP–GYF2 complex in which GYF2 interacts with the conserved tetraproline motifs of TTP.29
The Perturbed Balance and Activity of RBPs in Cancer
One can envision that any aberrations in many of the processes that comprise mRNA decay and translational repression can lead to the overproduction of gene products and thus their activity in cancer. ARE–mRNA expression is overrepresented in multiple cancer types.35 In cancer, several processes that change one or more of the following traits of ARE–RBPs occur: expression levels, compartment localization, and activity. In general, under‐expression or loss of the activity of mRNA decay‐promoting proteins, such as TTP or ZFP3L, will lead to the overexpression of cancer ARE genes. By contrast, overexpression or an increased activity of mRNA stability‐promoting proteins, such as HuR, is seen in cancer states. HuR expression and/or cytoplasmic preference are increased in many cancer types both in cell lines and in tumors, such as brain, colon, breast, and liver.30, 31, 32, 33 A higher HuR expression correlates with reduced patient survival in several cancers.34 However, TTP is deficient or inactive in cancer, and different reasons are outlined in Box 2. TTP deficiency leads to not only increased mRNA stabilization but also translation de‐repression, which results in the prolongation or overproduction of cancer genes that participate in different hallmarks of cancer. Thus, a perturbed balance of the levels or activity of mRNA stability‐ and mRNA decay‐promoting proteins is found in cancer. TTP controls HuR mRNA,12, 32 so the TTP–HuR axis is reversed in cancer because of both TTP deficiency and HuR overexpression/cytoplasmic preference. A large set of overexpressed cancer ARE genes was recently uncovered; this set correlated with the TTP/HuR mRNA ratio and was shown to be enriched in several processes, including mitotic cell cycle, cell motility, cell adhesion, and response to external stimuli.35
BOX 2. TTP DEFICIENCY IS A HALLMARK OF CANCER.
TTP repression is observed in many cancer types due to multiple processes that control different stages of TTP gene and protein expression. For the purpose of this review, the term TTP deficiency is applicable for both expression and activity of the protein. At transcriptional levels, no definite answer exists for the epigenetic silencing of the TTP promoter. However, methylation of the TTP promoter, specifically in a single CpG site located within a TGB1‐responsive region, has been pinpointed in liver cancer cells and tumor tissues where TTP is downregulated.36 Overexpression of histone deacetylases (HDACs), which leads to transcriptional repression through chromatin remodeling in cancer, may repress TTP promoter activity in colon cancer.37, 38 EGR1 can act as a transcriptional factor not only for TTP38 but also for other tumor suppressor genes, including PTEN, p53, and TGFβ1;39 thus, reduced levels of EGR1 in cancer40 may contribute to the deficiency of TTP and other tumor suppressors. Further studies are needed to establish the link between EGR1 and TTP in cancer. Another transcriptional mechanism for TTP deficiency is the failure of mutant p53, a frequent variant in cancer, to induce TTP transcription, particularly during the response of cancer cells to chemotherapeutic agents, such as doxorubicin.41 Cells with mutant or absent p53 have a lower TTP transcriptional activity than do p53‐proficient cells.41
At the mRNA level, miR‐29a, which is overexpressed in pancreatic and breast cancer, targets TTP 3′UTR for the mRNA degradation leading to decreased levels of TTP mRNA and protein.32, 42, 43 At the protein level, deficiency has been observed in breast cancer with a synonymous polymorphism (rs3746083) in the open reading frame of TTP, and as a result, TTP mRNA translation is reduced, but TTP mRNA stability is unaffected; this has been demonstrated in HER2‐positive breast cancer patients’ tumors and associated with a lack of response to the anti‐HER trastuzumab.44 How widespread this interesting mutation is in other types of cancers remains unknown but deserves in‐depth examination.
Another important cause of TTP deficiency is the aberrant signaling that occurs in cancer, such as during growth factor receptor over‐activity. Growth factor/cytokines bind their receptors to activate a series of signaling events leading to the phosphorylation of MEK3/MEK6 kinases that subsequently phosphorylate p38 MAP kinase and then MK2 kinase, which in turn phosphorylates TTP. As a result, its complex with the mRNA is sequestered by 14–3–3 binding proteins, and the phosphorylated TTP loses its ability to bind the AREs and recruit deadenylases and other effectors that cause ARE–mRNA stabilization and translational de‐repression.45 Other signals that are abnormal in cancer, such as Ras and the PI3K signaling pathway, may also participate in TTP phosphorylation and loss of its activity, which leads to ARE–mRNA stabilization.46
TTP becomes phosphorylated by several putative kinases and at multiple serine residues.47 High levels of phosphorylated, and thus inactive TTP has been demonstrated in several types of cancer cell lines and tissues such as those of head and neck and brain.48, 49 Normally, TTP protein is unstable and rapidly degraded by proteasome. Thus, a feature of the phosphorylated inactive form of TTP is that it becomes stable, during certain conditions such as p38/MK2 activation,50, 51 which further sustain the aberrant TTP–HuR axis. A recently studied player of TTP suppression is the pyruvate kinase M2 (PKM2), which is overexpressed in many cancers and participates in the increased glycolysis (Warbug effect). It is thought that PKM2 physically interacts with TTP and leads to its phosphorylation; however, it promotes its degradation via proteasome.52 The discrepancy in the relationship between phosphorylation and protein stability of TTP has been noted in literature. For example, the multiple eight serines mutant TTP form (thus unrphosphorylatable) was found stable when expressed in glioblastoma cells but still demonstrates increased ARE–mRNA instability compared to wild‐type TTP.48 Thus, TTP stability as affected by phosphorylation is dependent on the kinase type itself, the site stoichiometry, and probably the nature of interactions with 14–3–3 or other relevant proteins. Regardless of the protein stability outcome, phosphorylation of TTP in cancer states led to reduced mRNA decay promoting action of TTP and thus subsequently perturbed TTP–HuR balance.
TTP repression can lead to other abnormalities that are unrelated to its posttranscriptional function but are related to other unique functions. Specifically, TTP can act as a co‐repressor for estrogen factor (ERα) transactivation activity, so TTP repression contributes to sustained proliferation in ER‐positive cells.53 Another co‐repressor role is found, and it represses NF‐kB transcriptional activity.54, 55 Again, TTP repression can exaggerate the production of NF‐kB‐dependent cytokines that participate in the tumor microenvironment and the hallmarks of activating invasion and metastasis. In general, the NF‐kB pathway has been recognized as an important pathway in cancer maintenance.56
Many of the types of cancer involve TTP deficiency, and each type or subtype appears to have more than one cause of TTP repression. The outcome of TTP deficiency occurs in almost every hallmark of cancer, as discussed throughout this review.
Aberrant TTP–HuR axis in cancer cells can also occur as the result of changes in the phosphorylation status of each RPB (See Box 2 on TTP phosphorylation). Phosphorylated TTP becomes inactive and has reduced affinity toward AREs, and thus increased levels of ARE–mRNAs, including HuR mRNA, are observed in tumor cells. The function of HuR is highly dependent on the key phosphorylation site in the basic hinge region that harbors the nucleocytoplasmic shuttling sequence,17 which connects RRM2 and RRM3 domains.57 Since there are many kinases in which their activities are altered in cancer, they can affect the phosphorylation status of HuR and thus participate in the aberrant TTP–HuR axis. However, HuR phosphorylation is complex and dependent on the kinase and the phosphorylation site on HuR, meaning that they lead to either increased or decreased affinity of HuR to its mRNA targets. For example, in case of MK2, HuR phosphorylation shifts HuR toward cytoplasmic compartment and increase ARE–mRNA stability and translation.45 In contrast, cdk5, which is higher in cancer cells, can phosphorylate HuR, but leads to decreased ARE–mRNA stability.17 Thus, the role of HuR phosphorylation in cancer cells as opposed to normal cells remains elusive.
The deregulated expression of miRNAs is another important means through which cancer cells try to avoid mRNA decay‐promoting RBPs, or possibly; RBPs antagonize miRNA‐mediated mRNA decay and translation. A notable example is miR‐29a, which targets TTP 3′UTR and leads to its mRNA degradation (Box 2). HuR competes with certain miRNAs for nearly the same binding site on specific messages; a recent example is the ability of HuR to prevent miR‐21‐mediated repression of the tumor suppressor gene, programmed cell death 4 (PDCD4), which is under‐expressed in cancer. Other earlier examples of these HuR–miRNA interaction changes in cancer are miR‐122 and CAT‐1 mRNA, miR‐16 and COX‐2 mRNA, and miR‐331‐3p and ERBB2 (HER/neu) mRNA. The details of these examples can be found in previous reviews.4, 58
The following is an account of the involvement of AREs in each of the hallmarks of cancer. The hallmarks of cancer refer to a concept in which the authors of the original paper narrow cancer as a disease to six features, and later, these features were upgraded with two enabling and two hallmarks of cancer.59 To understand and appreciate the link between AREs in cancer, a brief introduction on each of the cancer hallmarks is presented in each section as well.
CANCER GROWTH AND DIVISION HALLMARKS AND AREs
Cellular growth and division are tightly regulated groups of transient processes that proceed whenever a need for them exists, such as during tissue regeneration, wound repair, blood cell formation, and immune cell expansion. When they are no longer required, various shut‐off mechanisms are in place to guarantee a healthy homeostasis. The ARE‐mediated pathway is one of these mechanisms that ensure the regulated half‐life of the mRNA and swift translation. An appreciable number of genes code for ARE–mRNAs that participate in cellular growth and division. In cancer cells, the following three hallmarks allow them to continue growing: sustained proliferative signaling, evading growth suppressors, and possessing a limitless replicative potential.
Sustaining Proliferative Signaling
Cancer cells tend to stimulate their own growth in a number of ways, so this hallmark is also called self‐sufficiency in growth signals. Over‐secretion of growth factors, abnormal mutant receptor activation, and aberrant signaling components can all promote the sustained growth of tumor cells. A number of gene products participating in this hallmark belong to the ARE transcriptome (Figure 1 and Table 1). A number of cell‐cycle regulators, notably cyclins, such as cyclin A1, cyclin B, and cyclin D, as well as c‐myc, c‐jun, and c‐fos, are coded by ARE–mRNAs and regulated by HuR; these are therefore overexpressed in cancer. MYC (class 1 ARE–mRNA), which encodes a nuclear phosphoprotein transcription factor that regulates the cell cycle, is amplified (DNA copy number) in many tumors and is one of the earliest well‐studied molecules in mRNA decay.126
Figure 1.
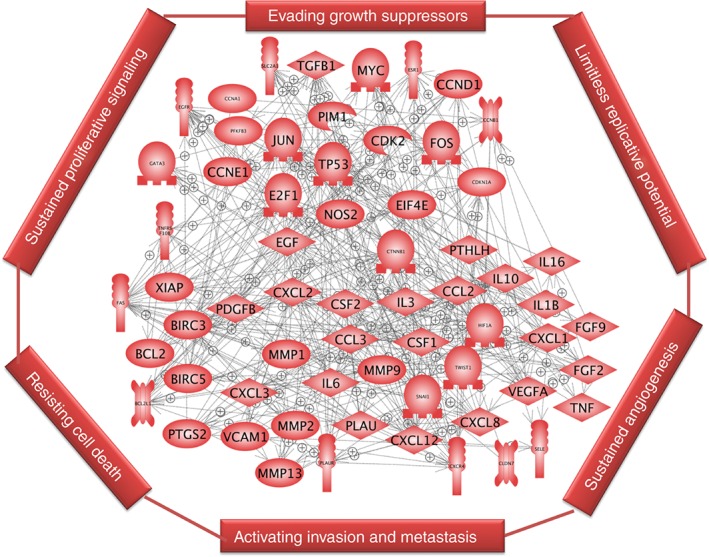
ARE gene product interactome. Gene products coded by ARE–mRNAs that participate in the hallmarks of cancer and their interactions are shown as an interactive map. Pathway Studio program (Elsevier) was used to create the interacting map. Because genes participating in cancer hallmarks overlap, they were not connected to the individual hallmarks.
Table 1.
ARE‐Genes and Hallmarks of Cancer
| ARE‐Gene/Other Name | Hallmarks of Cancer | C | RBP | Ref |
|---|---|---|---|---|
| BCL2 | Anti‐apoptosis | III | HuR, NCL, AUF1 | 60, 61 |
| BCL2L1 (Bcl‐X) | Anti‐apoptosis | U | NCL, HuR | 60, 62 |
| BIRC3 (cIAP2) | Anti‐apoptosis | III | TTP | 63 |
| BIRC5 (Suvivin) | Anti‐apoptosis | U | HuR, CELF1 | 64 |
| CCL2 | Metastasis | U | HuR, TTP | 65 |
| CCL3 | Metastasis | IV | TTP | 66 |
| PLK3 | Proliferation | V | TTP | 67 |
| CCL3 (MIP1α) | Metastasis | IV | TTP | 68 |
| CCNA1 (cyclin A1) | Proliferation | U | HuR, WTAP | 69, 70 |
| CCNB1 (cyclin B2) | Proliferation | U | HuR | 70 |
| CCND1 (cyclin D1) | Proliferation | V | HuR, AUF1, TTP | 71 |
| CCNE1 (cyclin E) | Proliferation | U | CERP, HuR, NF90 | 72 |
| CDK2 | Proliferation | V | HuR | 73 |
| CDKN1A (p21) | Proliferation | IV | AUF1, HuR, TTP, PCBP4 | 22, 74, 75 |
| CLDN7 (claudin‐1) | Invasion | U | HuR, TTP | 37 |
| SLC2A1/Glut1 | Proliferation | V | hnRNP A2 | 76, 77 |
| CSF1/M‐CSF | Invasion | U | TTP, GAPDH | 78 79 |
| CSF2/GMCSF | Angiogenesis | I | HuR, TTP, ZFP36, AUF1, NCL | 80 |
| CTNNB1 | Metastasis | U | KSRP | 81 |
| CXCL1 | Proliferation, angiogenesis | III | HuR, TTP | 82 |
| CXCL2 (MIP2a) | Metastasis | I | TTP, KSRP | 83 |
| CXCL3 (MIP‐2b) | Metastasis | III | TTP, KSRP | 84 |
| CXCL12 | Metastasis | V | — | 85 |
| CXCR4 | Metastasis | V | TTP, HuR | 86, 87 |
| EIF4E | Proliferation | U | HuR | 88 |
| EGF | Proliferation, angiogenesis, metastasis | U | HuR | 58 |
| EGFR | Proliferation | U | HuR | 89 |
| ESR1 (ERα) | Proliferation | V | TTP, HuR | 90 |
| E2F1 | Migration | AUF1, TTP | 71, 91 | |
| FGF2 | Proliferation, angiogenesis, metastasis | V | — | 92 |
| FGF9 | Invasion, anti‐apoptosis, angiogenesis | III | AUF1 | 93 |
| FOS | Proliferation | III | AUF1, HuR, KSRP, TTP | 94, 95 |
| HIF1A | Angiogenesis | III | TTP, HuR, NCL, PTB | 96, 97 |
| IL1B | Metastasis | II | TTP KSRP | 98 |
| IL3 | Angiogenesis | II | AUF1, HuR, TTP, ZFP36L1 | 99 |
| IL6 | Proliferation | IV | TTP, HuR, AUF1, KSRP | 49, 100, 101, 102 |
| IL8 | Angiogenesis, metastasis | III | HuR, TTP, KSRP | 84, 103 |
| IL10 | Evading immunity | V | AUF1, TTP, HuR | 104 |
| IL16 | Invasion, metastasis | U | TTP | 105 |
| NOS2 (iNOS) | Metastasis | V | AUF1, HuR, KSRP, TTP, TIAR, PTB | 106 |
| JUN | Proliferation | U | HuR, KSRP | 107 |
| MMP1 | Invasion | U | TTP | 32 |
| MMP2 | Invasion | U | TTP | 49 |
| MMP9 | Invasion | U | HuR, TTP | 49, 108 |
| MMP13 | Invasion | V | TTP | 32 |
| MYC | Proliferation, anti‐apoptosis | U | TTP, HuR, AUF1, TIAR | 109 |
| PDGF | Proliferation, angiogenesis | IV | HuR | 110 |
| PFKFB3 | Glucose metabolism | III | — | 111 |
| PIM1 | Proliferation, anti‐apoptosis | I | HuR, TTP | 112, 113 |
| PTGS/COX2 | Angiogenesis, anti‐apoptosis | III | HuR, TTP, KSRP, TIA | 31 |
| PTHLH | Metastasis | V | KSRP | 84 |
| PLAU (uPA) | Invasion | IV | HuR, TTP, | 114, 115 |
| PLAUR (uPAR) | Invasion | V | HuR, TTP | 114, 115 |
| SELE | Invasion, metastasis | III | — | |
| SNAI1 | Metastasis | U | HuR, TTP | 116, 117 |
| TWIST1 | Metastasis | III | TTP | 117 |
| TGFB1 | Evading immunity, metastasis | U | HuR | 33 |
| TNFRSF6/FAS | Anti‐apoptosis | III | HuR, NCL | 30, 118 |
| TNFRSF10B (DR5) | Apoptosis | III | HuR | 119 |
| TNF | Angiogenesis, invasion, metastasis | I | TTP, HuR, AUF1, KSRP, TIA, TIAR, ZFP26L1 | 120 |
| TP53 (p53) | Mutant P53 effects | U | HuR, NCL | 121 |
| VCAM1 | Metastasis | III | HuR | 122 |
| VEGF | Angiogenesis | III | AUF1, HuR, TTP | 123, 124 |
| XIAP (BIRC4) | Anti‐apoptosis | III | HuR, CELF1 | 64, 125 |
An example of an ARE–mRNA that codes for a protein involved in cancer growth is the Pim‐1 proto‐oncogene, serine/threonine kinase, which is targeted by TTP.127, 128 It act as an oncogene and is upregulated in several human tumors; it enables cell cycle progression and suppresses apoptosis. In cancer cells, pim‐1 mRNA has been shown to be functionally correlated with TTP expression in several cell lines and tumor tissues.112 Pim‐1 kinase phosphorylates different substrates, including Myc, p21/ Cip1/ WAF1, and p27K, and thus regulates cell cycle progression. TTP‐induced reduction of Pim‐1 thus leads to a decrease in the phosphorylation of p21 and p27; in turn, TTP deficiency can result in increased tumor cell growth.128 HuR overexpression, particularly with cytoplasmic localization, can increase pim‐1 mRNA stability, as shown in pancreatic cancer.113
Several tyrosine kinase growth factor receptor oncogenes, including ARE gene receptors EGFR and stem cell factor (kit, an ARE gene), in addition to other receptors, such as VEGFR, PDGFR, and hepatocyte growth factor (met), activate the PI3‐kinase/AKT pathway. The increased PI3‐kinase activity in cancer cells increases tumor growth and survival. The PI3 kinase/AKT kinase activity stabilizes cancer ARE–mRNAs, such as TNF, β‐catenin CCL2, CXCL3, CCND1, basic fibroblast growth factor (FGF2), and PLAU, by a process that involves phosphorylating and inactivating certain RPBs, including ZFP36L and KSRP modulation.129, 130 The response of EGFR to cancer‐associated EGF overexpression or constitutively active EGFR members leads to the activation of multiple signaling pathways, including PI3‐kinase, the Janus kinase/signal transducer and activator of transcription, and the RAS/extracellular signal‐regulated kinase (ERK) pathway. These pathways can lead to ARE–mRNA stabilization by a process that involves phosphorylation of ARE–RBPs. The mRNAs of EGF and EGFR, although not harboring strong AREs, such as class II AREs, can be both bound and upregulated by HuR.131 EGF can stabilize its own receptor, EGFR ARE–mRNA.132, 133 The alternative polyadenylation of HuR transcript has been characterized,12, 134 and changes in their patterns can affect cancer ARE–mRNA expression.35 In general, alternative polyadenylation that aberrantly occurs in cancer can lead to the overexpression of short 3′UTR transcripts, which escape repression by AREs and miRNAs.135, 136, 137
Evading Growth Suppressors
Normal cells possess regulated mechanisms that aim to stop cell growth when not needed, by developing negative feedback mechanisms, such as inhibitory signals. The checkpoints that are subject to inhibitory signals, e.g. are at the G1/Go and G2/M phases of the cell growth cycle. The most studied growth suppressors that are attenuated or inactivated in cancer are p53, retinoblastoma protein, phosphatase and tensin homolog (PTEN), and tumor growth factor β. The loss of PTEN function because of inactivating mutations that occur in tumors, such as those of prostate cancer, endometrial cancer, and glioblastoma, amplifies PI3K/AKT signaling and can subsequently lead to further ARE–mRNA stabilization of the cancer genes. p53 is a transcriptional factor that is induced by a variety of stresses, such as DNA damage, and it regulates a multitude of functions, including cell cycle control, senescence, apoptosis, DNA repair, and stem cell maintenance, in addition to various metabolic pathways.138 Mutations in p53 are a frequent feature in many types of cancer. Control of the p53 gene and protein expression is complex and involves both positive and negative regulators to ensure optimal levels. Not only is under‐expressed or inactivated p53 an unwanted event, but overexpressed p53 is also undesirable because it promotes aging and other abnormalities. Several mechanisms exist for the posttranscriptional control of p53, including those involving AREs, such as regulation of p53 mRNA stability and translation by a number of RBPs. The p53 3′UTR contain AU/U‐rich regions, and several RBPs have been shown to modulate p53; e.g., HuR has been shown to increase p53 translation during DNA damage. The double‐stranded‐RNA‐binding zinc finger protein, Wig1, in cooperation with hnRNP A2/B1, binds the p53 AU/U‐rich region and causes mRNA stability. Mutant p53 may not be able to participate in the transcriptional activity of the TTP promoter, which is thought to contain a p53 binding site. TTP itself plus ZFP36L can be considered tumor suppressors, in which cancer cells have developed evading mechanisms (Box 2 and Figure 2).
Figure 2.
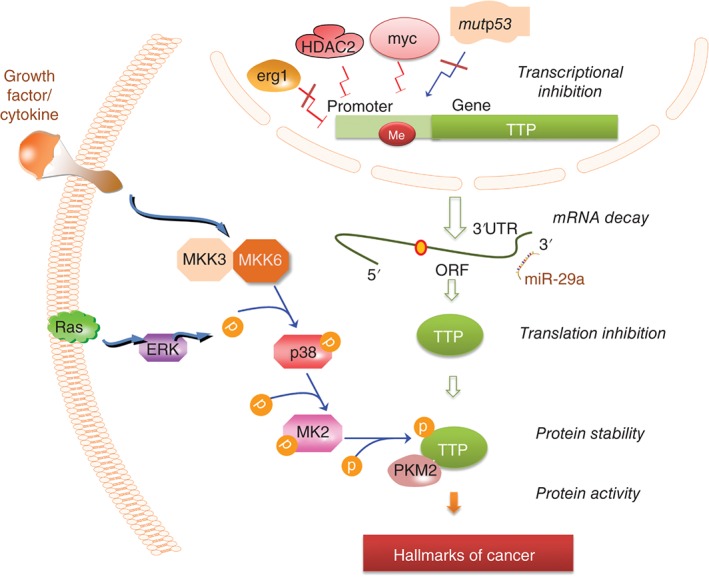
TTP repression in cancer. The various causes that lead to TTP repression in cancer cells: The details are found in Box 2 and throughout the review.
Limitless Replicative Potential
When undergoing a response to damage, such as in the case of wound damage or a response to a growth stimulus, normal cells divide in limited rounds. This process is largely governed by the telomeres, which are specialized structures with a conserved G‐rich sequence located at chromosomal ends. They consequentially become shortened as they divide until they become too short to support further division. Cancer cells have the potential to divide indefinitely by a process that either: reactivates the telomerase, the terminal transferase reverse transcriptase that adds ‘TTAGGG,’ or that maintains the telomere length by other activities.139
A posttranscriptional role in telomere maintenance has been observed, in which AUF1 binds to the G‐rich strand; this process leads to the destabilization of telomeric G‐rich tails and thus facilitates telomere extension.140 Although the telomerase catalytic subunit gene, TERT, does not code for ARE in its mRNA, it can be transcriptionally activated by AUF1. This finding was discovered from the observation that AUF1‐knockout mice exhibited accelerated aging and excessive telomere shortening.141 The work demonstrated that AUF1 is required for telomere maintenance in mouse through binding to and activation of TERT transcription.142
AVOID AREs, AVOID DEATH
Tumors need to resist apoptosis imparted by different stresses, such as chemotherapeutic cytotoxic drugs, so that these tumors have preferential growth. Morphologically, apoptosis or programmed cell death leads to a reduced cell size, followed by cell fragmentation and engulfment by phagocytic cells. Two main apoptotic pathways exist: one is the mitochondrial caspase (intrinsic) pathway, which is triggered by intracellular stress signals, such as DNA damage, and the extrinsic pathway, which is initiated by cell surface receptor engagement with their ligands. In the intrinsic pathway, the mitochondrial apoptosome consists of cytochrome c, dATP, and Apaf‐1, which recruit and activate the cysteinyl aspartate‐specific proteinase, caspase‐9; this process leads to the activation of other effector caspases and, finally, apoptosis.143
Several gene products that allow cancer cells to resist their programmed cell death are coded by ARE–mRNAs, such as the BCL2 family. The key B‐cell lymphoma 2 (BCL2) is an anti‐apoptotic protein that blocks cytochrome c release. It is overexpressed not only in certain types of lymphomas/leukemias because of chromosomal rearrangement but also in selected tumor types because of posttranscriptional mechanisms. BCL2 mRNA has a short half‐life due to the presence of cluster 3 ARE, which is a target for several RBPs, such as HuR, nucleolin (NCL), Ebp1, and AUF1. HuR and NCL synergistically stabilize BCL2 mRNA and enhance its translation by binding concurrently to BCL2 ARE, in which the NCL binding site is upstream from the HuR‐binding site.144 As a result of HuR overexpression or cytoplasmic active HuR, BCL2 and other anti‐apoptotic proteins in which their mRNAs harbor AREs, including BIRC5 (survivin) and the X‐linked inhibitor of apoptosis XIAP, contribute to resistance to apoptosis. The effect of HuR on BIRC5 appears to be conditional on p53 loss,64 which is encountered in most cancers by the mutant p53 status. The other anti‐apoptotic players in cancer are mutated p53 and amplified c‐myc, which can both be modulated by HuR because HuR stabilizes c‐myc and p53 mRNA and thus further amplifies the process.
The TTP/ZFP36 family members induce apoptosis through the intrinsic mitochondria pathway.145, 146, 147 Both TTP and ZFP36L1 interact with BCL2 ARE and cause mRNA destabilization, which leads to apoptosis; TTP also sensitizes cells to TNF‐induced apoptosis.145, 146 TTP sensitizes leukemic cells to anti‐CD20‐induced apoptosis in B‐cell lymphocytic cells and to cisplatin‐induced apoptosis in head and neck cancer cells.148, 149 Thus, TTP deficiency, which occurs in many tumor types, can lead to resistance to cellular death. Similarly, resistance to cell death by cytotoxic drugs can be affected by HuR overexpression or its cytoplasmic preference.
At least two gene members of the death receptor extrinsic pathway harbor AREs in their mRNA, including death receptor 5 (DR5) and the apoptosis‐mediating surface antigen FAS, which contains cluster 3 AREs in their 3′UTR. One key RBP that has been linked to this apoptotic pathway is HuR, and unlike its universal mRNA stabilization/translational role in other hallmarks of cancer, it has opposite functions in FAS‐mediated apoptosis. HuR has been shown to interact with ARE regions in the 3′UTR of FAS and thus inhibit its mRNA translation without affecting mRNA stability in hepatocellular carcinoma cell lines and clinically aggressive HCC; decreased resistance to Fas‐mediated apoptosis occurs as a result.30 Another interesting role of HuR is FAS exon 6 skipping, which leads to the synthesis of a soluble decoy Fas that inhibits apoptosis.150
SUSTAINED ANGIOGENESIS AND ARE–mRNA STABILITY
As the tumor grows, it consumes nutrients and oxygen leading to a state of oxygen deprivation (hypoxia) and nutrient deprivation. As a result, the malignant tumor induces new vessel formation (angiogenesis) and vascular hyper‐permeability to allow nutrients to the tumor microenvironment. Hypoxia triggers HIF1A, which is the master transcriptional factor for many genes that participate in angiogenesis and also in other processes needed for tumor growth and maintenance, such as the glycolytic pathway. HIF1A is regulated appreciably by the translational modification of its α subunit, which becomes stabilized during hypoxia, and HIF1A protein levels increase as a result. Additionally, with HIF1A being an ARE gene, mRNA stabilization and translation of HIF1A are augmented in cancer cells.151 HIF1A harbors cluster 3 (class II) AREs, which account for its labile mRNA behavior in a normal oxygen level environment. A number of RBPs, including TTP, HuR, and polypyrimidine tract‐binding protein (PTB), bind to U/AU‐rich sites in the HIF1A 3′UTR. Both HuR and BTP were shown to promote the translation of HIF1A mRNA.96 TTP has been shown to bind the AREs in the 3′UTR and accelerate HIF1A mRNA decay, whereas TTP deficiency (Box 2) leads to increased mRNA stabilization and protein levels of HIF1A, particularly during hypoxic conditions.97, 152 Because several cancer types are deficient in TTP, cancer cells have increased levels of HIF1A mRNA and protein, and subsequently increased levels of HIF1A target mRNAs in which their gene products participate in several hallmarks. Whether tumor endothelial cells per se are deficient in TTP or its function, it is unknown, but it is a possibility;152 in this case, as HIF1A increases, VEGF and VEGF receptors are upregulated, and, thus, the angiogenesis loop is enhanced.153 HIF1α also has a VEGF‐independent role in the glycolytic pathway in myeloid cells and their differentiation into macrophages and their survival in the tumor microenvironment.154
Hypoxia‐independent HIF1A can also be seen in other aberrant ARE‐mediated states; e.g., the myc oncoprotein, which is overexpressed in several cancers (Figure 2), can directly suppress TTP transcription155 and may therefore lead to HIF1A mRNA stability and high levels of HIF1A protein. During a screening of mediators for HIF1A‐mediated hypoxic response, USP52/PAN2, a component of P‐bodies, was found to promote the stability of HIF1A mRNA in 3′UTR‐dependent manner, as documented with the use of knockdown experiments.156 However, the direct binding of this protein to AREs was not studied, and interactions with de facto RPBs, such as TTP, can possibly happen in P‐bodies.
As HIF1A is triggered, in turn, it induces many HIF‐responsive genes, including those encoding for other ARE–mRNAs, notably, VEGFA, plus other growth factors, including PIGF, PDGF, COX2 (PTGS2), CXCL12, FGF2, HGF, and TGFA. The key angiogenesis mediator, VEGF, triggers a state called ‘angiogenic switch,’ which refers to the initiation from a dormant anti‐angiogenic state to an active phase of vascular growth to sustain tumor growth and maintenance.59, 157 VEGF can be released from the infiltrating immune cells in the microenvironment, and this contributes further to the sustained angiogenesis state.158 VEGF contains multiple U‐rich/ARE elements including cluster 3 (class II) AREs that cause strong mRNA destabilization under normal oxygen levels. Several RBPs, such as HuR, PTB, poly(A)‐binding protein‐interacting protein 2, AUF1, NF90, ZFP36L1, and TTP, bind to U‐rich/AREs in the VEGF 3′UTR and modulates its mRNA decay.159, 160 Hypoxia leads to significant VEGF mRNA stabilization independent of HIF1A, and it involves different assemblies of RBPs than those during normoxia.161 Another HIF1A‐indepdent mechanism could be the various kinases, such as Jun amino‐terminal kinase (JAK) and the p38 MAPK pathway,162 which are known to inactivate TTP by phosphorylation and thus leads to VEGF mRNA stabilization.
In cancer cells, HuR promotes the stability and translation of VEGF mRNA in several solid tumors, including colon, brain, and breast cancers.33, 163 In general, angiogenesis is associated with increased HuR expression and cytoplasmic localization. Hypoxia causes translocation of HuR from the nucleus to the cytoplasm164 to stabilize ARE–mRNAs and/or enhance their translation. On the other hand, as TTP destabilizes VEGF mRNAs,165 the aberrant TTP–HuR axis that is encountered in a number of cancers in principle contributes to sustained angiogenesis. Mice studies showed reduced microvessel density in transformed cell‐ derived xenografts in nude mice as a result of inducible TTP.166 The loss of the tumor suppressor Von Hippel‐Lindau (VHL), which is encountered in certain tumor types, such as renal carcinomas, can increase VEGF mRNA stability, which contributes to the sustained angiogenesis.167 This effect can be mediated by either the RRM1 domain of HuR, which interacts with VHL168 leading to the inhibition of its function, or by the fact that VHL decreases TTP expression.169 Another notable HIF1A‐inducible gene is FGF2, which is largely regulated by posttranscriptional mechanisms that involve alternative polyadenylation, 5′UTR‐mediated translation, and AREs.92 Thus, the aberrant TTP–HuR axis amplifies a network of angiogenesis through its direct effect on HIF1A mRNA, whose product is a transcriptional inducer of many ARE‐coding genes, including VEGF. VEGF itself can trigger the expression of other ARE genes, such as IL‐8 and other factors that can promote angiogenesis in a number of cancers.
ACTIVATING INVASION AND METASTASIS: THE ARE INTERACTOME
This hallmark of cancer is a prominent cause of cancer mortality. The process of metastatic cancer starts off with local invasion of the tumor tissues to the stroma, followed by intravasation, migration through the blood and lymphatic system, colonization, and growth at a distant organ. Many key pro‐invasion and pro‐metastasis genes code for ARE–mRNAs (see Table 1). Because of the importance of AREs in this cancer hallmark, the discussion in this section has been divided into several subsections.
Epithelial–Mesenchymal Transition
Epithelial–mesenchymal transition (EMT) is a multifactorial process in which cancer epithelial cells undergo further changes to acquire a mesenchymal phenotype that will allow them to invade local tissues. The cells undergo a cytoskeletal rearrangement and lose cell–cell adhesion structures and polarity. EMT is programmed through a number of specific transcriptional factors, notably the zinc finger Snail (SNAI1) and the basic helix‐loop‐helix factor, Twist1 (TWIST1). These factors inhibit epithelial cell features by repressing E‐cadherin transcription while activating N‐cadherin, a marker for mesenchymal cells. Cadherins, named after calcium‐dependent adhesion, are transmembrane proteins that form adherent junctions binding cells together. TTP has been recently shown to bind the 3′UTR, and promote the mRNA decay, of SNAIL and TWIST1 in an ARE‐dependent manner116, 117 and to inhibit EMT.117 Thus, TTP deficiency that occurs in cancer can be a plausible EMT factor, while the HuR overexpression encountered in cancer may promote EMT via enhancement of SNAIL mRNA stability or translation. This observation is evident from a work showing that loss of the tumor suppressor scribble, a component of a complex that participates in the polarity of epithelial cell structures, leads to HuR translocation to the cytoplasm and enhancement of SNAIL translation.116
An important mediator of EMT linked to AREs is the miR‐29a. Although it has been shown to have anti‐tumor activities, other evidence exists to show that miR‐29a promotes EMT and the invasion and metastatic potential of a number of cancers, including breast, colon, pancreatic, and nasopharyngeal cancers.32, 42, 43, 170, 171 Elevated levels of miR‐29a are found in these tumors and are associated with TTP suppression. miR‐29a suppresses TTP by targeting both mouse43 and human TTP 3′UTR in breast cancer32 and in pancreatic cancer.42 The ARE found within 100 bases downstream of the miR‐29a target sequence may cooperate in the accelerated mRNA decay of TTP.32 The TTP‐modulated effect of miR‐29a on EMT has been shown in breast cancer and also in pancreatic cells.42, 43 In a cell line model, TTP can induce EMT and in cooperation with oncogenic Ras signaling.43
Several pro‐tumor growth factors, including TGF‐β, FGF2, and PDGF, as well as signaling pathways, such as Notch and Hedgehog, and NF‐kB, participate in EMT. TGFB, a key regulator of EMT, is itself coded by an ARE–mRNA that is subject to HuR‐mediated mRNA stabilization.172 Another plausible player of EMT is AUF1. It is linked to tumorigenesis, as shown by studies on AUF1 p37 transgenic mice that developed sarcomas also involving high levels of cyclin D1.173 AUF1 may be overexpressed in cancer tissues compared with normal ones174 and promotes mesenchymal features in osteosarcoma cells.175 Notably, AUF1 can also act as either a destabilizing or stabilizing factor that interacts or acts in concert with other RBPs. For example, results from a global analysis of mRNA targets by AUF1 showed that AUF1 and HuR cooperate to promote the stabilization and translation of certain mRNAs.176 More studies are needed to further understand the role of AUF1 in cancer.
The Microenvironment
The tumor cells and stroma constitute an active microenvironment that promotes the processes underlying the hallmarks of cancer, particularly invasion. The tumor stroma comprises the basement membrane, extracellular matrix, and vasculature, together with a number of different cell types, including fibroblasts, pericyctes, immune cells, and endothelial cells. Two important and largely studied cells are the tumor‐associated macrophages (TAMs) and cancer‐associated fibroblasts (CAF). CAFs themselves have proinflammatory properties and secrete cytokines that promote TAM activity and macrophage infiltration. TAMs are key players of cancer‐related inflammation (an enabling hallmark of cancer) and constitute a large proportion of the inflammatory infiltrate in which tumor cells attract.177 They produce many cytokines, chemokines, and growth factors, which include many ARE‐coding gene products, such as EGF, CSF1, IL‐8 (CXCL8), CXL12, FGF, VEGF, PDGF, TGFB, and MMPs. TAMs assist in many of the hallmarks of cancers, including tumor growth, angiogenesis, and invasion. Together with myeloid‐derived suppressor cells, they can also suppress cytotoxic immune cells from killing the tumor cells, such as by secreting the immunosuppressive IL‐10.178
Cancer cells themselves also produce CSF‐1, which further amplifies the invasiveness of these cancer cells.179 Both CAFs and TAMs secrete large amounts of VEGF and FGF that contribute to the sustained angiogenesis hallmark. Furthermore, tumor‐associated endothelial cells overproduce VEGF receptors to activate angiogenesis. All these gene products are a part of the multiple growth factors that code ARE–mRNAs. In fact, cytokines and growth factors are overrepresented as a functional class in ARED. The aberrant TTP–HuR axis in the cells of the microenvironment, such as CAFs, should lead to the overproduction of these growth factors that feed the tumor.
Invasiveness
Invasion involves degradation of the extra‐cellular matrix, an important entry to surrounding tissues in cancer cells way to distant organs. Among the key players are gene products that are coded by ARE–mRNAs, such as matrix metalloproteases MMP1 and MMP13, in addition to serine proteases PLAU and PLAUR. The increased activities of these mediators have been associated with invasiveness in a number of tumors. uPA converts plasminogen to plasmin, which directly (or together with MMPs) degrades the extra‐cellular matrix. uPA has a well‐characterized ARE that binds to both TTP and HuR, and its expression has been linked to TTP deficiency and/or HuR overexpression.32, 114 The constitutive p38 MAPK activation that can occur in a number of cancers, such as breast, stomach cancer, and multiple myeloma,180, 181 has been linked to uPA and uPAR overexpression.114 The p38 MAP kinase pathway is activated during growth factor response, and this activation leads to the transient phosphorylation of TTP by the p38 MAPK‐activated protein kinase 2 (MK2); loss of binding to AREs results, and ARE–mRNA stability and translation are subsequently enhanced.50 These phosphorylation events decrease the affinity of TTP to the ARE and inhibit the ability to compete with HuR interaction with HuR 3′UTR; as a result, the TTP/HuR balance is switched toward ARE–mRNA stabilization and translation.45, 182 Thus, the pro‐invasive factors that are overproduced because of constitutive p38 MAPK are those regulated by TTP, such as uPA, uPAR, and MMP13.32 The pro‐invasion activity of miR‐29a has been shown to be mediated by the suppression of TTP, and it subsequently increases the HuR mRNA and protein levels because of the TTP–HuR axis.32
Migration and Metastasis
Metastasis is the major cause of mortality and morbidity in human cancer. Despite this fact, little progress has been achieved to considerably address this serious disease. One is expected to understand the intricate details of metastasis to thoroughly address the therapeutic possibilities of intervention. It is a complex process that allows cancer cells to seek refuge at distant organs, and it comprises several steps beginning with intravasation into the circulatory system, migration through circulation, and then seeding into the organs. Intravasation is the movement of cancer cells through the basal membrane into the circulatory system, both blood and lymphatic vessels, so that they can migrate to distant organs (metastasis). TAMs can help in the initial stages of metastasis by chemoattracting cancer cells toward blood vessels through secretion of pro‐cancer cytokines, such EGF and IL‐8. EGFR mRNA contains a 260 nt AU‐rich region which causes mRNA decay and becomes stabilized upon EGF response in breast cancer cell lines.133 Tumor cells also secrete TNF (cluster 1/class II ARE), which induces endothelial junction rearrangements that promote cancer cell transendothelial migration.183
In general, chemokine and specific growth factors facilitate metastatic processes. Important ARE–mRNAs that code for such factors are the components of the stromal cell‐derived factor‐1 (SDF‐1, CXCL12)–CXCR4 axis. CXCR4 is a G‐protein‐coupled chemokine receptor that promotes the chemotactic migration of breast cancer cells to distant organs along a gradient of its ligand.184 High levels of CXCR4 are associated with tumor aggressiveness in patients’ tissues.185 Recently, our work showed that CXCR4 3′UTR contains functional AREs that caused mRNA decay and bound both TTP and HuR.86 In cancer cells, where the TTP–HuR axis is aberrant, such as in invasive breast cancer cells, CXCR4 mRNA and protein are abundant and help cells migrate to their ligand CXCL12, which itself is coded by an ARE–mRNA. Several studies confirmed the role of TTP deficiency and HuR in promoting both the invasion and migration of cancer cells.32, 86, 115, 186 TTP and HuR regulate various ARE–mRNAs that participate in the ability of cancer cells to migrate (Table 1 and Figure 1). Mice models using xenografted tumors with altered expression in TTP or HuR demonstrate the vivo effects on invasion and metastasis. A notable example is tumor xenografts generated from TTP‐knockdown EMT cell line model, RasXT, that displays fewer metastases in the lung when compared to TTP intact cells.43 Silencing of HuR in a mouse model of glioblastoma using human xenograft cells proves attenuation of tumor invasion and migration as assessed with GFP‐labeled tumor cells.60 In patients, HuR overexpression and/or cytoplasmic localization have been shown to be associated with lymph node metastatic disease in several cancer types, such as breast and lung cancers.110, 187 An excellent coverage of the role of HuR in different cancer pathologies was given in this reference.188
CONCLUSIONS: RESTORING THE AREs ORDER
The TTP–HuR axis is aberrant in many cancer cells largely because of TTP deficiency, which causes ARE–mRNA stabilization and translational enhancement (Box 2 and Figure 2). Thus, a TTP activator would be a potential therapeutic option and may constitute a small molecule drug target that leads to upregulation or an increase in TTP activity (e.g., by dephosphorylation). Figure 3 depicts the process of normalization of the aberrant ARE‐mediated pathway in cancer cells if a TTP activator is used. Indeed, this ‘restoration’ has been experimentally validated in a highly invasive breast cancer cell line model with the use of a cell‐permeable peptide‐nucleic acid against miR‐29a. The inhibitor counteracted the miR‐29a repression of TTP mRNA, and the TTP–HuR axis normalized as a result, along with the mRNA levels for uPA, uPAR, MMP1, and CXCR4; the breast cancer cells became less invasive and displayed normalized actin cytoskeletal polymerization32, 86 (Figure 3, right panel).
Figure 3.
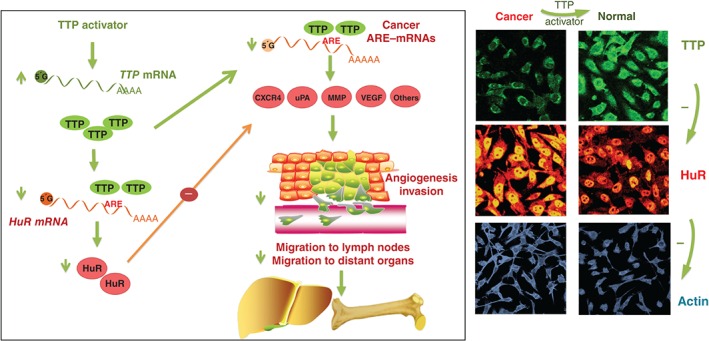
Restoring the perturbed ARE‐mediated pathway by a TTP activator. A graphical representation of how the aberrant TTP–HuR axis can be normalized by a TTP activator, which leads to a reduction of cancer ARE–mRNAs that participate in the hallmarks of cancer. The right panel shows confocal microscopy images for monitoring the changes in the expression levels of TTP and HuR, along with actin cytoskeleton visualization in normal and cancer cells.32 Further details are found under the various sections of this review. The images on the right were adopted from our previous work32 and reproduced from the Journal of Pathology by permission of the Pathological Society of Great Britain and Ireland.
Several groups have been attempting to find drugs that either stimulate TTP or repress HuR in cancer cells. Drugs that can activate TTP include the p38 MAP kinase inhibitor, SB 203580, which inhibits TTP phosphorylation and thus leads to active TTP189 that competes or ousts HuR in the quest for the cancer ARE–mRNA targets. Laboratory and mice studies have shown the benefits of using the p38 MAPK inhibitor in cancer models.190, 191 HDAC inhibitors, such as trichostatin, led to TTP de‐repression and increased its binding capacity to ARE–mRNAs, such as COX‐2 mRNA in colon cancer cells.37, 38 Because of the multifactorial nature of TTP repression, using a combination therapy to address the cancer may be necessary in the future. For HuR‐targeting drugs, drug screening approaches such as using HuR/ARE interaction assay identified several small molecule drugs were found to inhibit HuR mRNA stability‐promoting function through competitive binding to HuR.192, 193, 194
The restoration of the aberrant TTP–HuR axis by potential activators of TTP and the subsequent effects on the hallmarks of cancer in the laboratory prove the aberration of ARE‐mediated pathways in cancer. Several cancer genes that code for ARE–mRNAs were studied independent of ARE involvement or posttranscriptional control; or, several ARE–mRNAs were investigated for their posttranscriptional control but not in a cancer context. Thus, many opportunities exist for further investigating the role of AREs in the hallmarks of cancer. Importantly, in vivo work and preclinical models are now much needed to address the benefit of restoring the perturbed ARE‐mediated pathways (Box 3). Further work is required to find and assess kinase inhibitors or other small molecule drugs that override the various abnormalities encountered in cancer because of ARE dysregulation.
BOX 3. MOUSE MODELS AND OPPORTUNITIES IN CANCER RESEACH.
Although mouse models of TTP and HuR are generated in order to understand largely the immune and inflammatory modulating role of these RBPs, they constitute a significant and potential resource for cancer studies in future. Currently, the animal studies are immune‐deficient animals (e.g., nude mice) implanted with xenografted human tumors in which they were derived from cell lines with altered RBPs expression. In general, they confirm the in vivo activities of specific RBPs on the gross tumor growth. The increase or decrease of tumor volume is a result of the combined outcome of several hallmarks of cancer. Examples are xenograft tumors generated from TTP overexpressing tumor cell lines of pancreatic, gastric, breast, and lymphoma cancer types showing reduced volume of the xenografts.42, 105, 155, 165, 166, 195, 196 Silencing of HuR in several types of cancer cells such as those of brain, pancreatic and renal types, in tumor xenografts leads to significant reduction in the tumor volume in mice.60, 197, 198, 199 These results confirm the in vivo role of TTP and HuR as tumor suppressing and promoting factor, respectively. Other examples were mentioned throughout the review.
Because of the tremendous efforts of establishing TTP and other mouse models, it worth listing them here in order to stimulate the interest for their utilization in cancer. Being somewhat immunocompetent, they may not be entirely suitable with established human tumor xenograft models due to transplant rejection; however, syngeneic tumors can be used instead. Additionally, these models can be used to study the role of the RBP in the tumor microenvironment and for the emergence of spontaneous and/or carcinogen inducible tumors.
ZFP36 (TTP)‐knockout mice
This is first TTP mouse model, where TTP is knockdown by homologous recombination‐directed deletion in C57BL mice. Although appear normal at birth, they later develop arthritis, cachexia, dermatitis, autoimmunity, and myeloid hyperplasia in a manner that is largely dependent on increased TNF levels.200 This model can be used, e.g., to study carcinogen‐induced tumor development.
Myeloid‐specific ZFP36 knockout mice
The mice were created by taking the advantage of the M lysozyme promoter, specific for cells of the myeloid lineage, to express cre recombinase. The mice did not display the same systematic inflammatory patterns as the former. However, the monocytes secrete excessive TNF production in response to low doses of the gram‐negative bacterial endotoxin resulting in toxic shock and organ failure.201 Because the tumor associated macrophages play an important role in tumor maintenance (see Activating Invasion and Metastasis: The ARE Interactome section), this mouse model can be valuable in assessing the specific role of TTP in TAM‐assisted tumorigenesis.
ZFP36ΔARE mice
This is a latest laboratory variant in TTP models in which 136 bases of TTP 3′UTR, which harbors AREs, were deleted in the endogenous locus.202 TTP AREs are important in auto‐regulation of TTP mRNA stability.22, 203 Cells derived from the TPΔARE mice have stable TTP mRNA and higher levels of TTP protein levels, and they resist the typical inflammatory diseases seen in TTP‐knockout mice.202
ZFP36AA mice
These mice, instead of wild‐type TTP, express TTP that has alanine mutations in two prominent serine sites (ser52 and ser178 of murine ORF), so they will not be phosphorylated at these sites. The mice are normal and ARE–mRNAs such as TNF IL1, IL‐12, CXCL1, CXCL2, and PIM‐1 are reduced in the myeloid cells. Like the former model, they resist the systematic inflammatory response due to endotoxin.204 The previous two models may find benefit in cancer research to study the effect of TTP on cancer ARE–mRNAs in general particularly when using syngeneic (C57BL/6J) tumor xenograft or inducible tumors.
Both of the previous two models offer dual benefit, transgenic expression of TTP and elevated expression or increased activity of TTP, mimicking putative TTP‐inducing therapeutic drugs. This should allow, at least partially, assessment of the usefulness of TTP enhancing drugs in cancer.
Eμ‐ZFP36 and Eu‐ZFP36L1 mice
These are transgenic mice for either TTP or ZFP36L1 (Tis11b) which were utilized by creating double transgenics with Eμ‐myc transgenics, a mouse model of human lymphoma in which c‐myc is overexpressed from Ig enhancer promoter (thus, B cells). It was demonstrated that Eμ‐TTP/Eμ‐myc mice dramatically survived more than Eμ‐myc mice due to resistance to the development of aggressive lymphomas.155
ZFP36L1Δ/ZFP36L2Δ mice
The lymphocyte conditional Zfp36l1 and Zfp36l2 double knockout developed T‐lymphoblastic leukemia T‐ALL while the single knockouts were normal.205
Myeloid‐specific ZFP36L1Δ mice
Unlike myeloid ZFP36/TTP knockout mice, they do not appear to participate in the inflammatory response to endotoxin, possibly due to compensation from the TTP effect.206
ELAVL1 HuR mouse models
ELAVL1‐knockout mice display embryonic lethality, thus, a number of conditional HuR‐knockout or conditional HuR‐transgenic mice have been developed and largely for the study of the immune system.207, 208, 209, 210 These models have not been evaluated in cancer studies and may offer opportunities to study the effect specific immune cells as affected by HuR on cancer. It may be preferable to use the conditional knockout models as opposed to transgenic models to avoid opposite effects of HuR due to overexpression since HuR is abundantly expressed ubiquitous gene.
Other mouse models
Both KSRP‐ and AUF1‐knockout model are available in addition to AUF1‐transgenic mice.173, 211 Although the in vivo role of KSRP in cancer is not established, the latter model demonstrates that AUF1 overexpression promote development of sarcoma.173
ACKNOWLEDGMENT
The open access charge is supported by the intramural funding from King Faisal Specialist Hospital and Research Centre.
Conflict of interest: The author has declared no conflicts of interest for this article.
REFERENCES
- 1. Vlasova‐St Louis I, Bohjanen PR. Post‐transcriptional regulation of cytokine signaling by AU‐rich and GU‐rich elements. J Interferon Cytokine Res 2014, 34:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Halees AS, Hitti E, Al‐Saif M, Mahmoud L, Vlasova‐St Louis IA, Beisang DJ, Bohjanen PR, Khabar K. Global assessment of GU‐rich regulatory content and function in the human transcriptome. RNA Biol 2011, 8:681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moore AE, Young LE, Dixon DA. MicroRNA and AU‐rich element regulation of prostaglandin synthesis. Cancer Metastasis Rev 2011, 30:419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iadevaia V, Gerber AP. Combinatorial control of mRNA fates by RNA‐binding proteins and non‐coding RNAs. Biomolecules 2015, 5:2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerstberger S, Hafner M, Tuschl T. A census of human RNA‐binding proteins. Nat Rev Genet 2014, 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA‐binding protein HuR couples pre‐mRNA processing and mRNA stability. Mol Cell 2011, 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome‐wide analysis of regulatory interactions of the RNA‐binding protein HuR. Mol Cell 2011, 43:340–352. [DOI] [PubMed] [Google Scholar]
- 8. Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell‐permeable peptides. Science 2001, 294:1895–1901. [DOI] [PubMed] [Google Scholar]
- 9. Moore AE, Chenette DM, Larkin LC, Schneider RJ. Physiological networks and disease functions of RNA‐binding protein AUF1. Wiley Interdiscip Rev RNA 2014, 5:549–564. [DOI] [PubMed] [Google Scholar]
- 10. David Gerecht PS, Taylor MA, Port JD. Intracellular localization and interaction of mRNA binding proteins as detected by FRET. BMC Cell Biol 2010, 11:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kedar VP, Zucconi BE, Wilson GM, Blackshear PJ. Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins. J Biol Chem 2012, 287:5459–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Al‐Ahmadi W, Al‐Ghamdi M, Al‐Haj L, Al‐Saif M, Khabar KS. Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU‐rich elements and auto‐regulation. Nucleic Acids Res 2009, 37:3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dai W, Li W, Hoque M, Li Z, Tian B, Makeyev EV. A post‐transcriptional mechanism pacing expression of neural genes with precursor cell differentiation status. Nat Commun 2015, 6:7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Filippova N, Yang X, Nabors LB. Growth factor dependent regulation of centrosome function and genomic instability by HuR. Biomolecules 2015, 5:263–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr, Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev 2008, 22:1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doller A, Winkler C, Azrilian I, Schulz S, Hartmann S, Pfeilschifter J, Eberhardt W. High‐constitutive HuR phosphorylation at Ser 318 by PKC{delta} propagates tumor relevant functions in colon carcinoma cells. Carcinogenesis 2011, 32:676–685. [DOI] [PubMed] [Google Scholar]
- 17. Filippova N, Yang X, King P, Nabors LB. Phosphoregulation of the RNA‐binding protein Hu antigen R (HuR) by Cdk5 affects centrosome function. J Biol Chem 2012, 287:32277–32287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoon JH, Abdelmohsen K, Srikantan S, Guo R, Yang X, Martindale JL, Gorospe M. Tyrosine phosphorylation of HuR by JAK3 triggers dissociation and degradation of HuR target mRNAs. Nucleic Acids Res 2014, 42:1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Durie D, Lewis SM, Liwak U, Kisilewicz M, Gorospe M, Holcik M. RNA‐binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene 2011, 30:1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA‐stability factor HuR binds the 5'‐untranslated region of the human IGF‐IR transcript and differentially represses cap‐dependent and IRES‐mediated translation. Nucleic Acids Res 2005, 33:2962–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev 2002, 16:3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Al‐Haj L, Blackshear PJ, Khabar KS. Regulation of p21/CIP1/WAF‐1 mediated cell‐cycle arrest by RNase L and tristetraprolin, and involvement of AU‐rich elements. Nucleic Acids Res 2012, 40:7739–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al‐Ahmadi W, Al‐Haj L, Al‐Mohanna FA, Silverman RH, Khabar KS. RNase L downmodulation of the RNA‐binding protein, HuR, and cellular growth. Oncogene 2009, 28:1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brennan‐Laun SE, Li X‐L, Ezelle HJ, Venkataraman T, Blackshear PJ, Wilson GM, Hassel BA. RNase L attenuates mitogen‐stimulated gene expression via transcriptional and post‐transcriptional mechanisms to limit the proliferative response. J Biol Chem 2014, 289:33629–33643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rath S, Donovan J, Whitney G, Chitrakar A, Wang W, Korennykh A. Human RNase L tunes gene expression by selectively destabilizing the microRNA‐regulated transcriptome. Proc Natl Acad Sci USA 2015, 112:15916–15921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev 1999, 13:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tao X, Gao G. Tristetraprolin recruits eukaryotic initiation factor 4E2 to repress translation of AU‐rich element‐containing mRNAs. Mol Cell Biol 2015, 35:3921–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi MY, Wang ZZ, Zhang Z, Shao Q, Zeng A, Li XQ, Li WQ, Wang C, Tian FJ, Li Q, et al. AU‐rich‐element‐dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol Cell Biol 2012, 32:913–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu R, Olsen MT, Webb K, Bennett EJ, Lykke‐Andersen J. Recruitment of the 4EHP‐GYF2 cap‐binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU‐rich elements. RNA 2016, 22:373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu H, Berkova Z, Mathur R, Sehgal L, Khashab T, Tao RH, Ao X, Feng L, Sabichi AL, Blechacz B, et al. HuR suppresses Fas expression and correlates with patient outcome in liver cancer. Mol Cancer Res 2015, 13:809–818. [DOI] [PubMed] [Google Scholar]
- 31. Young LE, Sanduja S, Bemis‐Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 2009, 136:1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Al‐Ahmadi W, Al‐Ghamdi M, Al‐Souhibani N, Khabar KS. miR‐29a inhibition normalizes HuR over‐expression and aberrant AU‐rich mRNA stability in invasive cancer. J Pathol 2013, 230:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine‐ and uridine‐rich elements within the 3' untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res 2001, 61:2154–2161. [PubMed] [Google Scholar]
- 34. Kotta‐Loizou I, Giaginis C, Theocharis S. Clinical significance of HuR expression in human malignancy. Med Oncol 2014, 31:161. [DOI] [PubMed] [Google Scholar]
- 35. Hitti H, Bakheet T, Al‐Souhibani N, Moghrabi W, Al‐Yahya S, Al‐Ghamdi M, Al‐Saif M, Shoukri MM, Lánczk A, Grépin R, et al. Systematic analysis of AU‐rich element expression in cancer reveals common functional clusters regulated by key RNA‐binding proteins. Cancer Res. May 17. pii: canres.3110.2015. [Epub ahead of print] PMID: 27197193. [DOI] [PubMed] [Google Scholar]
- 36. Sohn BH, Park IY, Lee JJ, Yang SJ, Jang YJ, Park KC, Kim DJ, Lee DC, Sohn HA, Kim TW, et al. Functional switching of TGF‐beta1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology 2010, 138:1898–1908. [DOI] [PubMed] [Google Scholar]
- 37. Sharma A, Bhat AA, Krishnan M, Singh AB, Dhawan P. Trichostatin‐A modulates claudin‐1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis 2013, 34:2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sobolewski C, Sanduja S, Blanco FF, Hu L, Dixon DA. Histone deacetylase inhibitors activate tristetraprolin expression through induction of early growth response protein 1 (EGR1) in colorectal cancer cells. Biomolecules 2015, 5:2035–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53 and fibronectin: Egr1 is a potential target of gene therapy for prostate cancer. Cancer Gene Ther 2006, 13:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang H, Chen X, Wang J, Guang W, Han W, Tan X, Gu Y. EGR1 decreases the malignancy of human non‐small cell lung carcinoma by regulating KRT18 expression. Sci Rep 2014, 4:5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee JY, Kim HJ, Yoon NA, Lee WH, Min YJ, Ko BK, Lee BJ, Lee A, Cha HJ, Cho WJ, et al. Tumor suppressor p53 plays a key role in induction of both tristetraprolin and let‐7 in human cancer cells. Nucleic Acids Res 2013, 41:5614–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun XJ, Liu BY, Yan S, Jiang TH, Cheng HQ, Jiang HS, Cao Y, Mao AW. MicroRNA‐29a promotes pancreatic cancer growth by inhibiting tristetraprolin. Cell Physiol Biochem 2015, 37:707–718. [DOI] [PubMed] [Google Scholar]
- 43. Gebeshuber CA, Zatloukal K, Martinez J. miR‐29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 2009, 10:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Griseri P, Bourcier C, Hieblot C, Essafi‐Benkhadir K, Chamorey E, Touriol C, Pages G. A synonymous polymorphism of the Tristetraprolin (TTP) gene, an AU‐rich mRNA‐binding protein, affects translation efficiency and response to Herceptin treatment in breast cancer patients. Hum Mol Genet 2011, 20:4556–4568. [DOI] [PubMed] [Google Scholar]
- 45. Tiedje C, Holtmann H, Gaestel M. The role of mammalian MAPK signaling in regulation of cytokine mRNA stability and translation. J Interferon Cytokine Res 2014, 34:220–232. [DOI] [PubMed] [Google Scholar]
- 46. Kanies CL, Smith JJ, Kis C, Schmidt C, Levy S, Khabar KS, Morrow J, Deane N, Dixon DA, Beauchamp RD. Oncogenic Ras and transforming growth factor‐beta synergistically regulate AU‐rich element‐containing mRNAs during epithelial to mesenchymal transition. Mol Cancer Res 2008, 6:1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao H, Deterding LJ, Blackshear PJ. Identification of a major phosphopeptide in human tristetraprolin by phosphopeptide mapping and mass spectrometry. PLoS One 2014, 9:e100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suswam EA, Shacka JJ, Walker K, Lu L, Li X, Si Y, Zhang X, Zheng L, Nabors LB, Cao H, et al. Mutant tristetraprolin: a potent inhibitor of malignant glioma cell growth. J Neurooncol 2013, 113:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Tubergen EA, Banerjee R, Liu M, Vander Broek R, Light E, Kuo S, Feinberg SE, Willis AL, Wolf G, Carey T, et al. Inactivation or loss of TTP promotes invasion in head and neck cancer via transcript stabilization and secretion of MMP9, MMP2, and IL‐6. Clin Cancer Res 2013, 19:1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. Mitogen‐activated protein kinase‐activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine‐rich element. Mol Cell Biol 2006, 26:2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, Clark AR. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen‐activated protein kinase and extracellular signal‐regulated kinase pathways. Mol Cell Biol 2006, 26:2408–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang L, Yu Z, Zhang Z, Ma W, Song S, Huang G. Interaction with pyruvate kinase M2 destabilizes tristetraprolin by proteasome degradation and regulates cell proliferation in breast cancer. Sci Rep 2016, 6:22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barrios‐García T, Tecalco‐Cruz A, Gómez‐Romero V, Reyes‐Carmona S, Meneses‐Morales I, León‐Del‐Río A. Tristetraprolin represses estrogen receptor α transactivation in breast cancer cells. J Biol Chem 2014, 289:15554–15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA‐destabilizing factor tristetraprolin negatively regulates NF‐kappaB signaling. J Biol Chem 2009, 284:29383–29390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schichl YM, Resch U, Hofer‐Warbinek R, de Martin R. Tristetraprolin impairs NF‐kappaB/p65 nuclear translocation. J Biol Chem 2009, 284:29571–29581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tornatore L, Sandomenico A, Raimondo D, Low C, Rocci A, Tralau‐Stewart C, Capece D, D'Andrea D, Bua M, Boyle E, et al. Cancer‐selective targeting of the NF‐kappaB survival pathway with GADD45beta/MKK7 inhibitors. Cancer Cell 2014, 26:495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav‐like protein. J Biol Chem 1996, 271:8144–8151. [DOI] [PubMed] [Google Scholar]
- 58. Ciafrè SA, Galardi S. microRNAs and RNA‐binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol 2013, 10:934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000, 100:57–70. [DOI] [PubMed] [Google Scholar]
- 60. Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB. The RNA‐binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res 2011, 9:648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholanikunnel BG, Fernandes DJ, Spicer EK. Regulation of Bcl‐2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res 2009, 7:1354–1366. [DOI] [PubMed] [Google Scholar]
- 62. Zhang J, Tsaprailis G, Bowden GT. Nucleolin stabilizes Bcl‐X L messenger RNA in response to UVA irradiation. Cancer Res 2008, 68:1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim CW, Kim HK, Vo MT, Lee HH, Kim HJ, Min YJ, Cho WJ, Park JW. Tristetraprolin controls the stability of cIAP2 mRNA through binding to the 3'UTR of cIAP2 mRNA. Biochem Biophys Res Commun 2010, 400:46–52. [DOI] [PubMed] [Google Scholar]
- 64. Donahue JM, Chang ET, Xiao L, Wang PY, Rao JN, Turner DJ, Wang JY, Battafarano RJ. The RNA‐binding protein HuR stabilizes survivin mRNA in human oesophageal epithelial cells. Biochem J 2011, 437:89–96. [DOI] [PubMed] [Google Scholar]
- 65. Panganiban RP, Vonakis BM, Ishmael FT, Stellato C. Coordinated post‐transcriptional regulation of the chemokine system: messages from CCL2. J Interferon Cytokine Res 2014, 34:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kang J‐G, Amar MJ, Remaley AT, Kwon J, Blackshear PJ, Wang P‐y, Hwang PM. Zinc finger protein tristetraprolin interacts with CCL3 mRNA and regulates tissue inflammation. J Immunol 2011, 187:2696–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Horner TJ, Lai WS, Stumpo DJ, Blackshear PJ. Stimulation of polo‐like kinase 3 mRNA decay by tristetraprolin. Mol Cell Biol 2009, 29:1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sauer I, Schaljo B, Vogl C, Gattermeier I, Kolbe T, Müller M, Blackshear PJ, Kovarik P. Interferons limit inflammatory responses by induction of tristetraprolin. Blood 2006, 107:4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Horiuchi K, Umetani M, Minami T, Okayama H, Takada S, Yamamoto M, Aburatani H, Reid PC, Housman DE, Hamakubo T, et al. Wilms' tumor 1‐associating protein regulates G(2)/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci USA 2006, 103:17278–17283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 2000, 19:2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Al‐Khalaf HH, Colak D, Al‐Saif M, Al‐Bakheet A, Hendrayani SF, Al‐Yousef N, Kaya N, Khabar KS, Aboussekhra A. p16(INK4a) positively regulates cyclin D1 and E2F1 through negative control of AUF1. PLoS One 2011, 6:e21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guo X, Wu Y, Hartley RS. Cold‐inducible RNA‐binding protein contributes to human antigen R and cyclin E1 deregulation in breast cancer. Mol Carcinog 2010, 49:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Oda S, Nishida J, Nakabeppu Y, Sekiguchi M. Stabilization of cyclin E and cdk2 mRNAs at G1/S transition in Rat‐1A cells emerging from the G0 state. Oncogene 1995, 10:1343–1351. [PubMed] [Google Scholar]
- 74. Scoumanne A, Cho SJ, Zhang J, Chen X. The cyclin‐dependent kinase inhibitor p21 is regulated by RNA‐binding protein PCBP4 via mRNA stability. Nucleic Acids Res, 2010, 39:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 2000, 20:760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Griffin ME, Hamilton BJ, Roy KM, Du M, Willson AM, Keenan BJ, Wang XW, Nichols RC. Post‐transcriptional regulation of glucose transporter‐1 by an AU‐rich element in the 3'UTR and by hnRNP A2. Biochem Biophys Res Commun 2004, 318:977–982. [DOI] [PubMed] [Google Scholar]
- 77. Gantt KR, Cherry J, Richardson M, Karschner V, Atasoy U, Pekala PH. The regulation of glucose transporter (GLUT1) expression by the RNA binding protein HuR. J Cell Biochem 2006, 99:565–574. [DOI] [PubMed] [Google Scholar]
- 78. Bonafé N, Gilmore‐Hebert M, Folk NL, Azodi M, Zhou Y, Chambers SK. Glyceraldehyde‐3‐phosphate dehydrogenase binds to the AU‐rich 3′ untranslated region of colony‐stimulating factor‐1 (CSF‐1) messenger RNA in human ovarian cancer cells: possible role in CSF‐1 posttranscriptional regulation and tumor phenotype. Cancer Res 2005, 65:3762–3771. [DOI] [PubMed] [Google Scholar]
- 79. Chambers SK. Role of CSF‐1 in progression of epithelial ovarian cancer. Future Oncol 2009, 5:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ross HJ, Sato N, Ueyama Y, Koeffler HP. Cytokine messenger RNA stability is enhanced in tumor cells. Blood 1991, 77:1787–1795. [PubMed] [Google Scholar]
- 81. Ruggiero T, Trabucchi M, Ponassi M, Corte G, Chen C‐Y, al‐Haj L, Khabar KS, Briata P, Gherzi R. Identification of a set of KSRP target transcripts upregulated by PI3K‐AKT signaling. BMC Mol Biol 2007, 8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Qiu LQ, Lai WS, Bradbury A, Zeldin DC, Blackshear PJ. Tristetraprolin (TTP) coordinately regulates primary and secondary cellular responses to proinflammatory stimuli. J Leukoc Biol 2015, 97:723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jalonen U, Nieminen R, Vuolteenaho K, Kankaanranta H, Moilanen E. Down‐regulation of tristetraprolin expression results in enhanced IL‐12 and MIP‐2 production and reduced MIP‐3α synthesis in activated macrophages. Mediators Inflamm 2006, 2006:40691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Winzen R, Thakur BK, Dittrich‐Breiholz O, Shah M, Redich N, Dhamija S, Kracht M, Holtmann H. Functional analysis of KSRP interaction with the AU‐rich element of interleukin‐8 and identification of inflammatory mRNA targets. Mol Cell Biol 2007, 27:8388–8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nakayama T, Mutsuga N, Tosato G. FGF2 posttranscriptionally down‐regulates expression of SDF1 in bone marrow stromal cells through FGFR1 IIIc. Blood 2007, 109:1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Al‐Souhibani N, Al‐Ghamdi M, Al‐Ahmadi W, Khabar KS. Posttranscriptional control of the chemokine receptor CXCR4 expression in cancer cells. Carcinogenesis 2014, 35:1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Muralidharan R, Panneerselvam J, Chen A, Zhao YD, Munshi A, Ramesh R. HuR‐targeted nanotherapy in combination with AMD3100 suppresses CXCR4 expression, cell growth, migration and invasion in lung cancer. Cancer Gene Ther 2015, 22:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, Borden KL. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol 2009, 29:1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Licata L, Hostetter C, Keen J. The RNA binding protein HuR/ELAV1 regulates the estrogen receptor and epidermal growth factor receptor beta expression in BT 474 cells. Cancer Res 2009, 69:3076. [Google Scholar]
- 90. Barrios‐Garcia T, Tecalco‐Cruz A, Gomez‐Romero V, Reyes‐Carmona S, Meneses‐Morales I, Leon‐Del‐Rio A. Tristetraprolin represses estrogen receptor alpha transactivation in breast cancer cells. J Biol Chem 2014, 289:15554–15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lee HH, Lee SR, Leem SH. Tristetraprolin regulates prostate cancer cell growth through suppression of E2F1. J Microbiol Biotechnol 2014, 24:287–294. [DOI] [PubMed] [Google Scholar]
- 92. Touriol C, Morillon A, Gensac MC, Prats H, Prats AC. Expression of human fibroblast growth factor 2 mRNA is post‐transcriptionally controlled by a unique destabilizing element present in the 3'‐untranslated region between alternative polyadenylation sites. J Biol Chem 1999, 274:21402–21408. [DOI] [PubMed] [Google Scholar]
- 93. Chen T‐M, Hsu C‐H, Tsai S‐J, Sun HS. AUF1 p42 isoform selectively controls both steady‐state and PGE2‐induced FGF9 mRNA decay. Nucleic Acids Res 2010, 38:8061–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Winstall E, Gamache M, Raymond V. Rapid mRNA degradation mediated by the c‐fos 3' AU‐rich element and that mediated by the granulocyte‐macrophage colony‐stimulating factor 3' AU‐rich element occur through similar polysome‐associated mechanisms. Mol Cell Biol 1995, 15:3796–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Degese MS, Tanos T, Naipauer J, Gingerich T, Chiappe D, Echeverria P, LaMarre J, Gutkind JS, Coso OA. An interplay between the p38 MAPK pathway and AUBPs regulates c‐fos mRNA stability during mitogenic stimulation. Biochem J 2015, 467:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Galban S, Kuwano Y, Pullmann R Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA‐binding proteins HuR and PTB promote the translation of hypoxia‐inducible factor 1alpha. Mol Cell Biol 2008, 28:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kim TW, Yim S, Choi BJ, Jang Y, Lee JJ, Sohn BH, Yoo HS, Yeom YI, Park KC. Tristetraprolin regulates the stability of HIF‐1alpha mRNA during prolonged hypoxia. Biochem Biophys Res Commun 2010, 391:963–968. [DOI] [PubMed] [Google Scholar]
- 98. Kastelic T, Schnyder J, Leutwiler A, Traber R, Streit B, Niggli H, MacKenzie A, Cheneval D. Induction of rapid IL‐1 beta mRNA degradation in THP‐1 cells mediated through the AU‐rich region in the 3'UTR by a radicicol analogue. Cytokine 1996, 8:751–761. [DOI] [PubMed] [Google Scholar]
- 99. Stoecklin G, Gross B, Ming X‐F, Moroni C. A novel mechanism of tumor suppression by destabilizing AU‐rich growth factor mRNA. Oncogene 2003, 22:3554–3561. [DOI] [PubMed] [Google Scholar]
- 100. Van Tubergen E, Broek RV, Lee J, Wolf G, Carey T, Bradford C, Prince M, Kirkwood KL, D'Silva NJ. Tristetraprolin regulates IL‐6 which is correlated with tumor progression in head and neck squamous cell carcinoma. Cancer 2011, 117:2677–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shi JX, Su X, Xu J, Zhang WY, Shi Y. HuR post‐transcriptionally regulates TNF‐alpha‐induced IL‐6 expression in human pulmonary microvascular endothelial cells mainly via tristetraprolin. Respir Physiol Neurobiol 2012, 181:154–161. [DOI] [PubMed] [Google Scholar]
- 102. Kim Y, Noren Hooten N, Dluzen DF, Martindale JL, Gorospe M, Evans MK. Posttranscriptional regulation of the inflammatory marker C‐reactive protein by the RNA‐binding protein HuR and microRNA 637. Mol Cell Biol 2015, 35:4212–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bourcier C, Griseri P, Grepin R, Bertolotto C, Mazure N, Pages G. Constitutive ERK activity induces downregulation of tristetraprolin, a major protein controlling interleukin8/CXCL8 mRNA stability in melanoma cells. Am J Physiol Cell Physiol 2011, 301:C609–C618. [DOI] [PubMed] [Google Scholar]
- 104. Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome‐wide analysis identifies interleukin‐10 mRNA as target of tristetraprolin. J Biol Chem 2008, 283:11689–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Milke L, Schulz K, Weigert A, Sha W, Schmid T, Brune B. Depletion of tristetraprolin in breast cancer cells increases interleukin‐16 expression and promotes tumor infiltration with monocytes/macrophages. Carcinogenesis 2013, 34:850–857. [DOI] [PubMed] [Google Scholar]
- 106. Pautz A, Art J, Hahn S, Nowag S, Voss C, Kleinert H. Regulation of the expression of inducible nitric oxide synthase. Nitric Oxide 2010, 23:75–93. [DOI] [PubMed] [Google Scholar]
- 107. Gherzi R, Lee K‐Y, Briata P, Wegmüller D, Moroni C, Karin M, Chen C‐Y. A KH domain RNA binding protein, KSRP, promotes ARE‐directed mRNA turnover by recruiting the degradation machinery. Mol Cell 2004, 14:571–583. [DOI] [PubMed] [Google Scholar]
- 108. Akool el S, Kleinert H, Hamada FM, Abdelwahab MH, Forstermann U, Pfeilschifter J, Eberhardt W. Nitric oxide increases the decay of matrix metalloproteinase 9 mRNA by inhibiting the expression of mRNA‐stabilizing factor HuR. Mol Cell Biol 2003, 23:4901–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol 2007, 14:511–518. [DOI] [PubMed] [Google Scholar]
- 110. Luo NA, Qu YQ, Yang GD, Wang T, Li RL, Jia LT, Dong R. Post‐transcriptional up‐regulation of PDGF‐C by HuR in advanced and stressed breast cancer. Int J Mol Sci 2014, 15:20306–20320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chesney J, Mitchell R, Benigni F, Bacher M, Spiegel L, Al‐Abed Y, Han JH, Metz C, Bucala R. An inducible gene product for 6‐phosphofructo‐2‐kinase with an AU‐rich instability element: role in tumor cell glycolysis and the Warburg effect. Proc Natl Acad Sci USA 1999, 96:3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mahat DB, Brennan‐Laun SE, Fialcowitz‐White EJ, Kishor A, Ross CR, Pozharskaya T, Rawn JD, Blackshear PJ, Hassel BA, Wilson GM. Coordinated expression of tristetraprolin post‐transcriptionally attenuates mitogenic induction of the oncogenic Ser/Thr kinase Pim‐1. PLoS One 2012, 7:e33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, Meisner‐Kober N, Londin E, Rigoutsos I, Sawicki JA, et al. The mRNA‐binding protein HuR promotes hypoxia‐induced chemoresistance through posttranscriptional regulation of the proto‐oncogene PIM1 in pancreatic cancer cells. Oncogene 2015, 35:2529–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Montero L, Nagamine Y. Regulation by p38 mitogen‐activated protein kinase of adenylate‐ and uridylate‐rich element‐mediated urokinase‐type plasminogen activator (uPA) messenger RNA stability and uPA‐dependent in vitro cell invasion. Cancer Res 1999, 59:5286–5293. [PubMed] [Google Scholar]
- 115. Al‐Souhibani N, Al‐Ahmadi W, Hesketh JE, Blackshear PJ, Khabar KS. The RNA‐binding zinc‐finger protein tristetraprolin regulates AU‐rich mRNAs involved in breast cancer‐related processes. Oncogene 2010, 29:4205–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhou Y, Chang R, Ji W, Wang N, Qi M, Xu Y, Guo J, Zhan L. Loss of Scribble promotes Snail translation through translocation of HuR and enhances cancer drug resistance. J Biol Chem 2015, 291(1):291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yoon NA, Jo HG, Lee UH, Park JH, Yoon JE, Ryu J, Kang SS, Min YJ, Ju SA, Seo EH, et al. Tristetraprolin suppresses the EMT through the down‐regulation of Twist1 and Snail1 in cancer cells. Oncotarget 2016, 7:8931–8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wise JF, Berkova Z, Mathur R, Zhu H, Braun FK, Tao R‐H, Sabichi AL, Ao X, Maeng H, Samaniego F. Nucleolin inhibits Fas ligand binding and suppresses Fas‐mediated apoptosis in vivo via a surface nucleolin‐Fas complex. Blood 2013, 121:4729–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Pineda DM, Rittenhouse DW, Valley CC, Cozzitorto JA, Burkhart RA, Leiby B, Winter JM, Weber MC, Londin ER, Rigoutsos I, et al. HuR's post‐transcriptional regulation of death receptor 5 in pancreatic cancer cells. Cancer Biol Ther 2012, 13:946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU‐rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 1999, 19:4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Abdelmohsen K, Panda AC, Kang M‐J, Guo R, Kim J, Grammatikakis I, Yoon J‐H, Dudekula DB, Noh JH, Yang X, et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res 2014, 42:10099–10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Pietersma A, Tilly BC, Gaestel M, de Jong N, Lee JC, Koster JF, Sluiter W. p38 mitogen activated protein kinase regulates endothelial VCAM‐1 expression at the post‐transcriptional level. Biochem Biophys Res Commun 1997, 230:44–48. [DOI] [PubMed] [Google Scholar]
- 123. Zhou S, Gu L, He J, Zhang H, Zhou M. MDM2 regulates vascular endothelial growth factor mRNA stabilization in hypoxia. Mol Cell Biol 2011, 31:4928–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Fellows A, Griffin ME, Petrella BL, Zhong L, Parvin‐Nejad FP, Fava R, Morganelli P, Robey RB, Nichols RC. AUF1/hnRNP D represses expression of VEGF in macrophages. Mol Biol Cell 2012, 23:1414–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chang ET, Donahue JM, Xiao L, Cui Y, Rao JN, Turner DJ, Twaddell WS, Wang JY, Battafarano RJ. The RNA‐binding protein CUG‐BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. Biochem J 2012, 446:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Brewer G. An A + U‐rich element RNA‐binding factor regulates c‐myc mRNA stability in vitro. Mol Cell Biol 1991, 11:2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wingett D, Reeves R, Magnuson NS. Stability changes in pim‐1 proto‐oncogene mRNA after mitogen stimulation of normal lymphocytes. J Immunol 1991, 147:3653–3659. [PubMed] [Google Scholar]
- 128. Kim HK, Kim CW, Vo MT, Lee HH, Lee JY, Yoon NA, Lee CY, Moon CH, Min YJ, Park JW, et al. Expression of proviral integration site for Moloney murine leukemia virus 1 (Pim‐1) is post‐transcriptionally regulated by tristetraprolin in cancer cells. J Biol Chem 2012, 287:28770–28778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Graham JR, Hendershott MC, Terragni J, Cooper GM. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3‐kinase signaling. Mol Cell Biol 2010, 30:5295–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ruggiero T, Trabucchi M, Ponassi M, Corte G, Chen CY, al‐Haj L, Khabar KS, Briata P, Gherzi R. Identification of a set of KSRP target transcripts upregulated by PI3K‐AKT signaling. BMC Mol Biol 2007, 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sharma S, Verma S, Vasudevan M, Samanta S, Thakur JK, Kulshreshtha R. The interplay of HuR and miR‐3134 in regulation of AU rich transcriptome. RNA Biol 2013, 10:1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. McCulloch RK, Walker CE, Chakera A, Jazayeri J, Leedman PJ. Regulation of EGF‐receptor expression by EGF and TGF alpha in epidermoid cancer cells is cell type‐specific. Int J Biochem Cell Biol 1998, 30:1265–1278. [DOI] [PubMed] [Google Scholar]
- 133. Balmer LA, Beveridge DJ, Jazayeri JA, Thomson AM, Walker CE, Leedman PJ. Identification of a novel AU‐Rich element in the 3' untranslated region of epidermal growth factor receptor mRNA that is the target for regulated RNA‐binding proteins. Mol Cell Biol 2001, 21:2070–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Dai W, Zhang G, Makeyev EV. RNA‐binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res 2012, 40:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hitti E, Khabar KS. Sequence variations affecting AU‐rich element function and disease. Front Biosci (Landmark Ed) 2012, 17:1846–1860. [DOI] [PubMed] [Google Scholar]
- 136. An J, Zhu X, Wang H, Jin X. A dynamic interplay between alternative polyadenylation and microRNA regulation: implications for cancer (Review). Int J Oncol 2013, 43:995–1001. [DOI] [PubMed] [Google Scholar]
- 137. Khabar KS, Bakheet T, Williams BR. AU‐rich transient response transcripts in the human genome: expressed sequence tag clustering and gene discovery approach. Genomics 2005, 85:165–175. [DOI] [PubMed] [Google Scholar]
- 138. Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 2015, 16:393–405. [DOI] [PubMed] [Google Scholar]
- 139. Newbold RF. The significance of telomerase activation and cellular immortalization in human cancer. Mutagenesis 2002, 17:539–550. [DOI] [PubMed] [Google Scholar]
- 140. Eversole A, Maizels N. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol Cell Biol 2000, 20:5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Pont Adam R, Sadri N, Hsiao Susan J, Smith S, Schneider RJ. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol Cell 2012, 47:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Chen L‐Y, Lingner J. AUF1/HnRNP D RNA binding protein functions in telomere maintenance. Mol Cell 2012, 47:1–2. [DOI] [PubMed] [Google Scholar]
- 143. Woo M, Hakem R, Mak TW. Executionary pathway for apoptosis: lessons from mutant mice. Cell Res 2000, 10:267–278. [DOI] [PubMed] [Google Scholar]
- 144. Ishimaru D, Zuraw L, Ramalingam S, Sengupta TK, Bandyopadhyay S, Reuben A, Fernandes DJ, Spicer EK. Mechanism of regulation of bcl‐2 mRNA by nucleolin and A + U‐rich element‐binding factor 1 (AUF1). J Biol Chem 2010, 285:27182–27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Johnson BA, Blackwell TK. Multiple tristetraprolin sequence domains required to induce apoptosis and modulate responses to TNFalpha through distinct pathways. Oncogene 2002, 21:4237–4246. [DOI] [PubMed] [Google Scholar]
- 146. Zekavati A, Nasir A, Alcaraz A, Aldrovandi M, Marsh P, Norton JD, Murphy JJ. Post‐transcriptional regulation of BCL2 mRNA by the RNA‐binding protein ZFP36L1 in malignant B cells. PLoS One 2014, 9:e102625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Jackson RS 2nd, Cho YJ, Liang P. TIS11D is a candidate pro‐apoptotic p53 target gene. Cell Cycle 2006, 5:2889–2893. [DOI] [PubMed] [Google Scholar]
- 148. Park SB, Lee JH, Jeong WW, Kim YH, Cha HJ, Joe Y, Chung HT, Cho WJ, Do JW, Lee BJ, et al. TTP mediates cisplatin‐induced apoptosis of head and neck cancer cells by down‐regulating the expression of Bcl‐2. J Chemother 2015, 27:174–180. [DOI] [PubMed] [Google Scholar]
- 149. Baou M, Jewell A, Muthurania A, Wickremasinghe RG, Yong KL, Carr R, Marsh P, Murphy JJ. Involvement of Tis11b, an AU‐rich binding protein, in induction of apoptosis by rituximab in B cell chronic lymphocytic leukemia cells. Leukemia 2009, 23:986–989. [DOI] [PubMed] [Google Scholar]
- 150. Izquierdo JM. Cell‐specific regulation of Fas exon 6 splicing mediated by Hu antigen R. Biochem Biophys Res Commun 2010, 402:324–328. [DOI] [PubMed] [Google Scholar]
- 151. Galban S, Gorospe M. Factors interacting with HIF‐1alpha mRNA: novel therapeutic targets. Curr Pharm Des 2009, 15:3853–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Chamboredon S, Ciais D, Desroches‐Castan A, Savi P, Bono F, Feige JJ, Cherradi N. Hypoxia‐inducible factor‐1alpha mRNA: a new target for destabilization by tristetraprolin in endothelial cells. Mol Biol Cell 2011, 22:3366–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Lee SH, Jeong D, Han Y‐S, Baek MJ. Pivotal role of vascular endothelial growth factor pathway in tumor angiogenesis. Ann Surg Treat Res 2015, 89:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, et al. HIF‐1alpha is essential for myeloid cell‐mediated inflammation. Cell 2003, 112:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, Schaub FX, Sanduja S, Dixon DA, Blackshear PJ, et al. Tristetraprolin impairs myc‐induced lymphoma and abolishes the malignant state. Cell 2012, 150:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Bett John S, Ibrahim Adel FM, Garg Amit K, Kelly V, Pedrioli P, Rocha S, Hay RT. The P‐body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability. Biochem J 2013, 451:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Baeriswyl V, Christofori G. The angiogenic switch in carcinogenesis. Semin Cancer Biol 2009, 19:329–337. [DOI] [PubMed] [Google Scholar]
- 158. Scaldaferri F, Vetrano S, Sans M, Arena V, Straface G, Stigliano E, Repici A, Sturm A, Malesci A, Panes J, et al. VEGF‐A links angiogenesis and inflammation in inflammatory bowel disease pathogenesis. Gastroenterology 2009, 136:585–595.e5. [DOI] [PubMed] [Google Scholar]
- 159. Lejbkowicz F, Goldberg‐Cohen I, Levy AP. New horizons for VEGF: is there a role for nuclear localization? Acta Histochem 2005, 106:405–411. [DOI] [PubMed] [Google Scholar]
- 160. Griseri P, Pages G. Control of pro‐angiogenic cytokine mRNA half‐life in cancer: the role of AU‐rich elements and associated proteins. J Interferon Cytokine Res 2014, 34:242–254. [DOI] [PubMed] [Google Scholar]
- 161. Arcondeguy T, Lacazette E, Millevoi S, Prats H, Touriol C. VEGF‐A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post‐transcriptional level. Nucleic Acids Res 2013, 41:7997–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Wu MZ, Chen SF, Nieh S, Benner C, Ger LP, Jan CI, Ma L, Chen CH, Hishida T, Chang HT, et al. Hypoxia drives breast tumor malignancy through a TET‐TNFalpha‐p38‐MAPK signaling axis. Cancer Res 2015, 75:3912–3924. [DOI] [PubMed] [Google Scholar]
- 163. Yoo PS, Mulkeen AL, Silva T, Schmitz J, Tai N, Uchio EM, Chu E, Cha CH. RNA‐binding protein HUR regulates VEGF expression in human colorectal cancer cells. J Surg Res 2006, 130:219–220. [Google Scholar]
- 164. Goldberg‐Cohen I, Furneauxb H, Levy AP. A 40‐bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J Biol Chem 2002, 277:13635–13640. [DOI] [PubMed] [Google Scholar]
- 165. Lee HH, Son YJ, Lee WH, Park YW, Chae SW, Cho WJ, Kim YM, Choi HJ, Choi DH, Jung SW, et al. Tristetraprolin regulates expression of VEGF and tumorigenesis in human colon cancer. Int J Cancer 2010, 126:1817–1827. [DOI] [PubMed] [Google Scholar]
- 166. Essafi‐Benkhadir K, Onesto C, Stebe E, Moroni C, Pagès G. Tristetraprolin inhibits Ras‐dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol Biol Cell 2007, 18:4648–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Gnarra JR, Zhou S, Merrill MJ, Wagner JR, Krumm A, Papavassiliou E, Oldfield EH, Klausner RD, Linehan WM. Post‐transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci USA 1996, 93:10589–10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Datta K, Mondal S, Sinha S, Li J, Wang E, Knebelmann B, Karumanchi SA, Mukhopadhyay D. Role of elongin‐binding domain of von hippel lindau gene product on HuR‐mediated VPF//VEGF mRNA stability in renal cell carcinoma. Oncogene 2005, 24:7850–7858. [DOI] [PubMed] [Google Scholar]
- 169. Sinha S, Dutta S, Datta K, Ghosh AK, Mukhopadhyay D. Von Hippel‐Lindau gene product modulates TIS11B expression in renal cell carcinoma: impact on vascular endothelial growth factor expression in hypoxia. J Biol Chem 2009, 284:32610–32618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Tang W, Zhu Y, Gao J, Fu J, Liu C, Liu Y, Song C, Zhu S, Leng Y, Wang G, et al. MicroRNA‐29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E‐cadherin via KLF4. Br J Cancer 2014, 110:450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Qiu F, Sun R, Deng N, Guo T, Cao Y, Yu Y, Wang X, Zou B, Zhang S, Jing T, et al. miR‐29a/b enhances cell migration and invasion in nasopharyngeal carcinoma progression by regulating SPARC and COL3A1 gene expression. PLoS One 2015, 10:e0120969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Bai D, Gao Q, Li C, Ge L, Gao Y, Wang H. A conserved TGFbeta1/HuR feedback circuit regulates the fibrogenic response in fibroblasts. Cell Signal 2012, 24:1426–1432. [DOI] [PubMed] [Google Scholar]
- 173. Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res 2002, 62:1489–1495. [PubMed] [Google Scholar]
- 174. Zucconi BE, Wilson GM. Modulation of neoplastic gene regulatory pathways by the RNA‐binding factor AUF1. Front Biosci (Landmark Ed) 2011, 16:2307–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Al‐Khalaf HH, Aboussekhra A. MicroRNA‐141 and microRNA‐146b‐5p inhibit the prometastatic mesenchymal characteristics through the RNA‐binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT. J Biol Chem 2014, 289:31433–31447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Yoon J‐H, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJF, Martindale JL, et al. PAR‐CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun 2014, 5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Solinas G, Germano G, Mantovani A, Allavena P. Tumor‐associated macrophages (TAM) as major players of the cancer‐related inflammation. J Leukoc Biol 2009, 86:1065–1073. [DOI] [PubMed] [Google Scholar]
- 178. Sica A, Saccani A, Bottazzi B, Polentarutti N, Vecchi A, Damme JV, Mantovani A. Autocrine production of IL‐10 mediates defective IL‐12 production and NF‐kB activation in tumor‐associated macrophages. J Immunol 2000, 164:762–767. [DOI] [PubMed] [Google Scholar]
- 179. Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony stimulating factor‐1 receptor. Cancer Res 2009, 69:9498–9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Shin BA, Yoo HG, Kim HS, Kim MH, Hwang YS, Chay KO, Lee KY, Ahn BW, Jung YD. P38 MAPK pathway is involved in the urokinase plasminogen activator expression in human gastric SNU‐638 cells. Oncol Rep 2003, 10:1467–1471. [DOI] [PubMed] [Google Scholar]
- 181. Huang S, New L, Pan Z, Han J, Nemerow GR. Urokinase plasminogen activator/urokinase‐specific surface receptor expression and matrix invasion by breast cancer cells requires constitutive p38alpha mitogen‐activated protein kinase activity. J Biol Chem 2000, 275:12266–12272. [DOI] [PubMed] [Google Scholar]
- 182. Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. The p38/MK2‐driven exchange between tristetraprolin and HuR regulates AU‐rich element‐dependent translation. PLoS Genet 2012, 8:e1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Friedl J, Puhlmann M, Bartlett DL, Libutti SK, Turner EN, Gnant MF, Alexander HR. Induction of permeability across endothelial cell monolayers by tumor necrosis factor (TNF) occurs via a tissue factor‐dependent mechanism: relationship between the procoagulant and permeability effects of TNF. Blood 2002, 100:1334–1339. [PubMed] [Google Scholar]
- 184. Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410:50–56. [DOI] [PubMed] [Google Scholar]
- 185. Salvucci O, Bouchard A, Baccarelli A, Deschenes J, Sauter G, Simon R, Bianchi R, Basik M. The role of CXCR4 receptor expression in breast cancer: a large tissue microarray study. Breast Cancer Res Treat 2006, 97:275–283. [DOI] [PubMed] [Google Scholar]
- 186. Dormoy‐Raclet V, Ménard I, Clair E, Kurban G, Mazroui R, Di Marco S, von Roretz C, Pause A, Gallouzi I‐E. The RNA‐binding protein HuR promotes cell migration and cell invasion by stabilizing the β‐actin mRNA in a U‐rich‐element‐dependent manner. Mol Cell Biol 2007, 27:5365–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Lauriola L, Granone P, Ramella S, Lanza P, Ranelletti FO. Expression of the RNA‐binding protein HuR and its clinical significance in human stage I and II lung adenocarcinoma. Histol Histopathol 2012, 27:617–626. [DOI] [PubMed] [Google Scholar]
- 188. Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple functions of the RNA‐binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci 2013, 14:10015–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mitogen‐activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol Cell Biol 2001, 21:6461–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Leelahavanichkul K, Amornphimoltham P, Molinolo AA, Basile JR, Koontongkaew S, Gutkind JS. A role for p38 MAPK in head and neck cancer cell growth and tumor‐induced angiogenesis and lymphangiogenesis. Mol Oncol 2014, 8:105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Suswam E, Li Y, Zhang X, Gillespie GY, Li X, Shacka JJ, Lu L, Zheng L, King PH. Tristetraprolin down‐regulates interleukin‐8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res 2008, 68:674–682. [DOI] [PubMed] [Google Scholar]
- 192. Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, Roy A, Turner BA, McDonald P, Tunge JA, et al. Identification and validation of novel small molecule disruptors of HuR‐mRNA interaction. ACS Chem Biol 2015, 10:1476–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Meisner N‐C, Hintersteiner M, Mueller K, Bauer R, Seifert J‐M, Naegeli H‐U, Ottl J, Oberer L, Guenat C, Moss S, et al. Identification and mechanistic characterization of low‐molecular‐weight inhibitors for HuR. Nat Chem Biol 2007, 3:508–515. [DOI] [PubMed] [Google Scholar]
- 194. D'Agostino VG, Lal P, Mantelli B, Tiedje C, Zucal C, Thongon N, Gaestel M, Latorre E, Marinelli L, Seneci P, et al. Dihydrotanshinone‐I interferes with the RNA‐binding activity of HuR affecting its post‐transcriptional function. Sci Rep 2015, 5:16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Deng K, Wang H, Shan T, Chen Y, Zhou H, Zhao Q, Xia J. Tristetraprolin inhibits gastric cancer progression through suppression of IL‐33. Sci Rep 2016, 6:24505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Xu L, Ning H, Gu L, Wang Q, Lu W, Peng H, Cui W, Ying B, Ross CR, Wilson GM, et al. Tristetraprolin induces cell cycle arrest in breast tumor cells through targeting AP‐1/c‐Jun and NF‐kappaB pathway. Oncotarget 2015, 6:41679–41691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Jimbo M, Blanco FF, Huang Y‐H, Telonis AG, Screnci BA, Cosma GL, Alexeev V, Gonye GE, Yeo CJ, Sawicki JA, et al. Targeting the mRNA‐binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget 2015, 6:27312–27331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Danilin S, Sourbier C, Thomas L, Lindner V, Rothhut S, Dormoy V, Helwig J‐J, Jacqmin D, Lang H, Massfelder T. Role of the RNA‐binding protein HuR in human renal cell carcinoma. Carcinogenesis 2010, 31:1018–1026. [DOI] [PubMed] [Google Scholar]
- 199. López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA‐binding protein HuR in colon carcinogenesis. Oncogene 2003, 22:7146–7154. [DOI] [PubMed] [Google Scholar]
- 200. Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 1996, 4:445–454. [DOI] [PubMed] [Google Scholar]
- 201. Qiu LQ, Stumpo DJ, Blackshear PJ. Myeloid‐specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol 2012, 188:5150–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Patial S, Curtis AD 2nd, Lai WS, Stumpo DJ, Hill GD, Flake GP, Mannie MD, Blackshear PJ. Enhanced stability of tristetraprolin mRNA protects mice against immune‐mediated inflammatory pathologies. Proc Natl Acad Sci USA 2016, 113:1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Brooks SA, Connolly JE, Rigby WF. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal‐regulated kinase‐specific, AU‐rich element‐dependent, autoregulatory pathway. J Immunol 2004, 172:7263–7271. [DOI] [PubMed] [Google Scholar]
- 204. Ross EA, Smallie T, Ding Q, O'Neil JD, Cunliffe HE, Tang T, Rosner DR, Klevernic I, Morrice NA, Monaco C, et al. Dominant suppression of inflammation via targeted mutation of the mRNA destabilizing protein tristetraprolin. J Immunol 2015, 195:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, et al. Deletion of the RNA‐binding proteins Zfp36l1 and Zfp36l2 leads to perturbed thymic development and T‐lymphoblastic leukaemia. Nat Immunol 2010, 11:717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Hyatt LD, Wasserman GA, Rah YJ, Matsuura KY, Coleman FT, Hilliard KL, Pepper‐Cunningham ZA, Ieong M, Stumpo DJ, Blackshear PJ, et al. Myeloid ZFP36L1 does not regulate inflammation or host defense in mouse models of acute bacterial infection. PLoS One 2014, 9:e109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Ghosh M, Aguila HL, Michaud J, Ai Y, Wu M‐T, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T. Essential role of the RNA‐binding protein HuR in progenitor cell survival in mice. The Journal of Clinical Investigation 2009, 119:3530–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, Kontoyiannis DL. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell 2005, 19:777–789. [DOI] [PubMed] [Google Scholar]
- 209. Gubin MM, Techasintana P, Magee JD, Dahm GM, Calaluce R, Martindale JL, Whitney MS, Franklin CL, Besch‐Williford C, Hollingsworth JW, et al. Conditional knockout of the RNA‐binding protein HuR in CD4(+) T cells reveals a gene dosage effect on cytokine production. Mol Med 2014, 20:93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. DeMicco A, Naradikian MS, Sindhava VJ, Yoon JH, Gorospe M, Wertheim GB, Cancro MP, Bassing CH. B Cell‐intrinsic expression of the HuR RNA‐binding protein is required for the T cell‐dependent immune response in vivo. J Immunol 2015, 195:3449–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Lin WJ, Zheng X, Lin CC, Tsao J, Zhu X, Cody JJ, Coleman JM, Gherzi R, Luo M, Townes TM, et al. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol Cell Biol 2011, 31:3196–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]


