Summary
Agaricus bisporus is a secondary decomposer fungus and an excellent model for the adaptation, persistence and growth of fungi in humic‐rich environments such as soils of temperate woodland and pastures. The A. bisporus serine proteinase SPR1 is induced by humic acids and is highly expressed during growth on compost. Three Spr1 gene silencing cassettes were constructed around sense, antisense and non‐translatable‐stop strategies (pGRsensehph, pGRantihph and pGRstophph). Transformation of A. bisporus with these cassettes generated cultures showing a reduction in extracellular proteinase activity as demonstrated by the reduction, or abolition, of a clearing zone on plate‐based bioassays. These lines were then assessed by detailed enzyme assay, RT‐qPCR and fruiting. Serine proteinase activity in liquid cultures was reduced in 83% of transformants. RT‐qPCR showed reduced Spr1 mRNA levels in all transformants analysed, and these correlated with reduced enzyme activity. When fruiting was induced, highly‐silenced transformant AS5 failed to colonize the compost, whilst for those that did colonize the compost, 60% gave a reduction in mushroom yield. Transcriptional, biochemical and developmental observations, demonstrate that SPR1 has an important role in nutrient acquisition in compost and that SPR1 is a key enzyme in the adaptation of Agaricus to the humic‐rich ecological niche formed during biomass degradation.
Introduction
An ecosystem's carbon sequestration potential has gained increased interest due to current climate change concerns (Bailey et al., 2002; Strickland and Rousk, 2010). Forest ecosystems play a prominent role in global carbon cycling (Myneni et al., 2001), and understanding microbial involvement in decomposition in these ecosystems is required to estimate global carbon fluxes and their potential future changes (Zifcakova et al., 2011). Depolymerization of biopolymers is a key process in the cycling of carbon (Kahkonen and Hakulinen, 2011). With litter decomposition in temperate forests being mainly driven by fungal activity (Watling and Harper, 1998; Hattenschwiler et al., 2005; Steffen et al., 2007) these processes are likely to impact on the potential for carbon sequestration in such habitats. In terrestrial environments, basidiomycetes are probably the ecologically most significant group of fungi, involved in the breakdown and chemical conversion of litter components (Dix and Webster, 1995). They constitute a major fraction of the living biomass and are responsible for efficient degradation of many recalcitrant organic compounds in soil litter and the humic layer (Steffen et al., 2007). These humic substances are dark‐coloured products of biotransformation of plant material, chemically heterogeneous, stabilized physically by soil particle association and by micro‐aggregation into supramolecular associations (Piccolo, 2002; Six et al., 2002). Humic substances represent a significant component of stabilized carbon and nitrogen in soil and have important roles in global carbon and nitrogen cycling and in the regulation of the mobility and fate of plant nutrients and environmental contaminants (Murphy and Zachara, 1995; Weber, 1998; Christl et al., 2000; Zavarzina et al., 2004; Dong et al., 2006). Investigations into the mechanisms of humus biotransformation and the organisms responsible are particularly important under the changing environment and understanding of these processes may promote better predictions of the dynamics of soil organic matter.
Litter decomposing basidiomycetes are regarded as the key players in microbial lignin and humus degradation and produce a wide variety of oxidoreductases and hydrolytic enzymes capable of degrading all three principal litter components: cellulose, hemicellulose and lignin (Steffen et al., 2000, 2002; Martinez et al., 2005; Osono, 2007; Valaskova et al., 2007; Baldrian and Valaskova, 2008; Sinsabaugh, 2010; Snajdr et al., 2010; Burke et al., 2011). These extracellular enzymes can only be made if the fungus has an adequate nitrogen supply and so they are often accompanied by production of chitinases or proteases.
The basidiomycete Agaricus bisporus is a secondary decomposer fungus. Although it is not an effective competitor on un‐degraded plant debris, it grows well on partially‐decomposed plant material, hence is often abundant in the composted leaf and needle litter found in soils of temperate forests (Morin et al., 2012; Kerrigan et al., 2013). Given the wealth of information available concerning its commercial cultivation, coupled with the available genome data and transformation systems, A. bisporus is an ideal model for the study of fungal adaptation, persistence and growth in humic rich environments where nutrition is not readily available to primary degrading fungi (Burton et al., 1997a; Morin et al., 2012).
Serine proteinase activity has been identified as the major extracellular proteinase produced by A. bisporus mycelium growing in compost, in which nitrogen, a limiting factor of mushroom yield, is largely sequestered in the form of protein and microbial biomass, suggesting a nutritional role for this enzyme (Burton et al., 1997a). Extracellular mycelial serine proteinase is synthesized in response to humic‐associated protein rather than casein or glutamate as a nitrogen source (Burton et al., 1997a). High levels of serine proteinase activity are also produced in the stipe of A. bisporus mushrooms during post‐harvest senescence from which the SPR1 enzyme was purified and characterized biochemically (Burton et al., 1993, 1997b; Kingsnorth et al., 2001). Transgenic analysis of the Spr1 promoter elements confirmed serine proteinase production in mycelium is regulated in response to specific nitrogen sources, and clearly demonstrated expression patterns that are appropriate for nutrient acquisition in mycelia and transport through the stipe for fruiting body production (Heneghan et al., 2009).
Recent genomic research has revealed complexity in the mechanisms of adaptation of A. bisporus to the humic‐rich ecological niche. A detailed analysis of genomic sequence coupled with expression data showed that twenty‐two genes specifically expressed in humic‐rich environments share a specific promoter motif; these include 3 serine proteinases, 3 laccases, 2 cutinases and 10 carbohydrate‐active genes (Morin et al., 2012). The expression ratios of these three humic‐responsive serine proteinases in humic:non‐humic environments were 235 for Spr3, 107 for Spr4 and 103 for Spr1 (Morin et al., 2012). Serine proteinase genes Spr3 and Spr4 (see Supporting Information Table S1) have 34.4% and 24.3% pairwise identity, respectively, to Spr1. Two further serine proteinases Spr5 and Spr6 have high transcript levels in compost but lower humic:non‐humic expression ratios. Spr5 and Spr6 have 42.4% and 31.1% pairwise identity to Spr1. Heneghan and colleagues (2009) also described Spr2 and Spr2a, however although these have high pairwise identities to Spr1 (79.2% and 79.3%, respectively) and are within 20kb on the same scaffold, they are likely to play only minor roles in humic‐related nutrition as their transcript levels and humic:non‐humic expression ratios are much lower (Morin et al., 2012).
Whilst A. bisporus is well known for its commercial value as a cultivated mushroom, it is often overlooked that this and many other closely related species inhabit woodland and permanent pasture across most temperate environments, and thus there may be a vital role for proteases such as SPR1 in nutrient acquisition from humic associated material in soils and compost. Understanding the nutrient cycling role of fungi such as A. bisporus in ecosystems is a prerequisite to modelling and optimizing carbon management for sustainable forests. To fully elucidate the importance of the humic‐regulated serine proteinases in nutrient acquisition, a transgenic analysis of the role of Spr1 was conducted. We describe here the silencing of the Spr1 gene, the biochemical and molecular analysis of transformants and the monitoring of transformants through mushroom sporophore development.
Results
Transgenic analysis of SPR1was performed using three different silencing constructs engineered to contain the Spr1 cDNA in the sense (pGRsensehph) and anti‐sense (pGRantihph) orientations and as untranslatable‐sense (pGRstophph). Transformation of A. bisporus with these Hph‐Spr1 silencing cassettes resulted in the generation of 8 sense (S), 14 antisense (AS) and 17 stop (ST) stable transformants. Wild type A. bisporus (WT) was also transformed with GFP as a transformation control. Transformants were maintained on MMP plates supplemented with hygromycin. Transformants displayed normal morphology in plate cultures when compared to wild type A. bisporus.
Analysis of A. bisporus Spr1‐silenced transformants
A. bisporus Spr1‐silenced transformants, wild type and GFP control transformants were screened for proteinase activity using the plate‐based clearing zone assay. Average clearing zones for both non‐transformed control strain A15 and GFP transformants were similar: 3.16 mm for A15 and 3 mm for GFP transformants on SM medium with a gelatin overlay 17 day post‐inoculation (Supporting Information Fig. S2). Transformants exhibiting reduced or no proteinase activity were S1, S4, S5, AS1, AS3, AS5, AS10, AS13 ST4, ST6, ST8, ST9 and ST11. These were selected for further analysis along with three transformants exhibiting normal proteinase activity (S3, AS8, ST2).
Transformants were analysed by PCR and amplification using primers Hyg1 and Hyg2 resulted in the production of a 600 bp fragment in all A. bisporus transformants confirming transformation. The presence of an intact WT Spr1 gene was confirmed in the wild‐type and all transformants using primers SprpromFwd and Spr‐z which bind in the Spr1 promoter and gene to amplify a 1400 bp genomic DNA product showing that the Spr1 locus had not been disrupted as a result of transformation. The presence of the intact silencing cassettes in pGRsensehph, pGRantihph and pGRstophph transformants, was also assessed by PCR, screening for presence of the 5′ and 3′ regions in separate reactions to avoid cross‐reaction with the genomic Spr1 locus. Primers 004‐p1 and Spr‐z anneal within the A. bisporus gpdII promoter and Spr1 cDNA of pGRsensehph and pGRstophph plasmids to give a 1075 bp 5′ product. Primers SPR‐x and 004‐p1 anneal within the A. bisporus gpdII promoter and Spr1 cDNA of pGRantihph plasmid to yield a 1230 bp 5′ product. Primers Spr‐x and TrpCRev anneal within the Spr1 cDNA and A. nidulans TrpC terminator of pGRsensehph and pGRstophph plasmids giving a 1263 bp 3′ product. Primers Spr‐z and TrpCRev anneal within the Spr1 cDNA and A. nidulans TrpC terminator of pGRantihph plasmid resulting in a 1054 bp 3′ product. Amplification of these fragments confirmed the presence of the Spr1 silencing cassette in transformants.
Measurement of serine proteinase activity
Serine proteinase activity (normalized against dry weight) was assessed by inoculating sense‐transformants (S1, S3, S4, S5), antisense transformants (AS5, AS8, AS10, AS13) and sense‐stop transformants (ST2, ST6, ST9, ST11) along with wild type A15 and two transformed controls (ATA2, ATA4) into Treschow Media (TM) supplemented with humic acid (Fig. 1). Serine proteinase activity in control transformants (ATA2, ATA4) did not differ significantly from the non‐transformed control A15. The serine proteinase activities of five transformants, S4, S5, AS5, AS13 and ST6, were significantly reduced compared with the control strains (Fig. 1). The remaining transformants showed no significant difference to the control strains.
Figure 1.
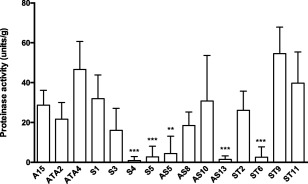
Normalized serine proteinase activity in 15 transformants growing in TM broth with humic fraction. A15 is the wild type strain, ATA2 and ATA4 are control transformants. Error bars are standard deviation of the mean of five biological replicates, with significance tested by Mann‐Whitney. A. bisporus transformants harbouring the Spr1 sense (S), anti‐sense (AS) and untranslatable sense (ST) constructs.
Induction of serine proteinase by nitrogen source
To determine the effect of induction of Spr1, six transformants (S4, S5, AS5, AS13, ST6 and ST9), wild‐type A15 and transformed control ATA4 were grown in TM supplemented with humic fraction or glutamate to compare induction and non‐induction of serine proteinase activity respectively. Serine proteinase enzyme activity was not induced in the presence of glutamate as sole nitrogen source for any of the strains tested (Fig. 2A). For growth in humic fraction as sole nitrogen source, control transformant ATA4 did not differ significantly from the wild‐type A15, both showing high levels of SPR1 activity. Serine proteinase activity in four transformants (S4, S5, AS13 and ST6) was significantly down‐regulated compared to the controls A15 and ATA4 in humic fraction. No significant differences in serine proteinase activity were found between the remaining transformants and the controls.
Figure 2.
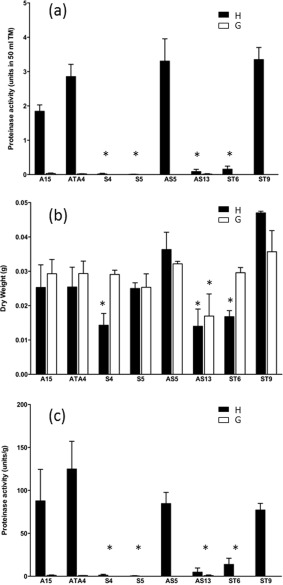
Induction of serine proteinase activity.
A. Serine protease activity in nine transformants growing in TM broth with humic acid (H) or glutamate (G) as sole nitrogen source.
B. Dry weights of transformants: A15 is the wild type strain, and ATA4 are control transformants.
C. Normalized serine proteinase activity with respect to biomass. Error bars are standard error of the mean of five biological replicates. A. bisporus transformants harbouring the Spr1 sense (S), anti‐sense (AS) and untranslatable sense (ST) constructs. Results were statistically analysed using t‐test and significant results are indicated by an asterisk (*).
The growth of the mycelial cultures was determined by measuring their dry weights at the end of the experiment (after 21 days growth). Control strains A15 and ATA4 and transformants S5, AS5 and AS13 had similar average dry weights for cultures grown on humic and glutamate as nitrogen source (Fig. 2B). However for AS13 both nutritional treatments resulted in lower dry weights than the controls. The dry weights of transformants S4 and ST6 were significantly reduced when grown on humic as sole nitrogen source while their growth on glutamate showed no difference compared with controls (Fig. 2B). Transformant ST9 had significantly increased dry weight when grown on humic fraction, compared with controls.
Normalized serine proteinase activity in media with either humic or glutamate nitrogen was significantly reduced in four transformants (S4, S5, AS13 and ST6) when compared with controls. In two transformants (AS5, ST9), there was no significant difference from non‐transformed and transformed controls (Fig. 2C).
Quantification of Spr1 transcripts
Serine proteinase transcript levels were quantified in transformants S4, S5, AS5, AS13, ST6, ST9, transformed control ATA4 and wild‐type A15 (Fig. 3) using RT‐qPCR analysis. Transcript levels in the control transformant ATA4 did not differ significantly from the wild‐type, however three transformants (S4, S5 and AS13) displayed significantly reduced levels of Spr1 transcripts.
Figure 3.
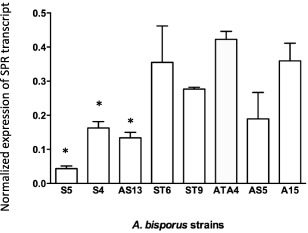
Quantification of serine proteinase transcripts determined by RT‐qPCR. Expression levels were normalized against 18S RNA. A15 is the wild type strain, ATA4 is a transformed control. Bars are means from SQRT scale. Error bars are +/− 0.5 standard error of the means of at least seven biological replicates A. bisporus transformants harbouring the Spr1 sense (S), anti‐sense (AS) and untranslatable sense (ST) constructs and significance assessed using Unpaired T‐test P < 0.05 indicated by an asterisk.
Correlation of serine proteinase activity and transcript levels
Serine proteinase transcript levels and enzyme activities of transformants (S4, S5, AS1, AS5, AS13, ST6 and ST9) and controls (A15 and ATA4) were compared. Linear regression analysis was performed on the square root of the averages of the Spr1 transcript levels and proteinase activities (Fig. 4). A significant correlation was found between Spr1 transcript levels and proteinase activities (correlation coefficient R 2 = 0.8355) (Fig. 4). The equation of the fitted line is: Serine proteinase activity = 20.7 x Spr1 transcript level–2.48. This suggests that a unit change in transcript level (increase or decrease) results in a 20‐fold change in enzyme activity. Transformants S4, S5 and AS13, exhibiting low level of Spr1 transcript, also had very low serine proteinase enzyme activity (Fig. 4). Transformant AS1 showed increased levels of both serine proteinase activity and transcript abundance.
Figure 4.
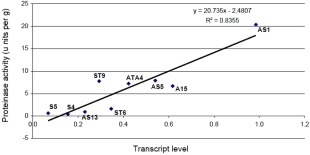
Linear regression analysis of the square root of the averages SPR1 transcript levels and proteinase activities for transformants S4, S5, AS1, AS5, AS13, ST6 and ST9 and controls (A15 and ATA4). The equation of the fitted line is: Serine proteinase activity = 20.7 x SPR1 transcript level–2.48.
Assessing the effect of Spr1 silencing on A. bisporus growth on compost and fruiting
The impact of Spr1 silencing on mycelial growth on compost and on mushroom production was determined for various selected transformants, non‐transformed control strain A15 and transformed control GFP1. Compost colonisation was assessed visually and subsequently, mushrooms were collected from 3 successive flushes and the weight of the mushrooms were recorded (Table 1). Transformant (AS5) failed to colonize the compost and hence produced no mushrooms. Four other transformants (S4, S5, ST4 and AS13) colonized the compost normally but gave significantly reduced yields of mushrooms (p<0.05) compared with the controls (non‐transformed A15 and transformed GFP1) (Table 1).
Table 1.
Mushroom fruiting results for transgenic strains.
| Average weight of mushrooms harvested (g) | ||||
|---|---|---|---|---|
| Strain/Transformant | Flush 1 | Flush 2 | Flush 3 | Total |
| A15 control | 614 | 256 | 71 | 941 |
| T‐ GFP control | 534 | 188 | 24 | 746 |
| S 1 | 509 | 159 | 83 | 751 |
| S 3 | 502 | 299 | 133 | 934 |
| S 4* | 113 | 0 | 30 | 143 |
| S 5* | 137 | 50 | 55 | 242 |
| AS 5* | 0 | 0 | 0 | 0 |
| AS 8 | 451 | 150 | 87 | 688 |
| AS 10 | 377 | 61 | 72 | 510 |
| AS 13* | 147 | 0 | 29 | 176 |
| ST 2 | 633 | 280 | 98 | 1012 |
| ST 4* | 205 | 145 | 30 | 381 |
| ST 6 | 354 | 253 | 57 | 664 |
| ST 9 | 648 | 226 | 6 | 880 |
| ST 11 | 644 | 195 | 71 | 909 |
Transgenic strains were grown in three replicate pots and mushrooms harvested at maturity stage 2–3 (Hammond and Nichols, 1976) over a period of 30 days. Mushrooms were collected from three successive flushes and the total number of mushrooms and weight of mushrooms recorded. Yields of Sense (S), Antisense (AS) and Sense‐Stop (ST) transformants of Spr1 were compared with two controls: non‐transformed A15 and GFP‐transformed T‐GFP. The LSD5% (Least Significant Difference) of the Total Yields was 288. The four transformants which were significantly different from the two controls are marked with an asterisk, *.
Discussion
SPR1 has previously been demonstrated to be the major extracellular protease produced by A. bisporus during growth on humic fractions and this protease is also abundantly expressed during post‐harvest senescence of the fruiting body (Burton et al., 1993, 1994, 1997a, 1997b). To further investigate the importance of SPR1 in nutrient acquisition and adaptation of Agaricus to humic environments, a transgenic analysis of Spr1 was conducted. A gene silencing approach was selected because it typically results in a diversity of transformants with varying silenced phenotypes, ranging from total inactivation of the gene to a small reduction in gene expression. Total inactivation, as achieved by gene disruption, may result in lethality to the fungus, whereas the suppression or downregulation mediated by gene silencing could permit a more practical functional analysis of critical genes (Heneghan et al., 2007; Kilaru et al., 2009). Furthermore, as A. bisporus is a multinucleate heterokaryon with two different nuclei, targeted gene knockouts of both alleles are likely to be difficult to achieve, whilst silencing should impact both as it is a dominant phenotype and should act similarly irrespective of the nuclear origin of the mRNA.
Three silencing strategies were employed using sense, antisense and sense‐stop constructs to trigger silencing of Spr1. Transformation of A. bisporus with the Hph‐Spr1 silencing cassettes (pGRsensehph, pGRantihph, and pGRstophph) resulted in the generation of a number of stable hygromycin resistant colonies; 8 sense (S), 14 antisense (AS) and 17 stop (ST) transformants. Molecular analysis confirmed the integrity of the wild type Spr1 gene in all transformants, demonstrating that gene knockouts had not occurred. The presence of a silencing cassette was confirmed in all transformants and given that previous work has established that only partial integration of the silencing cassette is necessary for silencing to occur (Namekawa et al., 2005; Heneghan et al., 2007) it was expected that amongst this population there would be some with a silencing phenotype. Of the 39 transformants obtained, 13 were impacted for protease production as indicated by reduction or abolition of clearing zones on plate‐based bioassay. Although these silencing frequencies were slightly lower than those reported for genes previously targeted in A. bisporus using complex hairpin‐based silencing constructs (Eastwood et al., 2008; Costa et al., 2009), it showed that these simpler cassettes were still effective in triggering silencing. The transformants with diminished clearing zones (S1, S4, S5, AS1, AS3, AS5, AS10, AS13, ST4, ST6, ST8, ST9 and ST11), indicative of a likely reduction in proteinase activity, along with three control transformants displaying normal proteinase activity, were selected for further analysis.
Whilst the plate‐based bioassay was indicative of reduced protease activity, it was not able to show which class of protease had been impacted. Therefore to specifically test for serine proteinase activity, a quantitative biochemical assay was employed using the serine protease‐specific substrate Succ‐Ala‐Ala‐Pro‐Phe‐pNA. This was conducted on 12 transformants (9 showing a reduction in proteinase activity and 3 demonstrating normal proteinase activity) grown in liquid culture supplemented with either humic or glutamic acid. Humic acid is known to induce SPR1 while nitrogen sources like glutamate do not induce expression (Burton et al., 1997a). Four of the transformants (S4, S5, AS13 and ST6) had significantly lower levels of serine proteinase activity than the controls when grown in humic fraction as sole nitrogen source. A strong correlation was found between Spr1 transcript level and serine protease enzyme activity using regression analyses. Given the almost complete down‐regulation of extracellular serine proteinase activity in some of these transformants despite the presence of the inducing humic fraction, this suggests SPR1 would normally account for the majority of the protease activity during growth on purified humic fraction.
The failure to colonize compost in AS5 and the reduced fruiting of other transformants highlights the role of SPR1 during growth on complex humic rich substrates. The three plasmids used in this study expressed the silencing cassette using the constitutive glyceraldehyde‐3‐phosphate dehydrogenase (gpdII) promoter from A. bisporus. Previous work has demonstrated the efficacy of this regulatory unit in transgene expression in basidiomycetes (Costa et al., 2008, 2009). The use of this constitutive promoter caused silencing of SPR1 throughout the life cycle of the basidiomycete, but this clearly impacted on normal growth on compost and on mushroom production. We note that SPR has been implicated as a detrimental enzyme impacting on both the shelf‐life and nutritional quality of mushrooms post‐harvest and, therefore, any silencing‐based approach to reduce the impact of SPR1 post‐harvest, would need to be designed incorporating the use of an appropriately regulated promoter to avoid impacting on SPR1 during cultivation.
We are aware that Morin and colleagues (2012) highlighted some 40 proteinases that might potentially be involved in the degradation of compost proteins, but of these genes, only three (Spr1, Spr3 and Spr4) were strongly up‐regulated during growth in mushroom compost, a complex humic‐rich substrate. Proteases Spr3, Spr4, Spr5 and Spr6 have 34%, 24%, 42.4% and 31.1% pair‐wise identities to Spr1, respectively, and this is below the degree of homology that would be required to facilitate cross‐silencing, so silencing of proteases other than SPR1 by our constructs is unlikely. Given that our experiments with purified humic fraction did not detect SPR3 or SPR4, it would suggest that they were not induced during growth in liquid culture on purified humic fraction. Their abundant transcript levels during growth on the more complex mushroom compost suggest that these other proteases are likely to be under a different regulatory pathway to SPR1. Even if other SPR proteases are expressed during growth on compost as inferred from the Morin and colleagues (2012) transcript data, the impact of silencing Spr1 alone was sufficient to reduce the ability of A. bisporus to colonize compost, showing that SPR1 plays a crucial role in supporting growth on such substrates. In addition the reduced numbers and mass of the resulting mushrooms highlights the role of this protease in supporting the production of fruiting bodies and, therefore, any impairments in SPR1 production would impact on the fecundity of A. bisporus.
Fungi such as A. bisporus were comparatively common in woodland and permanent pasture, but are becoming less abundant. The clear role of extracellular proteases such as SPR1 in allowing this fungus to access nitrogen from complex sources has been highlighted here, however, these proteases are tightly regulated and are likely to be impacted by changes in land management. We would speculate that some of the reduced occurrences of such fungi may well correlate with altered land management practises such as the application of fertilizers or manures which will impact on the regulation of these proteases. Negative impacts of nitrogen application has been observed particularly for basidiomycete fungi in other studies (Egli, 2011; Paungfoo‐Lonhienne et al., 2015). Given the roles of such basidiomycetes in nutrient cycling, this is likely to alter the C‐ and N‐cycles in soils and hence alter the fertility or carbon sequestration capacities of such soils. What this means for the long‐term health of such soil is difficult to predict.
The results presented here clearly demonstrate the critical role serine proteinases have in A. bisporus adaptation to humic rich substrates and a functional analysis of the organism's adaptation to its ecological niche. The silenced transformants either failed to colonize the compost, or resulted in a reduction in mushroom yield and hence fecundity, when compared to controls. Although three serine proteinases (Spr1, Spr3 and Spr4) have been identified showing up regulation on compost, indicating a possible role in nutrient acquisition, it would appear that SPR3 and SPR4 cannot compensate for silencing Spr1, assuming no cross silencing has occured. Biochemical analysis of enzyme activity levels and molecular analysis of transcript levels provide further evidence that SPR1 has been significantly down‐regulated in these transformants suggesting an essential role for this serine proteinase in nutrient acquisition from humic associated material in compost. SPR1 clearly plays a major role in allowing A. bisporus to access nitrogen, and hence also carbon sources, during growth on humic‐rich substrates and is, therefore, likely to be a determining factor in N‐ and C‐ cycles in the humic‐rich ecological niche formed during degradation of plant material in the soil.
Experimental procedures
Strains and culture maintenance
Escherichia coli strain DH5α was the host strain for plasmid construction and maintenance. Agrobacterium tumefaciens strains AGL1 and LBA1126 (Bundock and Hooykaas, 1996) were used for A. bisporus transformations and cultured as previously described (de Groot et al., 1998). The A. bisporus commercial strain Sylvan A15 (Sylvan, Kittanning, PA 16201, USA) was used for transformations. Mycelia were routinely maintained at 25°C on malt‐peptone (Leach et al., 2004) agar plates and supplemented with 25 μg ml−1 hygromycin B to select for transformants.
Construct design
Three different silencing constructs were designed and constructed to contain the Spr1 cDNA in the sense, anti‐sense and sense orientation with a stop codon to prevent translation (untranslatable stop). Spr1 cDNA was PCR amplified from pKing03 (contains Spr1 cDNA cloned into pBK‐CMV) using primers engineered to contain the appropriate restriction sites (Supporting Information Table S2). Spr1 cDNA in the sense orientation was amplified using primers Spr1‐p1 and Spr1‐p2 yielding a BspHI‐BamHI fragment, primers Spr1‐p4 and Spr1‐p5 resulted in the amplification of Spr1 cDNA in the antisense orientation as a AflIII‐BamHI fragment and Spr1 cDNA untranslatable stop was amplified using primers Spr1‐p3 and Spr1‐p2 yielding a BspHI‐BamHI fragment. Fragments were cloned into the Agaricus Molecular Toolkit (Burns et al., 2006) generating plasmids p004sense, p004anti‐sense and p004stop, respectively, (Supporting Information Fig. S1A). These plasmids contained the A. bisporus gpdII promoter and A. nidulans terminator as previously described (Burns et al., 2006). To introduce restriction sites, these plasmids were then cloned into a polylinker plasmid (pSL1180, Invitrogen) generating the Spr1 plasmids pSL004sense, pSL004anti‐sense and pSL004stop (Supporting Information Fig. S1B). The hygromycin cassette, hph, was isolated from phph004 (Burns et al., 2005) as a KpnI‐SacI fragment and cloned into pBluescript II (Stratagene). The hph cassette was then excised as a KpnI‐BssHII fragment and ligated to similarly digested pSL004Spr1 plasmids (pSL004sense, pSL004antisense and pSL004stop) thus linking the hph and Spr1 cassettes to create plasmids pSLsensehph, pSLantihph, and pSLstophph (Supporting Information Fig. S1C). The hph‐Spr1 cassettes were then excised as BglII‐SpeI fragments and ligated to pGREEN (Hellens et al., 2000) digested with BamHI and SpeI to form binary plasmids pGRsensehph, pGRantihph and pGRstophph (Supporting Information Fig. S1D). Correct assembly of all constructs was confirmed by sequence analysis.
Fungal transformations
Agaricus bisporus was transformed using Agrobacterium tumefaciens mediated transfection of gill tissue as previously described (Chen et al., 2000; Burns et al., 2005, 2006). Agaricus‐Agrobacterium co‐cultures were incubated at 20°C for 2–3 days before transferring the gill tissue to MMP agar containing 30 μg ml−1 hygromycin and 100 μg ml−1 cefotaxime. Selection plates were incubated at 25°C until transformed A. bisporus mycelia were visible (2–4 weeks) and transformants purified by three sequential subcultures under selection.
Genomic DNA was extracted from putative A. bisporus transformants and PCR screening was performed using Reddymix components (Abgene) with a general thermal cycling program of 95°C for 3 min, 30 cycles of (95°C for 30 sec, 50°C for 1 min, 72°C for 30 sec), and a final extension of 72°C for 10 min.
Fruiting studies
To cultivate mushroom fruiting bodies transgenic strains were grown on autoclaved rye grain to produce grain spawn (Elliott, 1985) which was used to inoculate compost substrate prepared according to commercial practice (Eastwood et al., 2008). Each transgenic strain was grown in three replicate pots each containing 3 kg compost. Mushrooms were harvested at maturity stage 2–3 and weighed (Hammond and Nichols, 1976) over a period of 30 days. The yield data was statistically evaluated by analysis of variants using Genstat (17.1).
Proteinase assays
Initial assessment of proteinase activity of wild type and transformed A. bisporus mycelia was screened by a clearing zone method. A. bisporus mycelia were inoculated onto SM medium with an overlay of 0.4% gelatin (w/v) in SM medium and examined after 17 days followed by staining with 0.1% (w/v) amido black in 7% (w/v) acetic acid and destained overnight using 7% acetic acid.
Expression of serine proteinase activity in liquid culture was determined by culturing A. bisporus mycelia of wild type, transformants, and transformed control in Treschow medium (TM; Treschow, 1944) supplemented with either humic fraction (0.94 g/l) or glutamate (5mM) and incubated at 25°C for 21 days. Dry weight was assessed and proteinase activity was measured spectrophotometrically using 0.15 mM Suc‐Ala‐Ala‐Pro‐Phe‐pNA substrate in 0.1 M sodium phosphate buffer (pH 7.0) with the OD measured continuously at 405 nm (Burton et al., 1997a). Five biological replicate cultures of each transformant or control were prepared. The protease activity per 50 ml of medium was determined and normalized to dry weight. One unit of proteinase activity was defined as an increase of one absorbance per minute.
Data of mycelial serine proteinase activities were subjected to square root (SQRT) transformations to stabilize the variance prior to analysis of variance (ANOVA). To analyse the correlation between serine proteinase enzyme activity and transcript levels, linear regression analysis was conducted on the square root of the averages of Spr1 transcripts and proteinase activities. Mushroom weight yield data were statistically analysed by ANOVA.
RT‐qPCR analysis
Total RNA was extracted from freeze‐dried mycelia using the method described by Sreenivasaprasad (2000). The RNA pellets were dried and dissolved in 100 μl of DMPC‐treated water. RNA was treated with DNase (Promega) prior to storage and preparations were confirmed free of contaminating genomic DNA by PCR amplification with primers SDH_Ab_F and SDH_Ab_R that span an intron of SDH gene of A. bisporus (Costa et al., 2009). Three independent RNA extractions were performed for each strain tested. cDNA synthesis was performed using 1 μg total RNA, with random hexamer primers and Thermoscript RT‐PCR System (Invitrogen).
The SPR 1 transcripts were assessed using Q‐PCR MasterMix Plus for SYBR Green I w/o UNG (Eurogentec) on the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) using primers qSPR1_F and qSPR1_R that amplify a 62 bp region from SPR1 cDNA. Expression levels of target genes were normalized against A. bisporus 18S rRNA using primers q18S_Ab_F and q18S_Ab_R, as described in Eastwood and colleagues (2008).
Two microliters of diluted cDNA products (1:3) was used in 15‐μL RT‐qPCR reactions with 500 nM of each primer and 2 μl Reaction buffer. Thermocycling parameters were: 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 1 min. Three replicate samples were run in 384‐well plates and sealed with optically clear heat seal film (Applied Biosystems). Standard curves were made using four serial dilutions (1, 1/4, 1/16, 1/64) of cDNA from the non‐silenced host. Negative controls (water, the non‐silenced host and a transformed control A. bisporus) were included in each experiment.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Construction of Spr1 silencing cassettes.
A. Spr1 cDNA was cloned into the Basidiomycete Molecular Toolkit (Burns et al., 2006) generating plasmids p004sense, p004stop and p004 anti‐sense.
B. To introduce restriction sites, these plasmids were then cloned into a polylinker plasmid (pSL1180, Invitrogen) generating the Spr1 plasmids pSL004sense, pSL004anti‐sense and pSL004stop.
C. The hygromycin cassette, hph, was isolated from phph004 (Burns et al., 2005) and cloned into pBluescript II (Stratagene). The hph cassette was then excised as a KpnI‐BssHII fragment and ligated to similarly digested pSL004Spr1 plasmids thus linking the hph and Spr1 cassettes to create plasmids pSLsensehph, pSLstophph, and pSLantihph.
D. The hph‐Spr1 cassettes were then excised as BglII‐SpeI fragments and ligated to pGREEN to form binary plasmids pGRsensehph, pGRstophph and pGRantihph.
Fig. S2. Proteinase clearing zone assays on milk or gelatin‐amended agar of non‐transformed control strain A15 (WT), GFP transformed Agaricus bisporus and hygromycin resistant sense (S), antisense (AS) and stop (ST) transformants. Colony and clearing zone diameters were measured 17 days after inoculation. (A) Control wild‐type A15 and GFP transformed Agaricus bisporus. (B) Spr1‐sense transformants. (C) Spr1‐antisense transformants. (D) Spr1‐stop transformants.
Table S1. Serine protease protein IDs and accession numbers.
Table S2. Sequence of primers used in the construction of molecular constructs, PCR analysis of transformants and other analysis. Incorporated restriction enzyme sites are underlined.
Acknowledgements
Research at Universities of Bristol, Warwick and East Malling Research was funded by grants from BBSRC (D19266) and DEFRA (HH2116SMU). We acknowledge the statistical advice by James Lynn and Phil Brain.
Contributor Information
Andy M. Bailey, Email: Andy.Bailey@bristol.ac.uk
Gary D. Foster, Email: Gary.Foster@bristol.ac.uk
References
- Bailey, V.L. , Smith, J.L. , and Bolton, H. (2002) Fungal‐to‐bacterial ratios in soils investigated for enhanced C sequestration. Soil Biol Biochem 34: 997–1007. [Google Scholar]
- Baldrian, P. , and Valaskova, V. (2008) Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 32: 501–521. [DOI] [PubMed] [Google Scholar]
- Bundock, P. , and Hooykaas, P.J.J. (1996) Integration of Agrobacterium tumefaciens T‐DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc Natl Acad Sci USA 93: 15272–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D.J. , Weintraub, M.N. , Hewins, C.R. , and Kalisz, S. (2011) Relationship between soil enzyme activities, nutrient cycling and soil fungal communities in a northern hardwood forest. Soil Biol Biochem 43: 795–803. [Google Scholar]
- Burns, C. , Gregory, K.E. , Kirby, M. , Cheung, M.K. , Riquelme, M. , Elliott, T.J. , et al (2005) Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol 42: 191–199. [DOI] [PubMed] [Google Scholar]
- Burns, C. , Leach, K.M. , Elliott, T.J. , Challen, M.P. , Foster, G.D. , and Bailey, A. (2006) Evaluation of Agrobacterium‐mediated transformation of Agaricus bisporus using a range of promoters linked to hygromycin resistance. Mol Biotechnol 32: 129–138. [DOI] [PubMed] [Google Scholar]
- Burton, K.S. , Wood, D.A. , Thurston, C.F. , and Barker, P.J. (1993) Purification and characterization of a serine proteinase from senescent sporophores of the commercial mushroom Agaricus bisporus . J Gen Microbiol 139: 1379–1386. [DOI] [PubMed] [Google Scholar]
- Burton, K.S. , Hammond, J.B.W. , and Minamide, T. (1994) Proteinase activity in Agaricus bisporus during periodic fruiting (flushing) and sporophore development. Curr Microbiol 28: 275–278. [Google Scholar]
- Burton, K.S. , Smith, J.F. , Wood, D.A. , and Thurston, C.F. (1997a) Extracellular proteinases from the mycelium of the cultivated mushroom Agaricus bisporus . Mycol Res 101: 1341–1347. [Google Scholar]
- Burton, K.S. , Partis, M.D. , Wood, D.A. , and Thurston, C.F. (1997b) Accumulation of serine proteinase in senescent sporophores of the cultivated mushroom, Agaricus bisporus . Mycol Res 101: 146–152. [Google Scholar]
- Chen, X. , Stone, M. , Schlagnhaufer, C. , and Romaine, C.P. (2000) A fruiting body tissue method for efficient Agrobacterium‐mediated transformation of Agaricus bisporus . Appl Environ Microbiol 66: 4510–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christl, I. , Knicker, H. , Kogel, I.K. , and Kretzschmar R. (2000) Chemical heterogeneity of humic substances: Characterization of size fractions obtained by hollow‐fibre ultrafiltration. Eur J Soil Sci 51: 617–625. [Google Scholar]
- Costa, A.M.S.B. , Mills, P.R. , Bailey, A.M. , Foster, G.D. , and Challen, M.P. (2008) Oligonucleotide sequences forming short self‐complimentary hairpins can expedite the down‐regulation of Coprinopsis cinerea genes. J Microbiol Methods 75: 205–208. [DOI] [PubMed] [Google Scholar]
- Costa, A.M.S.B. , Thomas, D.J.I. , Eastwood, D. , Cutler, S.B. , Bailey, A.M. , Foster, G.D. , et al (2009) Quantifiable down‐regulation of endogenous genes in Agaricus bisporus mediated by expression of RNA hairpins. J Microbiol Biotechnol 19: 271–276. [PubMed] [Google Scholar]
- de Groot, M.J.A. , Bundock, P. , Hooykaas, P.J.J. , and Beijersbergen, A.G.M. (1998) Agrobacterium tumefaciens‐mediated transformation of filamentous fungi. Nat Biotechnol 16: 839–842. [DOI] [PubMed] [Google Scholar]
- Dix, N.J. , and Webster, J. (1995) Fungal Ecology. Netherlands: Springer; [Google Scholar]
- Dong, L.H. , Yuan, Q. , and Yuan, H.L. (2006) Changes of chemical properties of humic acids from crude and fungal transformed lignite. Fuel 85: 2402–2407. [Google Scholar]
- Eastwood, D.C. , Challen, M.P. , Zhang, C.J. , Jenkins, H. , Henderson, J. , and Burton, K.S. (2008) Hairpin‐mediated down‐regulation of the urea cycle enzyme argininosuccinate lyase in Agaricus bisporus . Mycol Res 112: 708–716. [DOI] [PubMed] [Google Scholar]
- Egli, S. (2011) Mycorhizal mushroom diversity and productivity ‐ an indicator of forest health? Ann ForSci 68: 81–88. [Google Scholar]
- Elliott, T.J. (1985) Spawn‐making and spawns, The Biology and Technology of the Cultivated Mushroom, Chichester, England: John Wiley & Sons. [Google Scholar]
- Hammond, J.B.W. , and Nichols, R. (1976) Carbohydrate‐metabolism in Agaricus bisporus (Lange) sing ‐ changes in soluble carbohydrates during growth of mycelium and sporophore. J Gen Microbiol 93: 309–320. [DOI] [PubMed] [Google Scholar]
- Hattenschwiler, S. , Tiunov, A.V. , and Scheu, S. (2005) Biodiversity and litter decomposition interrestrial ecosystems. Annu Rev Ecol Evol Syst 36: 191–218. [Google Scholar]
- Hellens, R.P. , Edwards, E.A. , Leyland, N.R. , Bean, S. , and Mullineaux, P.M. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Mol Biol 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Heneghan, M.N. , Costa, A. , Challen, M.P. , Mills, P.R. , Bailey, A. , and Foster, G.D. (2007) A comparison of methods for successful triggering of gene silencing in Coprinus cinereus . Mol Biotechnol 35: 283–294. [DOI] [PubMed] [Google Scholar]
- Heneghan, M.N. , Porta, C. , Zhang, C. , Burton, K.S. , Challen, M.P. , Bailey, A.M. , and Foster, G.D. (2009) Characterization of serine proteinase expression in Agaricus bisporus and Coprinopsis cinerea by using green fluorescent protein and the A. bisporus SPR1 promoter. Appl Environ Microbiol 75: 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahkonen, M.A. , and Hakulinen, R. (2011) Hydrolytic enzyme activities, carbon dioxide production and the growth of litter degrading fungi in different soil layers in a coniferous forest in Northern Finland. Eur J Soil Biol 47: 108–113. [Google Scholar]
- Kerrigan, R.W. , Challen, M.P. , and Burton, K.S. (2013) Agaricus bisporus genome sequence: a commentary. Fungal Genet Biol 55: 2–5. [DOI] [PubMed] [Google Scholar]
- Kilaru, S. , Collins, C.M. , Hartley, A.J. , Bailey, A.M. , and Foster, G.D. (2009) Establishing molecular tools for genetic manip[ulaiton of the pleuromutilin‐producing fungus Clitopilus passeckerinus . Appl Environ Microbiol 75: 7196–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsnorth, C.S. , Eastwood, D.C. , and Burton, K.S. (2001) Cloning and postharvest expression of serine proteinase transcripts in the cultivated mushroom Agaricus bisporus . Fungal Genet Biol 32: 135–144. [DOI] [PubMed] [Google Scholar]
- Leach, K. , Odon, V. , Zhang, C. , Kim, H.K. , Henderson, J. , Warner, J. , et al (2004) Progress in Agaricus bisporus transformation: Agrobacterium methodologies and development of novel marker genes. Mushroom Sci 16: 93–102. [Google Scholar]
- Martinez, A.T. , Speranza, M. , Ruiz‐Duenas, F.J. , Ferreira, P. , Camarero, S. , Guillen, F. , et al (2005) Biodegradation of lignocellulosics: microbial chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol 8: 195–204. [PubMed] [Google Scholar]
- Morin, E. , Kohler, A. , Baker, A.R. , Foulongne‐Oriol, M. , Lombard, V. , Nagy, L.G. , et al (2012) Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic‐rich ecological niche. Proc Natl Acad Sci USA 109: 17501–17506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, E.M. , and Zachara, J.M. (1995) The role of sorbed humic substances on the distribution of organic and inorganic contaminants in groundwater. Geoderma 67: 103–124. [Google Scholar]
- Myneni, R.B. , Dong, J. , Tucker, C.J. , Kaufmann, R.K. , Kauppi, P.E. , Liski, J. , et al (2001) A large carbon sink in the woody biomass of Northern forests. Proc Natl Acad Sci USA 98: 14784–14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa, S.H. , Iwabata, K. , Sugawara, H. , Hamada, F.N. , Koshiyama, A. , Chiku, H. , et al (2005) Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double‐stranded RNA‐mediated gene silencing. Microbiol 151: 3669–3678. [DOI] [PubMed] [Google Scholar]
- Osono, T. (2007) Ecology of ligninolytic fungi associated with leaf litter decomposition. Ecol Res 22: 955–974. [Google Scholar]
- Piccolo, A. (2002) The supramolecular structure of humic substances. A novel understanding of humus chemistry and implications in soil science. Adv Agron 75: 57–134. [Google Scholar]
- Paungfoo‐Lonhienne, C. , Yeoh, Y.K. , Kasinadhuni, N.R.P. , Lonhienene, T.G.A. , Robinson, N. , Hugenholz, P. , et al (2015). Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci Rep 5: 8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsabaugh, R.L. (2010) Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol Biochem 42: 391–404. [Google Scholar]
- Six, J. , Conant, R.T. , Paul, E.A. , and Paustian, K. (2002) Stabilization mechanisms of soil organic matter: implications for C‐saturation of soils. Plant Soil 241: 155–176. [Google Scholar]
- Snajdr, J. , Steffen, K.T. , Hofrichter, M. , and Baldrian, P. (2010) Transformation of C‐14‐labelled lignin and humic substances in forest soil by the saprobic basidiomycetes Gymnopus erythropus and Hypholoma fasciculare . Soil Biol Biochem 42: 1541–1548. [Google Scholar]
- Sreenivasaprasad, S. (2000) Isolation of fungal nucleic acids In The Nucleic Acid Protocols Handbook. NJ, USA: Humana Press. [Google Scholar]
- Steffen, K.T. , Cajthaml, T. , Snajdr, J. , and Baldrian, P. (2007) Differential degradation of oak (Quercus petraea) leaf litter by litter‐decomposing basidiomycetes. Res Microbiol 158: 447–455. [DOI] [PubMed] [Google Scholar]
- Steffen, K.T. , Hofrichter, M. , and Hatakka, A. (2000)Mineralisation of C‐14‐labelled synthetic lignin and ligninolytic enzyme activities of litter‐decomposing basidiomycetous fungi. Appl Microbiol Biotechnol 54: 819–825. [DOI] [PubMed] [Google Scholar]
- Steffen, K.T. , Hatakka, A. , and Hofrichter, M. (2002) Degradation of humic acids by the litter‐decomposing basidiomycete Collybia dryophila . Appl Environ Microbiol 68: 3442–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland, M.S. , and Rousk, J. (2010) Considering fungal:bacterial dominance in soils ‐ Methods, controls, and ecosystem implications. Soil Biol Biochem 42: 1385–1395. [Google Scholar]
- Treschow, C. (1944) Nutrition of the cultivated mushroom. Dansk Botanisk Arkive 11: 1–180. [Google Scholar]
- Valaskova, V. , Snajdr, J. , Bittner, B. , Cajthaml, T. , Merhautova, V. , Hoffichter, M. , and Baldrian, P. (2007) Production of lignocellulose‐degrading enzymes and degradation of leaf litter by saprotrophic basidiomycetes isolated from a Quercus petraea forest. Soil Biol Biochem 39: 2651–2660. [Google Scholar]
- Watling, R. , and Harper, D.B. (1998) Chloromethane production by wood‐rotting fungi and an estimate of the global flux to the atmosphere. Mycol Res 102: 769–787. [Google Scholar]
- Weber, J.H. (1998) Binding and transport of metals by humic material In Humic Substances and their Role in the Environment. Frimmel F.H., and Christman R.F. (eds). Chichester, England: John Wiley and Sons, pp. 165–178. [Google Scholar]
- Zavarzina, A.G. , Leontievsky, A.A. , Golovleva, L.A. , and Trofimov, S.Y. (2004) Biotransformation of soil humic acids by blue laccase of Panus tigrinus 8/18: an in vitro study. Soil Biol Biochem 36: 359–369. [Google Scholar]
- Zifcakova, L. , Dobiasova, P. , Kolarova, Z. , Koukol, O. , and Baldrian, P. (2011) Enzyme activities of fungi associated with Picea abies needles. Fungal Ecol 4: 427–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Construction of Spr1 silencing cassettes.
A. Spr1 cDNA was cloned into the Basidiomycete Molecular Toolkit (Burns et al., 2006) generating plasmids p004sense, p004stop and p004 anti‐sense.
B. To introduce restriction sites, these plasmids were then cloned into a polylinker plasmid (pSL1180, Invitrogen) generating the Spr1 plasmids pSL004sense, pSL004anti‐sense and pSL004stop.
C. The hygromycin cassette, hph, was isolated from phph004 (Burns et al., 2005) and cloned into pBluescript II (Stratagene). The hph cassette was then excised as a KpnI‐BssHII fragment and ligated to similarly digested pSL004Spr1 plasmids thus linking the hph and Spr1 cassettes to create plasmids pSLsensehph, pSLstophph, and pSLantihph.
D. The hph‐Spr1 cassettes were then excised as BglII‐SpeI fragments and ligated to pGREEN to form binary plasmids pGRsensehph, pGRstophph and pGRantihph.
Fig. S2. Proteinase clearing zone assays on milk or gelatin‐amended agar of non‐transformed control strain A15 (WT), GFP transformed Agaricus bisporus and hygromycin resistant sense (S), antisense (AS) and stop (ST) transformants. Colony and clearing zone diameters were measured 17 days after inoculation. (A) Control wild‐type A15 and GFP transformed Agaricus bisporus. (B) Spr1‐sense transformants. (C) Spr1‐antisense transformants. (D) Spr1‐stop transformants.
Table S1. Serine protease protein IDs and accession numbers.
Table S2. Sequence of primers used in the construction of molecular constructs, PCR analysis of transformants and other analysis. Incorporated restriction enzyme sites are underlined.


