Abstract
Despite substantial achievements in elucidating the metabolic pathways of lipogenesis, a mechanistic representation of lipid accumulation and degradation has not been fully attained to-date. Recent evidence suggests that lipid accumulation can occur through increases of either the cytosolic copy-number of lipid droplets (LDs), or the LDs size. However, the prevailing phenotype, or how such mechanisms pertain to lipid degradation remain poorly understood. To address this shortcoming, we employed the–recently discovered–innate bioprocessing fluctuations in Yarrowia lipolytica, and performed single-cell fluctuation analysis using optical microscopy and microfluidics that generate a quasi-time invariant microenvironment. We report that lipid accumulation at early stationary phase in rich medium is substantially more likely to occur through variations in the LDs copy-number, rather than the LDs size. Critically, these mechanistics are also preserved during lipid degradation, as well as upon exposure to a protein translation inhibitor. The latter condition additionally induced a lipid accumulation phase, accompanied by the downregulation of lipid catabolism. Our results enable an in-depth mechanistic understanding of lipid biogenesis, and expand longitudinal single-cell fluctuation analyses from gene regulation to metabolism.
Introduction
Lipid droplets (LDs) are cytoplasmic emulsions, capable of storing neutral lipids such as triacylglycerols (TAG) and steryl esters (SE) at varying ratios [1–4]. By undergoing enzymatic hydrolysis, these stored compounds serve several cellular needs, such as membrane and lipoprotein biogenesis, as well as provide precursors towards oxidation mediated energy production. Similarly, autophagy may degrade LDs (i.e. lipophagy), whereas the LD content is directly released into lysosomes for further degradation [5]. LDs have attracted significant excitement in recent years as cost-effective biodiesel precursors in renewable energy production [6]. To this end, Yarrowia lipolytica has emerged as a model oleaginous yeast due to its genetic tractability, as well as enhanced lipid accumulation capability–most in the form of TAG [7–9]. In addition to industrial applications, more active roles of LDs have been recently recognized, such as their interactions with other organelles to coordinate immune responses [10], as well as cell protection against lipotoxicity [11].
Different pathways may induce lipid accumulation [12]. These include: (1) direct fatty acid internalization, esterification and incorporation to LDs [11]; and (2) de novo fatty acid synthesis through the mitochondrial TCA cycle and Kennedy pathway utilizing carbon precursors such as glucose and acetate [13]. According to the current consensus, the endoplasmic reticulum (ER) is the origin of LDs in most single-cell organisms [3, 4, 14]. This view is primarily based on the observation that essential enzymes to lipid biosynthesis reside in the ER [15], including diacylglycerol acyltransferase (e.g. DGAT1)–an enzyme involved in the final step in TAG biosynthesis. This LD biogenesis mechanism suggests that cytosolic lipid accumulation occurs primarily through the increase of the number of cytosolic LDs. More recently, an alternative mechanism of lipid accumulation was reported, evidencing that cytosolic LDs can also grow by size [16]. To this end, the glycerol-3-phospate acyltransferase (GPAT4), as well as diacylglycerol acyltransferase (DGAT2) were identified as essential components of those LDs that grow by size. Interestingly, the GPAT4 isoenzyme was not found to decorate all cytosolic LDs, but rather a smaller portion of them. This enzyme localization heterogeneity was identified as a mechanism generating two diverse LD populations: those that grow in size, and those remaining “static” [16].
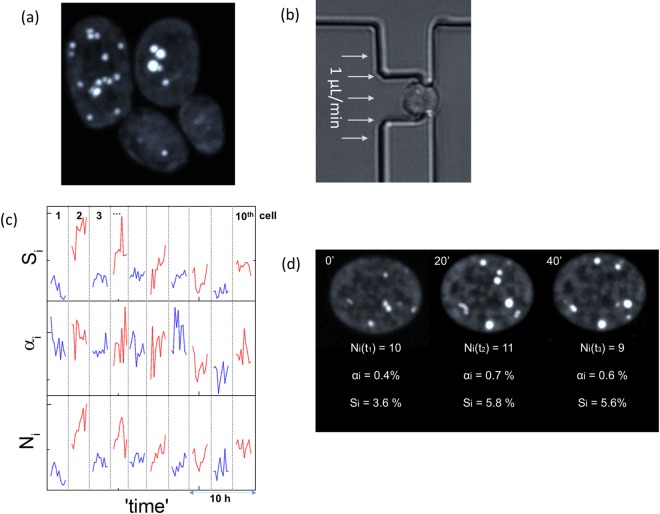
Another lipogenesis aspect that has also attracted substantial attention in recent years is the persistent cell-to-cell lipid content heterogeneity. A recent report identified this form of heterogeneity as a non-heritable trait, as well as its protection role against lipotoxicity [11]. To a similar end, we observed at the single-cell level that cytosolic lipid accumulation is far from monotonic with time [17]. We identified this form of bioprocessing noise as the origin of the cell-to-cell heterogeneity, confirmed its epigenetic origins and dependence on the extracellular environment [17]. In addition to the cell-to-cell lipid content heterogeneity, another form of phenotypic heterogeneity persists in clonal populations, whereas some cells contain large-but-fewer LDs, and others contain small-but-more LDs. A representative example of this innate phenotypic bistability is illustrated in Fig 1A for the Po1g strain of Yarrowia lipolytica [7–9]. While this form of phenotypic bistability has been appreciated since the first electron micrographs of yeast (see for example: [18]), they have yet to be extensively examined.
Fig 1.
(a) A maximum intensity projection of two budding Y. lipolytica Po1g cells, indicating two lipid-content phenotypes, namely: large-but-fewer LDs and small-but-more LDs. (b) A single Po1g cell trapped in the microfluidic system under continuous laminar flow at 1 μL/min. (c) Single-cell longitudinal time traces of the number of cytosolic LDs (Ni), their median area (αi) and the percentage of lipid content (Si); 10 single-cell traces are illustrated, in varying colors consecutively. (d) Time-lapse 3D imaging of neutral lipid fluctuations (Ni, αi, Si) of a single Po1g cell.
Despite substantial recent progress in identifying the different biochemical pathways of lipid accumulation [19], including the transcriptional regulatory changes under nitrogen starvation [20, 21], observations similar to those of Fig 1A still question the mechanistics of lipogenesis, namely: does lipid accumulation occurs primarily through the number of cytosolic LDs, or through their size? To answer this question, we explored neutral lipid expression at the single-cell level using microfluidics and optical microscopy. The approach was inspired by the plethora of single-cell analyses that have elucidated many features of gene regulation, which are otherwise hidden in bulk–population level–assays [22, 23]. Rather than gene expression, we probed the lipid content in Y. lipolytica, a model oleaginous organism [7–9, 13, 21, 24, 25]. Specifically, we selected the Po1g strain, which is transformed to overexpress the acetyl-CoA carboxylase (ACC) and diacylglycerol transferase (DGAT) genes, thereby allowing it to accumulate lipids [8]. By placing this strain within a microfluidic quasi-time invariant microenvironment–a crucial parameter in longitudinal analyses given the highly dynamic nature of the metabolome [26]–we previously reported the sporadic nature of lipid accumulation at early stationary phase [17]. Here, we employ these incessant fluctuations to statistically correlate the two phenotypes pertaining to large-but-few versus small-but-many LDs, and investigate the mechanistics of lipid accumulation and degradation under rich medium and protein translation inhibition steady-state conditions.
Results
Visualizing regulatory mechanistics
Following our previous analysis, the intracellular neutral lipid content (Si, namely total LD area normalized over the cell area) was observed to fluctuate strongly over time [17]. The Si fluctuations were manifested both through fluctuations of the number of LDs (Ni), as well as their average sizes (αi−normalized over the cell size)–illustrated the longitudinal traces for 10 Y. lipolytica Po1g cells in Fig 1C, as well as in the single-cell images of Fig 1D. To attain a deeper insight into both the synthesis and degradation of neutral lipids, we derived the instantaneous lipid flux in accordance to the equation:
| (1) |
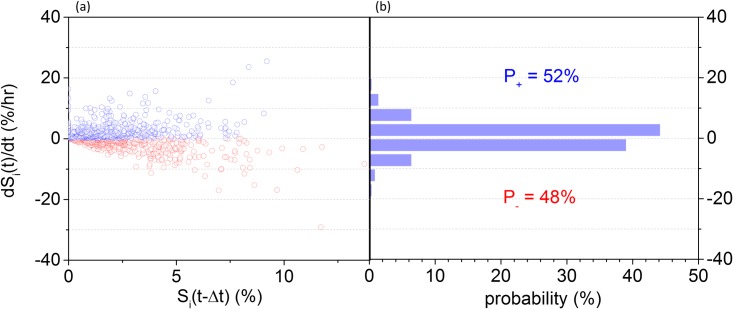
The instantaneous lipid flux can be either positive or negative, pertaining to a higher probability of lipid accumulation rather than degradation within the observation window (Δt). The latter was chosen to be Δt = 20 min (Fig 2A), limited by the sampling duration during 3D imaging, and the number of single-cells per chip. For Po1g, the positive and negative median fluxes were similar (within 6%), at 2.8%/hr (Fig 2B). This indicates that both lipid accumulation and degradation in Y. lipolytica occur at rates of comparable magnitudes. Despite their similar magnitudes, the positive lipid-flux observations during the 140 min of immobilization were marginally more likely (52% probability in total) than the negative ones (48% in total) as plotted in Fig 2B. This probability distribution indicates a moderately higher probability of de novo lipogenesis over degradation within the 140 min observation window.
Fig 2.
(a) A scatter plot of the lipid flux (dSi(t)/dt) as a function of the total lipid content (Si(t-Δt)); each point represents the instantaneous flux per cell, with positive ones plotted in blue, and negative in red. (b) A probability histogram of the lipid flux for all observations (n = 80 cells).
Additionally, the lipid flux (dSi/dt) is weakly correlated to its content (Si) at any instant (Fig 2A), with a Spearman correlation coefficient ρ = 0.23 (P = .001). To explain this, we developed a coarse-grain (deterministic) model, whereas the lipid flux depends on the activity levels and concentration of associated enzymes, as well as the substrate concentrations [27, 28]. This can be qualitatively described as follows (see S1 Document for a full derivation) [29]:
| (2) |
, where κ+ and κ- are the reaction rates of the enzyme manifolds associated with lipid accumulation (E+) and degradation (E-), and Ci is the available cytosolic carbon substrate dedicated to fatty acid synthesis and elongation.
Under the assumption that Ci is at equilibrium with the time-invariant extracellular supply, the weak Si−dSi/dt correlation suggests that the rate of de novo lipogenesis is primarily bounded by the rates of enzyme concentrations and activities and less by the intracellular substrate and product levels (Eq 2). Alternatively, the rate limiting step in expressing or degrading lipids rests with the enzyme reaction rates, rather than the cytosolic substrate or product concentrations. This conclusion is in agreement with our previous finding that bioprocessing noise depends inversely on the intracellular lipid content: in essence, the bounded lipid flux (dSi/dt) enforces stronger (weaker) lipid content (Si) fluctuations at low (high) cytosolic product concentrations [17].
LD copy-number or size?
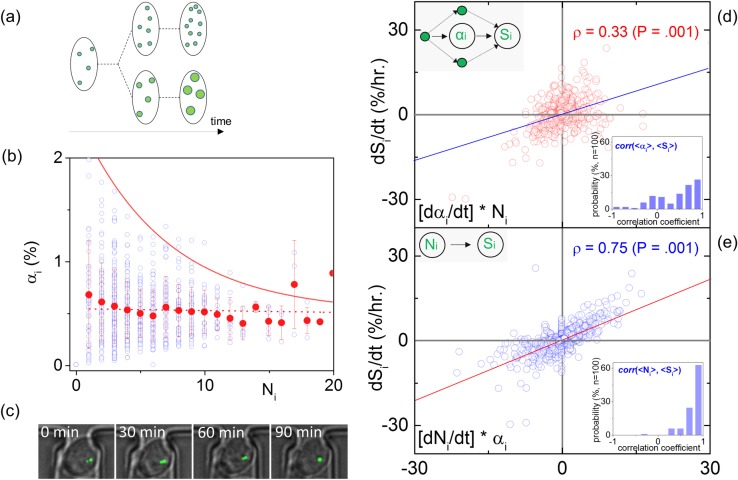
To investigate whether lipid accumulation occurs via the copy-number of cytosolic LDs versus the LDs size (Fig 3A) in the WT Y. lipolytica, we probed the relationship between the LDs copy-number with the median LD size–per cell at all instances. The scatter-plot of Fig 3B illustrates the inter-dependence between these variables for approximately n = 80 cells. It can be observed that Ni varies–as expected–in integer steps, while the αi values fluctuate for each Ni. Despite such fluctuations however, the median αi value remains relatively constant for all Ni. An exception to this apparent inter-dependence are the decreasing maximum values of αi for increasing Ni (solid red line–Fig 3B). This indicates while variable size LDs can be produced, their maximum size is ultimately bounded by the number of LDs. Namely, while Y. lipolytica cells can produce variable size LDs, their maximum size is ultimately limited by the number of intracellular LDs.
Fig 3.
(a) A schematic illustration of the two possible mechanistic pathways for lipid accumulation, namely by accumulating more LDs or larger LDs. (b) The dependence of Ni on αi (blue dots), including the mean values of αi per Ni (red dots–error bars denote the standard deviation), as well as the exponential decay fit of the αi maxima for increasing Ni (red solid line). (c) The dependence the lipid flux (dSi/dt) on [αi ∙ dNi/dt]; red circles denote observations per unit time and the solid blue line is the linear fit; upper inset depicts the cause-effect relationship between Si and αi; lower inset plots the histogram of the [αi—Si] correlation coefficient per single-cell. (d) Similar to (c), the dependence of dSi/dt on [Ni ∙ dαi/dt] is plotted, including the observations (blue circles), a linear fit (solid red line), the causality diagram (upper inset) and correlation coefficient per cell histogram during the observation window.
The weak correlation between the number of LDs (Ni) and their area (αi) for all cells at any given instance observed in Fig 3B questions which of the two variables is most integral to lipogenesis, and lipolysis. Does lipid accumulation occur through Ni in accordance to the consensus that LDs originate from the ER [4], or through αi in the presence of essential isoenzymes (Fig 3A) [16]? Additionally, is LD degradation a digital process through Ni, or can degradation occur through analogue decreases in αi?
To investigate the causality between αi and Ni with Si, we plotted the lipid flux (dSi/dt) as a function of its two weighted components, namely [Ni ∙ dαi/dt] and [αi ∙ dNi/dt] (see Eq 1), shown in Fig 3C and 3D. The lipid flux relationship with the rate of LD size change is complex and weakly correlated (Fig 3C) with a Spearman correlation coefficient of ρ = 0.33 (P = .001). This weak correlation suggests the presence of compensating mechanisms (see inset’s pathway diagram). Such compensating mechanisms dephase the Si - αi relationship, thereby limiting its causality [23]. This behavior is noted for both positive and negative instantaneous lipid fluxes (dSi/dt).
On the contrary, increased linearity between dSi/dt and dNi/dt is observed with a ρ = 0.75 Spearman correlation coefficient (P = .001) as illustrated in Fig 3D. This occurs for both positive and negative fluxes, indicating the causal relationship between Si and Ni (see inset in Fig 3D). The higher linearity between dSi/dt and dNi/dt (Fig 3C) than dSi/dt and dαi/dt (Fig 3D) manifests that intracellular lipid accumulation and degradation are substantially more likely to occur through fluctuations in the number of LDs (Ni) rather than their average size (αi). The outcome of the preceding instantaneous lipid flux analysis is in agreement with the [Ni—Si] and [αi—Si] correlations per single-cell time series, as shown by the inset histograms of Fig 3C and 3D.
Protein translation and lipogenesis
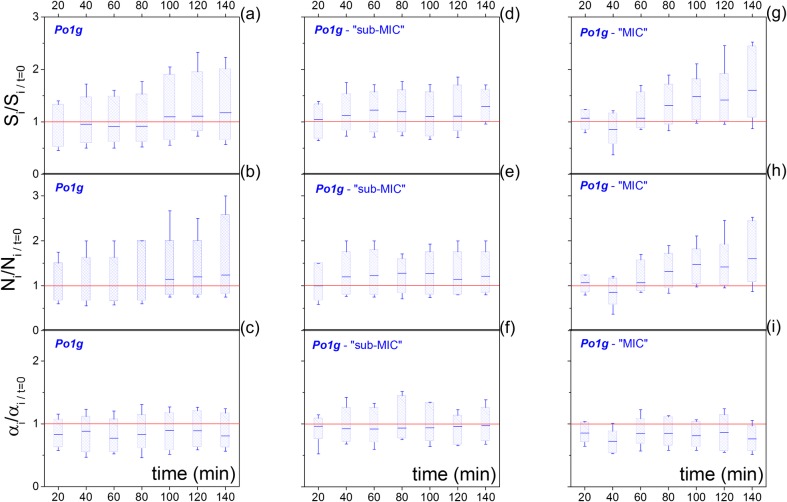
Our final investigation focused on lipogenesis under protein translation inhibition conditions. Under continuous (time-invariant) exposure to rich medium, the Po1g cells did not exhibit considerable neutral lipid increase during the microfluidic immobilization (140 min). This is illustrated in Fig 4A, where the lipid content (Si) is plotted as a box-chart time series, where each box-chart is normalized at the Si value of t = 0 for each individual cell lineage. During the first 100 minutes in YPD medium, on average lipid degradation took place (i.e. Si/normalized < 1 for 0min < t < 100min), followed by a 17% lipid increase at 140 min. This increase is probabilistic, and reflects the median response of the population, whereas 39% of the population exhibited lipid degradation, rather than accumulation. In agreement with the preceding analysis, lipid accumulation Si takes place primarily through the number of cytosolic LDs (Ni), rather than their area (αi). This is illustrated in the time dependent box-charts of Fig 4B and 4C, whereas–unlike αi−the median Ni is monotonic with Si.
Fig 4.
(a-c) Boxcharts of the longitudinal fluctuations of the lipid content (Si−a), number of LDs (Ni−b) and median LD size (αi−c). The boxcharts represent the median, 25% - 75% range, and the whiskers the 20% - 80% range. All single-cell observations are normalized to initial value at t = 0 for each cell-lineage. (d-f) and (g-i) illustrate the same longitudinal traces for the sub-MIC (d-f) and for the MIC (g-i) concentration of CHX.
Subsequently, we introduced cycloheximide (CHX) in the YPD medium flowing in the microfluidics. CHX acts on the E-site of the 60S ribosomal subunit, thus blocking the translocation step during elongation [30]. As a result, CHX inhibits protein translation, thereby suppressing growth and increasing the cytosolic concentration of free amino acids, which has been previously linked to the enhanced activity of the mTORC1 (Mammalian Target of Rapamycin) complex [31]. Therefore, by introducing CHX in the medium, protein translation and cell growth were artificially suppressed. Such ‘simulated’ growth suppression can also be achieved by modulating the nitrogen content of the nutrient supply. The latter would inevitably induce a transient period, during which the cells transition from growth phase to a lipid accumulation phase, thus adding an extra complication to the single-cell investigation.
Two CHX concentrations were employed. The first was below the minimum inhibitory concentration (“sub-MIC”) at 16 μg/ml [32], where cells still grow, albeit at longer lag-phases and slower growth-rates as determined in liquid batch cultures (S1 Fig). The second was the minimum inhibitory concentration (“MIC”) at 33 μg/ml, where the cells exhibit no/minimal growth, but rather a prolonged stationary phase (S1 Fig). For both CHX concentrations, the proportion of cells exhibiting net lipid degradation by the 140th min decreased substantially, to approximately 25% (45% in the absence of CHX–Fig 4A). Moreover, the sub-MIC concentration lead to a median 30% lipid content increase (Fig 4D), while the MIC concentration to a 60% median lipid content increase (Fig 4G) after 140 min of immobilization.
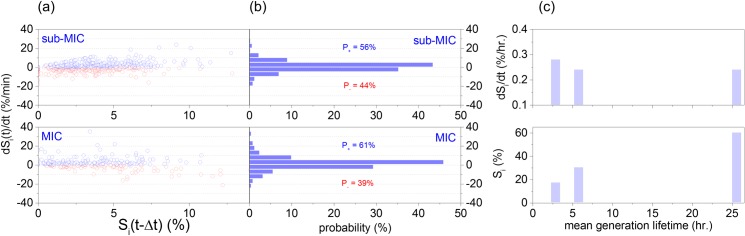
Regarding the mechanistics of lipid accumulation, both levels of protein translation inhibition (sub-MIC and MIC) induced lipid accumulation primarily through the number of cytosolic LDs rather than their area, as shown in Fig 4E and 4F, Fig 4H and 4I and S2 Fig. In addition, Fig 5A indicates that both lipid biogenesis and degradation are independent of the cytosolic lipid content and occur at comparable fluxes. Furthermore, complete inhibition of protein translation was accompanied by an enhanced probability of positive rather than negative lipid-flux observations, as illustrated in Fig 5B. Specifically, while either conditions of zero or sub-MIC concentrations of CHX resulted in equally distributed positive and flux observations, the MIC of CHX exhibited 22% higher positive flux observations than negative. The preceding analysis evidences that increasing the inhibition of protein translation and growth induce increasing cytosolic lipid levels (Fig 5C); nevertheless, this increase does not correlate with an increase in the instantaneous lipid flux (Fig 5C), but rather with a decreased probability of lipid degradation (Fig 5B).
Fig 5.
Similar to Fig 2, a scatter plot of the lipid flux (dSi/dt) as a function of the total lipid content (Si), and the associated probability histogram of the lipid flux for the sub-MIC (a) and for the MIC (b) concentration of CHX. (c) The dependence of the lipid content Si (in % per cell size by the 140th min. of observation) and the instantaneous lipid flux dSi/dt (%/hr) on the mean generation lifetime, controlled by different levels of CHX.
Discussion
Gene expression stochasticity imposes protein copy-number variability between individual cells in isogenic cultures. This form of phenotypic diversity was discovered decades ago [33], and can only be unmasked at the single-cell level [34]. Following substantial interest in the stochasticity of gene expression [35–40], infection [41], and cell growth [42–44], such stochastic fluctuations have been recently applied to the discovery of many unknown aspects of gene regulation [22, 23]. Here, we similarly employed the recently discovered innate fluctuations of lipid accumulation [17] (Fig 1C) to access the mechanistics of lipid regulation, including both de novo biogenesis and degradation.
Through single-cell microfluidics and optical microscopy, we determined the positive and negative instantaneous lipid flux for individual cells (Fig 2A). Under microfluidic immobilization and steady-state flow of rich medium, we observed that lipid accumulation and degradation occur at comparable fluxes. These fluxes also appear independent to the cytosolic concentration of product as shown in Fig 2B. Both the comparable accumulation and degradation fluxes and their independence to the cytosolic product concentration suggest that lipid accumulation and degradation undergo similar levels of regulation in rich medium as probed by way of example here for Y. lipolytica.
Two prominent mechanistic models characterize lipid accumulation. The first suggests that LDs originate in the endoplasmic reticulum (ER) [3, 4, 14]; the second indicates that LDs can also grow by size [16]. By performing longitudinal fluctuation analysis, we conclude that both mechanistic pathways are possible in Y. lipolytica. This is expected given that both glycerol-3-phospate acyltransferase isoenzyme (GPA) and diacylglycerol acyltransferase (DGA2) are conserved in Y. lipolytica [13]. However, lipid accumulation is more likely to occur through the number of LDs rather than their size, as evidenced by the correlation analysis presented in Fig 3D and Fig 3C. This evidences that lipid biogenesis at the ER is more probable than increases in the LD size. In regards to lipid degradation, Fig 3C indicates that it similarly occurs through decreases in the number of cytosolic LDs, rather than their size. As a result, we anticipate that the most dominant phenotype in wild-type Y. lipolytica to be the small-but-more LDs (Fig 3A).
The abovementioned behavior was confirmed for both rich medium and protein translation inhibition conditions (Fig 4E and 4F and Fig 4H–4L and S2 Fig). Additionally, the introduction of a protein translation inhibitor (CHX) at sub-MIC and MIC concentrations induced a 2- and 4-fold higher lipid content in comparison to the absence of protein translation inhibition, and correspond to a 2- and 8-fold decrease in growth-rates (Fig 5C) [45]. Finally, we observed that this increase in lipid accumulation was accompanied by a 20% higher probability of lipid accumulation rather than degradation (Fig 5B). While this observation cannot be immediately generalized to nitrogen limiting conditions, it is in agreement with reports on the transcriptional downregulation of β-oxidation in the oleaginous yeast Rhodosporidium toruloides [46] and Y. lipolytica grown in nitrogen limiting media [20].
To our knowledge, this is the first global investigation of the mechanistic regulation of lipid accumulation and degradation. Such mechanistic insight is critical towards engineering enhanced lipogenesis microorganisms as well as improved treatment of obesity and diabetes [47, 48]. In addition, the presented methodology expands upon single-cell investigations of gene regulatory networks to bioprocessing and metabolism, and therefore will provide an experimental platform towards similar investigations.
Materials and Methods
Microfluidics
To investigate the mechanistics of neutral lipid accumulation, we employed a single-cell assay, as previously described [17]. Briefly, microfluidics were used to isolate and immobilize individual cells in a microarray format under continuous laminar microflows, following the design by Tan et al. [49, 50]. Following cell immobilization (Fig 1B), we employed 1 μL/min flow rates to both sustain cell trapping and enable rapid replenishment of the environment, thereby giving rise to a quasi-time invariant extracellular environment. The latter is a crucial parameter in our time-dependent metabolic investigation, due to the highly dynamic character of the metabolome, which is known to react with the environment within very short time scales, in the order of a few seconds or less [26]. For this reason, the employed flow rates that are higher than typically employed in suspension cultures, but comparable to the flow rates typically employed in cell growth microfluidic chemostats [51], and substantially lower than those employed in flow cytometry. Under such conditions, we anticipate some minor loss of small molecular weight material from the cell, but given the low mechanical perturbation induced by these flow rates (see [17] for a more detailed discussion on yeast biomechanics under such flow-rates), cells are expected to be able to reach equilibrium under such conditions.
Cell and culture conditions
The immobilized cells were collected from a batch culture grown for 24h in rich YPD medium. Immobilized cells that exhibited no lipid content, or were actively dividing were not taken into consideration. Under microfluidic immobilization conditions, the neutral lipid content of the cells and the cell size was monitored every 20 min for approximately 140 min.
Microscopy
Cell size was monitored by bright field microscopy, while the lipid content was determined using Vesicle Photonics [52], coupled to a spin-disk confocal microscope (Fig 1A). For the latter, we employed the lipophilic fluorescent dye Bodipy (BODIPY® 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene, Molecular Probes), due to its enhanced specificity to LDs [53]. Cells were stained prior to microfluidic immobilization (250 ng/ml). To prolong the lipid content imaging, the Bodipy stain was included in the medium supplied during microfluidic immobilization (100 ng/ml). Under such conditions, a constant bodipy dye uptake was attained, as evidenced by the monotonic intracellular fluorescence increase shown in S3 Fig, contrary to the lipid content fluctuations shown in the same figure. It is worth adding, that similar dye uptake kinetics are observed in fixed cells; however, lipid content fluctuations are absent in this case, as illustrated in S3 Fig.
More detailed information on the employed cell strain, the procedures of cell growth, microfluidic fabrication, microscopy, image processing, and data analysis are included in the S1 Document.
Supporting Information
This section describes the derivation of Eq 2, as well as further details the materials and methods information employed in this work (including imaging, image/data analysis, sample preparation, and microfluidics fabrication).
(PDF)
Growth curves measured for Po1g in YPD medium at three concentrations of cycloheximide (CHX): 0 μg/ml (unperturbed), 16 μg/ml (sum-minimum inhibotory concentration–sub MIC), and 33 μg/ml (minimum inhibotory concentration–MIC). The measurements were performed in a 96-well plate using a Bioscreen C Pro instruement; each measurement represents an average of four independent wells at a 100x dilution.
(PDF)
The dependence the lipid flux (dSi/dt) on [αi ∙ dNi/dt] (red circles) and [Ni ∙ dαi/dt] (blue circles) for the Po1g strain under sub-minimum inhibitory concentrations (sub-MIC–left) and minimum inhibitory concentrations (MIC–right). Each data point denotes a single observation per unit time per cell, and the solid lines illustrate linear fits; insets include the Spearman correlation coefficient (ρ).
(PDF)
(a) A graph illustrating the dynamics of the intracellular fluorescence intensity (“fluorescence”) of the Bodipy dye, the total lipid content (Si in %), and the ratio of the fluorescence intensity of the propidium iodide (PI) dye (“PI ratio”). The latter denotes the ratio of the intracellular PI fluorescence over the extracellular fluorescence, which is less than 1 for live cells. (b) The same graph for fixed cells (PI ratio >1), where the lipid content fluctuations are reduced, despite the similar dye uptake kinetics.
(PDF)
Acknowledgments
AEV acknowledges support by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM104420. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. AEV also acknowledges funding from the Pacific Northwest National Laboratory (Linus Pauling Fellowship–PN12005/2406); GS acknowledges financial support from the US Department of Energy, Grant no. SC 0008744. Part of the research was performed using EMSL, a national user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research located at Pacific Northwest National Laboratory.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
AEV acknowledges support from an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P20 GM103408), as well as from the Pacific Northwest National Laboratory (Linus Pauling Fellowship – PN12005/2406); GS acknowledges financial support from the US Department of Energy, Grant no. SC 0008744. Part of the research was performed using EMSL, a national user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research located at Pacific Northwest National Laboratory.
References
- 1.Hashemi HF, Goodman JM. The life cycle of lipid droplets. Current Opinion in Cell Biology. 2015;33:119–24. 10.1016/j.ceb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nature Reviews Molecular Cell Biology. 2006;7(5):373–8. 10.1038/nrm1912 [DOI] [PubMed] [Google Scholar]
- 3.Ohsaki Y, Suzuki M, Fujimoto T. Open Questions in Lipid Droplet Biology. Chemistry & Biology. 2014;21(1):86–96. [DOI] [PubMed] [Google Scholar]
- 4.Thiam AR, Farese RV Jr., Walther TC. The biophysics and cell biology of lipid droplets. Nature Reviews Molecular Cell Biology. 2013;14(12):775–86. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. International journal of cell biology. 2012;2012:282041–. 10.1155/2012/282041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315(5813):801–4. 10.1126/science.1139612 [DOI] [PubMed] [Google Scholar]
- 7.Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nature Communications. 2014;5. [DOI] [PubMed] [Google Scholar]
- 8.Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metabolic Engineering. 2013;15:1–9. 10.1016/j.ymben.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 9.Beopoulos A, Mrozova Z, Thevenieau F, Le Dall M-T, Hapala I, Papanikolaou S, et al. Control of Lipid Accumulation in the Yeast Yarrowia lipolytica. Applied and Environmental Microbiology. 2008;74(24):7779–89. 10.1128/AEM.01412-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saka HA, Valdivia R. Emerging Roles for Lipid Droplets in Immunity and Host-Pathogen Interactions. In: Schekman R, editor. Annual Review of Cell and Developmental Biology, Vol 28. Annual Review of Cell and Developmental Biology. 282012. p. 411–37. [DOI] [PubMed]
- 11.Herms A, Bosch M, Ariotti N, Reddy BJN, Fajardo A, Fernandez-Vidal A, et al. Cell-to-Cell Heterogeneity in Lipid Droplets Suggests a Mechanism to Reduce Lipotoxicity. Current Biology. 2013;23(15):1489–96. 10.1016/j.cub.2013.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klug L, Daum G. Yeast lipid metabolism at a glance. Fems Yeast Research. 2014;14(3):369–88. 10.1111/1567-1364.12141 [DOI] [PubMed] [Google Scholar]
- 13.Silverman AM, Qiao K, Xu P, Stephanopoulos G. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Applied microbiology and biotechnology. 2016;100(8):3781–98. 10.1007/s00253-016-7376-0 [DOI] [PubMed] [Google Scholar]
- 14.Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. Journal of Cell Science. 2011;124(14):2424–37. [DOI] [PubMed] [Google Scholar]
- 15.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Progress in Lipid Research. 2001;40(5):325–438. [DOI] [PubMed] [Google Scholar]
- 16.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, et al. Triacylglycerol Synthesis Enzymes Mediate Lipid Droplet Growth by Relocalizing from the ER to Lipid Droplets. Developmental Cell. 2013;24(4):384–99. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasdekis AE, Silverman AM, Stephanopoulos G. Origins of Cell-to-Cell Bioprocessing Diversity and Implications of the Extracellular Environment Revealed at the Single-Cell Level. Scientific Reports. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osumi M. Visualization of yeast cells by electron microscopy. Journal of Electron Microscopy. 2012;61(6):343–65. 10.1093/jmicro/dfs082 [DOI] [PubMed] [Google Scholar]
- 19.Garay LA, Boundy-Mills KL, German JB. Accumulation of High-Value Lipids in Single-Cell Microorganisms: A Mechanistic Approach and Future Perspectives. Journal of Agricultural and Food Chemistry. 2014;62(13):2709–27. 10.1021/jf4042134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morin N, Cescut J, Beopoulos A, Lelandais G, Le Berre V, Uribelarrea J-L, et al. Transcriptomic Analyses during the Transition from Biomass Production to Lipid Accumulation in the Oleaginous Yeast Yarrowia lipolytica. Plos One. 2011;6(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerkhoven EJ, Pomraning KR, Baker SE, Nielsen J. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. npj Systems Biology and Applications. 2016;2:16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munsky B, Neuert G, van Oudenaarden A. Using Gene Expression Noise to Understand Gene Regulation. Science. 2012;336(6078):183–7. 10.1126/science.1216379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welf ES, Danuser G. Using Fluctuation Analysis to Establish Causal Relations between Cellular Events without Experimental Perturbation. Biophysical Journal. 2014;107(11):2492–8. 10.1016/j.bpj.2014.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wasylenko TM, Ahn WS, Stephanopoulos G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metabolic Engineering. 2015;30:27–39. 10.1016/j.ymben.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 25.Beopoulos A, Haddouche R, Kabran P, Dulermo T, Chardot T, Nicaud J-M. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Applied Microbiology and Biotechnology. 2012;93(4):1523–37. 10.1007/s00253-011-3506-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zenobi R. Single-Cell Metabolomics: Analytical and Biological Perspectives. Science. 2013;342(6163):1201–+. [DOI] [PubMed] [Google Scholar]
- 27.Stephanopoulos G. Metabolic Fluxes and Metabolic Engineering. Metabolic Engineering. 1999;1(1):1–11. 10.1006/mben.1998.0101 [DOI] [PubMed] [Google Scholar]
- 28.Nielsen J. It is all about metabolic fluxes. Journal of Bacteriology. 2003;185(24):7031–5. 10.1128/JB.185.24.7031-7035.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frenzen CL, Maini PK. Enzyne-Kinetics for a 2-Step Enzymatic Reaction with Comparable Initial Enzyme-Substrate Ratios Journal of Mathematical Biology. 1988;26(6):689–703. [DOI] [PubMed] [Google Scholar]
- 30.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nature Chemical Biology. 2010;6(3):209–17. 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewith R, Hall MN. Target of Rapamycin (TOR) in Nutrient Signaling and Growth Control. Genetics. 2011;189(4):1177–201. 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deris JB, Kim M, Zhang Z, Okano H, Hermsen R, Groisman A, et al. The Innate Growth Bistability and Fitness Landscapes of Antibiotic-Resistant Bacteria. Science. 2013;342(6162):1068–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick A, Weiner M. Enzyme Induction as an All or None Phenomenon. Proceedings of the National Academy of Sciences of the United States of America. 1957;43(7):553–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasdekis AE, Stephanopoulos G. Review of methods to probe single cell metabolism and bioenergetics. Metabolic Engineering. 2015;27:115–35. 10.1016/j.ymben.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–6. 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- 36.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nature Genetics. 2002;31(1):69–73. 10.1038/ng869 [DOI] [PubMed] [Google Scholar]
- 37.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427(6973):415–8. 10.1038/nature02257 [DOI] [PubMed] [Google Scholar]
- 38.Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, et al. Single-cell proteomic analysis of S-cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–6. 10.1038/nature04785 [DOI] [PubMed] [Google Scholar]
- 39.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: From theories to phenotypes. Nature Reviews Genetics. 2005;6(6):451–64. 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- 40.Li G-W, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475(7356):308–15. 10.1038/nature10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Read EL, Tovo-Dwyer AA, Chakraborty AK. Stochastic effects are important in intrahost HIV evolution even when viral loads are high. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(48):19727–32. 10.1073/pnas.1206940109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ. Stochasticity of metabolism and growth at the single-cell level. Nature. 2014;514(7522):376–+. 10.1038/nature13582 [DOI] [PubMed] [Google Scholar]
- 43.Labhsetwar P, Cole JA, Roberts E, Price ND, Luthey-Schulten ZA. Heterogeneity in protein expression induces metabolic variability in a modeled Escherichia coli population. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(34):14006–11. 10.1073/pnas.1222569110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Heerden JH, Wortel MT, Bruggeman FJ, Heijnen JJ, Bollen YJM, Planque R, et al. Lost in Transition: Start-Up of Glycolysis Yields Subpopulations of Nongrowing Cells. Science. 2014;343(6174):987–+. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Pan A, Spofford C, Zhou N, Alper HS. An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metab Eng. 2015;29:36–45. Epub 2015/03/01. 10.1016/j.ymben.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 46.Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nature Communications. 2012;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birch AM, Buckett LK, Turnbull AV. DGAT1 inhibitors as anti-obesity and anti-diabetic agents. Current Opinion in Drug Discovery & Development. 2010;13(4):489–96. [PubMed] [Google Scholar]
- 48.Walther TC, Farese RV, Jr. Lipid Droplets and Cellular Lipid Metabolism. In: Kornberg RD, editor. Annual Review of Biochemistry, Vol 81. Annual Review of Biochemistry. 812012. p. 687–714. [DOI] [PMC free article] [PubMed]
- 49.Tan W-H, Takeuchi S. A trap-and-release integrated microfluidic system for dynamic microarray applications. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(4):1146–51. 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasdekis AE. Single microbe trap and release in sub-microfluidics. RSC Advances. 2013;3(18):6343–6. [Google Scholar]
- 51.Zhang Z, Boccazzi P, Choi H-G, Perozziello G, Sinskey AJ, Jensen KF. Microchemostat-microbial continuous culture in a polymer-based, instrumented microbioreactor. Lab on a Chip. 2006;6(7):906–13. 10.1039/b518396k [DOI] [PubMed] [Google Scholar]
- 52.Vasdekis AE, Scott EA, Roke S, Hubbell JA, Psaltis D. Vesicle Photonics. Annual Review of Materials Research, Vol 43. 2013;43:283–305. [Google Scholar]
- 53.Gocze PM, Freeman DA. Factors Underlying the Variability of Lipid Droplet Fluorescence in MA-10 Leydig Tumor Cells. Cytometry. 1994;17(2):151–8. 10.1002/cyto.990170207 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This section describes the derivation of Eq 2, as well as further details the materials and methods information employed in this work (including imaging, image/data analysis, sample preparation, and microfluidics fabrication).
(PDF)
Growth curves measured for Po1g in YPD medium at three concentrations of cycloheximide (CHX): 0 μg/ml (unperturbed), 16 μg/ml (sum-minimum inhibotory concentration–sub MIC), and 33 μg/ml (minimum inhibotory concentration–MIC). The measurements were performed in a 96-well plate using a Bioscreen C Pro instruement; each measurement represents an average of four independent wells at a 100x dilution.
(PDF)
The dependence the lipid flux (dSi/dt) on [αi ∙ dNi/dt] (red circles) and [Ni ∙ dαi/dt] (blue circles) for the Po1g strain under sub-minimum inhibitory concentrations (sub-MIC–left) and minimum inhibitory concentrations (MIC–right). Each data point denotes a single observation per unit time per cell, and the solid lines illustrate linear fits; insets include the Spearman correlation coefficient (ρ).
(PDF)
(a) A graph illustrating the dynamics of the intracellular fluorescence intensity (“fluorescence”) of the Bodipy dye, the total lipid content (Si in %), and the ratio of the fluorescence intensity of the propidium iodide (PI) dye (“PI ratio”). The latter denotes the ratio of the intracellular PI fluorescence over the extracellular fluorescence, which is less than 1 for live cells. (b) The same graph for fixed cells (PI ratio >1), where the lipid content fluctuations are reduced, despite the similar dye uptake kinetics.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.