Summary
The oomycete pathogen Phytophthora infestans causes potato late blight, and as a potato and tomato specialist pathogen, is seemingly poorly adapted to infect plants outside the Solanaceae. Here, we report the unexpected finding that P. infestans can infect Arabidopsis thaliana when another oomycete pathogen, Albugo laibachii, has colonized the host plant. The behaviour and speed of P. infestans infection in Arabidopsis pre‐infected with A. laibachii resemble P. infestans infection of susceptible potato plants. Transcriptional profiling of P. infestans genes during infection revealed a significant overlap in the sets of secreted‐protein genes that are induced in P. infestans upon colonization of potato and susceptible Arabidopsis, suggesting major similarities in P. infestans gene expression dynamics on the two plant species. Furthermore, we found haustoria of A. laibachii and P. infestans within the same Arabidopsis cells. This Arabidopsis—A. laibachii—P. infestans tripartite interaction opens up various possibilities to dissect the molecular mechanisms of P. infestans infection and the processes occurring in co‐infected Arabidopsis cells.
Introduction
Plants have evolved diverse and effective mechanisms to protect against attack by microbial pathogens. Indeed, a central tenet of plant pathology is that resistance is the rule and disease the exception (Briggs, 1995). Although broad host‐range pathogens do occur, most plant pathogens are adapted to a limited number of taxonomically related host species and cause disease on only a few host plants. Those pathogens may not fare well on plants unrelated to their hosts because of adaptive evolution, which tends to drive organisms towards specialization, for example through the accumulation of mutations that enhance virulence on one host but impair it on another (Borhan et al., 2008; Dong et al., 2014; Dong et al., 2015; Ma et al., 2010; Raffaele et al., 2010; Tosa et al., 2006). In addition, nonhost resistance and species‐specific resistance serve to restrict the host range of plant pathogens (Lee et al., 2014; Schulze‐Lefert and Panstruga, 2011; Senthil‐Kumar and Mysore, 2013). Physical barriers, such as fortified cell walls and a waxy cuticle, production of antimicrobial secondary metabolites, and cell‐autonomous immunity all contribute to nonhost resistance (Bettgenhaeuser et al., 2014; Fellbrich et al., 2002; Miedes et al., 2014; Piasecka et al., 2015). Further, cell autonomous immunity is multi‐layered, involving pre‐invasive defences as well as cell surface and cytoplasmic immune receptors that perceive pathogens (Dodds and Rathjen, 2010; Win et al., 2012). Thus, a pathogen's ability to colonize a certain plant species includes its capacity to suppress or tolerate host immunity.
The oomycete plant pathogens comprise numerous host‐specific species (Fawke et al., 2015; Kamoun et al., 2015; Lamour and Kamoun, 2009; Thines and Kamoun, 2010). These filamentous microorganisms are some of the most destructive plant pathogens and remain persistent threats to both farmed and native plants (Akrofi et al., 2015; Enzenbacher et al., 2015; Hansen, 2015; Roy, 2015). For example, the Irish potato famine pathogen Phytophthora infestans, the causal agent of late blight, recurrently endangers global food security (Fisher et al., 2012; Fry et al., 2015). P. infestans is thought to have a relatively narrow host range, infecting a few wild Solanum species in their native habitats of central Mexico and the Andes, as well as cultivated potato and tomato in most regions where these crops are grown (Fry et al., 2009; Goss et al., 2014; Grunwald and Flier, 2005). In compatible hosts, P. infestans proliferates an extensive host‐intercellular hyphal network and projects digit‐like haustoria into single host cells (Blackwell, 1953). P. infestans can also infect other solanaceous plants, such as petunia and the experimental host Nicotiana benthamiana (Becktell et al., 2006; Chaparro‐Garcia et al., 2011). However, this pathogen is not known to complete its full infection cycle on plants outside the Solanaceae. For example, the model plant Arabidopsis thaliana, a member of the Brassicaceae family, is fully resistant to P. infestans and is considered a nonhost (Huitema et al., 2003; Lipka et al., 2005; Stein et al., 2006; Vleeshouwers et al., 2000).
On Arabidopsis leaves, as on other nonhost plants such as tobacco and parsley, P. infestans cysts germinate, form appressoria, and directly penetrate epidermal cells to form infection vesicles and occasionally secondary hyphae (Colon et al., 1992; Huitema et al., 2003; Naton et al., 1996; Schmelzer et al., 1995; Vleeshouwers et al., 2000). However, this early interaction is followed by the hypersensitive response, a localized cell death reaction of plants that restricts the spread of the pathogen (Huitema et al., 2003; Vleeshouwers et al., 2000). In the Arabidopsis pen2 mutant, which is deficient in the hydrolysis of 4‐methoxyindol‐3‐ylmethylglucosinolate (4MO‐I3M) into antimicrobial metabolites, the frequency of P. infestans penetration of epidermal cells increases, resulting in markedly enhanced hypersensitive cell death (Westphal et al., 2008). However, P. infestans does not complete its full infection cycle on pen2 mutants or pen2 mutants combined with mutations in other defence‐related genes (Kopischke et al., 2013; Lipka et al., 2005; Westphal et al., 2008). In these mutants, P. infestans hyphae fail to colonize the Arabidopsis mesophyll to the extent seen in compatible interactions and do not develop haustoria, the specialized hyphal extensions that project into host cells and are thought to be sites where the pathogen secretes virulence proteins (effectors) (Schornack et al., 2010; Whisson et al., 2007). To date, there are no published reports of Arabidopsis mutants that are fully deficient in nonhost resistance to P. infestans, and thus enable extensive biotrophic colonization and sporulation of this pathogen (Geissler et al., 2015; Stegmann et al., 2013).
One oomycete pathogen that can infect A. thaliana is A. laibachii, one of several specialist Albugo species that cause white blister rust disease (Kamoun et al., 2015; Kemen et al., 2011) by extensively colonizing host‐intercellular spaces, projection of knob‐like haustoria into host cells (Soylu, 2004), and by forming visible blister‐like dispersal pustules of sporangia on the lower side of leaves. Albugo spp. are obligate biotrophic parasites that are phylogenetically distinct from other oomycetes, such as Phytophthora, and thus have independently evolved the ability to colonize plants (Kemen and Jones, 2012; Thines and Kamoun, 2010). Albugo are widespread as endophytes in asymptomatic natural populations of Brassicaceae and likely influence the biology and ecology of their host species (Ploch and Thines, 2011). Remarkably, Albugo can suppress host immunity to enable colonization by other races of pathogens and subsequent genetic exchange between specialized genotypes with non‐overlapping host ranges (McMullan et al., 2015). Prior infection by Albugo enhances susceptibility to plant pathogens such as downy and powdery mildews (Bains and Jhooty, 1985; Cooper et al., 2008). For instance, pre‐infection with A. laibachii enables avirulent races of the Arabidopsis downy mildew Hyaloperonospora arabidopsidis to grow and sporulate on resistant Arabidopsis accessions (Cooper et al., 2008). A. laibachii suppresses the runaway cell death phenotype of the Arabidopsis lesion simulating disease1 mutant, further supporting the view that this pathogen is an effective suppressor of plant immunity (Cooper et al., 2008). The mechanisms by which Albugo spp. suppress immunity remain unknown, but probably involve suites of effector genes like those identified in the Albugo candida and A. laibachii genomes (Kemen et al., 2011; Links et al., 2011).
Here, we aimed to determine the degree to which A. laibachii would enable maladapted pathogens to colonize Arabidopsis. Pre‐infection with A. laibachii did not alter resistance of Arabidopsis to the Asian soybean rust pathogen (Phakopsora pachyrhizi) or the powdery mildew pathogen (Blumeria graminis f. sp. hordei (Bgh)). However, we discovered that pre‐infection with A. laibachii enables the potato pathogen P. infestans to fully colonize and sporulate on Arabidopsis, a plant that is considered to be a nonhost of this Solanaceae specialist. Our results show that when exposed to A. laibachii colonized tissues, P. infestans carries the potential to infect other plant species outside its natural host spectrum employing a conserved set of transcriptionally induced effector encoding genes. The interaction of Arabidopsis—A. laibachii—P. infestans will be an excellent model to examine how co‐infection of host cells enables infection by P. infestans.
Results
A. laibachii infection enables P. infestans colonization of the nonhost plant Arabidopsis
Previous work indicated that the potato late blight pathogen P. infestans can penetrate epidermal cells of its nonhost Arabidopsis resulting in hypersensitive cell death but little ingress beyond the infection site (Huitema et al., 2003; Vleeshouwers et al., 2000). To determine the extent to which A. laibachii alters the interaction between Arabidopsis and P. infestans, we carried out serial inoculations with the two pathogens. First, we infected rosette leaves of 5‐week‐old Arabidopsis Col‐0 with spores of A. laibachii strain Nc14 (Kemen et al., 2011). Successful infections were identified based on the formation of white sporangiophores on the abaxial side of rosette leaves 10 days after inoculation. At that stage, we detached the infected leaves, inoculated them with zoospores of P. infestans 88069 and monitored symptom development (Fig. 1A–B). Within 5 days after inoculation with P. infestans, we observed water‐soaked tissue, necrosis, and ultimately sporulation in co‐infected leaves (Fig. 1B). As controls we also applied P. infestans zoospores to uninfected leaves of A. thaliana Col‐0, and also monitored mock‐ and A. laibachii‐inoculated leaves (Fig. 1 A–B). No necrosis was observed in these negative controls (Fig. 1 A–B). To further investigate the degree to which P. infestans colonizes pre‐infected Arabidopsis leaves, we repeated the experiment with P. infestans 88069td, a transgenic strain that expresses the cytoplasmic red fluorescent protein (RFP) marker tandem dimer, and monitored pathogen ingress by microscopy. This revealed an extensive network of red fluorescent P. infestans hyphae and unlabelled A. laibachii hyphae in the co‐infected leaves that extended to most of the leaf within just 3 days after P. infestans inoculation and sharply contrasted with the P. infestans‐only treatment (Fig. 2 A–B). We also repeated the experiment with whole plants to ensure that the observed effect was not an artifact of the detached leaf assay. Here also, P. infestans triggered severe disease symptoms and formed an extensive hyphal network only in the mixed‐infection leaves (Fig. 1 C–E).
Figure 1.

Albugo laibachii enables Phytophthora infestans to colonize Arabidopsis on detached leaves and on whole plants.
A. Control leaves (Mock) or leaves from A. thaliana Col‐0 plants pre‐infected with A. laibachii were detached and droplets of water (H20) or P. infestans spore solution were applied to their abaxial sides and incubated for 4 days in high humidity.
B. A close‐up of (A) reveals P. infestans sporulation (arrowheads) as a dense cover of leaves pre‐infected by A. laibachii only.
B. Albugo laibachii enables P. infestans to colonize leaves infected on a whole plant. A. thaliana Col‐0 plants treated with water (H20) or pre‐infected with A. laibachii were inoculated with droplets of water (H20) or P. infestans spore solution and incubated in high humidity. Macroscopic observations of disease symptoms on whole plants at 3 days post inoculation.
D. A close‐up of (C) reveals P. infestans disease symptoms only on leaves pre‐colonized by A. laibachii (right panel).
E. The extent of P. infestans 88069td hyphal colonization (under RFP illumination) was assessed 3 days post inoculation using epifluorescence microscopy. Scale bar = 250 µm. All experiments were performed at least twice with similar results.
Figure 2.
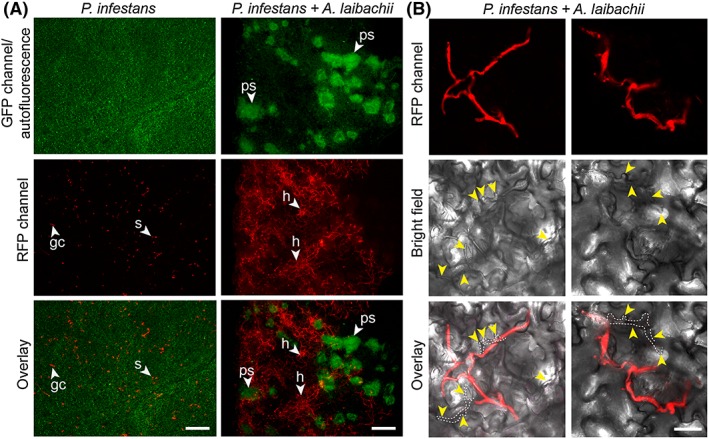
A. laibachii pre‐infection supports extensive hyphal growth of P. infestans in Arabidopsis.
A. Abaxial sides of control leaves of A. thaliana Col‐0 (left column) and leaves pre‐infected with A. laibachii (right column) have been infected with red fluorescent P. infestans 88069td. The extent of A. laibachii sporulation (under GFP illumination visible as green autofluorescent pustules, upper row) and P. infestans hyphal colonization (visible under RFP illumination as red hyphal network, middle row) was assessed 3 days post inoculation using epifluorescence microscopy. Bottom row represents merged fluorescence pictures.
B. Abaxial sides of co‐infected leaves at 2 dpi exhibiting dual colonization of P. infestans (in red) and by A. laibachii hyphae (not fluorescently labelled, indicated by yellow arrowheads) within the same area. All experiments were performed twice with similar results. Abbreviations: ps: pustules, h: hyphae; s: spores, gc: germinating cyst. Scale bars = 250 µm (A) or 50 µm (B).
Next, we quantified pathogen biomass during infection using kinetic PCR as previously described (Judelson and Tooley, 2000; Mauch et al., 2009) (Fig. 3). We amplified the P. infestans gene PiO8 to estimate relative levels of P. infestans DNA in infected plant tissue and observed a continuous increase over time in Arabidopsis leaves pre‐infected with A. laibachii (Fig. 3). We quantified A. laibachii biomass upon P. infestans co‐infection by using read data from our RNA sequencing experiment. We found numbers of reads matching to established constitutively expressed A. laibachii genes do not change upon infection with P. infestans (Fig. S1) suggesting that co‐colonization does not have a detrimental effect on A. laibachii colonization. Overall, the pathology, microscopy, and molecular biology experiments indicate that P. infestans becomes able to fully colonize nonhost Arabidopsis plants upon pre‐infection of those plants with A. laibachii (Figs 1, 2, 3, Fig. S1).
Figure 3.
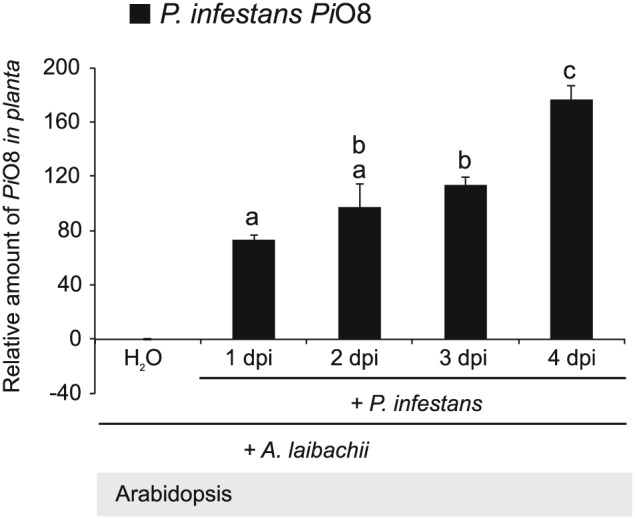
Quantification of P. infestans biomass upon infection of A. thaliana pre‐infected with A. laibachii.
Five‐week old leaves of A. thaliana Col‐0 pre‐infected with A. laibachii were detached and drop‐inoculated with a zoospore suspension of P. infestans isolate 06_3928A or mock‐treated with water applied to their abaxial sides and incubated for 4 days under high humidity. DNA was extracted at 0, 1, 2, 3, and 4 days post inoculation and used for quantitative PCR (qPCR) for PiO8 with gene‐specific primers for P. infestans. Pathogen DNA levels were normalized to the Arabidopsis SAND gene (At2g28390) and the relative amount of Pi08 was normalized to the DNA level in mock‐inoculated samples. Data are representative of one biological replicate with three technical replicates of qPCR reaction. Bars represent ratio between mean normalized expression of the infected samples with P. infestans and A. laibachii and the mock‐treated sample with A. laibachii and water (calibrator) (Mean ± SE). Data was analysed using repeated measures two way‐ANOVA (P < 0.0001). Letters indicate significant results of Fisher's LSD post‐hoc test. Experiment was performed twice with similar results.
Cellular dynamics of P. infestans colonization of pre‐infected Arabidopsis
To study the interaction between P. infestans and pre‐infected Arabidopsis in more detail, we performed confocal microscopy on leaves inoculated with P. infestans strain 88069td. We conducted side‐by‐side comparisons of the subcellular interactions of P. infestans in A. laibachii‐ and mock‐infected leaves. In both cases, we observed germinated P. infestans cysts on the leaf surface as well as appressoria (Fig. 4A, Fig. 4B) and infection vesicles within the plant epidermal cells (Fig. 4C, Fig. 4D). The difference between the two treatments became apparent at 1 day post infection (dpi) with the activation of host cell death (the hypersensitive response, HR) visible through accumulation of autofluorescent material in epidermal cell walls at sites of attempted infection by P. infestans in mock‐treated leaves only (Fig. 4E–F). Arabidopsis pre‐infected with A. laibachii did not display an HR at sites of penetration of P. infestans (Fig. 4G, Fig. 4H). To independently validate these data, we inoculated P. infestans strain 88069td on leaves that were mock treated or pre‐infected with A. laibachii, and then stained the leaves to quantify dead cells and monitor the invasion process at two different time points (6 h post infection, hpi, and 24 hpi) (Fig. 4I). This again confirmed that penetration of P. infestans was not associated with the HR in samples that were pre‐infected with A. laibachii, at both 6 and 24 hpi (Fig. 4I).
Figure 4.
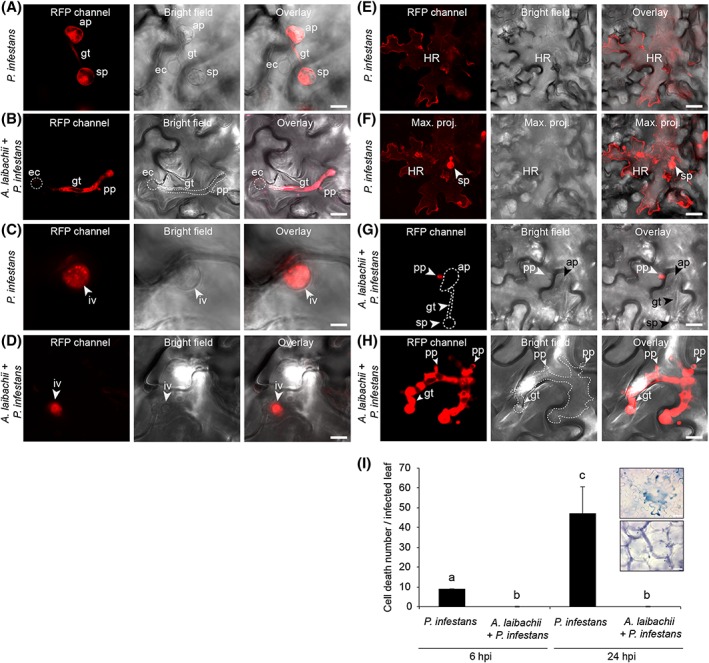
Hypersusceptibility of A. thaliana to P. infestans in leaves pre‐infected with A. laibachii is accompanied by a loss of the hypersensitive response Five‐week‐old leaves of A. thaliana Col‐0 mock‐treated or pre‐infected with A. laibachii were drop‐inoculated with a zoospore suspension of red fluorescent P. infestans 88069td Pathogen structures and autofluorescent dead epidermal cells were visualized with confocal laser scanning microscopy at 16 hpi (A–D) and at 24 hpi (E–H) in samples treated with P. infestans only (A, C, E, F) and in co‐infection experiments with A. laibachii (B, D, G, H). Panel F represents a maximum projection of images produced from 18 Z stacks showing a hypersensitive response of the same area as panel E. All experiments were performed twice with similar results. (I) Counts of dead cells per leaf after infection with P. infestans in the presence or absence of pre‐infection with A. laibachii. Data are representative of two biological replicates. Each replicate consists of counts from eight independent leaves. Bars represent mean ± SD. Data was analysed using one‐way‐ANOVA (P < 0.0001). Letters indicate significant results of Fisher's LSD post‐hoc test. The two light microscopy inserts show examples of an HR cell death in infected leaves with P. infestans only (top panel) and of absence of HR cell death in co‐infection experiments with A. laibachii and P. infestans (low panel) at 24 hpi. Abbreviations: sp: spores, gt: germ tube, ap: appressorium, ec: empty cyst, pp: penetration peg, iv: infection vesicle, HR: hypersensitive cell death, max. proj.: maximum projection. Scale bar = 25 µm (A–F) or 7.5 µm (G–H).
The ingress of P. infestans beyond its infection site became apparent starting at 36 hpi (1.5 dpi) in the A. laibachii pre‐infected leaves, with intercellular hyphae spreading from the penetration site (Fig. S2). In contrast to mock‐treated samples, the hyphae extended at 3 dpi to colonize the mesophyll and most of the leaf (Fig. S2). Branching hyphae with narrow, digit‐like haustoria expanded from the site of penetration to neighbouring cells through the intercellular space (Fig. S3). Starting at 3 dpi, the mycelium developed sporangiophores that released numerous sporangia to produce zoospores (Fig. S4). Thus, the P. infestans colonization of Arabidopsis pre‐infected with A. laibachii resembles, in behaviour and speed, the P. infestans infection reported on susceptible potatoes (Vleeshouwers et al., 2000).
A. laibachii and P. infestans haustoria within A single Arabidopsis cell
A. laibachii forms haustoria in Arabidopsis cells (Caillaud et al., 2012). Because we observed the formation of haustoria by P. infestans in Arabidopsis pre‐infected with A. laibachii, we searched for cells that harboured haustoria of both oomycetes. We recorded numerous events where single or multiple digit‐like P. infestans haustoria co‐occurred with multiple knob‐like A. laibachii haustoria in the same Arabidopsis cells (Fig. 5). In 45% of all assessed Arabidopsis mesophyll and epidermal cells with P. infestans haustoria we observed A. laibachii haustoria within the same confocal plane (Nobs = 17; 2 independent experiments). Thus, haustorium formation by A. laibachii or P. infestans does not trigger processes that prevent secondary penetration by another species. This observation will enable us to study how focal redirection of cellular compartments is affected by secondary penetration and how the two microbial pathogens vary in recruiting plant secretory processes to their haustoria.
Figure 5.
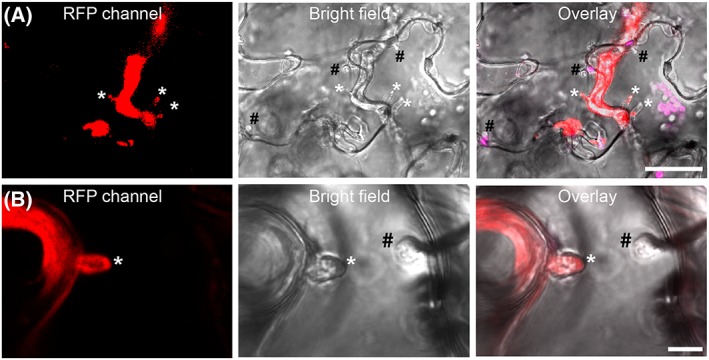
Phytophthora infestans and Albugo laibachii can form haustoria in the same Arabidopsis cell.
A. A. thaliana Col‐0 precolonized with A. laibachii was infected with red fluorescent P. infestans 88069td. Inspection by microscopy at 2 dpi revealed the presence of haustoria.
B. Frequently, plant cells were observed to harbour digit‐like, red fluorescent P. infestans 88069td haustoria as well as knob‐like A. laibachii haustoria. All experiments were performed twice with similar results. Abbreviations: #: haustoria of A. laibachii, *: haustoria of P. infestans. Scale bar = 25 µm (A) or 7.5 µm (B).
In planta expression dynamics of P. infestans secreted protein genes are similar on Arabidopsis and potato
Expression analyses have identified a significant set of P. infestans effector genes, which are transcriptionally induced during biotrophy in host–plant infections (Cooke et al., 2012; Haas et al., 2009; Pais et al., 2013). These studies have been limited to infections of potato and tomato, which both belong to the nightshade family (Solanaceae). To test whether the induced effector gene set is different in Arabidopsis pre‐infected with A. laibachii, we collected A. laibachii‐infected and mock‐infected Arabidopsis leaves at different time points following application of zoospores of P. infestans strain 06_3928A (13_A2 clonal lineage, (Cooke et al., 2012)). To compare sets of differentially regulated P. infestans genes in Arabidopsis with those differentially regulated in potato, we also infected and harvested potato leaves. Extracted RNA from all samples was subjected to Illumina RNA‐seq.
We found that during colonization of potato, the steady‐state transcript levels of 10 698 P. infestans genes were significantly altered. Of those, 7118 transcripts were also altered the same direction in A. laibachii pre‐infected Arabidopsis. In contrast, 776 transcripts were exclusively altered in the P. infestans—A. laibachii—Arabidopsis interaction (see Supplementary Table 1 for details).
We next examined changes in transcripts encoding secreted proteins and found 196 induced sequences, of which 136 (66%) were shared, 40 were uniquely induced in Arabidopsis/A. laibachii, and 20 uniquely induced in potato (Fig. S5A, Fig. 6A). We found a strong correlation between Arabidopsis/A. laibachii and potato in the degree of gene expression induction both at 2 and 3 dpi with P. infestans (Fig. 6B, Fig. S5). Out of a total of 96 induced effector gene transcripts, a common set of 78 (81%) were induced in both plant species, whereas 12 and 6 effector transcripts were induced in a host‐specific manner during colonization of Arabidopsis and potato respectively. Seven RXLR effector genes with known avirulence activity in specific potato cultivars were similarly induced in both host species (Fig. 6C). In summary, we conclude that the induction of secreted protein genes of P. infestans during colonization of potato and Arabidopsis/A. laibachii leaves do not greatly differ.
Figure 6.

Similar sets of effectors are induced during P. infestans colonization of potato (Solanum tuberosum) and Arabidopsis pre‐infected with A. laibachii.
A. Numbers of commonly and uniquely induced genes encoding secreted P. infestans proteins and effectors (a subset of the secreted proteins).
B. Dot blot comparing the transcript levels of P. infestans effector‐encoding genes between S. tuberosum and Arabidopsis pre‐infected with A. laibachii at 2 (left panel) and 3 days post infection (right panel).
C. Gene expression intensities relative to the average expression intensity in media (Rye sucrose) are shown for genes encoding avirulence proteins (Gene IDs in parentheses) during the interaction of P. infestans with A. laibachii pre‐infected Arabidopsis leaves (left panel) and S. tuberosum leaves (right panel) induced at 2 dpi and 3 dpi. Genes encoding ubiquitin ligases, Elongation factor 2, and Actin are shown as uninduced controls. All expression intensities are log2 transformed.
Arabidopsis leaves pre‐infected with A. laibachii do not become susceptible to barley powdery mildew fungus or Asian soybean rust fungus
To determine the degree to which the effect of A. laibachii on P. infestans extends to other maladapted pathogens, Arabidopsis leaves pre‐infected with A. laibachii were inoculated with the fungal pathogens B. graminis f. sp. hordei (Bgh) and P. pachyrhizi, the agents of barley powdery mildew and Asian soybean rust respectively. In both cases we observed no alteration of the interactions (Figs S6 and S7). Both of these fungal pathogens failed to penetrate leaves of both mock‐ and A. laibachii pre‐infected Arabidopsis plants.
Discussion
In this study, we demonstrated that the potato blight pathogen P. infestans becomes capable of colonizing Arabidopsis when this nonhost plant is pre‐infected by the obligate parasite A. laibachii. This is surprising, given that P. infestans is a Solanaceae specialist that is seemingly maladapted to plants from other botanical families. We took advantage of this tripartite interaction to perform comprehensive cellular and molecular analyses. On A. laibachii‐infected Arabidopsis, P. infestans goes through its full infection cycle to a degree that has not been observed to date with pre‐ and post‐invasive mutants (Geissler et al., 2015; Kobae et al., 2006; Lipka et al., 2005; Stegmann et al., 2013; Stein et al., 2006; Westphal et al., 2008). This includes the formation of haustoria, rapid hyphal proliferation, and profuse sporulation (Figs 1, 2, 3, 4, 5, Figs S1–S4). Expression dynamics of P. infestans genes encoding secreted proteins and effectors on susceptible (i.e., pre‐infected) Arabidopsis were generally similar to those on potato, indicating that this pathogen colonizes pre‐infected Arabidopsis in a similar manner as it colonizes its usual host plant (Fig. 6, Fig. S5B). In this study we used two different P. infestans isolates (88069td and 06_3928A) and obtained convergent results from transcriptome analysis and cell biological studies. Therefore, colonization of Albugo‐infected Arabidopsis is not strain specific.
P. infestans is a hemibiotroph and as such will initially colonize tissues biotrophically but will subsequently kill the tissue and feed on the remains. The initial biotrophic colonization including formation of haustoria is essential to colonization of host plants. We observed haustoria during colonization of A. laibachii‐infected Arabidopsis. Furthermore, autofluorescence monitoring using fluorescence microscopy and cell death trypan blue staining (Fig. 4) show presence of cell death only upon P. infestans singular infection, but not upon A. laibachii/P. infestans co‐infection. The induced expression of effector encoding genes previously associated with biotrophy in the P. infestans‐potato host system lends further support to an initial biotrophic growth. In conclusion, there are no data supporting an immediate necrotrophy when applying P. infestans spores to A. laibachii‐preinfected tissues. Instead, P. infestans exerts a hemibiotrophic lifestyle on potato as well as on A. laibachii colonized Arabidopsis.
The finding that P. infestans can fully colonize immunosuppressed plants distantly related to its hosts indicates that pathogen host range may not be fully determined by a lack of essential factors in the nonhost, one of several resistance mechanisms generally thought to determine host specificity (Agrios, 2005). Indeed, there is little evidence that nonhost resistance results primarily from the absence of taxon‐specific factors in the plant. For example, Garber's nutritional theory, which postulates that resistant plants provide a ‘nutritional environment that is inadequate for a parasite’ (Garber, 1956), has received little support over the years. By contrast, a greater understanding of the versatility and efficacy of the plant immune system has led to the view that active pre‐ and post‐invasive defences play a preponderant role in protecting most plants against most pathogens, and therefore in ultimately delimiting pathogen host range (Dodds and Rathjen, 2010; Jones and Dangl, 2006).
Our findings are consistent with the evolutionary history of the P. infestans lineage, which reflects significant plasticity in host range. This lineage, also known as clade 1c, consists of a tightknit group of closely related species that have specialized on host plants from four different botanical families as a consequence of a series of host jumps (Dong et al., 2014; Grunwald and Flier, 2005; Raffaele et al., 2010). This indicates that on a macroevolutionary scale, the P. infestans lineage has the capacity to generate variants that can infect divergent host plants (Dong et al., 2015). The split between P. infestans and its sister species P. mirabilis is estimated to have occurred relatively recently ~1300 years ago (Yoshida et al., 2013), providing some indication of the frequency of host jumps within the clade 1c lineage.
A. laibachii converts Arabidopsis into a fully susceptible host of P. infestans to a degree that has not been observed to date with genetic mutants. The pen2 and pen3 mutants, which are deficient in penetration resistance, display enhanced responses to P. infestans, exhibiting a macroscopically visible hypersensitive cell death that results from increased frequency of epidermal cell penetration (Kobae et al., 2006; Lipka et al., 2005; Stein et al., 2006). However, the extent to which Arabidopsis penetration resistance to P. infestans is effective at stopping pathogen ingress is debatable given that penetration events can also be observed on wild‐type Arabidopsis (Huitema et al., 2003; Vleeshouwers et al., 2000). In this study, we confirmed that penetration of Arabidopsis epidermal cells by P. infestans germinated cysts is commonly observed on wild‐type Arabidopsis (Fig. 4). Thus, although pen mutants enable increased plant cell penetration, pre‐invasive barriers do not fully block P. infestans infection, given that infection vesicles can be readily observed on mock‐treated wild‐type Arabidopsis at 16 hpi (Fig. 4). This view is consistent with the dramatic effect we observed on plants pre‐infected with A. laibachii, which did not display P. infestans‐triggered hypersensitivity probably as a consequence of post‐invasive immunosuppression. Consistent with a post‐invasive effect, A. laibachii did not alter Arabidopsis resistance to pathogens such as barley powdery mildew (Fig. S6) and Asian soybean rust fungi (Fig. S7), which cannot penetrate wild‐type Arabidopsis cells, in sharp contrast to P. infestans (Figs S2 and S3).
In P. infestans, as in many other filamentous pathogens, the expression of a subset of genes, notably secreted protein genes, is markedly induced during host infection (Cooke et al., 2012; Haas et al., 2009; Jupe et al., 2013; Pais et al., 2013). The mechanisms that underpin host signal perception by these pathogens, and the nature of these signals, remain largely unknown. We noted that the set of P. infestans effector genes induced on susceptible Arabidopsis largely overlaps with the genes induced in the host plant potato (Fig. 6). Patterns of effector gene expression displayed similar dynamics on both plants, with a peak during the biotrophic phase at 2 dpi. These results indicate that it is unlikely that P. infestans perceives a host‐specific plant signal to trigger in planta gene induction. One possibility is that as the pathogen progresses from host cell penetration to intercellular hyphal growth to haustorium formation, it undergoes a developmental programme that regulates gene expression.
Thines (2014) recently put forward the theory that Albugo‐infected plants could serve as a bridge that enables other oomycetes to shift from one host plant to another. Indeed, repeated cycles of co‐infection may facilitate the selection of genotypes of the maladapted pathogen that are virulent on the nonhost, eventually leading to a host jump. This scenario may have occurred with downy mildew species of the genus Hyaloperonospora, which tend to share Brassicaceae hosts with Albugo spp. (Thines, 2014). However, the degree to which Albugo has affected the ecological diversification of P. infestans and possibly other Phytophthora is unclear. First, it is not known whether the two pathogens are sympatric in central and south America, the natural geographic range of P. infestans and its sister species (Goss et al., 2014; Grunwald and Flier, 2005). Second, unlike P. infestans, most Phytophthora spp. are soil pathogens that do not spread aerially and are thus unlikely to colonize Albugo‐infected leaves. Nonetheless, the possibility that biotic agents, such as A. laibachii, have facilitated host jumps in the P. infestans lineage should not be disregarded and deserves to be studied, for example by genome sequencing of environmental leaf samples. Our study further highlights the importance of studying multitrophic interactions in order to fully understand the biology and ecology of plant pathogens (Kemen, 2014).
Few diseases rival the effect of P. infestans on humankind (Fisher et al., 2012; Yoshida et al., 2013). Long after it triggered the Irish potato famine, this pathogen is still regarded as a threat to global food security and is an active subject of research (Kamoun et al., 2015). To date, P. infestans research has focused mainly on its interaction with Solanacaeous plants. Little progress has been achieved using model systems such as A. thaliana, and work on Arabidopsis‐P. infestans has been limited to studies of nonhost resistance (Huitema et al., 2003; Kopischke et al., 2013; Lipka et al., 2005). Other Phytophthora spp., e.g. P. brassicae, P. cinnamomi, P. parasitica, and P. capsici, have been shown to infect Arabidopsis but they have been exploited to a lesser extent in research (Roetschi et al., 2001; Robinson and Cahill, 2003; Belhaj et al., 2009; Wang et al., 2011; Wang et al., 2013). The Arabidopsis—A. laibachii—P. infestans tripartite interaction opens up several new avenues of research: (i) to address the genetic diversity of Arabidopsis resistance towards P. infestans, (ii) to define the degree to which Albugo spp. have influenced the ecological diversification of P. infestans and enabled host jumps throughout evolution, and (iii) to dissect the molecular mechanisms, and focal retargeting of plant secretory pathways of co‐infected host cells, a situation that is likely to occur frequently under natural conditions.
Experimental procedures
Biological material
A. thaliana plants were grown on an ‘Arabidopsis mix’ (600 L F2 compost, 100 L grit, 200 g Intercept insecticide) in a controlled environment room (CER) with a 10 h day and a 14 h night photoperiod and at a constant temperature of 22 °C. A. thaliana Col‐0 ecotype was used for all experiments.
P. infestans isolates and 88069 expressing a cytosolic tandem RFP protein (88069td) and P. infestans strain 06_3928A (13_A2 clonal lineage) were cultured on rye sucrose agar at 18 °C in the dark as described earlier (Chaparro‐Garcia et al., 2011; Cooke et al., 2012). A. laibachii strain Nc14 was used in pre‐infection experiments in this study (Kemen et al., 2011). This strain was maintained on the A. thaliana Col‐5 line containing multiple insertions of the RPW8 powdery mildew resistance gene (Col‐gl RPW8.1 RPW8.2) (Xiao et al., 2001). The infected plants were kept overnight in a cold room (5 °C) then transferred to a growth cabinet under 10 h light and 14 h dark cycles with a 21 °C day and 14 °C night temperature as described (Kemen et al., 2011). Besides P. infestans and A. laibachii we used two obligate fungal parasites: B. graminis f. hordei CH4.8 (IPKBgh) and P. pachyrhizi isolate PPUFV02. A summary of fungal isolates used in this study and how they were maintained is provided in Supplementary Table 2.
Sequential infection assays
All infection assays were performed on four‐ or five‐week‐old Arabidopsis plants of ecotype Col‐0. Plants were pre‐inoculated with a zoospore suspension of A. laibachii (7.5 × 105 spores/ml) obtained from zoosporangia released from 14‐day‐old treated Col‐gl RPW8.1 RPW8.2 plants with A. laibachii isolate NC14 as described above. Briefly, whole Arabidopsis plants were sprayed with a zoospore suspension using a spray gun (1.25 ml/plant). They were incubated overnight in a cold room (5 °C) in the dark and transferred later to a growth cabinet under 10 h light and 14 h dark cycles with a temperature of 21 °C/14 °C per day/night. Control plants were mock‐treated with cold water. Plants pre‐infected with A. laibachii NC14 were then used for second infections eight to ten days after inoculation with the pathogens listed in Supplementary Table 2. Co‐infection assays with P. infestans were performed on detached leaves or whole plants as described earlier (Chaparro‐Garcia et al., 2011). Briefly, a zoospore suspension of P. infestans (1 × 105 spores ml−1) droplet was applied to the abaxial side of the leaf. Leaves were incubated on a wet paper towel in 100% relative humidity conditions with a 14 h/10 h day/night photoperiod and at a constant temperature of 18 °C.
Co‐infection assays with powdery mildew pathogen (B. graminis f. sp. hordei isolate CH 4.8) were performed on detached Arabidopsis leaves. Three‐centimetre leaf strips were cut from the cotyledon or first leaf of the barley cultivar and used as a control. Leaves were placed into agar plates containing 100 mg/l benzimidazole. Powdery mildew spores were collected from the barley‐infected leaves on a piece of paper. Infection was made in a settling tower by tapping and blowing the inoculum. Plates were allowed to settle for 10 min after infection in the tower before incubation in a growth cabinet at 15 °C (16 h light/8 h dark with 18 °C light/13 °C dark) (Brown and Wolfe, 1990).
Co‐infection assays with the Asian soybean rust were performed on detached leaves with P. pachyrhizi isolate PPUFV02 as described (Langenbach et al., 2013). Briefly, uredospores from P. pachyrhizi‐infected soybean leaves were collected at 14 days‐post inoculation (dpi), suspended in 0.01% (v/v) Tween‐20 at 1 mg/ml and used for inoculation. Spore suspension of P. pachyrhizi was sprayed on Arabidopsis leaves until the droplets covered the whole leaf surface. To allow fungal spore germination, infected leaves were maintained in moist conditions (100% humidity) and in the dark for the first 24 hpi.
Cytological analysis of infected material
Arabidopsis leaves infected with the red fluorescent P. infestans 88069td were visualized with a Fluorescent Stereo Microscope Leica M165 FC (Leica Microsystems Milton Keynes, UK) and an excitation wavelength for RFP: 510–560 nm. For confocal microscopy, patches of A. thaliana leaves were cut, mounted in water, and analysed with a Leica DM6000B/TCS SP5 confocal microscope (Leica Microsystems) with the following excitation wavelength for the GFP and the RFP channels: 458 nm and 561 nm respectively. Identical microscope settings were applied to all individuals.
To quantify the HR cell death response in infected samples, leaves were stained with lactoglycerol‐trypan blue and washed in chloral hydrate as described earlier (Belhaj et al., 2009). Specimens were mounted on microscope slides and analysed with a Leica DM2700 M microscope (Leica Microsystems).
Powdery mildew structures were stained with lactoglycerol‐trypan blue as described earlier (Vogel and Somerville, 2000). Briefly, excised leaves were destained in ethanol overnight than washed thoroughly with in water and placed in lactoglycerol (1:1:1 lactic acid:glycerol:water). Specimens were mounted on microscope slides with a few drops of 0.1% lactoglycerol‐trypan blue staining on top. Fungal structures were imaged with a Leica DM2700 M microscope (Leica Microsystems).
Asian soybean rust‐infected tissues were stained as described earlier in Ayliffe et al. (2011). Briefly, Arabidopsis leaf tissue was placed in 1 M KOH, then neutralized in 50 mM Tris, pH 7.0. The leaf was then stained with wheat germ agglutinin conjugated to fluorescein isothiocyanate (WGA‐FITC, Sigma‐Aldrich, UK) at 20 µg/ml. Specimens were mounted on a microscope slide and analysed with a Leica DM6000B/TCS SP5 confocal microscope (Leica Microsystems) with an excitation wavelength for GFP of 458 nm.
All microscopy images acquired for the various infections were analysed by using the Leica LAS AF software, ImageJ (2.0) and Adobe PHOTOSHOP CS5 (12.0).
Pathogen quantification
Genomic DNA was extracted from infected tissues using the DNeasy Plant Mini KIT (Qiagen, UK), following the manufacturer's protocol. Quantification of pathogen growth in planta was performed by quantitative PCR using a rotor gene 6000 apparatus (Corbett Research, UK) as previously described (Mauch et al., 2009). The PiO8 gene from P. infestans was used as a measure of in planta infection intensities of P. infestans with the following primers pair: PiO8‐3‐3F (5′‐CAATTCGCCACCTTCTTCGA‐3′) and PiO8‐3‐3R (5′‐GCCTTCCTGCCCTCAAGAAC‐3′) (Judelson and Tooley, 2000). SYBR Green (Qiagen, UK) was used as fluorescent reporter dye to amplify the PiO8 gene and was normalized to the Arabidopsis SAND gene (At2g28390) which was amplified with the following primer pairs SAND‐F (5′‐AACTCTATGCAGCATTTGATCCACT‐3′) and SAND‐R (5′‐TGATTGCATATCTTTATCGCCATC‐3′) (Mauch et al., 2009). The following LightCycler experimental protocol was used: denaturation at 95 °C for 15 min, amplification and quantification programme repeated 40 times (94 °C for 20s, 58 °C for 20s and 72 °C for 20s with two fluorescence measurements at 72 °C for 20 s (acquisition A on SYBR Green) and 77 °C for 15 s (acquisition B on SYBR Green)). A melting curve analysis was conducted from 60 °C to 95 °C in 0.5 °C steps and 5 s dwell time. Data were analysed with the Rotor‐Gene 4.4. Software package. The specificity of the amplification was confirmed by melting curve analysis. The amplification value (Efficiency) of each reaction was calculated. The ratio between P. infestans and Arabidopsis genomic DNA was calculated using the REST method as described (Pfaffl et al., 2002).
Statistical analysis
All data were analysed using the Prism software version 6.01 (GraphPad Software, USA). A one‐way ANOVA and repeated measures two‐way‐ANOVA were performed. Post hoc comparisons were conducted using the Fisher LSD test. A P value ≤ 0.001 or 0.05 was considered to be statistically significant.
RNA sequencing and analysis of the P. infestans and A. laibachii transcriptome
We sequenced the following samples: (i) 1 RNA sample from P. infestans isolate 06_3928A mycelia grown on RSA media, (ii) 2 RNA samples from the dual interaction of S. tuberosum (potato cv. Desiree) infected with P. infestans isolate 06_3928A and (iii) 3 RNA samples from the tripartite interaction of A. thaliana Col‐0 sequentially infected with A. laibachii isolate NC14 and P. infestans isolate 06_3928A (Supplementary Table 3). These samples were labelled as: (i) P. infestans isolate 06_3928A mycelia grown on rye sucrose agar RSA (Pinf_mycRSA), (ii) P. infestans isolate 06_3928A infecting Solanum tuberosum cv. Desiree and collected at 2 days post‐incoculation (dpi) (Pinf_Stub_2dpi), (iii) P. infestans isolate 06_3928A infecting S. tuberosum and collected at 3 dpi (Pinf_Stub_3dpi), (iv) A. laibachii isolate NC14 colonizing A. thaliana Col‐0 sequentially infected with P. infestans isolate 06_3928A and collected at 1 dpi (Alai_Atha_Pinf_1dpi), (v) A. laibachii isolate NC14 colonizing A. thaliana sequentially infected with P. infestans isolate 06_3928A and collected at 2 dpi (Alai_Atha_Pinf_2dpi), and (vi) A. laibachii isolate NC14 colonizing A. thaliana sequentially infected with P. infestans isolate 06_3928A and collected at 3 dpi (Alai_Atha_Pinf_3dpi). Mycelium was harvested after being grown in liquid Plich media for 15 days. It was washed with distilled water, vacuum dried, and ground in liquid nitrogen for RNA extraction. Detached leaves of both plant species were inoculated with 10 µl of a zoospore solution of P. infestans isolate 06_3928A at 1 × 105 spores ml−1. Leaf discs were collected at 2 and 3 days post inoculation (dpi) using a cork borer No. 4. Infected leaf samples were ground in liquid nitrogen until a fine powder was obtained and stored at −80 °C prior to RNA extraction. We used the RNeasy Plant Mini Kit (Qiagen, Cat No. 74904), following the manufacturer's instructions, to extract total RNA for all samples. cDNA libraries were prepared from total RNA using the TruSeq RNA sample prep kit v2 (Cat No. RS‐122‐2001). Library quality was confirmed before sequencing using the Agilent 2100 Bioanalyzer (Agilent Technologies). Sequencing was carried out using an Illumina Genome Analyzer II (Illumina Inc) with TruSeq Cluster generation kit v5 (Cat No. FC‐104‐5001) and TruSeq Sequencing kit v5 (Cat No. PE‐203‐5001). We performed read quality control by removing reads containing Ns and reads with abnormal read length (other than 76 bases) using FASTX‐Toolkit version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit). Total reads (76 bp paired‐end) that passed the parameters mentioned above for quality control were used for downstream analyses (Supplementary Table 3). All RNAseq reads are available at European Nucleotide Archive under the accession number PRJEB12248.
To describe the gene expression of coding genes of P. infestans isolate 06_3928A from the infected samples, we aligned each RNAseq experiment to the fasta nucleotide genome assembly of P. infestans strain T30‐4 version 2_2 (Haas et al., 2009) using TopHat software package version 2.0.6 (Kim et al., 2013) with 200 bp as the insertion length parameter. The alignments we obtained in sam format from TopHat software (Kim et al., 2013) were used for extract the expression of genes in P. infestans (Supplementary Table 3). A two‐stage analysis of the pathogen reads was applied to rescue multi‐mapped or ambiguous reads that cannot be uniquely assigned to groups of genes. First, we generated Reads Per Kilo Base per Million (RPKM) values for each gene by using the htseq‐count script that is part of the HTSeq python module (Anders et al., 2015). Next, we rescued reads that were enriched for gene families using multi‐map group (MMG) approach and customized perl scripts (Robert and Watson, 2015). In brief, we allocated multi‐mapped reads based on probability of multi‐mapped reads derived from particular locus that was calculated from RPKM, and then estimated final RPKM according to a published method (Mortazavi et al., 2008). The adjusted‐RPKM values of all reads after rescues were transformed into Log2 fold values by dividing the RPKM data to the RPKM values from mycelium of P. infestans isolate 06_3928A (Wagner et al., 2012). In planta‐induced genes exhibiting at least two‐fold gene induction between averaged media and infected sample (at 2 and/or 3 dpi) were considered induced during infection. Log2 values were loaded in Mev4_8 version 10.2 TM4 microarray software suite (Saeed et al., 2003) and analysed using hierarchical clustering method, gene tree selection, average linkage method, and Pearson correlation for distance metric selection. The gene expression heatmap obtained with Mev4_8 shows fold‐induction for P. infestans (PITG) genes with gene descriptions that are colour‐coded and highlight effector type custom annotations (Supplementary Fig. 5). We have also extracted read expression data of A. laibachii strain NC14 from the infected samples by aligning each time point to the A. laibachii NC14 reference genome (Kemen et al., 2011) using TopHat software v 2.0.6 and by generating RPKM values from read counts.
Conflict of interest
None of the authors has declared a conflict of interest.
Author contributions
K.B., S.S., and S.K. designed experiments. K.B., L.M.C., D.C.P., H.S., and H.P.v.E carried out experiments. K.B, L.M.C., K.Y., G.J.E., and Y.F.D. analysed data. D.C.P, A.K., H.S., J.D.G.J., and H. P.v.E. provided materials. K.B., S.K., and S.S. wrote the manuscript.
S.K. and S.S. contributed equally to this work.
Supporting information
Fig. S1. Gene expression as reads per kilobase per million mapped reads (RPKM) values for two ubiquitin control genes in P. infestans strain 3928A and Albugo laibachii strain NC14 during co‐infection in A. thaliana Col‐0. RPKM values were obtained from RNAseq reads counts using HTSeq programme. RPKM values show that the expression of two P. infestans ubiquitin control genes PITG_08025 and PITG_03199 increases over time but the expression of two other similar control genes in A. laibachii AlNc14C187G8343 and AlNc14C38G3321 are maintained. The expression of these ubiquitin genes was used as markers to measure accumulation of biomass of Phytophthora infestans and A. laibachii during co‐infection on A. thaliana.
Fig. S2. A. laibachii pre‐colonization supports formation of P. infestans infection structures in Arabidopsis beyond the surface penetration stage. Time course of red fluorescent P. infestans 88069td infection on A. thaliana Col‐0 pre‐infected with A. laibachii or mock‐treated with water. (A) A. laibachii pre‐colonized A. thaliana shows the first hyphae of P. infestans 88069td inside the leaf at 1.5 dpi and extensive intercellular colonization of A. thaliana Col‐0 leaf mesophyll at 2 and 3 dpi. (B) P. infestans does not grow on A. thaliana control leaves. Scale bars = 250 µm (A, C) or 50 µm (B). Experiment was performed twice with similar results. Abbreviations: h: hyphae of P. infestans; gc: germinating cyst of P. infestans; ps: pustules of A. laibachii.
Fig. S3. P. infestans can form haustoria in the nonhost plant Arabidopsis pre‐treated with A. laibachii. A. thaliana Col‐0 precolonized with A. laibachii was inoculated with red fluorescent P. infestans 88069td. Inspection by microscopy at 2 dpi revealed the presence of digit‐like haustoria of P. infestans independently of A. laibachii. Experiment was performed twice with similar results. Abbreviations: #: haustoria of A. laibachii, *: haustoria of P. infestans. Scale bar = 10 µm.
Fig. S4. P. infestans can extensively colonize and sporulate on nonhost Arabidopsis precolonized with A. laibachii. A. thaliana Col‐0 precolonized with A. laibachii was inoculated with red fluorescent P. infestans 88069td and imaged at 3 days post inoculation with confocal laser scanning microscopy. The upper panel shows confocal micrographs of hyphal extension and sporulation of P. infestans in A. laibachii pre‐treated Arabidopsis leaves. The lower panel is a close‐up of the region highlighted by the dotted square in the upper panel and shows P. infestans emerging sporangiophores from the leaf surface, giving rise to lemon‐shaped zoosporangia. Experiment was performed twice with similar results. Scale bar = 100 µm (upper panel) or 10 µm (lower panel).
Fig. S5. P. infestans genes encoding secreted proteins during infection of potato and Arabidopsis precolonized with A. laibachii. The heat map (A) illustrates 325 genes encoding secreted proteins; these genes were induced at least 2‐fold during the interaction of P. infestans with S. tuberosum leaves at 2 dpi and 3 dpi and during the interaction with Arabidopsis leaves colonized with A. laibachii at 1, 2, and 3 dpi. These genes were mean‐centred and hierarchically clustered by Euclidean distance. (B) Expression dynamics of selected P. infestans AVR genes and constitutively expressed control genes at 1, 2, and 3 days post inoculations (dpi).
Fig. S6. A. laibachii does not enable the nonhost powdery mildew pathogen to infect nonhost Arabidopsis. A. thaliana Col‐0 mock‐treated (control) or precolonized with A. laibachii was inoculated with Blumeria graminis f.sp hordei (Bgh) isolate CH4.8. The susceptible Barley cv. Golden Promise was used as a control for the infection. (A) Macroscopic phenotype of disease symptoms two weeks post inoculation with Bgh isolate CH4.8. (B) Maximum projection of images produced by light microscopy from 14 Z‐stacks from infected tissues. Micrographs show fungal structures 2 weeks post infection in both control (right panel) and samples pre‐colonized with A. laibachii (left panel) and reveal that the fungus was stopped at the penetration stage in both interactions. Experiment was performed twice with similar results. Abbreviations: c: conidium, pgt: primary germ tube, sgt: secondary germ tube, ap: appressorium, pa: papillae, ha: haustoria of A. laibachii, #: conidiospores of A. laibachii. Scale bar = 10 µm.
Fig. S7. A. laibachii does not enable the nonhost Asian soybean rust to infect nonhost Arabidopsis Five‐week‐old A. thaliana Col‐0 mock‐treated (control) or precolonized with A. laibachii was inoculated with spore suspension of Phakopsora pachyrhizi isolate PPUFV02 and incubated in high humidity. Pathogen structures were visualized with confocal laser scanning microscopy under GFP illumination at 6 days post inoculation. The micrographs show uredisniospores germinating on the leaf surface, producing an appressorium with no further growth in both control (upper panel) and pre‐colonized samples with A. laibachii (middle and lower panels). All experiments were performed twice with similar results. Abbreviations: u: urediniospore, ap: appressorium, st: stomata, #: conidiospores of A. laibachii. Scale bar = 10 µm.
Table S1. List of expressed genes in both potato/P. infestans and A. thaliana/A. laibachii‐P. infestans interactions. The file compiles three different worksheets containing lists (list 1‐3) of genes of P. infestans and their expression profiles during infection in potato (List 1) and/or in A. thaliana preinfected with A. laibachii (List 2‐3). List 1 comprises 10 698 coding genes of P. infestans on potato. List 2 comprises 7118 coding genes that are expressed in A. thaliana preinfected with A. laibachii only when compared to potato—P. infestans interaction. List 3 is a set of 325 in planta‐induced genes encoding secreted proteins in both treatments (potato and pre‐infected A. thaliana with A. laibachii). Abbreviations: GSR: gene sparse region, GDR: gene dense region, InBW: in between in between gene sparse and gene dense, NA: not applicable. If an NA is present in a column with annotations, this refers to no annotation to that type or description. If NA is present in a column with expression data this refers to a lower expression than 2‐fold (or log2 < 1) and in consequence it was not considered as in planta induced for that particular gene.
Table S2. Summary of pathogen isolates and media or susceptible plants used for maintaining pathogens.
Table S3. Alignment statistics of P. infestans isolate 06_3928A RNA sequences in infected materials. Pair‐end reads of 06_3928A isolate were aligned to the reference genome strain T30‐4 with the TopHat software package.
Supporting info item
Acknowledgements
We thank Stephen Whisson for providing the P. infestans 88069td strain, Francesca Stefanato for providing B. graminis f. sp. hordei CH4.8, Oliver Furzer and Wiebke Apel for supplying plant material, Matthew Moscow for providing WGA‐FITC, Jodie Pike for preparation of RNAseq libraries, and Joe Win for sequence data submission. This work was supported by the Gatsby Charitable Foundation, the European Research Council (ERC), and the Biotechnology and Biological Sciences Research Council (BBSRC). KY was supported by Japan Society of the Promotion of Science. H.S. was supported by the Institute Strategic Programme on Biotic Interactions for Crop Productivity BB/G042060/1.
Belhaj K., Cano L. M., Prince D. C., Kemen A., Yoshida K., Dagdas Y. F., Etherington G. J., Schoonbeek H.‐j., van Esse H. P., Jones J. D. G., Kamoun S., and Schornack S. (2017) Arabidopsis late blight: infection of a nonhost plant by Albugo laibachii enables full colonization by Phytophthora infestans , Cellular Microbiology, 19, e12628. doi: 10.1111/cmi.12628.
Contributor Information
Sophien Kamoun, Email: sophien.kamoun@tsl.ac.uk.
Sebastian Schornack, Email: sebastian.schornack@slcu.cam.ac.uk.
References
- Agrios, G.N. (2005) Plant Pathology. San Diego, CA: Elsevier. [Google Scholar]
- Akrofi, A.Y. , Amoako‐Atta, I. , Assuah, M. , and Asare, E.K. (2015) Black pod disease on cacao (Theobroma cacao, L) in Ghana: spread of Phytophthora megakarya and role of economic plants in the disease epidemiology. Crop Prot 72: 66–75. [Google Scholar]
- Anders, S. , Pyl, P.T. , and Huber, W. (2015) HTSeq—a Python framework to work with high‐throughput sequencing data. Bioinformatics btu638 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayliffe, M. , Jin, Y. , Kang, Z. , Persson, M. , Steffenson, B. , Wang, S. , and Leung, H. (2011) Determining the basis of nonhost resistance in rice to cereal rusts. Euphytica 179: 33–40. [Google Scholar]
- Bains, S. , and Jhooty, J. (1985) Association of Peronospora parasitica with Albugo candida on Brassica juncea leaves. J Phytopathol 112: 28–31. [Google Scholar]
- Becktell, M. , Smart, C. , Haney, C. , and Fry, W. (2006) Host–pathogen interactions between Phytophthora infestans and the Solanaceous hosts Calibrachoa × hybridus, Petunia × hybrida, and Nicotiana benthamiana . Plant Dis 90: 24–32. [DOI] [PubMed] [Google Scholar]
- Belhaj, K. , Lin, B. , and Mauch, F. (2009) The chloroplast protein RPH1 plays a role in the immune response of Arabidopsis to Phytophthora brassicae . Plant J 58: 287–298. [DOI] [PubMed] [Google Scholar]
- Bettgenhaeuser, J. , Gilbert, B. , Ayliffe, M. , and Moscou, M.J. (2014) Nonhost resistance to rust pathogens—a continuation of continua. Front Plant Sci 5 664: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell, E.M. (1953) Haustoria of Phytophthora infestans and some other species. Trans Br Mycol Soc 36: 138–IN135. [Google Scholar]
- Borhan, M.H. , Gunn, N. , Cooper, A. , Gulden, S. , Tör, M. , Rimmer, S.R. , and Holub, E.B. (2008) WRR4 encodes a TIR‐NB‐LRR protein that confers broad‐spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida . Mol Plant Microbe Interact 21: 757–768. [DOI] [PubMed] [Google Scholar]
- Briggs, S.P. (1995) Plant disease resistance. Grand unification theory in sight. Curr Biol 5: 128–131. [DOI] [PubMed] [Google Scholar]
- Brown, J. , and Wolfe, M. (1990) Structure and evolution of a population of Erysiphe graminis f. sp. hordei. Plant Pathol 39: 376–390. [Google Scholar]
- Caillaud, M.‐C. , Piquerez, S.J. , and Jones, J.D. (2012) Characterization of the membrane‐associated HaRxL17 Hpa effector candidate. Plant Signal Behav 7: 145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro‐Garcia, A. , Wilkinson, R.C. , Gimenez‐Ibanez, S. , Findlay, K. , Coffey, M.D. , Zipfel, C. , et al. (2011) The receptor‐like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen phytophthora infestans in Nicotiana benthamiana . PLoS One 6: e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon, L.T. , Eijlander, R. , Budding, D.J. , van Ijzendoorn, M.T. , Pieters, M.M.J. , and Hoogendoorn, J. (1992) Resistance to potato late blight (Phytophthora infestans (Mont.) de Bary) in Solanum nigrum, Solanum villosum and their sexual hybrids with Solanum tuberosum and Solanum demissum . Euphytica 66: 55–64. [Google Scholar]
- Cooke, D.E. , Cano, L.M. , Raffaele, S. , Bain, R.A. , Cooke, L.R. , Etherington, G.J. , et al. (2012) Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog 8: e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A.J. , Latunde‐Dada, A.O. , Woods‐Tor, A. , Lynn, J. , Lucas, J.A. , Crute, I.R. , and Holub, E.B. (2008) Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad‐spectrum suppression of innate immunity. Mol Plant Microbe Interact 21: 745–756. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Dong, S. , Stam, R. , Cano, L.M. , Song, J. , Sklenar, J. , Yoshida, K. , et al. (2014) Effector specialization in a lineage of the Irish potato famine pathogen. Science 343: 552–555. [DOI] [PubMed] [Google Scholar]
- Dong, S. , Raffaele, S. , and Kamoun, S. (2015) The two‐speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genet Dev 35: 57–65. [DOI] [PubMed] [Google Scholar]
- Enzenbacher, T. , Naegele, R.P. , and Hausbeck, M. (2015) Susceptibility of greenhouse ornamentals to Phytophthora capsici and P. tropicalis . Plant Dis 99: 1808–1815. [DOI] [PubMed] [Google Scholar]
- Fawke, S. , Doumane, M. , and Schornack, S. (2015) Oomycete interactions with plants: infection strategies and resistance principles. Microbiol Mol Biol Rev 79: 263–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellbrich, G. , Romanski, A. , Varet, A. , Blume, B. , Brunner, F. , Engelhardt, S. , et al. (2002) NPP1, a Phytophthora‐associated trigger of plant defense in parsley and Arabidopsis. Plant J 32: 375–390. [DOI] [PubMed] [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. , and Gurr, S.J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484: 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, W.E. , Grunwald, N.J. , Cooke, D.E.L. , McLeod, A. , Forbes, G.A. , and Cao, K. (2009) Population genetics and population diversity of Phytophthora infestans In Oomycete Genetics and Genomics. Lamoun K., and Kamoun S. (eds). Hobokem New Jersey: Wiley‐Blackwell. [Google Scholar]
- Fry, W. , Birch, P. , Judelson, H. , Grünwald, N.J. , Danies, G. , Everts, K.L. , et al. (2015) Five reasons to consider Phytophthora infestans a re‐emerging pathogen. Phytopathology 105: 966–981. [DOI] [PubMed] [Google Scholar]
- Garber, E. (1956) A nutrition‐inhibition hypothesis of pathogenicity. Amer Nat 90: 183–194. [Google Scholar]
- Geissler, K. , Eschen‐Lippold, L. , Naumann, K. , Schneeberger, K. , Weigel, D. , Scheel, D. , et al. (2015) Mutations in the EDR1 gene alter the response of Arabidopsis thaliana to Phytophthora infestans and the bacterial PAMPs flg22 and elf18. Mol Plant Microbe Interact 28: 122–133. [DOI] [PubMed] [Google Scholar]
- Goss, E.M. , Tabima, J.F. , Cooke, D.E. , Restrepo, S. , Fry, W.E. , Forbes, G.A. , et al. (2014) The Irish potato famine pathogen Phytophthora infestans originated in central Mexico rather than the Andes. Proc Natl Acad Sci U S A 111: 8791–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald, N.J. , and Flier, W.G. (2005) The biology of Phytophthora infestans at its center of origin. Annu Rev Phytopathol 43: 171–190. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. , et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- Hansen, E.M. (2015) Phytophthora species emerging as pathogens of forest trees. Curr Forest Res 1: 16–24. [Google Scholar]
- Huitema, E. , Vleeshouwers, V.G. , Francis, D.M. , and Kamoun, S. (2003) Active defence responses associated with non‐host resistance of Arabidopsis thaliana to the oomycete pathogen Phytophthora infestans . Mol Plant Pathol 4: 487–500. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. , and Dangl, J.L. (2006) The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. , and Tooley, P.W. (2000) Enhanced polymerase chain reaction methods for detecting and quantifying Phytophthora infestans in plants. Phytopathology 90: 1112–1119. [DOI] [PubMed] [Google Scholar]
- Jupe, J. , Stam, R. , Howden, A.J. , Morris, J.A. , Zhang, R. , Hedley, P.E. , and Huitema, E. (2013) Phytophthora capsici–tomato interaction features dramatic shifts in gene expression associated with a hemi‐biotrophic lifestyle. Genome Biol 14: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. , Furzer, O. , Jones, J.D. , Judelson, H.S. , Ali, G.S. , Dalio, R.J. , et al. (2015) The Top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol 16: 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen, E. (2014) Microbe–microbe interactions determine oomycete and fungal host colonization. Curr Opin Plant Biol 20: 75–81. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , and Jones, J.D. (2012) Obligate biotroph parasitism: can we link genomes to lifestyles? Trends Plant Sci 17: 448–457. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Gardiner, A. , Schultz‐Larsen, T. , Kemen, A.C. , Balmuth, A.L. , Robert‐Seilaniantz, A. , et al. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana . PLoS Biol 9: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. , and Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae, Y. , Sekino, T. , Yoshioka, H. , Nakagawa, T. , Martinoia, E. , and Maeshima, M. (2006) Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol 47: 309–318. [DOI] [PubMed] [Google Scholar]
- Kopischke, M. , Westphal, L. , Schneeberger, K. , Clark, R. , Ossowski, S. , Wewer, V. , et al. (2013) Impaired sterol ester synthesis alters the response of Arabidopsis thaliana to Phytophthora infestans . Plant J Cell Mol Biol 73: 456–468. [DOI] [PubMed] [Google Scholar]
- Lamour, K. , and Kamoun, S. (2009) Oomycete Genetics and Genomics: Diversity, Interactions and Research Tools. Hoboken, New Jersey: Wiley‐Blackwell, pp. 592 pp. [Google Scholar]
- Langenbach, C. , Campe, R. , Schaffrath, U. , Goellner, K. , and Conrath, U. (2013) UDP‐glucosyltransferase UGT84A2/BRT1 is required for Arabidopsis nonhost resistance to the Asian soybean rust pathogen Phakopsora pachyrhizi . New Phytol 198: 536–545. [DOI] [PubMed] [Google Scholar]
- Lee, H.A. , Kim, S.Y. , Oh, S.K. , Yeom, S.I. , Kim, S.B. , Kim, M.S. , et al. (2014) Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans . New Phytol 203: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Links, M.G. , Holub, E. , Jiang, R.H. , Sharpe, A.G. , Hegedus, D. , Beynon, E. , et al. (2011) De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics 12: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V. , Dittgen, J. , Bednarek, P. , Bhat, R. , Wiermer, M. , Stein, M. , et al. (2005) Pre‐ and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183. [DOI] [PubMed] [Google Scholar]
- Ma, L.‐J. , Van Der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.‐J. , Di Pietro, A. , et al. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch, F. , Torche, S. , Schlappi, K. , Branciard, L. , Belhaj, K. , Parisy, V. , and Si‐ammour, A. (2009) P. brassicae as a pathogen of Arabidopsis In Oomycete Genetics and Genomics: Diversity, Interactions and Research Tools. USA: Wiley‐Blackwell VWD, pp. 331. [Google Scholar]
- McMullan, M. , Gardiner, A. , Bailey, K. , Kemen, E. , Ward, B.J. , Cevik, V. , et al. (2015) Evidence for suppression of immunity as a driver for genomic introgressions and host range expansion in races of Albugo candida, a generalist parasite. eLife 4: e04550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes, E. , Vanholme, R. , Boerjan, W. , and Molina, A. (2014) The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5 358: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. , and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Naton, B. , Hahlbrock, K. , and Schmelzer, E. (1996) Correlation of rapid cell death with metabolic changes in fungus‐infected, cultured parsley cells. Plant Physiol 112: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais, M. , Win, J. , Yoshida, K. , Etherington, G.J. , Cano, L.M. , Raffaele, S. , et al. (2013) From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biol 14: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. , Horgan, G.W. , and Dempfle, L. (2002) Relative expression software tool (REST©) for group‐wise comparison and statistical analysis of relative expression results in real‐time PCR. Nucleic Acids Res 30: e36–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka, A. , Jedrzejczak‐Rey, N. , and Bednarek, P. (2015) Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol 206: 948–964. [DOI] [PubMed] [Google Scholar]
- Ploch, S. , and Thines, M. (2011) Obligate biotrophic pathogens of the genus Albugo are widespread as asymptomatic endophytes in natural populations of Brassicaceae. Mol Ecol 20: 3692–3699. [DOI] [PubMed] [Google Scholar]
- Raffaele, S. , Farrer, R.A. , Cano, L.M. , Studholme, D.J. , MacLean, D. , Thines, M. , et al. (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330: 1540–1543. [DOI] [PubMed] [Google Scholar]
- Robert, C. , and Watson, M. (2015) Errors in RNA‐Seq quantification affect genes of relevance to human disease. Genome Biol 16: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, L. , and Cahill, D. (2003) Ecotypic variation in the response of Arabidopsis thaliana to Phytophthora cinnamomi. Australas Plant Pathol 32: 53–64. [Google Scholar]
- Roetschi, A. , Si‐Ammour, A. , Belbahri, L. , Mauch, F. , and Mauch‐Mani, B. (2001) Characterization of an Arabidopsis‐Phytophthora Pathosystem: resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signalling. The Plant J 28: 293–305. [DOI] [PubMed] [Google Scholar]
- Roy, S.G. (2015) Phytophthora: a member of the sixth kingdom revisited as a threat to food security in the twenty‐first century In Value Addition of Horticultural Crops: Recent Trends and Future Directions. India: Springer, pp. 325–337. [Google Scholar]
- Saeed, A. , Sharov, V. , White, J. , Li, J. , Liang, W. , Bhagabati, N. , et al. (2003) TM4: a free, open‐source system for microarray data management and analysis. Biotechniques 34: 374. [DOI] [PubMed] [Google Scholar]
- Schmelzer, E. , Naton, B. , Freytag, S. , Rouhara, I. , Kuester, B. , and Hahlbrock, K. (1995) Infection‐induced rapid cell death in plants: a means of efficient pathogen defense. Can J Bot 73 (Suppl 1): S426–S434. [Google Scholar]
- Schornack, S. , van Damme, M. , Bozkurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. , et al. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci 107: 17421–17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze‐Lefert, P. , and Panstruga, R. (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci 16: 117–125. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. , and Mysore, K.S. (2013) Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu Rev Phytopathol 51: 407–427. [DOI] [PubMed] [Google Scholar]
- Soylu, S. (2004) Ultrastructural characterisation of the host–pathogen interface in white blister‐infected Arabidopsis leaves. Mycopathologia 158: 457–464. [DOI] [PubMed] [Google Scholar]
- Stegmann, M. , Anderson, R.G. , Westphal, L. , Rosahl, S. , McDowell, J.M. , and Trujillo, M. (2013) The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal Behav 8: e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, M. , Dittgen, J. , Sánchez‐Rodríguez, C. , Hou, B.‐H. , Molina, A. , Schulze‐Lefert, P. , et al. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, M. (2014) Phylogeny and evolution of plant pathogenic oomycetes—a global overview. Eur J Plant Pathol 138: 431–447. [Google Scholar]
- Thines, M. , and Kamoun, S. (2010) Oomycete‐plant coevolution: recent advances and future prospects. Curr Opin Plant Biol 13: 427–433. [DOI] [PubMed] [Google Scholar]
- Tosa, Y. , Tamba, H. , Tanaka, K. , and Mayama, S. (2006) Genetic analysis of host species specificity of Magnaporthe oryzae isolates from rice and wheat. Phytopathology 96: 480–484. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G. , van Dooijeweert, W. , Govers, F. , Kamoun, S. , and Colon, L.T. (2000) The hypersensitive response is associated with host and nonhost resistance to Phytophthora infestans . Planta 210: 853–864. [DOI] [PubMed] [Google Scholar]
- Vogel, J. , and Somerville, S. (2000) Isolation and characterization of powdery mildew‐resistant Arabidopsis mutants. Proc Natl Acad Sci 97: 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, G.P. , Kin, K. , and Lynch, V.J. (2012) Measurement of mRNA abundance using RNA‐seq data: RPKM measure is inconsistent among samples. Theory Biosci 131: 281–285. [DOI] [PubMed] [Google Scholar]
- Wang, Y.A.N. , Bouwmeester, K. , van de Mortel, J.E. , Shan, W. , and Govers, F. (2013) A novel Arabidopsis‐oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant, Cell Environ 36: 1192–1203. [DOI] [PubMed] [Google Scholar]
- Wang, Y.A.N. , Meng, Y. , Zhang, M. , Tong, X. , Wang, Q. , Sun, Y. , et al. (2011) Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol Plant Pathol 12: 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal, L. , Scheel, D. , and Rosahl, S. (2008) The coi1‐16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell 20: 824–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , et al. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118. [DOI] [PubMed] [Google Scholar]
- Win, J. , Chaparro‐Garcia, A. , Belhaj, K. , Saunders, D.G. , Yoshida, K. , Dong, S. , et al. (2012) Effector biology of plant‐associated organisms: concepts and perspectives. Cold Spring Harb Symp Quant Biol 77: 235–247. [DOI] [PubMed] [Google Scholar]
- Xiao, S. , Ellwood, S. , Calis, O. , Patrick, E. , Li, T. , Coleman, M. , and Turner, J.G. (2001) Broad‐spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120. [DOI] [PubMed] [Google Scholar]
- Yoshida, K. , Schuenemann, V.J. , Cano, L.M. , Pais, M. , Mishra, B. , Sharma, R. , et al. (2013) The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2: e00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Gene expression as reads per kilobase per million mapped reads (RPKM) values for two ubiquitin control genes in P. infestans strain 3928A and Albugo laibachii strain NC14 during co‐infection in A. thaliana Col‐0. RPKM values were obtained from RNAseq reads counts using HTSeq programme. RPKM values show that the expression of two P. infestans ubiquitin control genes PITG_08025 and PITG_03199 increases over time but the expression of two other similar control genes in A. laibachii AlNc14C187G8343 and AlNc14C38G3321 are maintained. The expression of these ubiquitin genes was used as markers to measure accumulation of biomass of Phytophthora infestans and A. laibachii during co‐infection on A. thaliana.
Fig. S2. A. laibachii pre‐colonization supports formation of P. infestans infection structures in Arabidopsis beyond the surface penetration stage. Time course of red fluorescent P. infestans 88069td infection on A. thaliana Col‐0 pre‐infected with A. laibachii or mock‐treated with water. (A) A. laibachii pre‐colonized A. thaliana shows the first hyphae of P. infestans 88069td inside the leaf at 1.5 dpi and extensive intercellular colonization of A. thaliana Col‐0 leaf mesophyll at 2 and 3 dpi. (B) P. infestans does not grow on A. thaliana control leaves. Scale bars = 250 µm (A, C) or 50 µm (B). Experiment was performed twice with similar results. Abbreviations: h: hyphae of P. infestans; gc: germinating cyst of P. infestans; ps: pustules of A. laibachii.
Fig. S3. P. infestans can form haustoria in the nonhost plant Arabidopsis pre‐treated with A. laibachii. A. thaliana Col‐0 precolonized with A. laibachii was inoculated with red fluorescent P. infestans 88069td. Inspection by microscopy at 2 dpi revealed the presence of digit‐like haustoria of P. infestans independently of A. laibachii. Experiment was performed twice with similar results. Abbreviations: #: haustoria of A. laibachii, *: haustoria of P. infestans. Scale bar = 10 µm.
Fig. S4. P. infestans can extensively colonize and sporulate on nonhost Arabidopsis precolonized with A. laibachii. A. thaliana Col‐0 precolonized with A. laibachii was inoculated with red fluorescent P. infestans 88069td and imaged at 3 days post inoculation with confocal laser scanning microscopy. The upper panel shows confocal micrographs of hyphal extension and sporulation of P. infestans in A. laibachii pre‐treated Arabidopsis leaves. The lower panel is a close‐up of the region highlighted by the dotted square in the upper panel and shows P. infestans emerging sporangiophores from the leaf surface, giving rise to lemon‐shaped zoosporangia. Experiment was performed twice with similar results. Scale bar = 100 µm (upper panel) or 10 µm (lower panel).
Fig. S5. P. infestans genes encoding secreted proteins during infection of potato and Arabidopsis precolonized with A. laibachii. The heat map (A) illustrates 325 genes encoding secreted proteins; these genes were induced at least 2‐fold during the interaction of P. infestans with S. tuberosum leaves at 2 dpi and 3 dpi and during the interaction with Arabidopsis leaves colonized with A. laibachii at 1, 2, and 3 dpi. These genes were mean‐centred and hierarchically clustered by Euclidean distance. (B) Expression dynamics of selected P. infestans AVR genes and constitutively expressed control genes at 1, 2, and 3 days post inoculations (dpi).
Fig. S6. A. laibachii does not enable the nonhost powdery mildew pathogen to infect nonhost Arabidopsis. A. thaliana Col‐0 mock‐treated (control) or precolonized with A. laibachii was inoculated with Blumeria graminis f.sp hordei (Bgh) isolate CH4.8. The susceptible Barley cv. Golden Promise was used as a control for the infection. (A) Macroscopic phenotype of disease symptoms two weeks post inoculation with Bgh isolate CH4.8. (B) Maximum projection of images produced by light microscopy from 14 Z‐stacks from infected tissues. Micrographs show fungal structures 2 weeks post infection in both control (right panel) and samples pre‐colonized with A. laibachii (left panel) and reveal that the fungus was stopped at the penetration stage in both interactions. Experiment was performed twice with similar results. Abbreviations: c: conidium, pgt: primary germ tube, sgt: secondary germ tube, ap: appressorium, pa: papillae, ha: haustoria of A. laibachii, #: conidiospores of A. laibachii. Scale bar = 10 µm.
Fig. S7. A. laibachii does not enable the nonhost Asian soybean rust to infect nonhost Arabidopsis Five‐week‐old A. thaliana Col‐0 mock‐treated (control) or precolonized with A. laibachii was inoculated with spore suspension of Phakopsora pachyrhizi isolate PPUFV02 and incubated in high humidity. Pathogen structures were visualized with confocal laser scanning microscopy under GFP illumination at 6 days post inoculation. The micrographs show uredisniospores germinating on the leaf surface, producing an appressorium with no further growth in both control (upper panel) and pre‐colonized samples with A. laibachii (middle and lower panels). All experiments were performed twice with similar results. Abbreviations: u: urediniospore, ap: appressorium, st: stomata, #: conidiospores of A. laibachii. Scale bar = 10 µm.
Table S1. List of expressed genes in both potato/P. infestans and A. thaliana/A. laibachii‐P. infestans interactions. The file compiles three different worksheets containing lists (list 1‐3) of genes of P. infestans and their expression profiles during infection in potato (List 1) and/or in A. thaliana preinfected with A. laibachii (List 2‐3). List 1 comprises 10 698 coding genes of P. infestans on potato. List 2 comprises 7118 coding genes that are expressed in A. thaliana preinfected with A. laibachii only when compared to potato—P. infestans interaction. List 3 is a set of 325 in planta‐induced genes encoding secreted proteins in both treatments (potato and pre‐infected A. thaliana with A. laibachii). Abbreviations: GSR: gene sparse region, GDR: gene dense region, InBW: in between in between gene sparse and gene dense, NA: not applicable. If an NA is present in a column with annotations, this refers to no annotation to that type or description. If NA is present in a column with expression data this refers to a lower expression than 2‐fold (or log2 < 1) and in consequence it was not considered as in planta induced for that particular gene.
Table S2. Summary of pathogen isolates and media or susceptible plants used for maintaining pathogens.
Table S3. Alignment statistics of P. infestans isolate 06_3928A RNA sequences in infected materials. Pair‐end reads of 06_3928A isolate were aligned to the reference genome strain T30‐4 with the TopHat software package.
Supporting info item


