Abstract
Microglia are brain resident macrophages important for brain development, connectivity, homeostasis and disease. However, it is still largely unclear how microglia functions and their identity are regulated at the molecular level. Although recent transcriptomic studies have identified genes specifically expressed in microglia, the function of most of these genes in microglia is still unknown. Here, we performed RNA sequencing on microglia acutely isolated from healthy and neurodegenerative zebrafish brains. We found that a large fraction of the mouse microglial signature is conserved in the zebrafish, corroborating the use of zebrafish to help understand microglial genetics in mammals in addition to studying basic microglia biology. Second, our transcriptome analysis of microglia following neuronal ablation suggested primarily a proliferative response of microglia, which we confirmed by immunohistochemistry and in vivo imaging. Together with the recent improvements in genome editing technology in zebrafish, these data offer opportunities to facilitate functional genetic research on microglia in vivo in the healthy as well as in the diseased brain. GLIA 2016;65:138–149
Keywords: microglia, zebrafish, RNA sequencing, transcriptome, neuronal cell death, proliferation
Introduction
Microglia are the resident macrophages of the central nervous system that serve important physiological functions related to neuronal plasticity and connectivity (Davalos et al., 2005; Li et al., 2012; Nimmerjahn, Kirchhoff, and Helmchen, 2005; Schafer et al., 2012; Stevens et al., 2007; Tremblay, Lowery, and Majewska, 2010). In addition, they play an important role in many neurodegenerative diseases, as scavengers of pathogens, debris and dead cells, and as regulators of immune responses (Hanisch and Kettenmann, 2007; Paloneva et al., 2002; Prinz et al., 2011). Moreover, several genetic neurological diseases have been found to be caused by microglial defects (Prinz and Priller, 2014). Nonetheless, the exact mechanisms by which microglia regulate brain homeostasis and contribute to disease are still unclear. Recently, genome wide gene expression analyses of acutely isolated microglia from mouse brains have revealed many of the genes and pathways that distinguish microglia from other brain and immune cell types (Butovsky et al., 2014; Chiu et al., 2013; Hickman et al., 2013; Matcovitch‐Natan et al., 2016; Zhang et al., 2014). However, the role and significance of many of these genes for microglial function remains to be elucidated. Microglial identity is induced by interplay of their developmental ontogeny and their position in the heterogeneous brain tissue, and therefore functional analysis of microglia in healthy and diseased brain is best addressed in vivo (Gosselin et al., 2014; Lavin et al., 2014; Sica and Mantovani, 2012; Xue et al., 2014).
Zebrafish share high similarity in embryonic development, cell biology and genetics with mammals and they are transparent at larval stages, which makes them highly suitable for non‐invasive imaging in vivo (Howe et al., 2013; Oosterhof, Boddeke, and van Ham, 2015; Vacaru et al., 2014). Analogous to mammalian microglia development, the first zebrafish microglia develop from a subset of early macrophages in the rostral blood island on the embryonic yolk sac that migrate into the brain (Ginhoux et al., 2010; Gomez Perdiguero et al., 2015; Herbomel, Thisse, and Thisse, 2001; Kierdorf et al., 2013; Matcovitch‐Natan et al., 2016; Xu et al., 2015). Functions described in vivo for zebrafish microglia include the clearance of dead brain cells and debris, the detection and removal of invading pathogens and regulation of neuronal activity (Herbomel, Thisse, and Thisse, 2001; Li et al., 2012; Peri and Nusslein‐Volhard, 2008; van Ham, Kokel, and Peterson, 2012). Phenotype driven genetic screens for microglial defects in zebrafish have already yielded new insight in microglial biology (Meireles et al., 2014; Shen, Sidik, and Talbot, 2016). Advances made in genome editing technology in zebrafish have now made it possible to perform reverse genetic screens in zebrafish (Burger et al., 2016; Hruscha et al., 2013; Hwang et al., 2013; Schmid and Haass, 2013; Shah et al., 2015). Therefore, the zebrafish appears to be an excellent model to further elucidate in vivo microglia gene function in development and in a disease context in a systematic manner by using reverse genetics.
However, as only a handful of zebrafish microglial genes are currently known, it is unknown how zebrafish microglia compare with mammalian microglia at the gene expression level (Herbomel, Thisse, and Thisse, 2001; Rossi et al., 2015; Shiau et al., 2015, 2013; Xu et al., 2016). To identify genome‐wide gene expression in microglia we optimized acute isolation of microglia from zebrafish brains by FACS and used RNA sequencing to compare their gene expression signature to the expression profile of other brain cells. Here, we identified the zebrafish microglia transcriptome, including many orthologs of mammalian microglia‐specific genes, indicating conservation of microglia gene expression across vertebrate classes. In addition, we applied RNA sequencing to study how microglia respond to induced neuronal cell death, and identified that neuronal death induces extensive local proliferation of microglia. These findings will facilitate investigating the genetics of microglial biology and their role in disease.
Materials and Methods
Animals
For all experiments in adult fish we used neuronal nitroreductase (NTR)‐mCherry expressing zebrafish incrossed with mpeg1‐GFP transgenic zebrafish as described previously that were kept on a 14 h/10 h light–dark cycle at 28°C (van Ham et al., 2014). They were fed brine shrimp twice a day. During experiments animals were kept in system water under standard water quality parameters. For in vivo imaging we used 6 dpf larvae expressing neuronal NTR‐mCherry in addition to expression of mpeg1‐ or apoeb‐driven GFP expression. Animal experiments were approved by the Animal Experimentation Committee of the Erasmus MC, Rotterdam and UMCG, Groningen.
Acute Isolation Microglia
For microglial isolation 3‐month‐old neuro‐NTR/mpeg1‐GFP zebrafish were euthanized in ice water according to animal welfare regulations. The heads were severed behind the gills, followed by removal of the gills, lower jaw and eyes using a watchmaker's forceps. The brains (5 per sample) were taken out of the skull after removal of the skull base and collected in ice cold PBS. Subsequently, the brains were cut using scalpels followed by dissociation in 0.25% trypsin‐0.1% EDTA in PBS for approximately 2 h at 4°C, while re‐suspending regularly. Upon complete dissociation of the brain, trypsin was inactivated by adding 1/6 volume of a 6 mM CaCl2 solution in PBS. The cell suspension was run through a 70 µm cell strainer and collected in a 22% Percoll solution (Schaafsma et al., 2015). Ice cold PBS was placed on top of the cell suspension while avoiding mixing of the layers, followed by centrifugation at 1,000 rcf at 4°C for 45 min. The remaining cell pellet was re‐suspended in suspension solution (high‐glucose DMEM without phenol red, 0.8 mM CaCl2, 1% v/v Penicillin/Streptomycin). The suspension was transferred to FACS tubes with 35 µm cell strainer caps, immediately followed by FACS sorting using a MoFlo Astrios cell sorter (Beckman Coulter, Fullerton, USA). DAPI was added to label and exclude dead cells.
RNA Extraction and Library Synthesis
Total RNA extraction was performed using the Qiagen miRNeasy kit according to the manufacturer's instructions (Qiagen, Hilden, Germany) and RNA sample quality was determined using on an Agilent Bioanalyzer 2100 total RNA 6000 Pico series chip (Agilent, Santa Clara, USA). Subsequently, cDNA libraries were created using the SMARTer Ultra Low Input RNA Kit for Sequencing – v3 (Clontech, Mountain View, USA). Illumina RNAseq libraries were prepared from cDNA using the Illumina TruSeq™ RNA Sample Prep Kit v2 according to the manufacturer's instructions (Illumina, Inc., San Diego, USA). In all libraries 50 nucleotide single‐end reads (SR50) were sequenced on an Illumina HiSeq2500 sequencer according to the manufacturer's protocol, obtaining 10–20 million reads per sample library. Image analysis and base calling were done using the Illumina pipeline. Reads were aligned to the zebrafish genome (GRCz10) using TopHat (version 2.0.5) (Li et al., 2009). The resulting files were filtered using SAMtools (version 0.1.18) to exclude secondary alignment of reads.
Bioinformatics
The aligned and filtered data was quantified with the Bioconductor package Genomic Ranges (Lawrence et al., 2013). Differential gene expression analysis was performed with Bioconductor package EdgeR (Robinson, McCarthy, and Smyth, 2010). The differentially expressed gene lists were functionally annotated using Qiagen's Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Data were inspected using MultiDimensional Scaling (MDS) plots, Principal Component Analysis (PCA), and inter‐sample correlation plots. The neuronal ablation‐associated microglia, 24 h and 48 h after treatment, had very similar transcriptional profiles and were grouped together as neuronal ablation‐associated microglia. The RNA‐seq data is available via GEO (www.ncbi.geo, accession number: GSE86921) and via the Glia Open Access Database (www.goad.education) (Holtman et al., 2015a). The Biomart Bioconductor Package was used to annotate the genes, and to identify mouse orthologs. Heatmaps were generated with heatmap.2 of Bioconductor package Gplots (Warnes et al., 2009). The zebrafish microglia expression profile was compared with recently reported pure mouse microglia expression profiles (Butovsky et al., 2014; Hickman et al., 2013; Zhang et al., 2014). With the Biomart Bioconductor tool, mouse orthologs, with their corresponding gene symbols, were identified for all zebrafish genes. This gene symbol list was intersected with the gene symbol list of genes expressed in mouse microglia and only high‐confidence orthologs were selected. For several mouse genes, multiple zebrafish high‐confidence orthologs were identified.
Neuronal Cell Ablation
For neuronal ablation neuro‐NTR transgenic zebrafish were used as described previously (van Ham et al., 2014). 3‐Month‐old zebrafish were placed in system water containing either 0.47% DMSO (control) or 5 mM MTZ for 48 h or 0.46% DMSO for 24 h followed by 5 mM MTZ for 24 h. The medium was refreshed after 24 h of treatment. Fish were kept under 14 h light/10 h dark cycles in a temperature controlled incubator (28°C). Fish were fed brine shrimp twice a day during the 48 h treatment. All experiments were performed according to the animal welfare regulations.
Immunofluorescence Staining
Immunohistochemistry was performed as described (van Ham et al., 2014; van Ham, Kokel, and Peterson, 2012). Briefly, fish were euthanized in ice water, followed by fixation of the brain inside the skull in 4% PFA at 4°C. Subsequently, the brains were carefully removed from the skulls and dehydrated with a 25%, 50%, 75%, 100% MeOH series and stored at −20°C for at least 12 h. After rehydration, brains were embedded in 4% w/v low melting point agarose in PBS and cut into 80 µm sections using a Microm HM 650V vibratome (Thermo Scientific, Waltham, USA). Immunostainings on free‐floating sections were performed as described (Adolf et al., 2006). Primary antibodies: PCNA (1:250, Dako, Glostrup, Denmark), l‐plastin (1:1,000). Secondary antibodies: DyLight Alexa 488 (1:500), DyLight Alexa 647 (1:500). For nuclear staining Hoechst was used. Sections were mounted in Vectashield mounting medium H1000 (Vector Laboratories, Burlingame, USA).
Combined TUNEL/Antibody Staining in Whole Mount Brain
For TUNEL staining the Click‐iT TUNEL Alexa Fluor 647 Kit (Invitrogen, Carlsbad, USA) was used. Fish were euthanized in ice water, followed by fixation of the brain inside the skull in 4% PFA at 4°C. Subsequently, the brains were carefully removed from the skulls and incubated in 11 µg/ml Proteinase K in PBST (PBS containing 0.2% Triton X‐100) at room temperature for 40 min. Then the brains were incubated in fixative again (4% PFA) at room temperature for 20 min. After washing with PBST the brains were incubated in the reaction buffer at room temperature for 30 min, followed by overnight incubation in the reaction cocktail at room temperature, according to the manufacturer's instructions. After washing with 3% w/v BSA in PBST the brains were incubated in the Click‐iT reaction cocktail at room temperature for 3 h, followed by washing with 3% BSA in PBST. Then the brains were incubated in PBST containing 1% v/v DMSO and 1% BSA at room temperature for 2 h. The brains were incubated with primary antibody (l‐plastin, gift from Yi Feng, University of Edinburgh, 1:500) in 5% BSA in PBST at 4°C for 72 h. Subsequently, brains were incubated overnight at 4°C with fluorescently labeled secondary antibody (DyLight Alexa 488 1:250) and Hoechst in PBST containing 2% BSA. Brains were sections as described and sections were mounted in Vectashield mounting medium.
Imaging and Quantification
Mounted sections were imaged on a LSM 700 Zeiss confocal system using a 20x dry objective (PlanApo, NA = 0.8) using 405, 488, 555 and 633 laser lines. Confocal z‐stack images were acquired. Images were processed with Zen 2012 (Zeiss, Oberkochen, Germany) and ImageJ/FIJI software. Quantifications were performed on 3 individual 4.0 × 10−4 mm3 volumes within the olfactory bulb in 3 animals. These volumes were chosen in the central areas of the olfactory bulb that contained a similar density of Hoechst‐positive nuclei across all imaged brain slices. Average numbers of three individual volumes per fish were quantified. Student's t‐tests were carried out on the averages of measurements in at least 3 individual animals (n = 3 or n = 4) were used to determine P‐values.
Intravital Imaging
Intravital imaging in zebrafish brains was largely performed as previously described (van Ham et al., 2014). Briefly, zebrafish larvae were mounted in 1.8% low melting point agarose containing 0.016% MS‐222 as sedative and anesthetic in HEPES‐buffered E3. The imaging dish containing the embedded larva was filled with HEPES‐buffered E3 containing 0.016% MS‐222. Imaging was performed using an Lcl Plan‐Neofluar 63×/1.3 lens on the Zeiss LSM780 system. For two‐photon excitation of red (mCherry) and green (GFP) fluorophores the laser (Coherent, Santa Clara, CA) was tuned to ∼990 nm.
Results
Acute Isolation and RNA Sequencing of Zebrafish Microglia
To identify microglial gene expression in zebrafish, we performed RNA sequencing on acutely isolated adult zebrafish microglia. To label microglia, we used transgenic mpeg1‐GFP zebrafish expressing GFP specifically in cells of the macrophage lineage, including microglia (Ellett et al., 2011; Svahn et al., 2013). Approximately 100,000 GFP+ cells were isolated from 5 pooled zebrafish brains, using fluorescence activated cell sorting (FACS) (Fig. 1A and B, Supporting Information Fig. S1A). The GFP− cellular fraction, representing neurons and other glial cells, was used to determine genome wide gene expression in non‐microglial brain cells. RNA sequencing was carried out on 3 biological replicates. Principal component analysis based on the expression profile showed that the GFP+ and GFP− samples formed separate distant clusters (Supporting Information Fig. S1B), indicating highly distinctive gene expression patterns.
Figure 1.
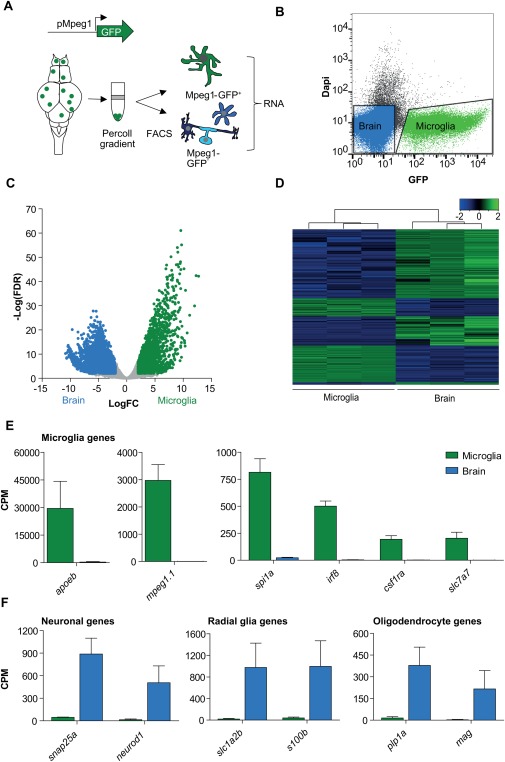
Sequencing of the zebrafish microglia transcriptome. (A) Schematic representation of acute isolation of zebrafish microglia from mpeg1‐promoter driven GFP transgenic zebrafish (mpeg1‐GFP). (B) FACS plot showing isolated populations for RNAseq in zebrafish microglia (green) and other brain cells (blue). (C) Differential gene expression (Volcano plot) showing genes significantly higher expressed in microglia (green) and other brain cells (blue). FDR <0.01, LogFC > |2|, n = 3. (D) Heatmap showing Z‐score values of all genes differentially expressed between microglia and other brain cells (6511 genes) (FDR <0.01, LogFC > |2|). (E, F) Expression values (CPM) for known microglial, neuronal, radial glial and oligodendrocyte genes in GFP+ (microglia) and GFP− (brain) cells. Values in (E) and (F) represent means of three independent experiments; Error bars in (E,F) represent standard deviation. FDR, False discovery rate; CPM, counts per million.
To confirm that isolated mpeg‐GFP+ cells are microglia and express previously identified zebrafish microglial genes, we investigated differentially expressed genes in the isolated GFP+ cell fraction. We identified a total of 6,511 differentially expressed genes (FDR <0.01, LogFC > |2|) of which 2,411 genes showed significantly higher expression in the GFP+ fraction (Fig. 1C and D, Supporting Information Table S1). These 2,411 genes included mpeg1, on which FACS sorting was based, and other genes previously described in zebrafish microglia including apoeb, csfr1a, spi1a, slc7a7 and irf8 (Fig. 1E, Supporting Information Table S1) (Herbomel, Thisse, and Thisse, 2001; Rossi et al., 2015; Shiau et al., 2015; Svahn et al., 2013). In contrast, genes mostly expressed in neurons (snap25a, neurod1), oligodendrocytes (plp1a, mag) and radial glia (slc1a2b, s100b) were significantly higher expressed in the GFP− neuronal and glia fraction (Fig. 1F). Altogether, we identified previously known, and many novel, zebrafish microglia genes in the isolated mpeg1‐GFP+ microglia population.
Functional Conservation of Microglia
To determine whether zebrafish microglia express typical vertebrate macrophage genes and genes related to immune function, we investigated the differentially expressed genes in more detail. Myeloid transcription factors that are essential for macrophage identity and immune function such as Irf8, Pu.1, Mafb, Cebp/α, and Jun showed high expression in zebrafish microglia and were hardly detectable in other brain cells (Supporting Information Table S1). Moreover, Ingenuity Pathway Analysis (IPA) revealed that genes with a significantly higher expression in microglia compared with other brain cells are mainly associated with immune responses, including production of reactive oxygen species (ROS) in macrophages and monocytes, NF‐κB and interleukin signaling (Fig. 2A). Zebrafish microglia also showed high expression of several Toll like receptors (TLRs) (e.g., tlr1, tlr7, tlr21), chemokine receptors (CR) (e.g., cxcr5, ccr12a, ccr9a), purinergic receptors (PR) (e.g., p2rx3a, p2rx7, p2ry12) and components of the mhc class II complex (e.g., cd74a, cd74b, mhc2dab), that were also hardly detectable in other brain cells (Fig. 2B, Supporting Information Table S1). Last, we found high microglia‐specific expression of components of the complement system, including C1q homologs C1qa and C1qb, components of the complement receptor 3 (CR3) complex, and progranulin, all of which have been shown to be involved in synaptic pruning in mice (Hong et al., 2016; Lui et al., 2016). Taken together, zebrafish microglia express many of the transcriptional regulators, immune and pathogen recognition receptor repertoire and pruning‐associated genes found in mammals, indicating that zebrafish microglia show similar functionality as found in mammals.
Figure 2.
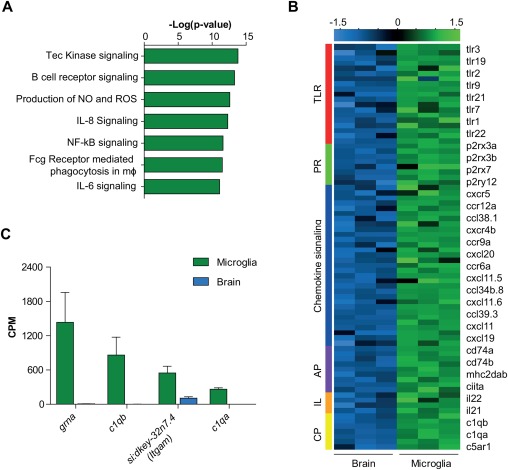
Conserved microglia functions in the zebrafish. (A) Ingenuity pathway analysis (IPA) canonical pathways most significantly enriched in zebrafish microglia. (B) Heatmap showing expression Z‐scores of immune genes significantly higher expressed in microglia than other brain cells (FDR <0.01 and logFC >1). (C) CPM values of zebrafish orthologs of genes involved in synaptic pruning in microglia and brain samples. Values in (C) represent mean of three independent experiments; Error bars in (C) represent standard deviation. TLR, Toll‐like receptors; PR, purinergic receptors; AP, antigen presentation; IL, interleukins + interleukin receptors; CP, complement.
Identification of a Conserved Zebrafish Microglia Gene Expression Signature
To investigate to what extent the microglial gene expression profile is conserved between zebrafish and mammals, we compared the zebrafish dataset to several previously published mouse microglia transcriptomes (Butovsky et al., 2014; Hickman et al., 2013; Zhang et al., 2014). Zhang et al., (2014) used RNA sequencing and identified 500 significantly enriched genes in mouse microglia compared with other cell types in the brain, including neurons, astrocytes and oligodendrocytes. For these genes we found 361 annotated zebrafish orthologs, of which 163 orthologs showed significantly higher expression in adult zebrafish microglia (e.g., c1qb, cd68) compared with other brain cells (FDR <0.01, logFC > 2) (Fig. 3A and B, Supporting Information Table S2). A second study applied direct RNA sequencing on mouse microglia and reported 100 genes encoding mainly cell surface molecules with significantly higher expression in microglia compared with whole brain (Hickman et al., 2013). Comparison with zebrafish microglial genes expression showed that out of 66 identified zebrafish orthologs, 42 orthologs (e.g., slco2b1 and gpr84) are significantly higher expressed in the zebrafish microglia transcriptome (FDR <0.01, logFC > 2) (Fig. 2A and B, Supporting Information Table S2). In a third study, Butovsky et al. (2014) performed quantitative mass spectrometry and gene expression profiling on isolated microglia and showed 106 genes with significantly higher expression in microglia than in whole brain samples. We found 101 zebrafish orthologs of which 44 are significantly higher expressed in zebrafish microglia compared with other brain cells, including cmklr1 and entpd1 (FDR <0.01, logFC > 2) (Fig. 3A and B, Supporting Information Table S2). Taken together, we identified at least 213 mouse genes for which microglia‐specific expression is conserved in the zebrafish (Fig. 3C). In all, a large fraction of the mouse microglia‐specific gene expression signature is conserved in the zebrafish, suggesting evolutionary conservation of processes regulated by these genes across vertebrates from fish to mammals.
Figure 3.
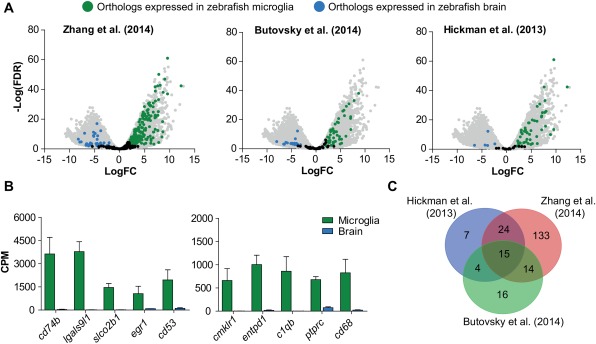
Conserved microglia gene expression in the zebrafish. (A) Volcano plot showing expression of zebrafish orthologs of genes found to be enriched in microglia compared with Zhang et al. (2014), Butovsky et al. (2014), and Hickman et al. (2013), respectively. All zebrafish genes are shown in grey. Differentially expressed orthologs (FDR <0.01, LogFC > |2|) are shown in green (microglia) or blue (other brain cells). (B) Expression values (CPM) of orthologs for genes found by one or more of the above mentioned studies. (C) Venn diagram showing overlap of microglia‐specific orthologs of genes found in three transcriptomic studies (Butovsky et al., 2014; Hickman et al., 2013; Zhang et al., 2014). Values in (B) represent means of three independent experiments; Error bars in (B) represent standard deviation.
RNAseq Reveals Proliferation As an Acute Transcriptional Microglia Response to Neuronal Cell Death
Microglia are involved in many age‐related neurodegenerative diseases and there is a widely held view that the microglia state can influence disease outcome. Therefore, transcriptome studies on microglia acquired from mouse models including amyotrophic lateral sclerosis (ALS) and aging have been carried out to identify disease and aging specific signatures. Processes that were identified to be differentially regulated, although highly dependent on the nature of the model or insult, involve inflammation, phagocytosis, lysosomal processing, priming, and the inflammasome (Chiu et al., 2013; Hickman et al., 2013).
As virtually all neurodegenerative diseases show extensive degeneration and death of neurons, we wanted to study the microglial transcriptional response specifically to acute degeneration of neurons by conditional neuronal ablation (van Ham et al., 2014; van Ham, Kokel, and Peterson, 2012). This could allow us to isolate the microglial processes mostly affected by neuronal cell death in vivo. We have shown previously that addition of the ligand metronidazole (MTZ) to zebrafish larvae expressing a nitroreductase (NTR)‐mCherry (neuro‐NTR) fusion protein mainly in neurons of the olfactory bulb effectively kills only the transgene expressing cells. Neuronal ablation in zebrafish is accompanied by an increase in number of highly phagocytic microglia, characterized by an amoeboid morphology (Kuipers et al., 2016).
To determine whether NTR‐mediated neuronal ablation can be used in adult zebrafish to induce neuronal cell death accompanied by microglia activation, we performed immunofluorescence (microglia) and TUNEL (apoptotic cells) labeling in brains of adult Neuro‐NTR expressing zebrafish after treatment with MTZ. MTZ‐treated animals showed high numbers of TUNEL+ cells (314.8 ± 44.1) in the olfactory bulb, whereas TUNEL+ cells were rarely observed in DMSO‐treated controls (0.6 ± 0.7) (Fig. 4A), showing that NTR‐mediated neuronal ablation induces extensive neuronal cell death in the olfactory bulb of adult zebrafish. In addition, the olfactory bulbs of MTZ‐treated animals showed abundant amoeboid microglia, with a significant increase in microglia numbers (DMSO: 7.8 ± 2.0; MTZ: 44.8 ± 3.1) (Fig. 4A). The NTR‐mCherry fusion transgene in neurons allowed us to track the fate of ablated neurons because of persisting mCherry fluorescence, even after engulfment by microglia (van Ham et al., 2014; van Ham, Kokel, and Peterson, 2012). This revealed accumulation of ablated neurons in microglia of MTZ‐treated zebrafish, showing that the induction of neuronal cell death causes microglia to become highly phagocytic (Fig. 4A, Supporting Information Fig. S2A). These data show that our non‐invasive NTR/MTZ‐mediated conditional neuronal ablation is an effective strategy to induce neuronal cell death and subsequent microglia activation in adult zebrafish.
Figure 4.
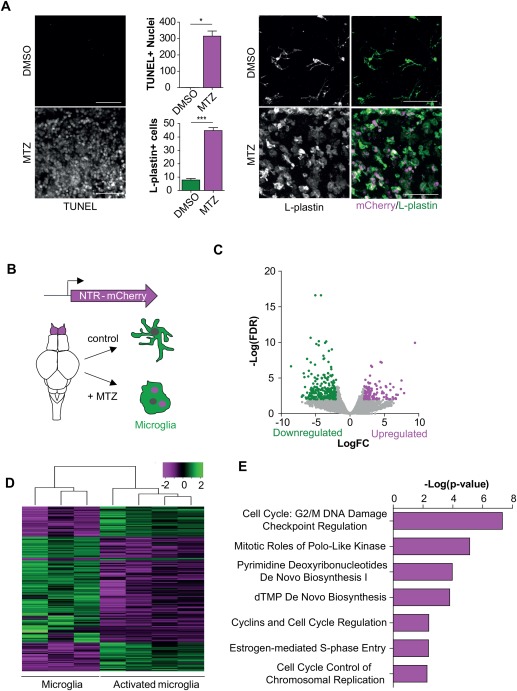
Identification of gene expression changes upon neuronal cell death. (A) TUNEL and L‐plastin staining showing increased neuronal cell death upon treatment with MTZ for 48 hours accompanied by microglia activation. mCherry signal represents engulfed neurons. (B) Schematic representation of cells isolated for RNA sequencing on activated microglia. (C) Volcano plot showing differentially expressed genes upon the activation of microglia (FDR <0.01; LogFC > |2|). (D) Heatmap showing Z‐score values of all genes differentially expressed between activated microglia and control microglia (367 genes) (FDR <0.01, LogFC > |2|).(E) IPA canonical pathway analysis on significantly upregulated genes in microglia upon NTR‐mediated ablation (FDR <0.01;LogFC > |2|). Scale bar = 40 µm in (A). For quantification in (A) cells were counted in 3 selected volumes in the olfactory bulb (4.0 × 10 − 4 mm3) per fish (n = 3). Error bars represent standard deviation, *P < 0.05, ***P < 0.001 (Student's t‐test).
To compare genome‐wide transcriptional changes accompanying microglial activation upon induced neuronal death, we performed RNA sequencing on isolated GFP+ microglia from Neuro‐NTR expressing zebrafish undergoing conditional neuronal ablation for 24 or 48 h (Fig. 4B). Principal component analysis (PCA) and differential gene expression analysis of the transcriptome revealed that the expression profiles of microglia 24 or 48 h after neuronal ablation are very similar and showed only 2 differentially expressed genes (FDR <0.01, LogFC > |2|) (Supporting Information Fig. S1B, data not shown). Therefore, subsequent analysis was performed on pooled data from 24 and 48 h treated animals, 2 biological replicates each, to increase the statistical power. Differential gene expression analysis of microglia gene expression in fish undergoing conditional neuronal ablation and in control animals revealed 367 differentially expressed genes (FDR <0.01, LogFC > |2|) of which 125 genes showed increased expression upon NTR‐mediated ablation (Fig. 4C and D, Supporting Information Table S3). IPA analysis revealed that the upregulated genes upon neuronal death are mostly associated with cell cycle control and DNA replication (Fig. 4E). Interestingly, increased expression of genes previously identified in disease models involved in processes such as phagocytosis and inflammation did not show significantly increased expression (Supporting Information Fig. S2B, data not shown). This suggests that the microglial signature upon detection of extensive neuronal cell death is characterized primarily by upregulation of genes involved in molecular processes related to proliferation.
Microglia Proliferate upon the Induction of Neuronal Death
Proliferation of microglia could explain the increase in numbers we observed upon conditional neuronal ablation. However, the expression markers we used to isolate and label microglia, mpeg1 and lplastin, do not distinguish microglia from potentially infiltrating macrophages (Ellett et al., 2011). Therefore, we aimed to address whether infiltration of peripheral monocytes and/or macrophages could explain part of the observed increase in microglia numbers and proliferation. From mouse studies it is known that infiltrating macrophages express high levels of genes encoding MHC class II components, CD45, CD40 and CD44 (Mildner et al., 2009; Vainchtein et al., 2014). We reasoned that if peripheral macrophages were infiltrating in large numbers this would have been detectable as an increased expression of these genes. However, differential gene expression analysis did not show increased expression of orthologs for these genes and even showed a significantly reduced expression of MHC class II molecules (e.g., mhc2dab). Similarly, a large contribution of infiltrating macrophages would likely yield a dilution of microglia, which would be reflected by a decrease in the expression of microglial specific genes. We could not detect a reduction in expression of microglial specific genes. Therefore, these findings suggest that there is no major contribution of macrophages from the periphery (Supporting Information Fig. S3).
To investigate whether the increased expression of cell cycle genes is indeed followed by increased proliferation of microglia, we performed immunofluorescence staining for PCNA (dividing cells) after the induction of neuronal ablation. This revealed a significant increase in the number of PCNA+ microglia (DMSO: 3.3 ± 2.7; MTZ: 17.8 ± 8.5) as well as increase in the PCNA+ microglia fraction (DMSO: 0.4 ± 0.2; MTZ: 0.7 ± 0.02) in the olfactory bulbs of MTZ‐treated animals compared with DMSO‐treated controls (Fig. 5A). We conclude that proliferation of microglia is an acute response to extensive neuronal death.
Figure 5.
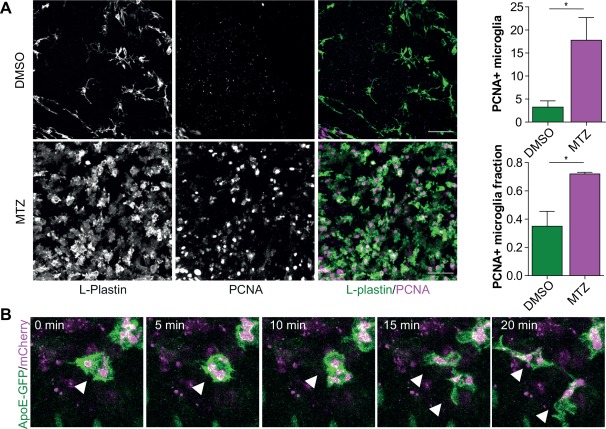
Microglia proliferation upon the induction of neuronal cell death. (A) Immunofluorescence staining in the olfactory bulbs of 3‐month‐old treated MTZ‐treated and DMSO‐treated (control) fish. (B) Intravital imaging in 7 dpf zebrafish larvae with Apoe‐driven GFP undergoing NTR‐mediated neuronal cell death, showing the presence of dividing zebrafish microglia upon neuronal death. n = 3. Scale bar = 40 µm in (A). For quantification in (A) cells were counted in 3 selected volumes in the olfactory bulb (4.0 × 10 − 4 mm3) per fish (n = 3). Error bars represent standard deviation, *P < 0.05 (Student's t‐test).
To study in vivo whether microglia proliferate locally, we performed long‐term intravital imaging in transgenic zebrafish expressing apoeb‐driven GFP after neuronal ablation. Transgenic zebrafish larvae expressing GFP under the apoeb promoter, show high GFP expression in microglia, but not in peripheral macrophages (Peri and Nusslein‐Volhard, 2008; van Ham et al., 2014). We observed occasional mitosis of phagocytic microglia, showing that microglia proliferate locally upon induced neuronal cell death (Fig. 5B, Supporting Information Movie S1).Taken together, our transcriptomic, immunohistochemistry and in vivo imaging data indicate that neuronal death induces an immediate proliferative response of microglia apparent at the transcriptional as well as cellular level.
Discussion
Zebrafish are highly suitable for in vivo microscopic imaging and because of recent advances in genome editing could be an ideal model organism for functional genetic studies of microglia development and function. In this study we used RNA sequencing to map the zebrafish microglia transcriptome, which should aid in elucidating basic microglia biology. We found that, many of the genes expressed in mouse microglia are also expressed in microglia in the zebrafish. In addition, by using RNA sequencing on microglia in the first stages following neuronal cell death, we showed that proliferation is a very first, and major transcriptional response of microglia to dying neurons. As several neurological diseases are caused by genetic defects of microglia for which the pathogenic mechanisms are currently unknown, it is important to better understand how microglia development and function are regulated genetically. Our gene expression datasets will be a useful tool to facilitate the elucidation of microglia genetic mechanisms relevant to brain development, aging and disease.
The majority of mouse genes for which we could not find homologs encode chemokines, chemokine receptors and genes involved in adaptive immunity (data not shown). It is thought that in general in fish species innate immunity is highly evolved, which may compensate for a less sophisticated adaptive immune system when compared with mammals (Lieschke and Trede, 2009). In addition, as chemokines and their receptors are amongst the most rapidly evolving gene clusters, it is difficult to compare them across species (DeVries et al., 2006; Nomiyama et al., 2003, 2008). Therefore it is not surprising to find non‐overlapping gene expression in these particular processes between zebrafish and mouse. However, the differences in species specific microglial gene expression between fish and mammals will be interesting to analyze as this may reveal genetic mechanisms underlying potential microglial adaptations to increasing brain complexity.
The specific identity and gene expression signature, which microglia adopt in the brain, is controlled by environmental factors in conjunction with developmental ontogeny (Gosselin et al., 2014; Lavin et al., 2014). Recently, it has been shown in mice that TGFβ is a main driver of microglia‐specific gene expression (Butovsky et al., 2014). Interestingly, upstream regulator prediction analysis on our zebrafish microglia gene expression data, predicted TGFb1 to be one of the main upstream regulators (Supporting Information Table S4). This indicates that beyond transcriptome similarities also the upstream regulation shows important similarities across species, and zebrafish may serve as a powerful model system to address the steps involved in acquiring the unique microglial identity.
Nitroreductase‐mediated ablation allowed us to distinguish the very first microglial response specifically to dying brain cells. We did not find increased expression of classes identified in previous studies on microglia gene‐expression in mouse models for chronic neurodegenerative diseases. Instead we identified extensive proliferation of microglia as a primary response to induced neuronal death, apparent at both the transcriptional and cellular level. One explanation for this difference could be that previous microglial gene expression studies were performed in models that show a more chronic, gradually developing neurodegenerative process, whereas we observed microglia immediately following neuronal cell death (Chiu et al., 2013; Holtman et al., 2015b). This is corroborated by microglia labeling at 24 h after treatment which already shows an increase in proliferative gene expression, when they have not all adopted an amoeboid morphology yet or increased in numbers (data not shown). Another possible explanation is that neurodegenerative diseases are associated with a combination of disease‐related cues including misfolded proteins and/or dying neurons causing altered immunological activity of microglia. In contrast, NTR‐mediated ablation causes a very clean insult consisting only of programmed cell death in the absence of other factors (Davison et al., 2007; van Ham, Kokel, and Peterson, 2012).
It is clear from several studies including the current that microglia express many genes involved in pathogen recognition, phagocytosis and lysosomal processing of phagocytic cargo (Butovsky et al., 2014; Chiu et al., 2013; Hickman et al., 2013; Zhang et al., 2014). Therefore, it may not be surprising that the first upregulated genes are not related to these basic microglia functions. Instead, microglia appear to prepare to counter an immense phagocytic task by locally increasing their numbers by self‐renewal as previously described in a mouse model for amyotrophic lateral sclerosis (Ajami et al., 2007). Although it is as yet unclear whether and when microglial proliferation is beneficial or detrimental in disease, it would be interesting to identify the cues that drive proliferation under these circumstances. In fact, strategies to manipulate microglia production are currently under investigation as potential treatments for patients suffering from neurodegenerative disease (Olmos‐Alonso et al., 2016).
In conclusion, with recent advances in scalable genome editing, we anticipate that our study will prove an important guide for functional genetic dissection of microglia activity and behavior, critical to understanding the role of microglia in physiology and brain disease. Our data provide novel insight into the microglial response to dying neurons in vivo and, together with the identified zebrafish microglia transcriptome, may accelerate the pace of elucidating molecular mechanisms involved in basic microglia function in vivo relevant to brain homeostasis.
Contributions
NO and TVH conceived the study, HVDL, NO, LK, TVH performed experiments, IH, NO, and TVH collected and analyzed data, EB and BJLE gave technical support and conceptual advice, NO and TVH wrote the manuscript with contributions from all authors.
Supporting information
Fig. S1. High‐throughput sequencing of the zebrafish microglia transcriptome. (A) Gating strategy for FACS‐sorting of GFP+ (microglia) and GFP− (brain) cell fractions. (B) PCA analysis showing GFP− brain cell samples (blue), GFP+ control microglia samples (green) and GFP+ activated microglia samples (magenta).
Fig. S2. Microglia become phagocytic upon neuronal cell death. (A) FACS data showing that many microglia are also positive for mCherry signal. mCherry is expressed in NTR‐expressing neurons. Therefore, increased numbers of mCherry positive microglia suggest increased phagocytic activity of microglia. (B) CPM values of genes involved in phagocytosis in activated and control microglia. No significant difference in the expression of genes involved in phagocytosis. Values in (B) represent means of at least three independent experiments. Error bars represent standard deviation.
Fig. S3. No increased expression of genes associated with infiltrating macrophages. CPM values of genes expected to increase in expression upon infiltration of monocyte‐derived macrophages. Values represent means of at least three independent experiments. Error bars represent standard deviation.
TABLE S1. Microglial Enriched Genes in the Zebrafish. (FDR < 0.01; LogFC >|2|)
TABLE S2. Zebrafish Orthologs for Mouse Microglia‐Specific Genes. (FDR < 0.01; LogFC > 2)
TABLE S3. Differentially Expressed Genes in Microglia Upon Neuronal Cell Death. (FDR < 0.01; LogFC >|2|)
TABLE S4. Upstream Regulators Zebrafish Microglia Transcriptome. 15 Most Significant Upstream Regulators Identified by the IPA Software
MOVIE S1. Proliferation of ApoE+ Microglia Intravital Imaging in 7 dpf Zebrafish Larvae Undergoing NTR‐Mediated Neuronal Cell Death, Showing Dividing Microglia in Apoe‐Driven Microglia GFP Expression
Supplemental material
Acknowledgment
We thank Annemarie Meijer (Leiden University), Nieske Brouwer, Jeroen Kuipers and Ben Giepmans (UMCG), for advice on the project, and Rob Willemsen (Erasmus MC) and Emma de Pater (Erasmus MC) for comments on the manuscript. We thank Ron Dirks (ZF‐screens) for help with RNA sequencing. The members of the Erasmus MC Optical Imaging Center (OIC) are acknowledged for assistance on microscopy. Grant sponsor: ZonMW; Grant number: 016.136.150; Grant sponsor: Marie Curie Career Integration; Grant number: 322368; Grant sponsor: Alzheimer Nederland fellowship; Grant number: WE.15‐2012‐01; Grant sponsor: UMCG Research School of Behavioural and Cognitive Neurosciences (BCN).
References
- Adolf B, Chapouton P, Lam CS, Topp S, Tannhauser B, Strahle U, Gotz M, Bally‐Cuif L. 2006. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev Biol 295:278–293. [DOI] [PubMed] [Google Scholar]
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. 2007. Local self‐renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci 10:1538–1543. [DOI] [PubMed] [Google Scholar]
- Burger A, Lindsay H, Felker A, Hess C, Anders C, Chiavacci E, Zaugg J, Weber LM, Catena R, Jinek M, MD Robinson, C Mosimann. 2016. Maximizing mutagenesis with solubilized CRISPR‐Cas9 ribonucleoprotein complexes. Development 143:2025–2037. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Z Fanek, L Liu, Z Chen, JD Rothstein, RM Ransohoff, SP Gygi, JP Antel, HL Weiner. 2014. Identification of a unique TGF‐beta‐dependent molecular and functional signature in microglia. Nat Neurosci 17:131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O'Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, RM Myers, T Maniatis. 2013. A neurodegeneration‐specific gene‐expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep 4:385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. 2005. ATP mediates rapid microglial response to local brain injury in vivo . Nat Neurosci 8:752–758. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. 2007. Transactivation from Gal4‐VP16 transgenic insertions for tissue‐specific cell labeling and ablation in zebrafish. Dev Biol 304:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries ME, Kelvin AA, Xu L, Ran L, Robinson J, Kelvin DJ. 2006. Defining the origins and evolution of the chemokine/chemokine receptor system. J Immunol 176:401–415. [DOI] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. 2011. mpeg1 promoter transgenes direct macrophage‐lineage expression in zebrafish. Blood 117:e49–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, IM Samokhvalov, M Merad. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, HR Rodewald. 2015. Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 518:547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, CK Glass. 2014. Environment drives selection and function of enhancers controlling tissue‐specific macrophage identities. Cell 159:1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch UK, Kettenmann H. 2007. Microglia: Active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 10:1387–1394. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. 2001. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M‐CSF receptor‐dependent invasive process. Dev Biol 238:274–288. [DOI] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. 2013. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci 16:1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtman IR, Noback M, Bijlsma M, Duong KN, van der Geest MA, Ketelaars PT, Brouwer N, Vainchtein ID, Eggen BJ, Boddeke HW. 2015a. Glia open access database (GOAD): A comprehensive gene expression encyclopedia of glia cells in health and disease. Glia 63:1495–1506. [DOI] [PubMed] [Google Scholar]
- Holtman IR, Raj DD, Miller JA, Schaafsma W, Yin Z, Brouwer N, Wes PD, Moller T, Orre M, Kamphuis W, EM Hol, EW Boddeke, BJ Eggen. 2015b. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: A co‐expression meta‐analysis. Acta Neuropathol Commun 3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja‐Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, CA Lemere, DJ Selkoe, B Stevens. 2016. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352:712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, S McLaren, I Sealy, M Caccamo, C Churcher, C Scott, JC Barrett, R Koch, GJ Rauch, S White, W Chow, B Kilian, LT Quintais, JA Guerra‐Assuncao, Y Zhou, Y Gu, J Yen, JH Vogel, T Eyre, S Redmond, R Banerjee, J Chi, B Fu, E Langley, SF Maguire, GK Laird, D Lloyd, E Kenyon, S Donaldson, H Sehra, J Almeida‐King, Loveland J, S Trevanion, M Jones, M Quail, D Willey, A Hunt, J Burton, S Sims, K McLay, B Plumb, J Davis, C Clee, K Oliver, R Clark, C Riddle, D Elliot, G Threadgold, G Harden, D Ware, S Begum, B Mortimore, G Kerry, P Heath, B Phillimore, A Tracey, N Corby, M Dunn, C Johnson, J Wood, S Clark, S Pelan, G Griffiths, M Smith, R Glithero, P Howden, N Barker, C Lloyd, C Stevens, J Harley, K Holt, G Panagiotidis, J Lovell, H Beasley, C Henderson, D Gordon, K Auger, D Wright, J Collins, C Raisen, L Dyer, K Leung, L Robertson, K Ambridge, D Leongamornlert, S McGuire, R Gilderthorp, C Griffiths, D Manthravadi, S Nichol, G Barker, S Whitehead, M Kay, J Brown, C Murnane, E Gray, M Humphries, N Sycamore, D Barker, D Saunders, J Wallis, A Babbage, S Hammond, M Mashreghi‐Mohammadi, L Barr, S Martin, P Wray, A Ellington, N Matthews, M Ellwood, R Woodmansey, G Clark, J Cooper, A Tromans, D Grafham, C Skuce, R Pandian, R Andrews, E Harrison, A Kimberley, J Garnett, N Fosker, R Hall, P Garner, D Kelly, C Bird, S Palmer, I Gehring, A Berger, CM Dooley, Z Ersan‐Urun, C Eser, H Geiger, M Geisler, L Karotki, A Kirn, J Konantz, M Konantz, M Oberlander, S Rudolph‐Geiger, M Teucke, C Lanz, G Raddatz, K Osoegawa, B Zhu, A Rapp, S Widaa, C Langford, F Yang, SC Schuster, NP Carter, J Harrow, Z Ning, J Herrero, SM Searle, A Enright, R Geisler, RH Plasterk, C Lee, M Westerfield, PJ de Jong, LI Zon, JH Postlethwait, C Nusslein‐Volhard, TJ Hubbard, H Roest Crollius, J Rogers, DL Stemple. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. 2013. Efficient CRISPR/Cas9 genome editing with low off‐target effects in zebrafish. Development 140:4982–4987. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. 2013. Efficient genome editing in zebrafish using a CRISPR‐Cas system. Nat Biotechnol 31:227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, DN Muller, B Luckow, T Brocker, K Debowski, G Fritz, G Opdenakker, A Diefenbach, K Biber, M Heikenwalder, F Geissmann, F Rosenbauer, M Prinz. 2013. Microglia emerge from erythromyeloid precursors via Pu.1‐ and Irf8‐dependent pathways. Nat Neurosci 16:273–280. [DOI] [PubMed] [Google Scholar]
- Kuipers J, Kalicharan RD, Wolters AH, van Ham TJ, Giepmans BN. 2016. Large‐scale scanning transmission electron microscopy (nanotomy) of healthy and injured zebrafish brain. J Vis Exp e53635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher‐Gonen R, David E, Keren‐Shaul H, Merad M, Jung S, Amit I. 2014. Tissue‐resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159:1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Proc GPD. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Du XF, Liu CS, Wen ZL, Du JL. 2012. Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo . Dev Cell 23:1189–1202. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Trede NS. 2009. Fish immunology. Curr Biol 19:R678–R682. [DOI] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, G Coppola, SA Sloan, CL Hsieh, CC Kim, EH Bigio, S Weintraub, MM Mesulam, R Rademakers, IR Mackenzie, WW Seeley, A Karydas, BL Miller, B Borroni, R Ghidoni, RV Farese Jr., JT Paz, BA Barres, EJ Huang. 2016. Progranulin deficiency promotes circuit‐specific synaptic pruning by microglia via complement activation. Cell 165:921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch‐Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben‐Yehuda H, David E, Zelada Gonzalez F, Perrin P, H Keren‐Shaul, M Gury, D Lara‐Astaiso, CA Thaiss, M Cohen, K Bahar Halpern, K Baruch, A Deczkowska, E Lorenzo‐Vivas, S Itzkovitz, E Elinav, MH Sieweke, M Schwartz, I Amit. 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353:aad8670. [DOI] [PubMed] [Google Scholar]
- Meireles AM, Shiau CE, Guenther CA, Sidik H, Kingsley DM, Talbot WS. 2014. The phosphate exporter xpr1b is required for differentiation of tissue‐resident macrophages. Cell Reports 8:1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner A, Mack M, Schmidt H, Bruck W, Djukic M, Zabel MD, Hille A, Priller J, Prinz M. 2009. CCR2+Ly‐6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain 132:2487–2500. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. 2005. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo . Science 308:1314–1318. [DOI] [PubMed] [Google Scholar]
- Nomiyama H, Egami K, Tanase S, Miura R, Hirakawa H, Kuhara S, Ogasawara J, Morishita S, Yoshie O, Kusuda J, K Hashimoto. 2003. Comparative DNA sequence analysis of mouse and human CC chemokine gene clusters. J Interferon Cytokine Res 23:37–45. [DOI] [PubMed] [Google Scholar]
- Nomiyama H, Hieshima K, Osada N, Kato‐Unoki Y, Otsuka‐Ono K, Takegawa S, Izawa T, Yoshizawa A, Kikuchi Y, Tanase S, R Miura, J Kusuda, M Nakao, O Yoshie. 2008. Extensive expansion and diversification of the chemokine gene family in zebrafish: Identification of a novel chemokine subfamily CX. BMC Genomics 9:222:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos‐Alonso A, Schetters ST, Sri S, Askew K, Mancuso R, Vargas‐Caballero M, Holscher C, Perry VH, Gomez‐Nicola D. 2016. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer's‐like pathology. Brain 139:891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhof N, Boddeke E, van Ham TJ. 2015. Immune cell dynamics in the CNS: Learning from the zebrafish. Glia 63:719–735. [DOI] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, L Tranebjaerg, Y Konttinen, L Peltonen. 2002. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet 71:656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri F, Nusslein‐Volhard C. 2008. Live imaging of neuronal degradation by microglia reveals a role for v0‐ATPase a1 in phagosomal fusion in vivo . Cell 133:916–927. [DOI] [PubMed] [Google Scholar]
- Prinz M, Priller J. 2014. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat Rev Neurosci 15:300–312. [DOI] [PubMed] [Google Scholar]
- Prinz M, Priller J, Sisodia SS, Ransohoff RM. 2011. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci 14:1227–1235. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, Casano AM, Henke K, Richter K, Peri F. 2015. The SLC7A7 transporter identifies microglial precursors prior to entry into the brain. Cell Rep 11:1008–1017. [DOI] [PubMed] [Google Scholar]
- Schaafsma W, Zhang X, van Zomeren KC, Jacobs S, Georgieva PB, Wolf SA, Kettenmann H, Janova H, Saiepour N, Hanisch UK, P Meerlo, PJ van den Elsen, N Brouwer, HW Boddeke, BJ Eggen. 2015. Long‐lasting pro‐inflammatory suppression of microglia by LPS‐preconditioning is mediated by RelB‐dependent epigenetic silencing. Brain Behav Immun 48:205–221. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. 2012. Microglia sculpt postnatal neural circuits in an activity and complement‐dependent manner. Neuron 74:691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Haass C. 2013. Genomic editing opens new avenues for zebrafish as a model for neurodegeneration. J Neurochem 127:461–470. [DOI] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. 2015. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 12:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Sidik H, Talbot WS. 2016. The Rag‐Ragulator complex regulates lysosome function and phagocytic flux in microglia. Cell Rep 14:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau CE, Kaufman Z, Meireles AM, Talbot WS. 2015. Differential requirement for irf8 in formation of embryonic and adult macrophages in zebrafish. PLoS One 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau CE, Monk KR, Joo W, Talbot WS. 2013. An anti‐inflammatory NOD‐like receptor is required for microglia development. Cell Rep 5:1342–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Mantovani A. 2012. Macrophage plasticity and polarization: In vivo veritas. J Clin Invest 122:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, A Sher, AM Litke, JD Lambris, SJ Smith, SW John, BA Barres. 2007. The classical complement cascade mediates CNS synapse elimination. Cell 131:1164–1178. [DOI] [PubMed] [Google Scholar]
- Svahn AJ, Graeber MB, Ellett F, Lieschke GJ, Rinkwitz S, Bennett MR, Becker TS. 2013. Development of ramified microglia from early macrophages in the zebrafish optic tectum. Dev Neurobiol 73:60–71. [DOI] [PubMed] [Google Scholar]
- Tremblay ME, Lowery RL, Majewska AK. 2010. Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8:e1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacaru AM, Unlu G, Spitzner M, Mione M, Knapik EW, Sadler KC. 2014. In vivo cell biology in zebrafish—Providing insights into vertebrate development and disease. J Cell Sci 127:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainchtein ID, Vinet J, Brouwer N, Brendecke S, Biagini G, Biber K, Boddeke HW, Eggen BJ. 2014. In acute experimental autoimmune encephalomyelitis, infiltrating macrophages are immune activated, whereas microglia remain immune suppressed. Glia 62:1724–1735. [DOI] [PubMed] [Google Scholar]
- van Ham TJ, Brady CA, Kalicharan RD, Oosterhof N, Kuipers J, Veenstra‐Algra A, Sjollema KA, Peterson RT, Kampinga HH, Giepmans BN. 2014. Intravital correlated microscopy reveals differential macrophage and microglial dynamics during resolution of neuroinflammation. Dis Model Mech 7:857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham TJ, Kokel D, Peterson RT. 2012. Apoptotic cells are cleared by directional migration and elmo1‐dependent macrophage engulfment. Curr Biol 22:830–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S. 2009. gplots: Various R programming tools for plotting data. R package Version 2.6.0. [Google Scholar]
- Xu J, Wang T, Wu Y, Jin W, Wen Z. 2016. Microglia colonization of developing zebrafish midbrain is promoted by apoptotic neuron and lysophosphatidylcholine. Dev Cell 38:214–222. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhu L, He S, Wu Y, Jin W, Yu T, Qu JY, Wen Z. 2015. Temporal‐spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev Cell 34:632–641. [DOI] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, H Ganesan, A Nino‐Castro, MR Mallmann, L Labzin, H Theis, M Kraut, M Beyer, E Latz, TC Freeman, T Ulas, JL Schultze. 2014. Transcriptome‐based network analysis reveals a spectrum model of human macrophage activation. Immunity 40:274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, S Deng, SA Liddelow, C Zhang, R Daneman, T Maniatis, BA Barres, JQ Wu. 2014. An RNA‐sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. High‐throughput sequencing of the zebrafish microglia transcriptome. (A) Gating strategy for FACS‐sorting of GFP+ (microglia) and GFP− (brain) cell fractions. (B) PCA analysis showing GFP− brain cell samples (blue), GFP+ control microglia samples (green) and GFP+ activated microglia samples (magenta).
Fig. S2. Microglia become phagocytic upon neuronal cell death. (A) FACS data showing that many microglia are also positive for mCherry signal. mCherry is expressed in NTR‐expressing neurons. Therefore, increased numbers of mCherry positive microglia suggest increased phagocytic activity of microglia. (B) CPM values of genes involved in phagocytosis in activated and control microglia. No significant difference in the expression of genes involved in phagocytosis. Values in (B) represent means of at least three independent experiments. Error bars represent standard deviation.
Fig. S3. No increased expression of genes associated with infiltrating macrophages. CPM values of genes expected to increase in expression upon infiltration of monocyte‐derived macrophages. Values represent means of at least three independent experiments. Error bars represent standard deviation.
TABLE S1. Microglial Enriched Genes in the Zebrafish. (FDR < 0.01; LogFC >|2|)
TABLE S2. Zebrafish Orthologs for Mouse Microglia‐Specific Genes. (FDR < 0.01; LogFC > 2)
TABLE S3. Differentially Expressed Genes in Microglia Upon Neuronal Cell Death. (FDR < 0.01; LogFC >|2|)
TABLE S4. Upstream Regulators Zebrafish Microglia Transcriptome. 15 Most Significant Upstream Regulators Identified by the IPA Software
MOVIE S1. Proliferation of ApoE+ Microglia Intravital Imaging in 7 dpf Zebrafish Larvae Undergoing NTR‐Mediated Neuronal Cell Death, Showing Dividing Microglia in Apoe‐Driven Microglia GFP Expression
Supplemental material


