Summary
Impairment of neurocognitive functioning is a common consequence of cerebral neoplasms and treatment, though considerable heterogeneity exists in the pattern and severity of problems across individuals and tumor types. While the influence of numerous clinical and patient characteristics have been documented, relatively little research has been devoted to understanding the influence of genetic variation upon neurocognitive outcomes in patients with brain tumors. This review highlights preliminary evidence associating genes from diverse pathways with risk of adverse neurocognitive outcomes in brain tumor patients, including genes specific to neuronal function and those involved in more systemic cellular regulation. Related literature involving other disease populations is also briefly surveyed, pointing to additional candidate genes. Methodological considerations are also discussed and the need for future research integrating novel investigative techniques is emphasized.
Keywords: brain tumor, cognition, genetics, genomics, individual differences
Introduction
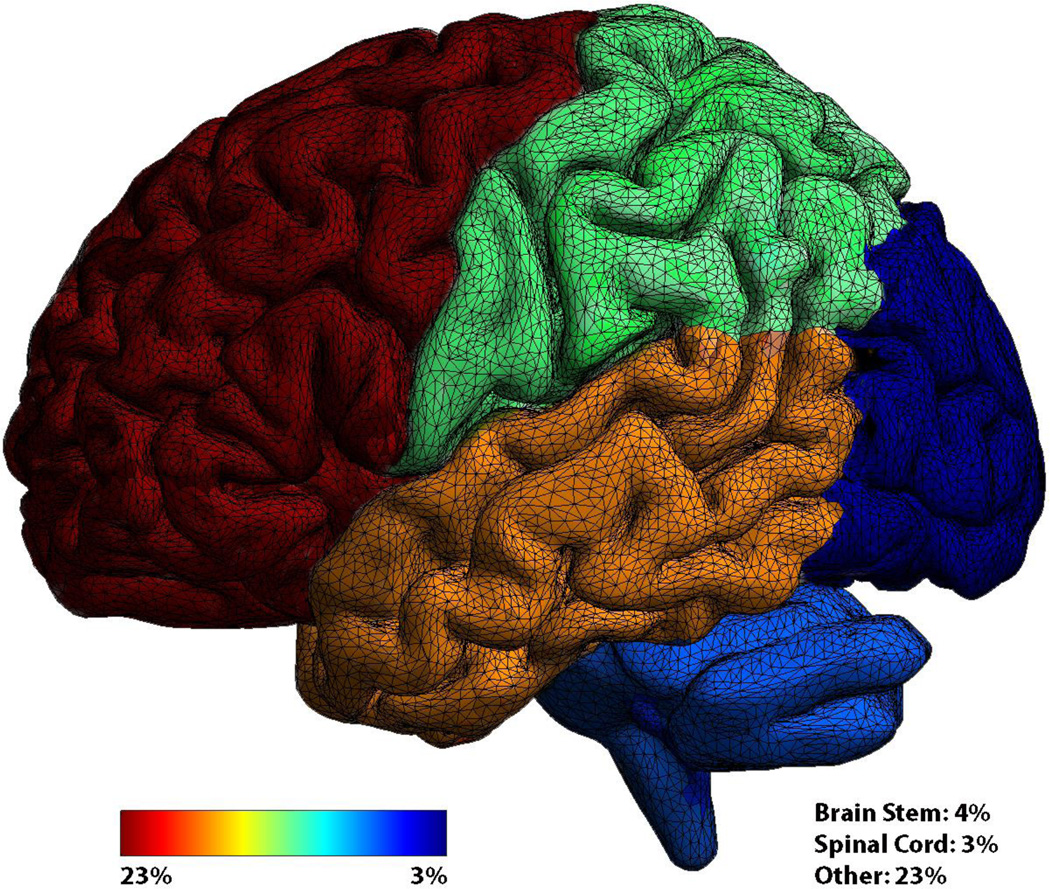
Primary brain tumors are relatively rare neoplasms, with an incidence of 21.4 cases per 100,000, comprising 1.4% of all cancers.1 These tumors may arise from the meninges or brain parenchyma, including neuronal and supportive glial cells. In adults, brain tumors are most frequently located in the frontal and temporal lobes (Figure 1), while childhood brain tumors are more often infratentorial. Much research has been devoted to glial tumors, or glioma, which represent nearly 30% of all cerebral tumors and 80% of all malignant brain tumors. Of all glioma, WHO grade IV glioblastoma represent the most common (54%) and most aggressive, with a median overall survival of 14.6 months despite surgical resection and standard adjuvant treatment consisting of concurrent chemoradiation with temozolomide, followed by single agent temozolomide.2 This is in stark contrast to low grade glioma (WHO grade II), which have a median overall survival of 13.8 years when treated with fractionated radiotherapy followed by procarbazine, lomustine, and vincristine. Despite survival differences across histological subtypes and pediatric versus adult populations,3 all cerebral tumors and treatments can have profound consequences upon patient neurocognitive functioning and health-related quality of life.
Figure 1.
Frequency of cerebral tumors by region. Frontal (23%), Temporal (17%), Parietal (11%), Occipital (3%), Cerebellum (5%).1
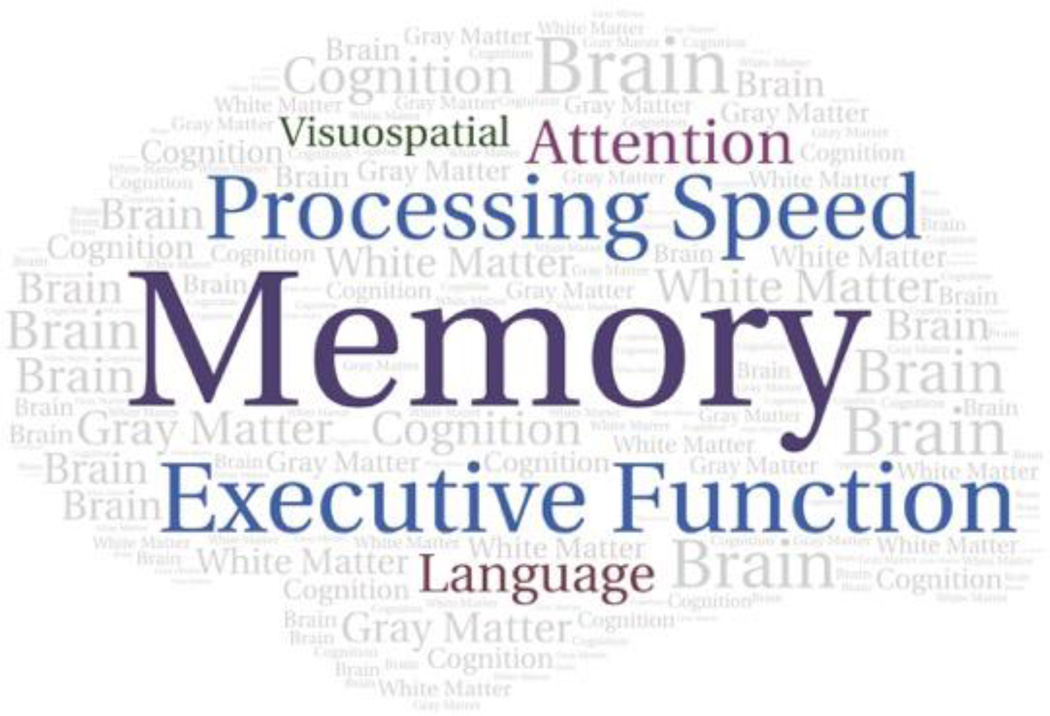
Most patients with brain tumors exhibit neurocognitive impairment at some point in the disease course.4,5 Impairment may be found within a variety of domains, with memory and executive functioning comprising the most frequently impacted (Figure 2). Impairment often presents early in the disease and can be progressive in nature, with significant consequences upon social and occupational functioning. Additionally, evidence indicates that neurocognitive impairment impacts health-related quality of life more than physical or other neurological symptoms.6 Nonetheless, much variability exists in the pattern and severity of impairment across individuals.
Figure 2.
Word cloud representing frequently impaired domains of neurocognitive functioning. Larger font indicates greater frequency of impairment observed on formal neuropsychological testing.4,5
Numerous clinical and demographic factors are known to contribute to inter-individual differences in neurocognitive outcomes in patients with brain tumors. These include lesion size and location,7 tumor histology,8 effects of treatment,9 and cognitive reserve or level of pre-illness functioning.10 Germline and tumor genetic factors are also believed to influence neurocognitive functioning in patients with glioma, both at the time of diagnosis and in response to treatment. Unfortunately, very little research has been devoted to understanding the contribution of genetic variability to neurocognitive functioning in patients with brain tumors. At present, behavioral genetics research in neuro-oncology is limited by the relatively few neuropsychological investigations assaying both genes and cognition, in addition to the fact that most brain tumors represent orphan diseases necessitating long study durations and/or multi-institutional collaborations to achieve adequate sample sizes. As such, the neurocognitive endophenotype of patients with brain tumors has not been well-characterized.11
This review summarizes the current adult and pediatric literature regarding genetic variability and neurocognition in patients with primary brain tumors, both at the individual and tumor-specific levels. To provide a broad perspective on this issue, we also incorporate selected findings from studies regarding genetics and neurocognitive functioning in non-central nervous system (non-CNS) cancer, non-cancer neuromedical populations, healthy individuals, and animal models. These studies point to potential molecular pathways that may contribute to variability in neurocognitive outcomes and provide direction for future research. Important methodological considerations are also discussed, including difficulties inherent to the study of neurocognitive functioning and contrasts between common genetics approaches. Of the genetic methods, focus is placed upon genome-wide association studies (GWAS) and targeted approaches. That is, GWAS provides an agnostic approach to discovering associations across genome-wide assays that may not have been hypothesized a priori, whereas targeted approaches (e.g., candidate variant, gene, or pathway) investigate specific variants thought to be more likely associated with the outcome of interest. Strengths and weaknesses of these methods are briefly addressed.
Individual Genetic Characteristics and Cognition
Adults
Decades of research have identified strong germline genetic influences on general cognition (e.g., intelligence), in addition to some genes implicated in more specific neurocognitive processes such as memory, processing speed, and attention/working memory.12,13 While general intelligence tends to be robust throughout adulthood, inter-individual differences in more specific neurocognitive processes tend to be large and increase with age. Explanations for this variability include gene-gene and gene-environment interactions, with some evidence suggesting that genetic effects upon neurocognition may become magnified in older adults.14 However, when investigating genetic influences upon neurocognitive functioning in medical populations, one must also consider the possibility that stochastic events (e.g., disease and treatment) may have greater effects on neurocognition than genetic variability, obscuring inter-individual differences in relationships between genes and neurocognition. Indeed, this has been observed in patients with Alzheimer’s disease, in which heritability of neurocognitive function is less than that of their unaffected family members.15 Nonetheless, it is almost certainly true that our genes interact with and have some influence upon the impact of cancer and treatment on brain activity and neurocognitive functioning.
As summarized by McAllister et al.,16 early efforts to identify genetic modulators of cancer survivors’ neurocognitive functioning employed targeted approaches focusing on single nucleotide polymorphisms (SNPs) of genes believed to have a direct effect on neurobiological processes. Perhaps the most studied gene associated with neurocognitive functioning is apolipoprotein E (ApoE), a glycoprotein important to neural repair.17 Associations between ApoE ε4 allele polymorphisms and increased risk of late-onset Alzheimer’s disease are well-established,18 and ApoE status is known to impact neurocognitive outcome in numerous other populations, including traumatic brain injury19 and stroke.20 Worse neurocognitive outcome in patients with at least one ApoE ε4 allele has been attributed to increased cerebral amyloid burden, reduced cholinergic function, dysregulation of neuronal repair mechanisms, and alterations in glucose and phospholipid metabolism.21 Evidence also suggests that ApoE genotype influences development of neurocognitive impairment following chemotherapy in patients with non-CNS cancers, such as breast carcinoma.22–24
Recently, associations between neurocognitive functioning and ApoE status have been documented in adults with brain tumors. Correa and colleagues investigated the association of ApoE genotype with neurocognitive performances in a large cohort of patients with brain tumors at approximately 4 years after completion of treatment.25 Over 90% of patients had previously received chemoradiation or chemotherapy alone. The authors found that those with at least one ε4 allele exhibited significantly lower performances on tests of learning, memory, and executive functioning than those without the allele. The authors also identified 9 other ApoE SNPs associated with attention, executive functioning, and memory. While informative, this investigation was not longitudinal and thus was not able to demonstrate that differences in trajectories of neurocognitive changes were attributable to ApoE genotype. Additionally, considerable heterogeneity in tumor grades and treatment received limit potential inferences regarding relationships between specific clinical characteristics and ApoE status.
Despite the limitations of the Correa et al. study,25 very similar findings have been reported by Ahles and colleagues in survivors of breast cancer and lymphoma.22,24 Additionally, both Correa et al.25 and Ahles and colleagues24 found that ApoE ε4 carriers with a history of cigarette smoking exhibited better neurocognitive performances than those without a smoking history. Accordingly, the observed relationships between ApoE status and neurocognitive function may not be specific to CNS-tumors but more generally relevant to the impact of cancer therapies. Specifically, the ε4 polymorphism may limit neuronal repair following radiation and/or chemotherapy, treatments known to contribute to vascular damage, oxidative stress and inflammation, white matter changes, and disruption of hippocampal neurogenesis. Further, smoking may attenuate some of the risk associated with the ε4 polymorphism, perhaps by counteracting cholinergic dysfunction known to be associated with the at-risk allele.14
In a more recent study published in abstract form, Correa et al. focused on other candidate genes with known importance to neuronal functioning, including catechol-O methyltransferase (COMT), brain-derived neurotrophic factor (BDNF), and dystrobrevin binding protein 1 (DTNBP1).26 While detailed results were not reported, the authors found associations between SNPs in these genes and aspects of attention, executive functioning, and memory in a sample of brain tumor patients, most of whom completed treatment with chemotherapy and/or radiation. Such relationships are consistent with prior studies investigating the role of these genes in the neurocognitive functioning of other populations, including chemotherapy treated breast cancer survivors.27
COMT is an enzyme involved in the degradation of catecholamine neurotransmitters, synaptic plasticity, and regulation of dopamine in the frontal lobes, all of which support attention and executive abilities.28 In chemotherapy-treated breast cancer patients, Val carriers of the COMT Val158Met variant perform more poorly on tests of attention, verbal fluency, and motor speed relative to COMT Met homozygotes.27 The COMT gene has also been implicated in the neurocognitive functioning of patients with other neurological and psychiatric conditions, including schizophrenia,29 Parkinson’s disease,30 and Alzheimer’s disease, though the Val allele is not always associated with worse neurocognitive outcomes.31 Indeed, Correa and colleagues reported that although Val homozygotes showed reduced working memory in their sample of brain tumor patients, Val/Met heterozygotes actually performed better on measures of executive function and memory.26 Accordingly, future work is needed to determine precisely what risks and/or protection are associated with the Val158Met variants of the COMT gene, and within which cognitive domains.
Neurotrophic factors, including BDNF, are proteins found in both central and peripheral nervous systems, which promote the growth and integrity of neurons and synapses.32 Alterations in the expression of BDNF can have a significant impact on neurocognition, with reductions of BDNF mRNA being found in Alzheimer’s disease and other neurodegenerative conditions.33 The DTNBP1 gene encodes dystrobrevin-binding protein, which is expressed downstream in the synaptic terminals of numerous cortical and subcortical regions.34 Polymorphisms of this gene have been implicated in susceptibility to schizophrenia and bipolar disorder, both of which are often accompanied by alterations of neurocognitive functioning.35 In light of the findings of Correa et al.,26 variation in these neuronal processes attributable to genetic polymorphisms may contribute to the pattern and severity of neurocognitive impairment in patients with brain tumors, though further investigation is clearly needed.
In contrast to the more selective and targeted approaches discussed so far, Liu and colleagues utilized a broader pathway-based approach in a recent study of glioma patients.36 Specifically, they examined 10,967 SNPs mapping to 580 genes involving inflammatory, metabolism, and DNA repair pathways in a large cohort of newly diagnosed glioma patients. They identified associations between SNPs in each of these pathways and the processing speed and executive function domains of neurocognitive functioning. They further reported a linear relationship between severity of neurocognitive impairment and the number of at-risk SNPs in these pathways, suggestive of potential gene-gene interactions.
Regarding the inflammatory pathway, Liu et al.36 found the strongest associations between processing speed and the insulin receptor substrate-1 (IRS-1) rs6725330 SNP, as well as executive functioning and the nitric oxide synthase 1 (NOS1) rs11611788 and interleukin-6 (IL-6) rs1912124 SNPs. These associations are consistent with known associations between inflammatory markers and neurocognitive functioning in other neuromedical populations. For instance, cytokines (e.g., interleukins and tumor necrosis factor alpha) help regulate several aspects of normal brain function, including neuronal inflammation, repair, and metabolism of neurotransmitters important to neurocognitive function.37 Levels of circulating cytokines have been implicated in the development of neurotoxicity and neurological disease.38 Evidence also suggests that reduced neurocognitive functioning in patients with non-CNS cancer is attributable in part to elevations of circulating pro-inflammatory markers, both prior to and following initiation of therapies.39 Additionally, carriers of the tumor necrosis alpha (TNF-alpha) 308A promoter allele show significantly lower mean age at onset of Alzheimer’s disease compared to non-carriers,40 and genetic deletion of the p55 or p75 TNF-alpha receptor has been linked to exacerbation of neurobehavioral deficits and neural damage in animal models of traumatic brain injury.41 Accordingly, these genes may also be important to the development of neurocognitive impairment in patients with brain tumors. However, relationships between genetics and cytokines are complex and the presence of at-risk alleles does not necessarily entail elevations in pro-inflammatory markers.42 Accordingly, future work incorporating nongenetic factors and gene-environment interactions are needed.
In addition to the inflammatory pathway, Liu et al.36 found associations between DNA repair pathway genes and neurocognition in their brain tumor population. Specifically, excision repair cross-complementation group 4 (ERCC4) rs1573638 was associated with processing speed, and polymerase (DNA directed) epsilon catalytic subunit (POLE) rs5744761 was associated with executive functioning. These genes encode proteins and enzymes involved in DNA repair and combination. Interestingly, known molecular effects of chemotherapeutic agents include causing DNA strand breaks and shortening of telomere length.43,44 Taken together, these findings suggest that certain individuals may be particularly vulnerable to inflammatory cascades with reduced DNA repair capabilities, increasing susceptibility to the effects chemotherapy and brain tumor itself upon neurocognitive functioning.
Other candidate genes
Through both GWAS and targeted approaches, a number of other genes have been implicated in the neurocognitive functioning of healthy adults that may be relevant to the study of neurocognition in patients with brain tumors. For instance, neurotransmitter receptor genes are known to play important roles in facilitating neurocognition. The dopamine receptor d2 (DRD2) gene is involved in dopamine signaling, and variation in its expression may impact neurocognitive functioning in healthy adults.34 For instance, carriers of the DRD2 Taq1 allele show worse non-verbal intellectual functioning,45 memory,46 and executive functioning.47 Various SNPs of the muscarinic acetylcholine receptor M2 (CHRM2) gene have also been associated with aspects of intelligence in healthy adults, though effect sizes are relatively small.34 The CHRM2 gene encodes the cholinergic/muscarinic type 2 autoreceptor, which modulates neurotransmitter release. Given the importance of acetylcholine to memory functioning,48 which is frequently impaired following cancer diagnosis and treatment, this gene may play an important role in the neurocognitive function of brain tumor patients. Another candidate is the 5-hydroxytryptamine transporter (5-HTT) gene involved in modulating re-uptake of serotonin from the synapse. Individuals with the s-allele exhibit reduced performance on list learning and memory tasks,49 the same measures on which patients with brain tumors frequently show impairment. While intuitively promising, future work is needed to extend these findings to brain tumor populations.
In addition to the pathways noted above, genes directly involved in cell cycle regulation may contribute to susceptibility to neurocognitive impairment in patients with cerebral tumors. For instance, the cyclin D1 (CD1) gene is known to moderate neuronal damage following traumatic brain injury.50 Specifically, ablation of CD1 appears to cause reduced cell cycle activation, attenuated neurodegeneration and hippocampal cell loss, smaller lesion size, and lower cortical microglial activation in mice following brain trauma. Given that brain tumor treatment often involves acquired damage to healthy tissue, CD1 might also contribute to the etiology of impaired neurocognitive functioning in patients with central nervous system neoplasms.
Pediatrics
As many as half of pediatric brain tumor patients develop progressive neurocognitive deficits.51,52 Many of these children also fail to fully attain their academic and professional potential as they do not develop at the same rate as their peers.53 Specifically, IQ scores decline an average of 2.2 points per year, with younger patients showing a more immediate degradation than older children.53,54 In addition to age of onset, cranial radiation dose and female gender are also associated with increased risk for these adverse neurocognitive outcomes.51,52, 54–59 Despite these known risk factors, a great amount of inter-individual difference exists in the development of neurocognitive impairment in pediatric brain tumor populations. Increasing evidence suggests that such differences may relate to host germline variability, moderating the biological consequences of cancer and treatment.60
Most of the literature specifically examining neurocognitive functioning in pediatric brain tumor patients involves children with medulloblastoma. In addition to radiation therapy and neurosurgery, these tumors are often treated with alkylating agents, nitrosoureas, and platinum containing compounds.61 While use of these chemotherapies have contributed to extended survival, they are often accompanied by considerable toxicities, including peripheral neuropathy, renal failure, cardiomyopathy, hearing loss, and neurocognitive impairment.38,62,63 Evidence suggests that development of such toxicities may depend in part upon genetic polymorphisms of genes of the glutathione S-transferase (GST) enzyme family.64,65 Genes of this family encode GSTs, isoenzymes important to the catalysis of detoxification of alkylating and platinum containing agents, as well as oxygen free radicals that accumulate with chemotherapy and radiation treatment. Barahmani and colleagues investigated relationships between GST genes and development of toxicities, including impairment on intelligence testing, in a relatively small cohort of children with medulloblastoma.66 Most patients were treated with craniospinal radiation and high-dose platinum containing agents. Patients with at least one null GSTM1 or GSTT1 genotype had greater risk of toxicities and exhibited lower performances on tests of general, verbal, and non-verbal intelligence.
Bracket and colleagues also investigated the role of enzyme polymorphisms in neurocognitive outcomes of medulloblastoma survivors.67 They found that homozygous GSTM1 deletion was related to greater anxiety, depression, and global distress compared to those with the non-null genotypes. Additionally, survivors reported significantly greater problems with processing efficiency and memory compared to their healthy siblings. However, self-reported cognitive functioning did not vary by GSTM1 genotype. While this contrasts with the findings of Barahmani et al.,66 differences between studies may relate to variation in neurocognitive assessment techniques (i.e., performance vs. self-report measures) and domains assessed (i.e., global vs. specific processes).. Accordingly, it remains unclear if GST genes relate to more domain-specific functions in this population, such as memory, visuospatial functioning, and executive abilities.
Similar to findings in adults with brain tumors, polymorphisms of the COMT gene have been implicated in the neurocognitive functioning of pediatric brain tumor populations. In a study of non-medulloblastoma pediatric brain tumor survivors, those with the Met allele of the COMT Val158Met variant showed better performance on a self-ordered verbal task assessing executive functions compared to those who were homozygous for the Val allele.68 This is in line with the role of COMT in regulating frontal lobe dopamine activity and facilitation of attention and executive abilities as previously discussed.
Other candidate genes
A majority of childhood non-CNS cancer survivors will have one or more late effects of therapy, including neurocognitive impairment. Most studies of neurocognitive functioning and non-CNS pediatric cancer focus on leukemia. In a study of children diagnosed with acute lymphoblastic leukemia (ALL), Krajinovic and colleagues found that nitric oxide synthase 3 (NOS3) 894T homozygosity was associated with longitudinal decline in intellectual functioning only among patients treated with cranial radiation.69 This provides further evidence implicating inflammatory pathways and gene-environment interactions in the development of neurocognitive decline related to cancer and treatment, particularly regarding neurotoxicity associated with radiotherapy.
The folate pathway has also been implicated in the neurocognitive functioning of childhood ALL survivors.70 After adjusting for age at diagnosis and gender, Krull and colleagues reported that patients with the methylene tetrahydrofolate reductase (MTHFR) 1298AC or 1298CC genotype were 7.4 times more likely to develop attention deficit hyperactivity disorder than those with the 1298AA variant. In an expanded sample of ALL survivors from the same center,71 survivors with the MTHFR 1298AC/CC genotypes showed reduced executive functioning compared to those with the MTHFR AA genotype. Further, survivors with enhancer region of the thymidylate synthase (TSER) 2R/3R and 3R/3R genotypes also exhibited worse executive function performances than survivors with the TSER 2R/2R genotypes. Gene-gene interactions were also evident, as survivors with adverse alleles in six or more folate pathway variants demonstrated greater reductions in processing speed and executive functioning compared to those with less than six adverse alleles. Interestingly, folate deficiency has been linked to impaired brain development and intellectual deficiency in children,72 as well as age-related cognitive decline in older adults.73 Animal models provide some insight into the mechanism by which variation in folate regulation may impact processes underlying neurocognitive functions. Specifically, reduced folate levels appear to elevate plasma homocysteine levels, alter brain monoamine metabolism, and inhibit hippocampal neurogenesis.74 Accordingly, abnormalities in folate pathways that contribute to oxidative stress may impact neurocognition following cancer treatment.
Further evidence of the importance of oxidative stress pathways comes from another investigation by Krull and colleagues regarding genetic influences upon neurocognitive functioning in survivors of childhood ALL.75 Survivors with the A2756G polymorphism in the 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR) gene were more likely to have deficits in attentiveness and response speed. MTR encodes methionine synthase, which converts homocysteine to methionine. Dysregulation of the homocysteine-methionine cycle can lead to an excess of homocysteine, or hyperhomocysteinenemia, which can cause endothelial cell damage, disruption of the blood-brain-barrier, and ischemic injury.76,77 Additional genes implicated in neurocognition following childhood ALL, include GST, COMT, ApoE and NOS discussed above, as well as polymorphisms of the monoamine oxidase A gene (MAOA), involved in the regulation of amine neurotransmitters. Patients with at-risk polymorphisms of these genes tend to exhibit greater impairment following treatment of childhood ALL, particularly with attention and concentration.78 Accordingly, similar pathways and genes appear to contribute to susceptibility of neurocognitive impairment in both children and adults following cancer and treatment—namely, those involved with metabolic regulation, neurotransmitter synthesis and degradation, oxidative stress, inflammatory cascades, neural repair, and DNA maintenance.
Brain Tumor Genetics and Cognition
Evidence suggests that higher grade tumors entail more aggressive growth and greater neurocognitive impairment.8 However, advances in molecular profiling of brain tumors indicate that tumor genetic markers are more accurate indicators of growth kinetics than histological grading.79 While little work has investigated relationships between brain tumor genetics and neurocognition, preliminary evidence suggests that patients with malignant glioma harboring mutation of the isocitrate dehydrogenase one gene (IDH1) exhibit less impairment than their wild type counterparts.80 This has been attributed to the more aggressive proliferation characteristic of the IDH1 wild type tumors, allowing for less neuroplastic cerebral reorganization of function. While this work is promising, numerous other tumor markers exist with known impact on prognosis, including 1p/19q co-deletion, O6-Methylguanine-DNA methyltransferase (MGMT) promoter methylation, alpha-thalassemia/mental retardation syndrome X-linked (ATRX) loss, among others. Accordingly, these markers may interact and share relationships with neurocognitive outcomes in patients with primary brain tumors. Future work is needed that includes multiple genetic pathways that may more accurately determine prognosis and risk of neurocognitive impairment.
Methodological Considerations
Study Design
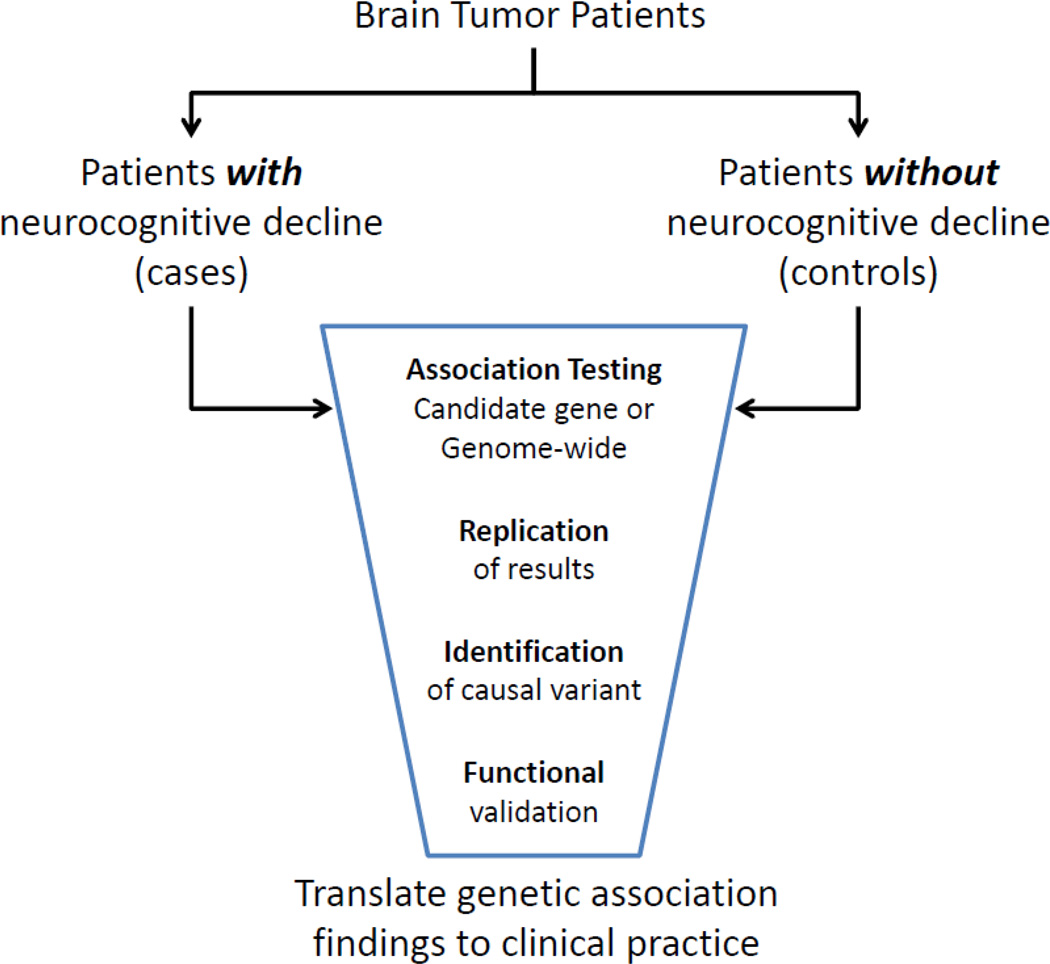
Case-control studies are one of the most common epidemiologic study designs used in genetic association studies (Figure 3). In a case-control study where SNPs are evaluated as genetic risk factors, the odds of having a particular SNP are compared between cases and controls. This study design is particularly useful when investigating rare outcomes. In studies assessing neurocognitive functioning in brain tumor patients one can still utilize the case-control design despite all of the subjects having cancer. In this scenario, cases refer to those with an adverse treatment-related effect (neurocognitive impairment), whereas the controls are those without the effect. This should not be confused with the “case-only” design used in the assessment of gene-gene and gene-environment interactions, in which all the subjects have the outcome of interest.
Figure 3.
Flow diagram outlining the case-control genetic association method.
Improving upon the case-control design are longitudinal cohort studies. While particularly useful in the context of examining adverse outcomes in patients with brain tumors, they have been less frequently employed secondary to logistical and practical limitations inherent to closely following patients with a rare disease over time. Nonetheless, these studies enable the definitive demonstration of a temporal relationship between risk factors and outcome. Additionally, as time-to-event data are collected as part of cohort studies, one can answer questions about which SNPs are most likely to contribute to the development or resolution of adverse effects and at what rate compared to other SNPs.
Measuring Neurocognitive Outcomes
As the biological gap between genes and outcomes of interest widen (e.g., cellular function versus behavior or neurocognitive function) it becomes increasingly difficult to detect genetic effects. Attempts to optimize signal to noise are thus critical in such studies. Utilizing tests of neurocognitive function with adequate psychometric properties including good test-retest reliability and sensitivity to the outcome of interest is of paramount importance. Fortunately, the use of mental status screening instruments such as the Mini Mental State Examination (MMSE) in brain tumor trials has given way to incorporation of more comprehensive, psychometrically robust, and sensitive neuropsychological tests,81,82 allowing for more powerful correlative biological questions to be asked in these studies. There has also been some preliminary consensus on a core set of neuropsychological tests83–85 that are being increasingly incorporated into trials by a diverse array of investigators, allowing data to be more readily pooled across studies to answer gene-neurocognition association questions. This is essential as such questions frequently require larger sample sizes that are difficult to obtain in rare disease populations where neuropsychological expertise is less readily available.
Another complicating issue in the study of genetic variability and neurocognitive functioning in brain tumor patients involves collapsing histopathological groups. That is, many of the extant studies of genetic influences upon cognition include patients with tumors of varying histologies, despite the fact that neurocognitive functioning tends to vary from low to high grade.8 Fortunately, modern brain tumor clinical trials are now routinely gentotyping both tumor and patient while incorporating neurocognitive outcomes, which will enable the longitudinal study of gene-neurocognition associations in more homogeneous clinical samples of large size. Studies associating genes and neurocognition in brain tumor patients will also need to address several other variables that may cloud the interpretation of such association studies, including the potential impact of concomitant medications that can impact neurocognitive function (e.g., steroids, psychostimulants, psychotropics, pain meds, etc.), the timing and nature of tumor progression, and the impact of multiple lines of therapy many brain tumor patients are exposed to during the course of their illness.
Statistical Models
There are several considerations when determining the most appropriate statistical model for genetic association studies of treatment-related adverse effects. Two important factors are the study design (e.g., case-control versus cohort discussed above) and how the outcome of interest is measured. For instance, when evaluating a measurement where many values are possible within a range (i.e., a continuous variable) an investigator may opt to use a linear regression model. However, one could also define the adverse outcome in a binary fashion (i.e., present vs. absent) and use logistic regression to determine the association between candidate SNPs and the outcome. If investigators utilize a cohort study design when assessing treatment-related or other adverse outcomes and have time-to-event data, the model most commonly used is Cox proportional hazards regression.
As many investigators opt to build predictive statistical models of adverse outcomes in adults and children treated for cancer, it is important to evaluate the performance and predictive ability of those models. This is often done by constructing receiver operating characteristic (ROC) curves and calculating the area under the curve (AUC) statistic. However, the ROC curve may not be sensitive to differences in probabilities between models. In light of this limitation, new metrics are being used to compare nested models. For instance, two newer approaches for assessing the predictive ability of SNPs are the integrated discrimination improvement (IDI) and the net reclassification improvement (NRI) approaches.86 While many additional options are available suited to various behavioral genetics research questions, the methods and designs briefly discussed represent some of the more commonly used statistical methods and are meant to constitute only a cursory introduction to the field.
Selecting Genes and SNPs of Interest
A key question in conducting a genetic association study of treatment-related effects is deciding what SNPs to include in the analysis. Early on, investigators examined candidate SNPs with a known or putative function of interest—so-called targeted approaches as mentioned earlier. For instance, as neurocognitive impairment in childhood ALL survivors is potentially mediated by folate depletion and homocysteine elevation following methotrexate treatment, a study discussed above focused exclusively on SNPs of genes known to alter folate levels.71 However, a more extensive approach could have been chosen, in which several SNPs of a number of genes were selected across broader pathways of interest (e.g., drug metabolism or DNA repair) to gain greater coverage.
With more recent advances in DNA-sparing genotyping methods, GWAS have been used in the context of genetic epidemiology allowing a more “agnostic” evaluation of SNPs across the entire human genome. The number of SNPs in a GWAS ranges from 100,000 to approximately 5 million on newer SNP arrays. This raises obvious questions related to multiple comparisons; however, these studies are particularly valuable for generating hypotheses. Furthermore, these investigations utilize a much more stringent p-value for determining statistical significance (typically p < 10−8). Finally, with newer technologies that allow inexpensive whole exome or genome sequencing, researchers are now able to compare the entire DNA sequence of individuals with an adverse outcome to those without the outcome. It is hoped that this information will be valuable in elucidating new variants associated with treatment-related effects, including changes in neurocognitive functioning.
Validation of Findings
Another important consideration in conducting genetic association studies is the replication and validation of findings in independent populations. This can be difficult when studies are small and all available individuals must be pooled for a study, as is frequently the case in brain tumor investigations. Another option is using cross-validation, where a single sample is broken into a “training” (or discovery) subset and a “validation” subset. However, the power to detect genetic associations is influenced by several factors, including sample size, the minor allele frequency, and the strength of the association. While brain tumor studies often suffer from small sample sizes, strong associations have been documented,67,71 suggesting that larger studies may not be needed to discover clinically significant findings. However, regardless of the sample size, if the genetic variant under investigation is rare (i.e., has a low minor allele frequency), very large sample sizes would be required. Investigators should be cognizant of this issue when examining genetic effects in their clinical populations of limited size. Additionally, GWAS tends to require larger sample sizes and may not detect biologically relevant variants, leaving much functional work to be done to determine the exact variant leading to the effect on biologic activity. On the other hand, targeted approaches may increase power, though this is often accompanied by lost opportunity to examine previously unstudied variants.
A final consideration in genetic association studies, as with all epidemiologic studies, is accounting for bias and confounding factors. An important source of bias in genetic association studies is referred to as population stratification, which can lead to a false association or the masking of a genetic effect. Specially, population stratification arises when subgroups within a population (e.g., different race/ethnicity or age groups) have different genetic profiles and/or different frequencies of the outcome of interest.87 However, the use of ancestry informative markers (AIMs) and other techniques (e.g., principal components analysis) can better identify an individual’s ancestry in order to control for these effects. Another source of bias is in defining relatively homogeneous populations in terms of diagnosis and treatment. It is important to consider all of these factors not only at the analysis phase, but also during the design of a genetic association study.
Future Directions
While there is a growing body of work concerning both gene-gene and gene-environment interactions in other cognate fields of neuroscience, such studies remain rare in patients with brain tumors. A detailed discussion of the methods being used in this area and the caveats specific to conducting interaction analyses are outside the scope of this review. However, readers are referred to a recent review by Dick et al. for suggestions on how best to interpret the results from such studies and recommendations on providing more robust data to this conversation with brain tumor populations.88 The effects of epigenetic mechanisms (e.g., changes to the DNA that are not represented by changes in the genetic sequence), such as methylation, micro-RNAs, or histone modifications, are also being increasingly recognized as important factors in many disorders, including neurocognition dysfunction. An important factor with epigenetic changes is that, once identified, they can potentially be modified.89 However, these effects can be more difficult to detect as they are often tissue or cell specific and can change over time or with different exposures, such as nutrition or medications.
Conclusion
This review integrates the current corpus of literature regarding genetic influences upon neurocognition in adult and pediatric patients with brain tumors. The studies reviewed point to a number of candidate genes within varying pathways, including those that modulate cellular metabolism, DNA maintenance and repair, neural growth and repair, neurotransmitter synthesis and degradation, as well as oxidative stress and inflammation. In addition to germline mutations, evidence also suggests that tumor genetic variation may play an important role in the development of neurocognitive impairment in patients with cerebral neoplasms. Nonetheless, the paucity of existing research regarding genetic variation and neurocognition in patients with brain tumors is striking, particularly when contrasted with the vast behavioral genetics literature accumulating regarding non-central nervous system cancer, neurological, and psychiatric populations. Further studies are obviously needed, not only to validate preliminary work but also to explore potential targets identified from cognate populations with novel methodologies.
Identification of genetic markers that accurately predict development of neurocognitive impairment related to brain tumor and treatment are necessary for two primary reasons: 1) to better tailor cancer treatment for maximal antineoplastic effect while simultaneously minimizing risk of neurocognitive decline, and 2) to identify potential targets for the development of interventions to ameliorate or prevent neurocognitive problems.90,91 For instance, those at risk for neurocognitive impairment could potentially receive modified radiation schedules or chemotherapy regimens or be offered cognitive remediation, medication, and behavioral modification for intervention or prevention of impairment. Additionally, populations with minimal genetic risk may be able to receive intensified therapy in the hopes of better tumor control without increased toxicity. In other words, better understanding of the modulatory effect of genetic background on brain tumor and treatment-related alterations of neurocognitive functioning may lead to further opportunities to personalize cancer therapy through risk adapted therapy or pharmacogenetically-determined therapy selection, prognostication of neurocognition, triage of patients for surveillance purposes, and identification of targets for the development of therapeutics.
Search Strategy and Selection Criteria
References for the Adult section of this review were identified through searches of PubMed with the search terms “brain tumor” AND "cognitive” or “neuropsychology” AND “genetics” or “genes” from 1990 until May 2015. Given limited results, the “brain tumor” term was omitted to obtain articles addressing cognition and genetics more generally. References for the pediatrics section of this review were identified through searches of PubMed with the search terms "childhood" or “pediatric” AND “brain tumor” or “medulloblastoma” AND "cognitive” or “neuropsychology” AND “genetics” or “genes” from 1990 until May 2015. Results were again limited and the search strategy was expanded to include the broader term “cancer” over the more specific “brain tumor” or “medulloblastoma” terms. Articles were also identified through Google Scholar searches and review of the reference sections of obtained articles. Only papers published in English were reviewed. The final reference list was generated on the basis of originality and relevance to the broad scope of this review.
Table 1.
Studies investigating relationships between genetics and neurocognitive functioning in adult and pediatric CNS and other cancer populations
| Authors | Population and Design | Genotyping | Findings |
|---|---|---|---|
| Correa et al.25 |
|
|
|
| Correa et al.26 (Abstract) |
|
|
|
| Liu et al.36 |
|
|
|
| Wefel et al.80 (Abstract) |
|
|
|
| Ahles et al.22 |
|
|
|
| Ahles et al.24 |
|
|
|
| Koleck et al.23 |
|
|
|
| Small et al.27 |
|
|
|
| Bahramani et al.66 |
|
|
|
| Brackett et al.67 |
|
|
|
| Howarth et al.68 |
|
|
|
| Krajinovic et al.69 |
|
|
|
| Krull et al.70 |
|
|
|
| Kamdar et al.71 |
|
|
|
| Krull et al.75 |
|
|
|
| Cole et al.78 |
|
|
|
ATTN = attention, COMP = comprehension, CONST = construction, EF = executive function, FSIQ = full scale IQ, IQ = intelligence quotient, LRN = learning, MEM = memory, MOT = motor, NCF = neurocognitive function, PIQ = performance IQ, PS = processing speed, WM = working memory, VF = verbal fluency, VIQ = verbal IQ.
Acknowledgments
This publication was supported by the National Institute of Nursing Research of the National Institutes of Health under award number R01NR014195 (J.S.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank Shelli Kesler, Ph.D., for her assistance with creation of the figures.
All authors have read and approved the manuscript. The manuscript has not been previously published in whole or in part, or submitted elsewhere for review.
Declaration of interests
The authors report receipt of funds from F. Hoffman La Roche (J.S.W.) and Angiochem (J.S.W.), as well as personal fees from AbbVie (J.S.W.) and Genentech (J.S.W.).
Footnotes
Contributors
All authors contributed to the conception and writing of the manuscript.
References
- 1.Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(suppl 4):iv1–iv63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran B, Rosenthal M. Survival comparison between glioblastoma multiforme and other incurable cancers. J Clin Neurosci. 2010;17(4):417–421. doi: 10.1016/j.jocn.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Pignatti F, van den Bent M, Curran D, et al. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20(8):2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- 4.Habets EJ, Kloet A, Walchenbach R, Vecht CJ, Klein M, Taphoorn MJ. Tumour and surgery effects on cognitive functioning in high-grade glioma patients. Acta Neurochir. 2014;156(8):1451–1459. doi: 10.1007/s00701-014-2115-8. [DOI] [PubMed] [Google Scholar]
- 5.Tucha O, Smely C, Preier M, Lange KW. Cognitive deficits before treatment among patients with brain tumors. Neurosurgery. 2000;47(2):324–334. doi: 10.1097/00006123-200008000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Henriksson R, Asklund T, Poulsen HS. Impact of therapy on quality of life, neurocognitive function and their correlates in glioblastoma multiforme: a review. J Neurooncol. 2011;104(3):639–646. doi: 10.1007/s11060-011-0565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheibel RS, Meyers CA, Levin VA. Cognitive dysfunction following surgery for intracerebral glioma: influence of histopathology, lesion location, and treatment. J Neurooncol. 1996;30(1):61–69. doi: 10.1007/BF00177444. [DOI] [PubMed] [Google Scholar]
- 8.Noll KR, Sullaway C, Ziu M, Weinberg JS, Wefel JS. Relationships between tumor grade and neurocognitive functioning in patients with glioma of the left temporal lobe prior to surgical resection. Neuro Oncol. 2015;17(4):580–587. doi: 10.1093/neuonc/nou233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu AS, Witgert ME, Lang FF, et al. Neurocognitive function before and after surgery for insular gliomas: Clinical article. J Neurosurg. 2011;115(6):1115–1125. doi: 10.3171/2011.8.JNS11488. [DOI] [PubMed] [Google Scholar]
- 10.Dwan TM, Ownsworth T, Chambers S, Walker DG, Shum DH. Neuropsychological assessment of individuals with brain tumor: comparison of approaches used in the classification of impairment. Front Oncol. 2015;5 doi: 10.3389/fonc.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman II, Gould TD. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. Am J Psychiatry. 2003;160(4):636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 12.Deary IJ, Spinath FM, Bates TC. Genetics of intelligence. Eur J Human Genet. 2006;14(6):690–700. doi: 10.1038/sj.ejhg.5201588. [DOI] [PubMed] [Google Scholar]
- 13.Vogler C, Gschwind L, Coynel D, et al. Substantial SNP-based heritability estimates for working memory performance. Transl Psychiatry. 2014;4(9):e438. doi: 10.1038/tp.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papenberg G, Lindenberger U, Bäckman L. Aging-related magnification of genetic effects on cognitive and brain integrity. Trends Cogn Sci. 2015;19(9):506–514. doi: 10.1016/j.tics.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Wilson RS, Barral S, Lee JH, et al. Heritability of different forms of memory in the Late Onset Alzheimer’s Disease Family Study. J Alzheimers Dis. 2011;23(2):249. doi: 10.3233/JAD-2010-101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAllister TW, Ahles TA, Saykin AJ, et al. Cognitive effects of cytotoxic cancer chemotherapy: predisposing risk factors and potential treatments. Curr Psychiatry Rep. 2004;6(5):364–371. doi: 10.1007/s11920-004-0023-y. [DOI] [PubMed] [Google Scholar]
- 17.Mahley RW, Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012;76(5):871–885. doi: 10.1016/j.neuron.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genin E, Hannequin D, Wallon D, et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry. 2011;16(9):903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandy S, DeKosky ST. APOE ε4 status and traumatic brain injury on the gridiron or the battlefield. Sci Transl Med. 2012;(4):134ed4. doi: 10.1126/scitranslmed.3004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagle J, Farner L, Flekkøy K, et al. Association between ApoE epsilon4 and cognitive impairment after stroke. Dement Geriatr Cogn Disord. 2008;27(6):525–533. doi: 10.1159/000223230. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Boyle PA, Leurgans S, Schneider JA, Bennett DA. Disentangling the effects of age and APOE on neuropathology and late life cognitive decline. Neurobiol Aging. 2014;35(4):819–826. doi: 10.1016/j.neurobiolaging.2013.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahles TA, Saykin AJ, Noll WW, et al. The relationship of APOE genotype to neuropsychological performance in long-term cancer survivors treated with standard dose chemotherapy. Psychooncology. 2003;12(6):612–619. doi: 10.1002/pon.742. [DOI] [PubMed] [Google Scholar]
- 23.Koleck TA, Bender CM, Sereika SM, et al. Apolipoprotein E genotype and cognitive function in postmenopausal women with early-stage breast cancer. Oncol Nurs Forum. 2014:E313–E325. doi: 10.1188/14.ONF.E313-E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahles TA, Li Y, McDonald BC, et al. Longitudinal assessment of cognitive changes associated with adjuvant treatment for breast cancer: the impact of APOE and smoking. Psychooncology. 2014;23(12):1382–1390. doi: 10.1002/pon.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Correa DD, Satagopan J, Baser RE, et al. APOE polymorphisms and cognitive functions in patients with brain tumors. Neurology. 2014;83(4):320–327. doi: 10.1212/WNL.0000000000000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Correa D, Satagopan J, Baser R, Cheung K, DeAngelis L, Orlow I. NC-03 Polymorphisms in the COMT, BDNF, and DTNBP1 genes and cognitive functions in patients with brain tumors. Neuro Oncol. 2014;16(suppl 5):v134. doi: 10.1093/neuonc/now057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Small BJ, Rawson KS, Walsh E, et al. Catechol-O-methyltransferase genotype modulates cancer treatment-related cognitive deficits in breast cancer survivors. Cancer. 2011;117(7):1369–1376. doi: 10.1002/cncr.25685. [DOI] [PubMed] [Google Scholar]
- 28.Stefanis NC, van Os J, Avramopoulos D, Smyrnis N, Evdokimidis I, Stefanis CN. Effect of COMT Val158Met polymorphism on the Continuous Performance Test, Identical Pairs Version: tuning rather than improving performance. Am J Psychiatr. 2014;162(9):1752–1754. doi: 10.1176/appi.ajp.162.9.1752. [DOI] [PubMed] [Google Scholar]
- 29.Green MJ, Chia T-Y, Cairns MJ, et al. Catechol-O-methyltransferase (COMT) genotype moderates the effects of childhood trauma on cognition and symptoms in schizophrenia. J Psychiatr Res. 2014;49:43–50. doi: 10.1016/j.jpsychires.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Williams-Gray CH, Hampshire A, Barker RA, Owen AM. Attentional control in Parkinson's disease is dependent on COMT val158met genotype. Brain. 2008;131(2):397–408. doi: 10.1093/brain/awm313. [DOI] [PubMed] [Google Scholar]
- 31.Lanni C, Garbin G, Lisa A, et al. Influence of COMT Val158Met polymorphism on Alzheimer's disease and mild cognitive impairment in Italian patients. J Alzheimers Dis. 2012;32(4):919–926. doi: 10.3233/JAD-2012-120358. [DOI] [PubMed] [Google Scholar]
- 32.Waterhouse EG, An JJ, Orefice LL, et al. BDNF promotes differentiation and maturation of adult-born neurons through GABAergic transmission. J Neurosci. 2012;32(41):14318–14330. doi: 10.1523/JNEUROSCI.0709-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu B, Nagappan G, Guan X, Nathan PJ, Wren P. BDNF-based synaptic repair as a disease-modifying strategy for neurodegenerative diseases. Nat Rev Neurosci. 2013;14(6):401–416. doi: 10.1038/nrn3505. [DOI] [PubMed] [Google Scholar]
- 34.Sabb F, Burggren A, Higier R, et al. Challenges in phenotype definition in the whole-genome era: multivariate models of memory and intelligence. Neuroscience. 2009;164(1):88–107. doi: 10.1016/j.neuroscience.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fett A-KJ, Viechtbauer W, Penn DL, van Os J, Krabbendam L. The relationship between neurocognition and social cognition with functional outcomes in schizophrenia: a meta-analysis. Neurosci Biobehav Rev. 2011;35(3):573–588. doi: 10.1016/j.neubiorev.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Zhou R, Sulman EP, et al. Genetic modulation of neurocognitive function in glioma patients. Clin Cancer Res. 2015;21(14):3340–3346. doi: 10.1158/1078-0432.CCR-15-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson CJ, Finch CE, Cohen HJ. Cytokines and cognition—the case for a head-to-toe inflammatory paradigm. J Am Geriatr Soc. 2002;50(12):2041–2056. doi: 10.1046/j.1532-5415.2002.50619.x. [DOI] [PubMed] [Google Scholar]
- 38.Mandelblatt JS, Hurria A, McDonald BC, et al. Cognitive effects of cancer and its treatments at the intersection of aging: what do we know; what do we need to know? Semin Oncol. 2013;40(6):709–725. doi: 10.1053/j.seminoncol.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wefel JS, Kesler SR, Noll KR, Schagen SB. Clinical characteristics, pathophysiology, and management of noncentral nervous system cancer-related cognitive impairment in adults. CA Cancer J Clin. 2014;65(2):123–138. doi: 10.3322/caac.21258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lio D, Annoni G, Licastro F, et al. Tumor necrosis factor-α− 308A/G polymorphism is associated with age at onset of Alzheimer's disease. Mech Ageing Dev. 2006;127(6):567–571. doi: 10.1016/j.mad.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Longhi L, Perego C, Ortolano F, et al. Tumor necrosis factor in traumatic brain injury: effects of genetic deletion of p55 or p75 receptor. J Cereb Blood Flow Metab. 2013;33(8):1182–1189. doi: 10.1038/jcbfm.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bower JE, Ganz PA, Irwin MR, Castellon S, Arevalo J, Cole SW. Cytokine genetic variations and fatigue among patients with breast cancer. J Clin Oncol. 2013;31(13):1656–1661. doi: 10.1200/JCO.2012.46.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell J, Bennett M, Waters R, Reed S. A novel global genome method to measure and map DNA damage: application for chemotherapy treatment stratification. Lancet. 2014;383:S83. [Google Scholar]
- 44.Benitez-Buelga C, Sanchez-Barroso L, Gallardo M, et al. Impact of chemotherapy on telomere length in sporadic and familial breast cancer patients. Breast Cancer Res Treat. 2014;149(2):385–394. doi: 10.1007/s10549-014-3246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai S-J, Yu Y, Lin C-H, Chen T-J, Chen S-P, Hong C-J. Dopamine D2 receptor and N-methyl-D-aspartate receptor 2B subunit genetic variants and intelligence. Neuropsychobiology. 2001;45(3):128–130. doi: 10.1159/000054951. [DOI] [PubMed] [Google Scholar]
- 46.Bartrés-Faz D, Junqué C, Serra-Grabulosa JM, et al. Dopamine DRD2 Taq I polymorphism associates with caudate nucleus volume and cognitive performance in memory impaired subjects. Neuroreport. 2002;13(9):1121–1125. doi: 10.1097/00001756-200207020-00010. [DOI] [PubMed] [Google Scholar]
- 47.Barnes JJ, Dean AJ, Nandam LS, O'Connell RG, Bellgrove MA. The molecular genetics of executive function: role of monoamine system genes. Biol Psychiatry. 2011;69(12):e127–e143. doi: 10.1016/j.biopsych.2010.12.040. [DOI] [PubMed] [Google Scholar]
- 48.De Jaeger X, Cammarota M, Prado MA, Izquierdo I, Prado VF, Pereira GS. Decreased acetylcholine release delays the consolidation of object recognition memory. Behav Brain Res. 2013;238:62–68. doi: 10.1016/j.bbr.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Strange BA, Kroes MC, Roiser JP, Tan GC, Dolan RJ. Emotion-induced retrograde amnesia is determined by a 5-HTT genetic polymorphism. J Neurosci. 2008;28(28):7036–7039. doi: 10.1523/JNEUROSCI.0834-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabadi SV, Stoica BA, Loane DJ, et al. Cyclin D1 gene ablation confers neuroprotection in traumatic brain injury. J Neurotrauma. 2012;29(5):813–827. doi: 10.1089/neu.2011.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulhern RK, Palmer SL, Merchant TE, et al. Neurocognitive consequences of risk-adapted therapy for childhood medulloblastoma. J Clin Oncol. 2005;23(24):5511–5519. doi: 10.1200/JCO.2005.00.703. [DOI] [PubMed] [Google Scholar]
- 52.Ris MD, Packer R, Goldwein J, Jones-Wallace D, Boyett JM. Intellectual outcome after reduced-dose radiation therapy plus adjuvant chemotherapy for medulloblastoma: a Children’s Cancer Group study. J Clin Oncol. 2001;19(15):3470–3476. doi: 10.1200/JCO.2001.19.15.3470. [DOI] [PubMed] [Google Scholar]
- 53.Palmer SL, Gajjar A, Reddick WE, et al. Predicting intellectual outcome among children treated with 35–40 Gy craniospinal irradiation for medulloblastoma. Neuropsychology. 2003;17(4):548. doi: 10.1037/0894-4105.17.4.548. [DOI] [PubMed] [Google Scholar]
- 54.Palmer SL, Goloubeva O, Reddick WE, et al. Patterns of intellectual development among survivors of pediatric medulloblastoma: a longitudinal analysis. J Clin Oncol. 2001;19(8):2302–2308. doi: 10.1200/JCO.2001.19.8.2302. [DOI] [PubMed] [Google Scholar]
- 55.Copeland DR, Moore BD, III, Ater JL. Neurocognitive development of children after a cerebellar tumor in infancy: a longitudinal study. J Clin Oncol. 1999;17(11):3476–3486. doi: 10.1200/JCO.1999.17.11.3476. [DOI] [PubMed] [Google Scholar]
- 56.Moore BD, Ater JL, Copeland DR. Improved neuropsychological outcome in children with brain tumors diagnosed during infancy and treated without cranial irradiation. J Child Neurol. 1992;7(3):281–290. doi: 10.1177/088307389200700308. [DOI] [PubMed] [Google Scholar]
- 57.Moore BD. Neurocognitive outcomes in survivors of childhood cancer. J Pediatr Psychol. 2005;30(1):51–63. doi: 10.1093/jpepsy/jsi016. [DOI] [PubMed] [Google Scholar]
- 58.Lähteenmäki P, Harila-Saari A, Pukkala E, Kyyrönen P, Salmi T, Sankila R. Scholastic achievements of children with brain tumors at the end of comprehensive education: a nationwide, register-based study. Neurology. 2007;69(3):296–305. doi: 10.1212/01.wnl.0000265816.44697.b4. [DOI] [PubMed] [Google Scholar]
- 59.Mulhern RK, Kepner JL, Thomas PR, Armstrong FD, Friedman HS, Kun LE. Neuropsychologic functioning of survivors of childhood medulloblastoma randomized to receive conventional or reduced-dose craniospinal irradiation: a Pediatric Oncology Group study. J Clin Oncol. 1998;16(5):1723–1728. doi: 10.1200/JCO.1998.16.5.1723. [DOI] [PubMed] [Google Scholar]
- 60.Brouwers P. Commentary: study of the neurobehavioral consequences of childhood cancer: entering the genomic era? J Pediatr Psychol. 2005;30(1):79–84. doi: 10.1093/jpepsy/jsi018. [DOI] [PubMed] [Google Scholar]
- 61.Sorrentino BP. Chemoprotection in Brain Tumor Patients: Another Success for Stem Cell Gene Therapy. Mol Ther. 2012;20(8):1485–1487. doi: 10.1038/mt.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dietrich J, Monje M, Wefel J, Meyers C. Clinical patterns and biological correlates of cognitive dysfunction associated with cancer therapy. Oncologist. 2008;13(12):1285–1295. doi: 10.1634/theoncologist.2008-0130. [DOI] [PubMed] [Google Scholar]
- 63.Ahles TA, Saykin A. Cognitive effects of standard-dose chemotherapy in patients with cancer. Cancer Invest. 2001;19(8):812–820. doi: 10.1081/cnv-100107743. [DOI] [PubMed] [Google Scholar]
- 64.Cho H-J, Eom H-S, Kim H-J, Kim I-S, Lee GW, Kong S-Y. Glutathione-S-transferase genotypes influence the risk of chemotherapy-related toxicities and prognosis in Korean patients with diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2010;198(1):40–46. doi: 10.1016/j.cancergencyto.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Lecomte T, Landi B, Beaune P, Laurent-Puig P, Loriot M-A. Glutathione S-transferase P1 polymorphism (Ile105Val) predicts cumulative neuropathy in patients receiving oxaliplatin-based chemotherapy. Clin Cancer Res. 2006;12(10):3050–3056. doi: 10.1158/1078-0432.CCR-05-2076. [DOI] [PubMed] [Google Scholar]
- 66.Barahmani N, Carpentieri S, Li X-N, et al. Glutathione S-transferase M1 and T1 polymorphisms may predict adverse effects after therapy in children with medulloblastoma. Neuro Oncol. 2009;11(3):292–300. doi: 10.1215/15228517-2008-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brackett J, Krull KR, Scheurer ME, et al. Antioxidant enzyme polymorphisms and neuropsychological outcomes in medulloblastoma survivors: a report from the Childhood Cancer Survivor Study. Neuro Oncol. 2012:nos123. doi: 10.1093/neuonc/nos123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howarth RA, Adamson AM, Ashford JM, et al. Investigating the relationship between COMT polymorphisms and working memory performance among childhood brain tumor survivors. Pediatr Blood Cancer. 2014;61(1):40–45. doi: 10.1002/pbc.24649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krajinovic M, Robaey P, Chiasson S, et al. Polymorphisms of genes controlling homocysteine levels and IQ score following the treatment for childhood ALL. Pharmacogenomics. 2005;6(3):293–302. doi: 10.1517/14622416.6.3.293. [DOI] [PubMed] [Google Scholar]
- 70.Krull KR, Brouwers P, Jain N, et al. Folate pathway genetic polymorphisms are related to attention disorders in childhood leukemia survivors. J Pediatr. 2008;152(1):101–105. doi: 10.1016/j.jpeds.2007.05.047. [DOI] [PubMed] [Google Scholar]
- 71.Kamdar KY, Krull KR, El-Zein RA, et al. Folate pathway polymorphisms predict deficits in attention and processing speed after childhood leukemia therapy. Pediatr Blood Cancer. 2011;57(3):454–460. doi: 10.1002/pbc.23162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strand TA, Taneja S, Ueland PM, et al. Cobalamin and folate status predicts mental development scores in North Indian children 12–18 mo of age. Am J Clin Nutr. 2013;97(2):310–317. doi: 10.3945/ajcn.111.032268. [DOI] [PubMed] [Google Scholar]
- 73.Bowman G, Silbert L, Howieson D, et al. Nutrient biomarker patterns, cognitive function, and MRI measures of brain aging. Neurology. 2012;78(4):241–249. doi: 10.1212/WNL.0b013e3182436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kronenberg G, Harms C, Sobol RW, et al. Folate deficiency induces neurodegeneration and brain dysfunction in mice lacking uracil DNA glycosylase. J Neurosci. 2008;28(28):7219–7230. doi: 10.1523/JNEUROSCI.0940-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krull KR, Bhojwani D, Conklin HM, et al. Genetic mediators of neurocognitive outcomes in survivors of childhood acute lymphoblastic leukemia. J Clin Oncol. 2013;31(17):2182–2188. doi: 10.1200/JCO.2012.46.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamath AF, Chauhan AK, Kisucka J, et al. Elevated levels of homocysteine compromise blood-brain barrier integrity in mice. Blood. 2006;107(2):591–593. doi: 10.1182/blood-2005-06-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Collaboration HS. Homocysteine and risk of ischemic heart disease and stroke: a meta-analysis. JAMA. 2002;288(16):2015–2022. doi: 10.1001/jama.288.16.2015. [DOI] [PubMed] [Google Scholar]
- 78.Cole P, Neuberg DS, Silverman LB, et al. Polymorphisms in genes related to oxidative stress are associated with inferior cognitive function after therapy for childhood ALL. Blood. 33(19):2205–2211. doi: 10.1200/JCO.2014.59.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baldock AL, Yagle K, Born DE, et al. Invasion and proliferation kinetics in enhancing gliomas predict IDH1 mutation status. Neuro Oncol. 2014;16(6):779–786. doi: 10.1093/neuonc/nou027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wefel J, Noll K, Rao G, Cahill D. NC-15 Relationships between neurocognitive functioning and IDH1 genetic mutation status in malignant astrocytoma. Neuro Oncol. 2014;16(suppl 5):v137. [Google Scholar]
- 81.Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown PD, Pugh S, Laack NN, et al. Memantine for the prevention of cognitive dysfunction in patients receiving whole-brain radiotherapy: a randomized, double-blind, placebo-controlled trial. Neuro Oncol. 2013;15(10):1429–1437. doi: 10.1093/neuonc/not114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin NU, Lee EQ, Aoyama H, et al. Challenges relating to solid tumour brain metastases in clinical trials, part 1: patient population, response, and progression. A report from the RANO group. Lancet Oncol. 2013;14(10):e396–e406. doi: 10.1016/S1470-2045(13)70311-5. [DOI] [PubMed] [Google Scholar]
- 84.Lin NU, Wefel JS, Lee EQ, et al. Challenges relating to solid tumour brain metastases in clinical trials, part 2: neurocognitive, neurological, and quality-of-life outcomes. A report from the RANO group. Lancet Oncol. 2013;14(10):e407–e416. doi: 10.1016/S1470-2045(13)70308-5. [DOI] [PubMed] [Google Scholar]
- 85.Wefel JS, Vardy J, Ahles T, Schagen SB. International Cognition and Cancer Task Force recommendations to harmonise studies of cognitive function in patients with cancer. Lancet Oncol. 2011;12(7):703–708. doi: 10.1016/S1470-2045(10)70294-1. [DOI] [PubMed] [Google Scholar]
- 86.Pencina MJ, D'Agostino RB, Demler OV. Novel metrics for evaluating improvement in discrimination: net reclassification and integrated discrimination improvement for normal variables and nested models. Stat Med. 2012;31(2):101–113. doi: 10.1002/sim.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Campbell CD, Ogburn EL, Lunetta KL, et al. Demonstrating stratification in a European American population. Nature Genet. 2005;37(8):868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 88.Dick DM, Agrawal A, Keller MC, et al. Candidate gene–environment interaction research: reflections and recommendations. Perspect Psychol Sci. 2015;10(1):37–59. doi: 10.1177/1745691614556682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Day JJ. New approaches to manipulating the epigenome. Dialogues Clin Neurosci. 2014;16(3):345. doi: 10.31887/DCNS.2014.16.3/jday. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thompson SJ, Leigh L, Christensen R, et al. Immediate neurocognitive effects of methylphenidate on learning-impaired survivors of childhood cancer. J Clin Oncol. 2001;19(6):1802–1808. doi: 10.1200/JCO.2001.19.6.1802. [DOI] [PubMed] [Google Scholar]
- 91.Mulhern RK, Khan RB, Kaplan S, et al. Short-term efficacy of methylphenidate: a randomized, double-blind, placebo-controlled trial among survivors of childhood cancer. J Clin Oncol. 2004;22(23):4795–4803. doi: 10.1200/JCO.2004.04.128. [DOI] [PubMed] [Google Scholar]