Abstract
The “cancer immunogenomics” paradigm has facilitated the search for tumor-specific antigens over the last 4 years by applying comprehensive cancer genomics to tumor antigen discovery. We applied this methodology to identify tumor-specific “neoantigens” in the C57BL/6-derived GL261 and VM/Dk-derived SMA-560 tumor models. Following DNA whole exome and RNA sequencing, high-affinity candidate neoepitopes were predicted and screened for immunogenicity by ELISPOT and tetramer analyses. GL261 and SMA-560 harbored 4,932 and 2,171 non-synonymous exome mutations, respectively, of which less than half were expressed. To establish the immunogenicities of H-2Kb and H-2Db candidate neoantigens, we assessed the ability of the epitopes predicted in silico to be the highest affinity binders to activate tumor-infiltrating T cells harvested from GL261 and SMA-560 tumors. Using IFNγ ELISPOT, we confirmed H-2Db–restricted Imp3D81N (GL261) and Odc1Q129L (SMA-560) along with H-2Kb–restricted E2f8K272R (SMA-560) as endogenous tumor-specific neoantigens that are functionally immunogenic. Furthermore, neoantigen-specific T cells to Imp3D81N and Odc1Q129L were detected within intracranial tumors as well as cervical draining lymph nodes by tetramer analysis. By establishing the immunogenicities of predicted high-affinity neoepitopes in these models, we extend the immunogenomics-based neoantigen discovery pipeline to glioblastoma models and provide a tractable system to further study the mechanism of action of T cell–activating immunotherapeutic approaches in preclinical models of glioblastoma.
Keywords: Neoantigen, immunogenomics, TIL, glioblastoma, GL261, SMA-560
Introduction
Glioblastoma is the most common and lethal malignancy of the central nervous system (CNS) in adults. Despite multimodality standard-of-care treatment involving surgery followed by concurrent chemoradiation, patients will eventually relapse or progress with a median overall survival of 12–15 months (1). The unmet need for new treatments has been a strong stimulus to better understand the molecular basis of glioblastoma in order to identify new therapeutic targets. However, although we now have deep insights into the genomic landscapes of these tumors (2, 3), the “mutation-to-targeted drug” paradigm based on genomically-guided precision medicine has not emerged as an effective treatment option as it has for other solid tumors (4). Due to the successes of immunotherapies in other cancer types (5), combined with the realization that the CNS is not an hermetically immunoprivileged site (6), enthusiasm is growing regarding the use of immune-based treatments for patients with glioblastoma. Indeed, vaccine approaches targeting EGFRvIII (7) and other shared epitopes (8), autologous dendritic cell (9) and heat shock protein (10) vaccines, and other modalities including the checkpoint blockade agents are in clinical trials for glioblastoma. However, our understanding of how these immune-based strategies, designed to enhance tumor-specific T cell recognition and effector function, control glioblastoma progression has been limited by our ability to identify and monitor tumor-specific T cell responses.
To this end, the “cancer immunogenomics” concept has greatly enabled the search for tumor mutation–specific antigens over the last 4 years by applying cancer genomics information in a new way (11–13). The aim of this approach is to identify expressed tumor-specific, missense mutations that are predicted to bind with high affinity to an individual patient’s MHC molecules for presentation as “neoantigens” to host effector T cells. Thus, this view prioritizes the antigenic potential of a somatic mutation in cancer cells over the more traditional “driver” versus “passenger” hierarchy. Since its initial conception and application to neoantigen discovery in murine sarcomas (13, 14), this approach has been employed to identify antigenic targets of tumor-specific T cells arising spontaneously or as the result of checkpoint blockade in other preclinical (15–17) and human (18–21) settings.
To better understand the endogenous T cell immune response to brain tumor antigens, we applied the cancer immunogenomics methodology to the study of two mouse models of glioblastoma. The GL261 model was derived from an intracranially-induced methylcholanthrene tumor in C57BL/6 mice (22), while the SMA-560 model was derived from a spontaneous astrocytoma that developed in VM/Dk mice (23). Although both models grow progressively when transplanted into syngeneic hosts, both GL261 and SMA-560 are responsive to checkpoint blockade immunotherapy (24–28). To identify candidate targets of the host anti-tumor CD8 T cell response, we performed whole exome DNA and RNA sequencing of both GL261 and SMA-560 gliomas to characterize expressed tumor-specific mutations. We then applied multiple in silico MHC class I binding prediction algorithms to identify putative high-affinity H-2Db and H-2Kb–restricted neoepitopes and then assessed the immunogenicities of these predicted neoantigens. Using IFNγ ELISPOT assays and tetramer analysis, we confirmed the presence of an endogenous CD8 T cell response specific to the H-2Db–restricted neoantigens, Imp3D81N (GL261) and Odc1Q129L (SMA-560). Additionally, we identified reactivity to the H-2Kb–restricted SMA-560-derived neoantigen, E2f8K272R. Furthermore, endogenous neoantigen-specific T cell populations in the brain and draining lymph nodes to Imp3D81N and Odc1Q129L were detected. By characterizing the neoepitope profile and using this information to identify and monitor neoantigen-specific host anti-glioma T cell responses in these preclinical models, we extend the cancer immunogenomics approach to glioblastoma and provide a genomics-based system to further explore the mechanisms of action of immunotherapeutics in glioblastoma.
Materials and Methods
Animals and Cells
Animal studies were approved by the Animal Studies Committee at Washington University. C57BL/6 were purchased from Taconic Biosciences (Hudson, NY), and VM/Dk mice were obtained from Dr. John Sampson (Duke University). Mice were housed in specific pathogen-free conditions. GL261 was obtained from the NCI Tumor Repository (Frederick, MD) in 2014 and fully characterized by DNA whole exome and RNA sequencing. SMA-560 was obtained from Drs. John Sampson and Peter Fecci (Duke University) and fully characterized by DNA whole exome and RNA sequencing. Either 1×106 (subcutaneous) or 5×104 (intracranial) GL261 or SMA-560 cells were implanted into 6–10 week old naïve syngeneic C57BL/6 mice or VM/Dk, respectively. For intracranial experiments, tumor cells were resuspended in 5 µL PBS and injected into the right striatum of anesthetized syngeneic mice in a stereotactic frame. Subcutaneously implanted tumors were harvested when approximately 10 mm in greatest diameter (approximately 2 weeks for SMA-560 and 3 weeks for GL261). Intracranially implanted tumors were harvested when mice became moribund.
Lymphocyte isolation
Subcutaneous or intracranial tumors were minced into 1–2 mm chunks, plated in 12-well plates, and incubated at 37°C in culture media (RPMI-1640, L-glutamine, penicillin/streptomycin, β-mercaptoethanol, MEM, 10% FBS) with 100 U/mL recombinant human IL-2. After 2–5 days, TIL were harvested and passed through a 70 micron cell strainer. Draining lymph nodes and spleens were mechanically dissociated and filtered through a 70-micron cell strainer. Lymphocytes were purified usisng the Dead Cell Removal kit (Miltenyi Biotec, Auburn, CA). For ELISPOT assays, mononuclear cells were isolated from splenocytes by Ficoll-Paque PLUS density gradient (GE Healthcare Life Sciences, Pittsburgh, PA).
DNA whole exome and RNA sequencing
Libraries were captured using the Agilent Mouse Exome reagent. Sequencing was performed on an Illumina HiSeq2000 (Illumina Inc, San Diego, CA). Sequence coverage was as follows: C57BL/6 normal (92.1X), GL261 tumor (76.7X), VM/Dk normal (86.0X), SMA-560 tumor (82.4X). Data were aligned to reference sequence using bwa(29) version 0.5.9 then merged and deduplicated using picard version 1.46 (https://broadinstitute.github.io/picard/). SNVs were detected using the union of three callers: 1) samtools(30) version r963 intersected with Somatic Sniper (31) version 1.0.2 and processed through false-positive filter v1, 2) VarScan (32) version 2.2.6 filtered by varscan-high-confidence filter version v1 and processed through false-positive filter v1, 3) Strelka (33) version 0.4.6.2. Indels were detected using the union of three callers: 1) GATK (34) somatic-indel version 5336 pindel version 0.5 filtered with pindel false- positive and VAF filters (params: --variant-freq-cutoff=0.08), 2) VarScan (32) version 2.2.6 filtered by varscan-high-confidence-indel version v1 and 3) Strelka (33) version 0.4.6.2. SNVs and Indels were further filtered using a bayesian classifier (https://github.com/genome/genome/blob/master/lib/perl/Genome/Model/Tools/Validation/IdentifyOutliers.pm), retaining variants classified as somatic with a binomial log-likelihood of at least 3. Results were filtered to require expression of the mutant allele (FPKM >1 and at least one variant-supporting read in the RNA) and variant allele frequency (VAF) >1%.
MHC class I binding prediction
The potential for GL261 and SMA-560 missense mutations to bind to H-2Db or H-2Kb molecules was predicted using multiple pipelines: Stabilized Matrix Method (SMM) algorithm, the Stabilized Matrix Method with a Peptide:MHC Binding Energy Covariance algorithm (SMMPMBEC), Artificial Neural Network (ANN) algorithm, and NetMHCpan algorithm provided by the Immune Epitope Database and Analysis Resource (http://www.immuneepitope.org) as well as the Position Specific Scoring Matrices (PSSM). Results were expressed as affinity values (1/IC50 × 100; where IC50 is the half-maximum inhibitory concentration [nM]). The mean affinity value was calculated from predicted binding affinities of all five algorithms. A binding cut-off of IC50 < 500 nM was empirically applied as an additional filter.
Enzyme linked immunospot (ELISPOT) assay
Naïve splenocytes were plated at a concentration of 100,000–200,000 cells/well in 100 µL serum-free C.T.L. media (Cellular Technology, Ltd., Shaker Heights, OH) on precoated murine IFNγ ELISPOT plates (Cellular Technology, Ltd., Shaker Heights, OH). Harvested TIL were added at a final concentration of 25,000 cells/well in 200 µL with peptide (10 µM) (Peptide 2.0 Inc., Chantilly, VA). Concavalin A (1 µg/well) was a positive control. Plates were incubated overnight at 37°C and analyzed using the C.T.L. ImmunoSpot kit (Cellular Technology, Ltd., Shaker Heights, OH).
Tetramers and Flow cytometry
Recombinant H-2Db heavy chains and human β2-microglobulin were produced in BL21-CodonPlus (DE3)-RIPL Escherichia coli (Agilent, Santa Clara, CA) and purified from inclusion bodies. Purification of MHC class I heavy and light chain by size-exclusion FPLC was performed as described (35). Peptide-specific monomers were generated by UV-mediated exchange as described (36). MHC class I multimers were generated using streptavidin-conjugated PE or APC (Invitrogen, Burlington, ON). For TIL and lymph nodes, 1×106 cells were dual stained with PE- and APC-peptide:MHC class I tetramers for 15 minutes at 37°C. Cells were then stained with CD8α-FITC, Thy1.2-PE/Cy7, and Zombie NIR Fixable Viability Kit (BioLegend, San Diego, CA) and analyzed on a BD Fortessa flow cytometer.
Statistical analysis
The intergroup difference in mean number of spots on ELISPOT was evaluated using a Student’s t-test with p <0.05 as statistically significant.
Results
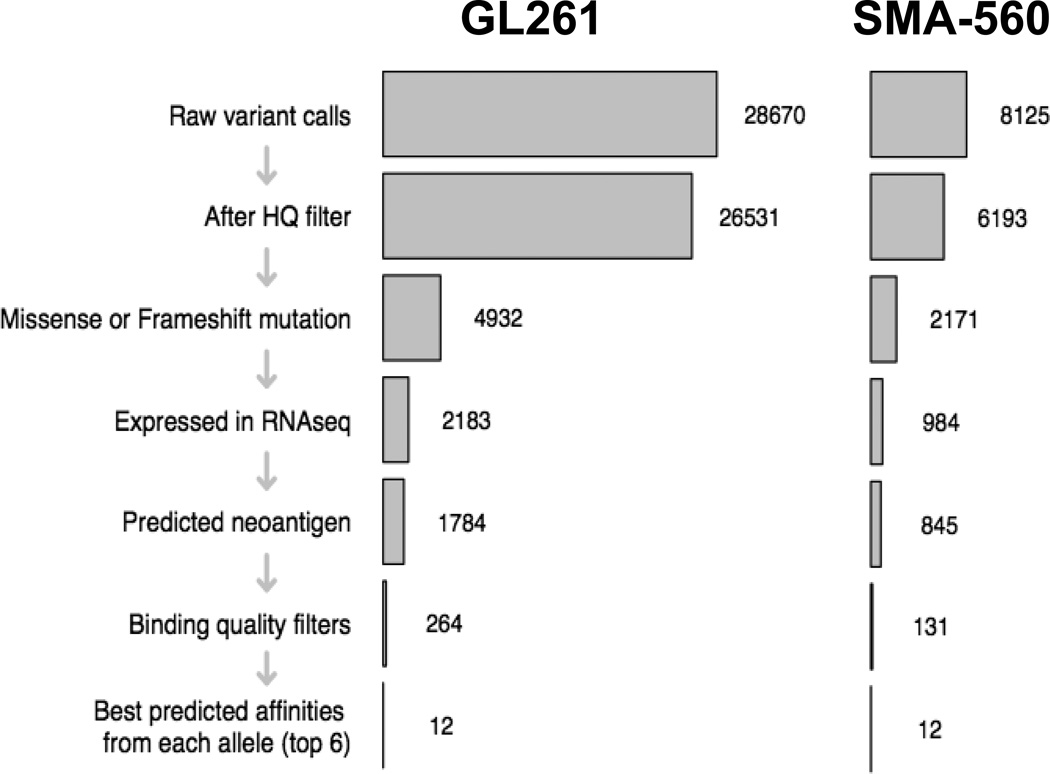
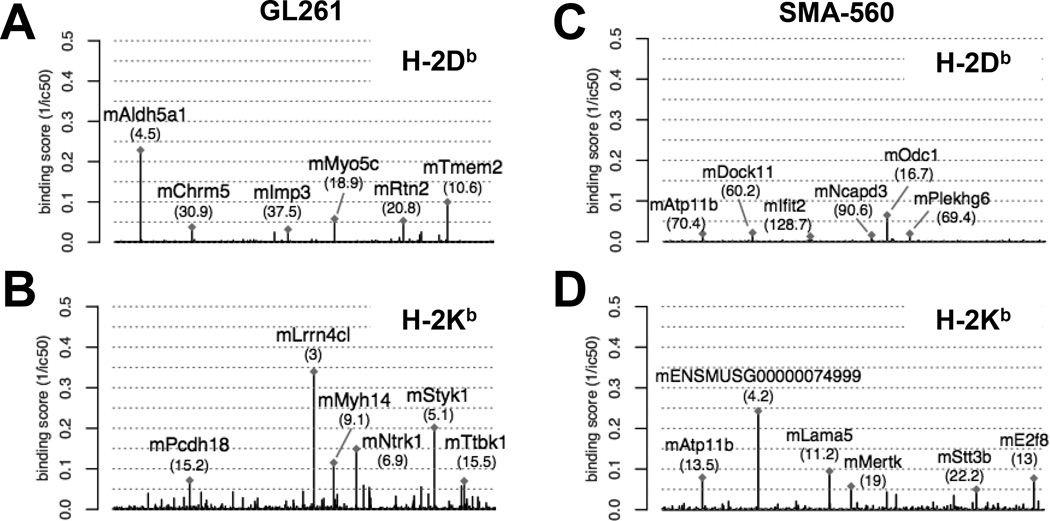
To identify candidate neoantigens within GL261 and SMA-560, we applied a cancer immunogenomics discovery pipeline similar to the approach that has been described previously (11, 14, 15). Briefly, unbiased genomic characterization is performed to detect expressed tumor-specific variants, and existing in silico algorithms are used to determine the predicted affinity strength with which translated peptide sequences bind to particular MHC class I molecules. DNA whole exome sequencing was employed to identify tumor-specific, somatic missense single nucleotide variants (SNVs). GL261 harbored a total of 26,531 somatic mutations compared to the syngeneic C57BL/6 genome, of which 4,932 were missense or frameshift variants (Fig. 1; Supplementary Table 1). Conversely, SMA-560 harbored 6,193 somatic mutations compared to the syngeneic VM/Dk exome, of which 2,171 were missense or frameshift variants (Fig. 1; Supplementary Table 2). To determine which of the identified SNVs were expressed and could be translated into candidate neoepitopes, RNA-sequencing was performed on both tumors. These data showed that fewer than half of somatic SNVs were expressed at the transcript level; GL261 and SMA-560 expressed 2,183 and 984 mutations, respectively (Fig. 1; Supplementary Tables 1 and 2). Because both C57BL/6 and VM/Dk genetic backgrounds possess the H-2b haplotype, we used in silico computational approaches to determine which mutant proteins represented candidate neoepitopes that were predicted to bind with high affinity to the associated class I major histocompatibility complexes (MHC), H-2Db and H-2Kb. We calculated the median binding affinities from a combination of five MHC class I binding prediction algorithms, and after applying an affinity cutoff of ic50 < 500 nM, we identified 181 H-2Db and 1,599 H-2Kb predicted high affinity neoepitopes in GL261 (Fig. 2a,b; Supplementary Table 1). Similarly, there were 77 H-2Db and 647 H-2Kb predicted high affinity neoepitopes in SMA-560 (Fig. 2c–d; Supplementary Table 2). The top six predicted binding affinities for each allele were prioritized for further evaluation (Tables 1, 2). These data showed that both GL261 and SMA-560 harbored a high mutational load and expressed potentially high-affinity candidate neoantigens.
Fig. 1.
Mutational burden of GL261 and SMA-560. Schematic of workflow for GL261 and SMA-560 neoantigen discovery. Numbers to right of bar graph indicate number of mutations identified following application of indicated filter on left side of bar graph.
Fig. 2.
Neoantigen landscape of GL261 and SMA-560. Manhattan plot of mean binding affinity (1/ic50) of putative candidate GL261-derived neoantigens for H-2Db (a) and H-2Kb (b). Manhattan plot of mean binding affinity (1/ic50) of putative candidate SMA-560-derived neoantigens for H-2Db (c) and H-2Kb (d). Labeled are the six highest predicted binding affinity candidate neoantigens. Numbers in parentheses represent calculated ic50 (nM) for each candidate neoantigen.
Table 1.
Top 6 in silico predicted H-2Db and H-2Kb candidate GL261-derived neoantigens with the corresponding amino acid sequence. The mutated amino acid is denoted by bold and underlining.
| H-2Db Restriction | |
|---|---|
| Amino acid mutation | Neoantigen sequence |
| mAldh5a1 V444F | FAIANAAEV |
| mTmem2 K1042N | VMLENGYTI |
| mMyo5c L822M | YMVRNLYQL |
| mRtn2 L405F | GAIFNGFTL |
| mChrm5 R503W | YALCNRTFW |
| mImp3 D81N | AALLNKLYA |
| H-2Kb Restriction | |
| Amino acid mutation | Neoantigen sequence |
| mLrrn4cl V200L | VTLVYAAL |
| mStyk1 L429I | ISYSFSVI |
| mNtrk1 H470Q | MSLQFMTL |
| mMyh14 G135V | LIYTYSVL |
| mPcdh18 Q1012R | MSSVFRRL |
| mTtbk1 C450R | RSLRYRRV |
Table 2.
Top 6 in silico predicted H-2Db and H-2Kb candidate SMA-560-derived neoantigens with the corresponding amino acid sequence. The mutated amino acid is denoted by bold and underlining.
| H-2Db Restriction | |
|---|---|
| Amino acid mutation | Neoantigen sequence |
| mOdc1 Q129L | YAASNGVLM |
| mDock11 G1958V | SVQVNAVPL |
| mPlekhg6 G10C | FGPPNECPL |
| mAtp11b K884M | FFYMNVCFI |
| mNcapd3 V510L | NTVLNPSPL |
| mIfit2 V60I | ATMCNILAYI |
| H-2Kb Restriction | |
| Amino acid mutation | Neoantigen sequence |
| mENSMUSG00000074999 C15F | SSFIYAYI |
| mLama5 G897V | VRFVFNPL |
| mE2f8 K272R | MSQRFVML |
| mAtp11b K884M | VQYFFYMNV |
| mMertk R682P | TFLLYSPL |
| mStt3b G323V | AAVVFALL |
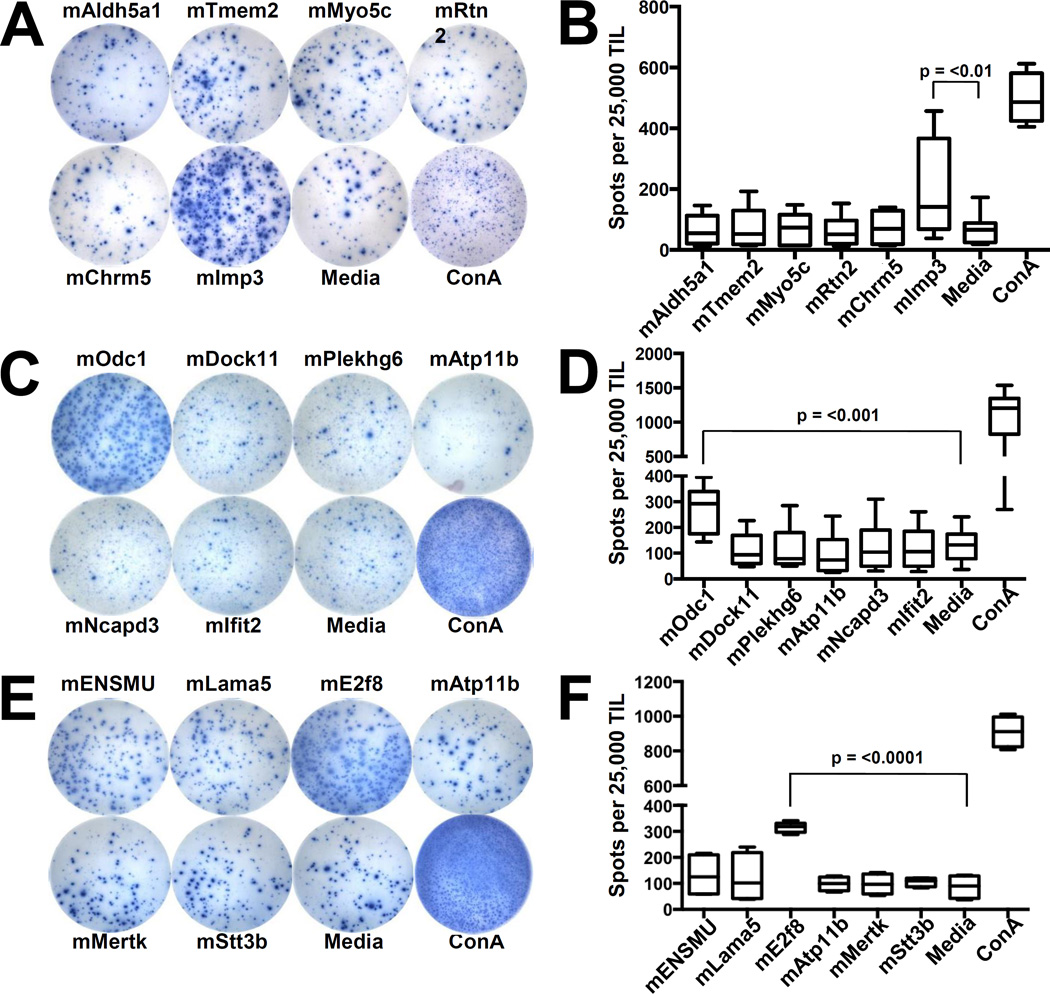
To determine the immunogenicities of the top predicted neoantigen candidates, we isolated and cultured TIL from established subcutaneously implanted tumors and performed IFNγ ELISPOT assays to assess the presence of neoantigen-specific T cells within intratumoral TIL populations. GL261-derived TIL demonstrated increased activation following stimulation with the H-2Db–restricted peptide, Imp3D81N (referred to as mImp3; Fig. 3a,b). SMA-560 TIL stimulated with the H-2Db–restricted candidate neoantigen, Odc1Q129L (referred to as mOdc1), as well as the H-2Kb–restricted epitope, E2f8K272R (referred to as mE2f8), had significantly increased IFNγ (Fig. 3c–f). Of note, none of the top predicted H-2Kb–restricted GL261 neoantigens induced increased IFNγ production from GL261-derived TIL (data not shown).
Fig. 3.
Identification of neoantigen-reactive TIL in GL261 and SMA-560. a) Representative images from IFNγ ELISPOT of GL261 TIL stimulated with H-2Db–restricted candidate neoantigens. TIL were isolated from established subcutaneously implanted GL261 at day 21 and incubated for 4 days in IL-2 (50 U/mL). Cultured TIL (25,000 TIL/well) were then incubated overnight with the indicated peptide (10 µM) and assessed for IFNγ production the following day by ELISPOT. b) Bar graph quantifying number of IFNγ spots per well. c) Representative wells from IFNγ ELISPOT of SMA-560 TIL stimulated with H-2Db–restricted candidate neoantigens. d) Bar graph quantifying number of IFNγ spots per well. e) Representative wells from IFNγ ELISPOT of SMA-560 TIL stimulated with H-2Kb–restricted candidate neoantigens. f) Bar graph quantifying number of IFNγ spots per well. Presented data depict pooled results from at least 3 experiments with 2–3 mice per experiment.
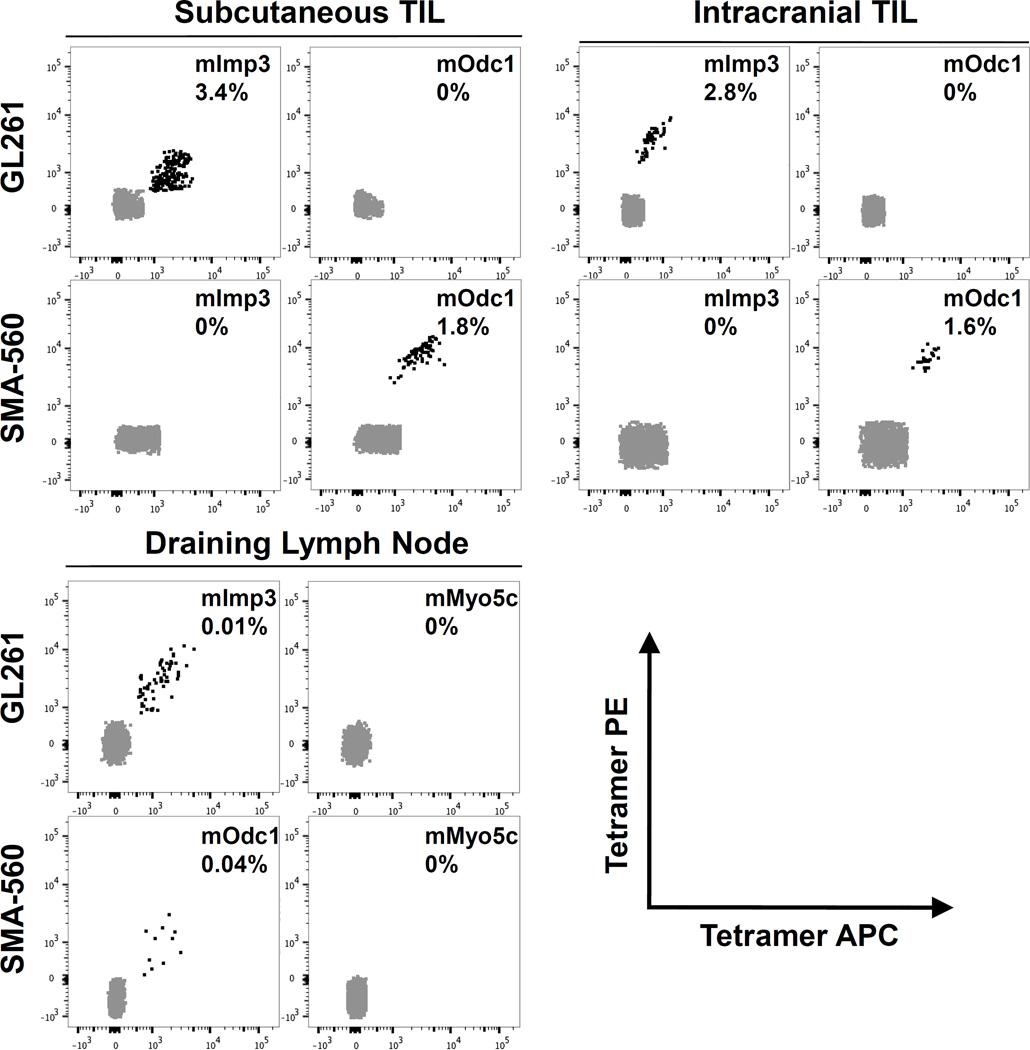
Having demonstrated the functional immunogenicities of candidate neoantigens by ELISPOT, we next asked whether we could directly detect of the presence of TIL-derived CD8 T cells specific to the cognate neoantigen:MHC complex using tetramer analysis. For these experiments, we focused solely on the H-2Db–restricted neoantigens. In TIL derived from established GL261 subcutaneously implanted tumors, approximately 3.4% (range 2.5 to 3.9%) of short-term cultured CD8 T cells were specific for the H-2Db–mImp3 tetramer, whereas there were no cells that stained with the negative control H-2Db–mOdc1 tetramer (Fig. 4, top left graph). Likewise, approximately 1.84% (range 1.1% to 3.1%) of CD8 T cells in TIL derived from established subcutaneously implanted SMA-560 tumors were found to be tetramer positive for mOdc1 (Fig. 4, top left graph). Of note, no detectable tetramer-positive CD8+ T cell populations specific to other predicted GL261 or SMA-560 H-2Db–restricted putative neoantigens were appreciated (data not shown). Together with the ELISPOT data, these results demonstrate the presence of functional, endogenous neoantigen-specific CD8 T cells in each of these transplantable glioblastoma models.
Fig. 4.
Detection of neoantigen-specific CD8 T cells by tetramer within TIL and draining lymph node. TIL isolated from subcutaneously implanted (top left graph) or intracranially implanted (top right graph) GL261 (top row) and SMA-560 (bottom row) tumors were cultured for 4 days in IL-2 (50 U/mL) prior to tetramer staining. Indicated peptides were loaded into H-2Db–restricted tetramers and dual labeled with PE and APC fluorochromes. Tetramer positive cells are identified as double positive populations. Draining cervical lymph nodes were surgically removed at time of sacrifice and stained directly with indicated tetramers (bottom left graph). Representative FACS plots of at least three experiments containing pooled TIL from 2–5 mice with similar results are shown. Number in each FACS plot represents average percentage of tetramer positive cells within CD8+ population of cells.
Finally, due to the distinct features of immune responses in the CNS compared to those in extracranial sites, we asked whether we could detect evidence of endogenous host immune responses to brain tumor neoantigens following orthotopic intracerebral implantation. We isolated TIL from intracranial GL261 and SMA-560 tumors and assessed for the presence of mImp3- and mOdc1-specific CD8 T cells, respectively, by tetramer analysis. Both mImp3- and mOdc1-specific CD8 T cells were detectable at comparable frequencies (GL261 – 2.8%, range 1.7% to 4.3%; SMA-560 – 1.6%, range 0.8% to 3.2%) (Fig. 4, top right graph). Because it has been shown that there may be a physical lymphatic connection between the brain and the cervical lymph nodes (37, 38), we next asked whether we could identify neoantigen-specific T cells within the draining cervical lymph nodes in mice orthotopically transplanted with GL261 or SMA-560 tumors. Surprisingly, a small but discrete population of tetramer-positive, neoantigen-specific CD8 T cells were detectable within the cervical lymph nodes of both C57BL/6 mice harboring intracranial GL261 tumors and VM/Dk mice harboring intracranial SMA-560 mice (Fig. 4, bottom right graph). Taken together, these data show endogenous neoantigen-specific CD8 T cell responses are detectable in both orthotopically-transplanted brain tumors as well as cervical lymph nodes.
Discussion
We employed a cancer immunogenomics approach to characterize the neoantigen landscape of two distinct, well-studied, murine orthotopic transplant models of glioblastoma. Our work represents the first application of this approach for neoantigen discovery in preclinical brain tumor model systems. The presence of H-2Db–restricted Imp3D81N (GL261) and Odc1Q129L (SMA-560) as well as H-2Kb–restricted E2f8K272R (SMA-560) neoantigen-reactive TIL was detected by screening candidate neoantigens with the highest predicted binding affinities using a functional IFNγ ELISPOT assay. H-2Db–restricted tetramer analysis validated the presence of mImp3 and mOdc1-specific CD8 T cell populations both within intracranial tumors as well as cervical lymph nodes.
The growing use of T cell–activating immunotherapies in cancer treatment has stimulated further studies into the identities of the antigens recognized by this effector subset. More broadly, we have had a longstanding interest in how the concept of cancer immunoediting applies to malignant glioma (39). Although there are several types of tumor antigens that T cells may recognize (40), neoantigens derived from somatic mutations are tumor-restricted in most cases and therefore harbor a lower likelihood of inducing tolerance or autoimmunity. Especially in the brain, it is important to limit the cross-reactivity of induced immune responses against normal tissue. The identities of the neoantigens we characterized underscore the emphasis of the cancer immunogenomics perspective on prioritizing somatic mutant proteins as antigens rather than as obvious drivers or passengers. Although Imp3, a small ribonucleoprotein, and Odc1, ornithine decarboxylase 1, have been implicated in cancer in some studies (41, 42), neither gene is recurrently mutated. Moreover, the contributions of mutant E2f8, an atypical E2F family repressor, to oncogenesis remain to be clarified.
Although our results provide evidence that predicted neoantigens can be validated as immunogenic in brain tumor models, the functional validation of only a subset of the highest predicted binding candidates suggests that additional work is needed to address further refinements to the in silico approach in order to increase the concordance between neoantigenicity and immunogenicity. Furthermore, the identified neoantigen-specific T cells represented a small subset of the total CD8 TIL population suggesting the presence of yet to be identified neoantigens. Although there exists substantial experience in prediction algorithm development (43–45), further refinements are necessary to enhance our ability to predict immunogenicity. Specifically, further work is focusing on the incorporation of other parameters—such as mutant transcript expression levels, IC50 thresholds of neoantigen binding to MHC molecules, comparative binding affinities of mutant peptides to wild type counterparts, and location of amino acid substitutions within the TCR/peptide:MHC topography—to enhance the predictive potential of the neoantigen discovery pipeline. Moreover, ongoing work is directed at determining the extent of neoantigen screening needed to capture the entire immunogenic landscape, especially in tumors with high mutational burdens such as these preclinical models, and how best to exploit neoantigen identification for therapeutic approaches. Nevertheless, the cancer immunogenomics approach to neoantigen detection has become a powerful method to probe the cancer immunome (11).
Our identification of the H-2Db–restricted, GL261-derived immunodominant neoantigen, Imp3D81N, supports the findings of a previous study (46), providing further validity to our immunogenomics approach. Specifically, an attenuated oncolytic strain of HSV-1 was injected into an established subcutaneously implanted tumor, and a GL261-specific cytotoxic T cell line was derived from the isolated splenocytes. Using a cDNA expression cloning approach, an antigen encoded by the 1190002L16Rik gene was identified and termed “GARC-1.” Strikingly, during the preparation of our manuscript, we determined that the amino acid sequence of mutated GARC-1 recognized by this GL261 T cell clone is identical to the Imp3D81N 9-mer we identified, AALLNKLYA. Thus, these data provide independent validation of the immunogenomics pipeline and further corroboration of the immunogenicity of the Imp3D81N antigen in the GL261 model.
Our observations of neoantigen-specific CD8 T cells within brain tumors and cervical lymph nodes provide further opportunities to study the cellular immunobiology of spontaneously arising immune responses to endogenous glioma antigens. Although it is acknowledged at this point that the brain is not “immunoprivileged” (6), it is clearly immunologically specialized. A number of studies have provided evidence that there may be a connection between the cerebrum and secondary lymphoid tissues, particularly the cervical lymph nodes (38). Recent work identified a dural lymphatic network that may represent the physical conduit by which transit of antigens from the brain to the extracranial secondary lymph nodes takes place (37, 47). Of note, we did not detect tumor cells within the lymph nodes we analyzed (data not shown). Additional work will be necessary to clearly establish the cellular basis for the anti-glioma immune response both in naturally occurring immunosurveillance and following vaccination or other therapeutic interventions.
Our results point to several other clinically relevant topics. First, personalized vaccination targeting mutated antigens is an active area of investigation (11, 12, 48) and clinical trials testing this possibility in the treatment of patients with glioblastoma are presently accruing patients (http://clinicaltrials.gov: NCT02287428, NCT02510950). The cancer immunogenomics neoantigen discovery pipeline employed in the current study therefore establishes a preclinical setting in which various methods of vaccination against genomically-identified glioma neoantigens can be rigorously tested. Secondly, the mutational load of both the GL261 and SMA-560 tumors is much higher than is typically observed in primary human glioblastoma in which the tumor exome mutational load is usually less than 100 (2). Thus, the number of potentially targetable neoantigens may be more limited in contexts harboring this mutational burden. However, work over the last 10 years has identified that approximately 20–30% of recurrent glioblastomas harbor a “hypermutator” phenotype (49–53) in which the number of exome mutations are significantly higher than at presentation and similar to the number of mutations seen in these preclinical tumors representing a subpopulation of glioblastoma patients that may benefit from neoantigen-based personalized vaccination. Moreover, due to the growing literature that tumors with hypermutated phenotypes may be more sensitive to checkpoint blockade immunotherapy (54), preclinical models with similar genomic landscapes may represent valuable models to explore the immunologic basis for these observations and whether they are generalizable to the CNS.
Supplementary Material
Acknowledgments
Funding Support: This study was funded by National Institutes of Health grant K08NS092912 (G.P.D.), American Cancer Society-Institutional Research grant (G.P.D), and the Physician-Scientist Training Program at Washington University School of Medicine (T.M.J., J.P.W.).
The authors thank Dr. Ravi Uppaluri for reviewing this manuscript.
Footnotes
Disclosures: The current Editor-in-Chief of Cancer Immunology Research (Robert D. Schreiber) is an author on this article. Otherwise, no potential conflicts of interest exist.
Compliance with Ethical Standards
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted. This article does not contain any studies with human participants performed by any of the authors.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Witt Hamer PC. Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol. 2010;12:304–316. doi: 10.1093/neuonc/nop068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan MK, Postow MA, Wolchok JD. Targeting T Cell Co-receptors for Cancer Therapy. Immunity. 2016;44:1069–1078. doi: 10.1016/j.immuni.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Dunn GP, Okada H. Principles of immunology and its nuances in the central nervous system. Neuro Oncol. 2015;17:vii3–vii8. doi: 10.1093/neuonc/nov175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Herndon JE, 2nd, Lally-Goss D, et al. An epidermal growth factor receptor variant III-targeted vaccine is safe and immunogenic in patients with glioblastoma multiforme. Mol Cancer Ther. 2009;8:2773–2779. doi: 10.1158/1535-7163.MCT-09-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phuphanich S, Wheeler CJ, Rudnick JD, Mazer M, Wang H, Nuno MA, et al. Phase I trial of a multi-epitope-pulsed dendritic cell vaccine for patients with newly diagnosed glioblastoma. Cancer Immunol Immunother. 2013;62:125–135. doi: 10.1007/s00262-012-1319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antonios JP, Everson RG, Liau LM. Dendritic cell immunotherapy for brain tumors. J Neurooncol. 2015;123:425–432. doi: 10.1007/s11060-015-1830-1. [DOI] [PubMed] [Google Scholar]
- 10.Bloch O, Crane CA, Fuks Y, Kaur R, Aghi MK, Berger MS, et al. Heat-shock protein peptide complex-96 vaccination for recurrent glioblastoma: a phase II, single-arm trial. Neuro Oncol. 2014;16:274–279. doi: 10.1093/neuonc/not203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125:3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacohen N, Fritsch EF, Carter TA, Lander ES, Wu CJ. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer Immunol Res. 2013;1:11–15. doi: 10.1158/2326-6066.CIR-13-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 17.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 18.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30:2394–2400. [PubMed] [Google Scholar]
- 23.Fraser H. Astrocytomas in an inbred mouse strain. J Pathol. 1971;103:266–270. doi: 10.1002/path.1711030410. [DOI] [PubMed] [Google Scholar]
- 24.Fecci PE, Ochiai H, Mitchell DA, Grossi PM, Sweeney AE, Archer GE, et al. Systemic CTLA-4 blockade ameliorates glioma-induced changes to the CD4+ T cell compartment without affecting regulatory T-cell function. Clin Cancer Res. 2007;13:2158–2167. doi: 10.1158/1078-0432.CCR-06-2070. [DOI] [PubMed] [Google Scholar]
- 25.Reardon DA, Gokhale PC, Klein SR, Ligon KL, Rodig SJ, Ramkissoon SH, et al. Glioblastoma Eradication Following Immune Checkpoint Blockade in an Orthotopic, Immunocompetent Model. Cancer Immunol Res. 2016;4:124–135. doi: 10.1158/2326-6066.CIR-15-0151. [DOI] [PubMed] [Google Scholar]
- 26.Wainwright DA, Chang AL, Dey M, Balyasnikova IV, Kim C, Tobias AL, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4 and PD-L1 in mice with brain tumors. Clin Cancer Res. 2014;20:5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng J, See AP, Phallen J, Jackson CM, Belcaid Z, Ruzevick J, et al. Anti-PD-1 blockade and stereotactic radiation produce long-term survival in mice with intracranial gliomas. Int J Radiat Oncol Biol Phys. 2013;86:343–349. doi: 10.1016/j.ijrobp.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belcaid Z, Phallen JA, Zeng J, See AP, Mathios D, Gottschalk C, et al. Focal radiation therapy combined with 4-1BB activation and CTLA-4 blockade yields long-term survival and a protective antigen-specific memory response in a murine glioma model. PLoS One. 2014;9:e101764. doi: 10.1371/journal.pone.0101764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 34.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, et al. Design and use of conditional MHC class I ligands. Nat Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 36.Bakker AH, Hoppes R, Linnemann C, Toebes M, Rodenko B, Berkers CR, et al. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc Natl Acad Sci U S A. 2008;105:3825–3830. doi: 10.1073/pnas.0709717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523:337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cserr HF, Harling-Berg CJ, Knopf PM. Drainage of brain extracellular fluid into blood and deep cervical lymph and its immunological significance. Brain Pathol. 1992;2:269–276. doi: 10.1111/j.1750-3639.1992.tb00703.x. [DOI] [PubMed] [Google Scholar]
- 39.Dunn GP, Fecci PE, Curry WT. Cancer immunoediting in malignant glioma. Neurosurgery. 2012;71:201–222. doi: 10.1227/NEU.0b013e31824f840d. discussion 22–3. [DOI] [PubMed] [Google Scholar]
- 40.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 41.Hogarty MD, Norris MD, Davis K, Liu X, Evageliou NF, Hayes CS, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68:9735–9745. doi: 10.1158/0008-5472.CAN-07-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samanta S, Sun H, Goel HL, Pursell B, Chang C, Khan A, et al. IMP3 promotes stem-like properties in triple-negative breast cancer by regulating SLUG. Oncogene. 2016;35:1111–1121. doi: 10.1038/onc.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lundegaard C, Lund O, Kesmir C, Brunak S, Nielsen M. Modeling the adaptive immune system: predictions and simulations. Bioinformatics. 2007;23:3265–3275. doi: 10.1093/bioinformatics/btm471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC immunology. 2008;9:8. doi: 10.1186/1471-2172-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Udaka K, Mamitsuka H, Zhu S. Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools. Briefings in bioinformatics. 2012;13:350–364. doi: 10.1093/bib/bbr060. [DOI] [PubMed] [Google Scholar]
- 46.Izuka Y, Kojima H, Kobata T, Kawase T, Kawakami Y, Toda M. Identification of a glioma antigen, GARC-1, using cytotoxic T lymphocytes induced by HSV cancer vaccine. Int J Cancer. 2006;118:942–949. doi: 10.1002/ijc.21432. [DOI] [PubMed] [Google Scholar]
- 47.Aspelund A, Antila S, Proulx ST, Karlsen TV, Karaman S, Detmar M, et al. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J Exp Med. 2015;212:991–999. doi: 10.1084/jem.20142290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Thuijl HF, Mazor T, Johnson BE, Fouse SD, Aihara K, Hong C, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129:597–607. doi: 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28:318–328. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.