Abstract
Cancer cell vascular invasion is a crucial step in the malignant progression towards metastasis. Here we used a genome-wide RNAi screen with E0771 mammary cancer cells to uncover drivers of endothelial monolayer invasion. We identified keratin-associated protein 5-5 (Krtap5-5) as a candidate. Krtap5-5 belongs to a large protein family that is implicated in crosslinking keratin intermediate filaments during hair formation, yet these keratin-associated proteins have no reported role in cancer. Depletion of Krtap5-5 from cancer cells led to cell blebbing and a loss of keratins 14 and 18, in addition to the upregulation of vimentin intermediate filaments. This intermediate filament subtype switching induced dysregulation of the actin cytoskeleton and reduced the expression of hemidesmosomal α6/β4-integrins. We further demonstrate that knockdown of keratin 18 phenocopies the loss of Krtap5-5, suggesting that Krtap5-5 crosstalks with keratin 18 in E0771 cells. Disruption of the keratin cytoskeleton by perturbing Krtap5-5 function broadly altered the expression of cytoskeleton regulators and the localization of cell surface markers. Krtap5-5 depletion did not impact cell viability but reduced cell motility and extracellular matrix invasion, as well as extravasation of cancer cells into tissues in zebrafish and mice. We conclude that Krtap5-5 is a previously unknown regulator of cytoskeletal function in cancer cells that modulates motility and vascular invasion. Thus, in addition to its physiologic function, a keratin-associated protein can serve as a switch towards malignant progression.
Keywords: cancer, cytoskeleton, vascular invasion, keratin, KRTAP5-5
INTRODUCTION
Metastatic disease is the major cause of cancer mortality1 and there is a pressing need to unravel the mechanisms that govern cancer cell spread from a primary tumor. Invasive behavior is governed by the cytoskeleton, a structure essential for cell motility2,3 that also demarcates the boundary between cancer cells and their surrounding microenvironment4,5. The cytoskeleton is comprised of actin, microtubules, and intermediate filaments6,7, and a key example of metastatic behavior involving the cytoskeleton is vascular invasion, during which cancer cells bind to, separate, and then migrate through intact endothelial layers8,9. Since vascular invasion is a significant feature of cancer progression, we sought to evaluate drivers of this process with an RNA interference (RNAi) screen. In a previous experimental series, cancer cell attachment to an endothelial monolayer was used as a functional approach. Using a kinome-wide screen we identified the LATS1 kinase in the Hippo pathway as a crucial modulator of the vascular invasive phenotype of cancer cells10. In the present study, we utilized this approach in an unbiased, genome-wide RNAi screen to identify drivers of cancer cell vascular invasion outside the kinome. From this screen we identified keratin-associated protein 5-5 (Krtap5-5), a gene with no previously known role in cancer.
Krtap5-5 belongs to a large superfamily of over 100 genes coding for keratin-associated proteins (Krtap, or Kap) that are unique to mammals and involved in hair growth11–13, where Krtap mRNA comprises ~58% of all transcripts in the developing hair shaft14. Krtaps are subdivided into 3 major categories: high glycine/tyrosine, high sulfur, and ultrahigh sulfur15. Krtap5-5 is a member of the ultrahigh sulfur group and cysteine residues comprise >30% of its amino acid content. This cysteine richness is thought to impart structural rigidity upon hair by facilitating the crosslinking of keratin intermediate filaments16,17, a function that was very recently teased apart by atomic force microscopy of the hair follicle fiber18. The relationship between keratin-associated proteins and hair stiffness is also corroborated by the whole-genome mapping of the domestic goat, Capra hircus, where it was found that ultrahigh sulfur Krtaps were upregulated in the hair follicles that produce cashmere19, a hair fiber known for its structural resilience. In mice, the Krtap5 family consists of 5 members on the same chromosomal locus, and in humans this locus is duplicated to generate a total of 11 Krtap5 family genes20. Because Krtaps are expressed at low levels outside the hair follicle, distinct functions for the individual genes have not been reported and these genes have thus evaded study and characterization.
Of the three major cytoskeletal components in epithelial cells, keratin intermediate filaments are largely considered to have a structural purpose despite serving as a bridge to a variety of cell signaling pathways21,22. The broad influence of keratins is described in the current study, where we report that small hairpin RNA (shRNA)-mediated knockdown of Krtap5-5 in murine mammary E0771 carcinoma cells elicits an unexpected cellular switching of keratin intermediate filaments to vimentin intermediate filaments. The cytoskeletal crisis that follows does not impact on cell viability but leads to a reduction in cell motility, invasion from a spheroid into a 3D matrix, as well as extravasation in zebrafish and mice. Taken together, these surprising results provide new insights into how cells respond to the changes in molecules impacting their keratin cytoskeleton and suggest that Krtap5-5 is a keratin 18 regulator that can modulate cancer cell vascular invasion.
RESULTS
A genome-wide RNAi screen implicates Krtap5-5 in cancer cell interaction with endothelium
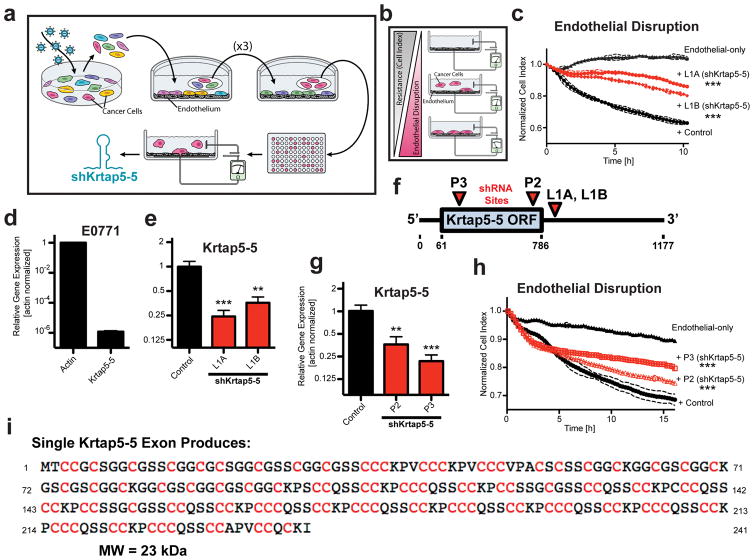
We used an unbiased, genome-wide RNAi screen to identify driver genes of cancer cell interactions with endothelia (Figure 1a). The murine mammary cancer cell line E0771 was chosen for the experiments because it generates hematogenous organ metastases from a local implantation site in immunologically intact mice23. We hypothesized that shRNAs preventing cancer cell interaction with endothelial monolayers would also interfere with vascular invasion as a first step in the metastatic cascade. A pool of cancer cells transduced with a genome-wide lentiviral RNAi library was subjected to serial rounds of selection by panning the cells on an intact endothelial monolayer and harvesting unattached cancer cell subpopulations. These subpopulations of cells were expanded between pannings by growth in regular culture dishes to avoid selection for shRNAs that negatively impact general cell attachment, growth, or viability. The selection procedure enriched for cell populations that represented a functionally distinct pool (Supplementary Figures S1A and B). To identify targeted genes, we evaluated the pooled cell population as well as clonal cell lines derived from the pool through limiting dilution. From shRNAs most enriched in the cell pool we identified four candidate genes Axin1, Pafr, Nxph1 and Snx5 by an Affymetrix array that recognizes the specific bar codes contained in the shRNA expression vector (Supplementary Figure S1C and D). Unfortunately, attempts to validate these four candidate genes using independent shRNAs were not successful (Supplementary Figures S1E and F).
Figure 1.
Genome-wide RNAi screen implicates Krtap5-5 in cancer cell interaction with an endothelial monolayer. (a) Schematic of RNAi screen. E0771 transduced with a genome-wide RNAi library were subjected to Human Umbilical Vein Endothelial Cell (HUVEC) monolayer attachment. Unattached cells were collected and expanded over three rounds. After the 4th round, clonal cells were generated by limiting dilution and tested functionally, and the inserted shRNA was identified by sequencing. (b) Electric Cell-substrate Impedance Sensing (ECIS) monitors real-time disruption of an endothelial monolayer by invading cancer cells10. (c) Clonal E0771 lines derived from the RNAi library screen (L1A, L1B) show reduced endothelial monolayer disruption relative to control cells with a non-silencing shRNA. The plot depicts real-time disruption of endothelial monolayer after addition of cancer cells (data point intervals, 15 min). *** p<0.001, vs control; mean ± SD. (d) Expression of Krtap5-5 mRNA in E0771 cells relative to actin. Mean ± SEM. (e) Krtap5-5 mRNA in RNAi library screen-derived L1A and L1B cells. ** p<0.01, *** p<0.001, vs controls; mean ± SEM. (f) shRNA targeting sites relative to the Krtap5-5 mRNA. P2, P3 shRNAs were used to generate de novo pooled knockdown cell lines. ORF = open reading frame. (g) Krtap5-5 mRNA in P2 and P3 cells. ** p<0.01, *** p<0.001, vs controls; mean ± SEM. (h) P2 and P3 cell disruption of an endothelial monolayer as in panel c. *** p<0.001, mean ± SD. (i) Krtap5-5 amino acid sequence highlighting cysteine residues.
Alternatively, direct sequencing of shRNAs expressed in the clonal cell lines identified Fam73b and Krtap5-5 as potential candidate genes, neither of which had a previously known function in cancer. The Fam73b shRNA-containing cell lines showed a reduced ability to disrupt an endothelial monolayer relative to a control transduced with non-silencing shRNA (Supplementary Figure S1G). However, we were unable to validate a gene knockdown in those cells (Supplementary Figure S1H) and concluded that the phenotype was likely due to an off-target effect of the shRNA. Beyond these five false-positive candidate genes, we identified a Krtap5-5 targeting shRNA in two separate clonal cell lines selected after the panning (L1A, L1B). These cell lines showed reduced endothelial invasion when compared with the non-silencing shRNA control in an assay that monitors the real-time disruption of an endothelial monolayer (Figures 1b and c), and Krtap5-5 mRNA expression was also decreased (Figures 1d and e). Thus, preliminary data indicated that Krtap5-5 might be required for vascular invasion by E0771 cells, and we sought to further verify that this dependency was not an artifact of cloning lines from the shRNA screen. Indeed, Krtap5-5 knockdown with independent shRNAs also mitigated the cells’ ability to disrupt an endothelial monolayer (P2, P3; Figures 1f–h; Supplementary Table S1). The Krtap5-5 gene codes for a cysteine-rich protein (Figure 1i) that is highly homologous to other Krtap proteins20. Therefore, quantitation of the endogenous Krtap5-5 protein by mass spectrometry or by antibody-based approaches was not possible. Still, distinct expression of specific Krtaps in the hair follicle has been shown by detection of the respective mRNAs15 and here we thus utilized real-time RT-PCR for quantitation. Overall, we conclude from the above data that Krtap5-5 is a candidate gene that impacts cancer cell interaction with endothelia in vitro. We decided to study Krtap5-5 in detail because the function of a keratin-associated protein had yet to be described within the context of cancer.
Depletion of Krtap5-5 reduces cancer cell motility via disruption of the keratin cytoskeleton
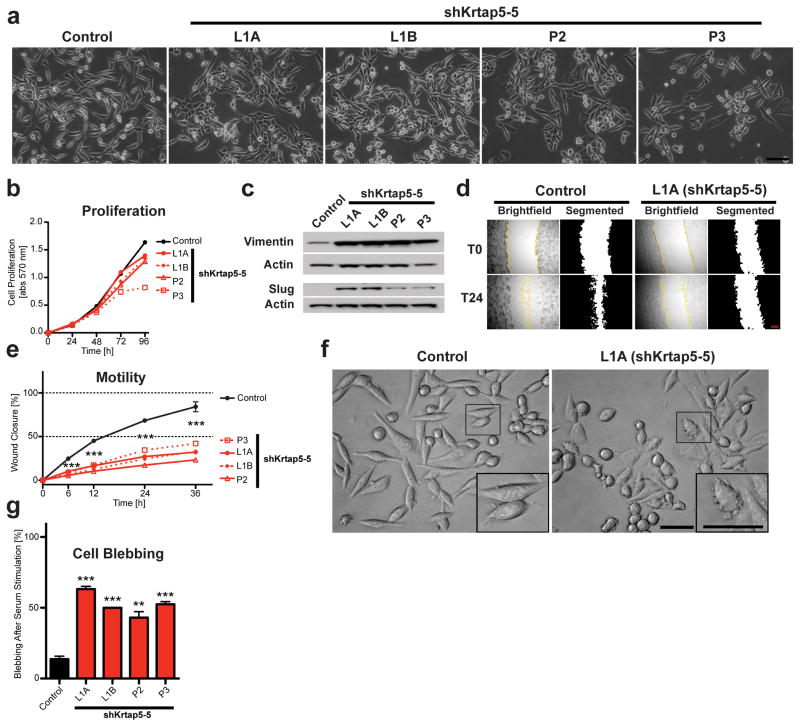
A comparison of cell morphology revealed that the Krtap5-5 knockdown cells tended to aggregate into clusters (Figure 2a), which impacted cell growth only at high cell density (Figure 2b). This morphological change suggested a more epithelial characteristic, implying that shRNA-mediated knockdown of Krtap5-5 could be causing a mesenchymal-epithelial transition. However, the Krtap5-5 knockdown cells exhibited a concomitant increase in the expression of vimentin and Slug, both of which are associated with a mesenchymal phenotype (Figure 2c; Supplementary Figures S2A and B). Additionally, reduced motility of the Krtap5-5 knockdown cell lines in scratch-wound assays matched with the clustered cell growth (Figures 2d and e).
Figure 2.
Krtap5-5 is required for cell motility and morphology. (a) Phase contrast images of control and Krtap5-5 knockdown cells. Scale bar, 100 μm. (b) Cell proliferation. Mean ± SEM; n=3. (c) Western blot for vimentin and slug. (d) Example images of scratch-wound assays at 0 and 24 hrs. Scale bar, 0.25 mm. (e) Quantitation of scratch-wound closure (with 5 μg/ml mitomycin C). n = 12 / cell line. *** p<0.001, vs control; mean ± SEM. (f) Example of cell blebbing in a Krtap5-5 knockdown line after serum addition. Inset: Magnified individual cells. Scale bar, 50 μm. (g) Quantitation of cell blebbing. Cells with blebs were counted in 15 independent, 40x microscopy fields per line. ** p<0.01, *** p<0.001, vs control; mean ± SEM.
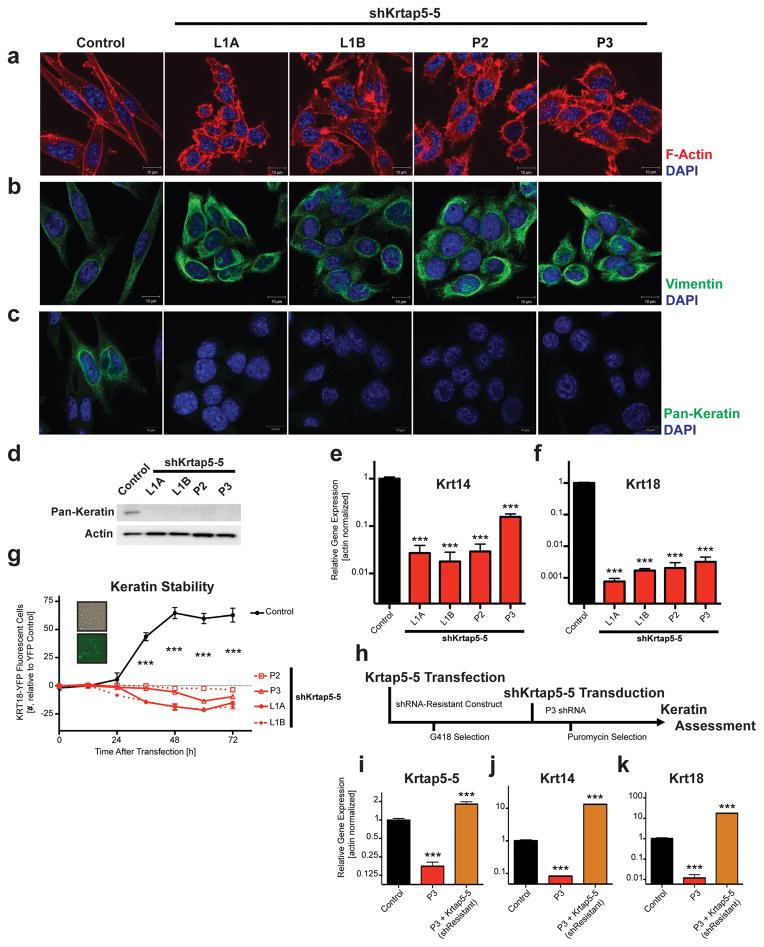
Upon closer inspection of cells after serum stimulation, we saw significantly increased blebbing in the Krtap5-5 knockdown cells (Figures 2f and g), suggesting cytoskeletal effects of the knockdown24. Indeed, confocal microscopy showed altered F-actin organization in the Krtap5-5 knockdown lines (Figure 3a). While actin stress fibers were readily observable in the control, Krtap5-5 knockdown lines demonstrated an increased amount of filopodial protrusions (Supplementary Figure S3A), despite tubulin being unaffected (Supplementary Figures S3B and C). The increase in vimentin protein in the knockdown cell lines (see Figure 2c) was also confirmed via immunofluorescence (Figure 3b; Supplementary Figure S3D). Because Krtap5-5 is a keratin-associated protein, we conjectured that the increase in vimentin intermediate filaments could be cellular compensation for a perturbation of keratin intermediate filaments. Consistent with this hypothesis, pan-keratin staining was below detection in Krtap5-5 knockdown cells (Figure 3c; Supplementary Figure S3E). Western blot analysis showed that total keratin protein was also below detection, corroborating the immunofluorescence observations (Figure 3d; Supplementary Figure S3F). We sought to further validate these findings at the mRNA level, focusing on keratins 5/14 and 8/18 because they are expressed by both normal and cancerous mammary epithelia25–27. We found that the mRNA levels for both Krt14 and Krt18 were significantly downregulated in Krtap5-5 knockdown cells (Figures 3e and f; Supplementary Figure S3G–J). Since depletion of Krtap5-5 led to the loss of key keratins, we hypothesized that Krtap5-5 function was necessary for the stability of keratin intermediate filaments in these cells. To address this, the Krtap5-5 knockdown lines were transiently transfected with a fluorescently tagged human KRT18 expression vector28 to provide an excess of dimerization partner for the endogenous Krt8, whose level was unaffected by the Krtap5-5 shRNA (Figure S3I). Exogenously introduced KRT18 was readily detectable in control cells between 36 and 72 hours after transfection (Figure 3g). However, only minimal expression of the protein was observable in Krtap5-5 knockdown cells, suggesting that they were unable to process keratins into intermediate filaments, and that delivery of exogenous KRT18 could not rescue the phenotype (Figure 3g; Supplementary Figure S3K).
Figure 3.
Krtap5-5 stabilizes the keratin cytoskeleton. (a) Phalloidin staining for F-actin reveals cytoskeletal dynamics in Krtap5-5 knockdown lines. (b) Confocal imaging of vimentin intermediate filaments. (c) Confocal imaging of keratin intermediate filaments. Scale bars are 10 μm. (d) Western blot for pan-keratin in the control vs Krtap5-5 knockdown cell lines. (e, f) Keratin 14 (e) and Keratin 18 (f) mRNA expression in Krtap5-5 knockdown lines. *** p<0.001, vs control; mean ± SEM. (g) Time-course of expression of exogenous, fluorescently tagged human keratin 18 in control and Krtap5-5 knockdown cells. Fluorescent cells were quantitated in twelve microscopy fields. The fluorescent cell number was derived after normalization to a YFP transfection control. *** p<0.001, vs control; mean ± SEM. (h) Schematic depicting the generation of a cell line resistant to the Krtap5-5 shRNA. Expression of (i) Krtap5-5, (j) Krt14, and (k) Krt18 mRNA in E0771 cells relative to actin after transfection with the shRNA-resistant Krtap5-5 expression vector. *** p<0.001, vs control; mean ± SEM.
Since these data indicated that Krtap5-5 function was required for Krt14 and Krt18 expression, we next determined if exogenous expression of an shRNA-resistant Krtap5-5 (Figure 3h) could reverse the cytoskeletal alterations and cellular phenotype. Indeed, the shRNA-resistant Krtap5-5 expression vector restored Krtap5-5 expression (Figure 3i), normalized cell morphology, reverted vimentin protein expression to control levels (Supplementary Figures S3L and M) and restored expression of Krt14 and Krt18 (Figures 3j and k). However, in the Krtap5-5 rescue cells the expression of exogenously introduced shRNA-resistant Krtap5-5 was 2-fold above control and endogenous Krt14 and Krt18 were raised to >10-fold of control (Fig. 3i, j and k). The high level of keratin expression in the rescued cells can induce stiffening of the cytoskeleton as reported recently18 and thus impact cellular behavior beyond the mere rescue from the Krtap5-5 knockdown. In summary, Krtap5-5 regulates keratin intermediate filaments and can thus modulate cancer cell motility.
Krtap5-5 impacts integrin signals and cell invasion through extracellular matrix
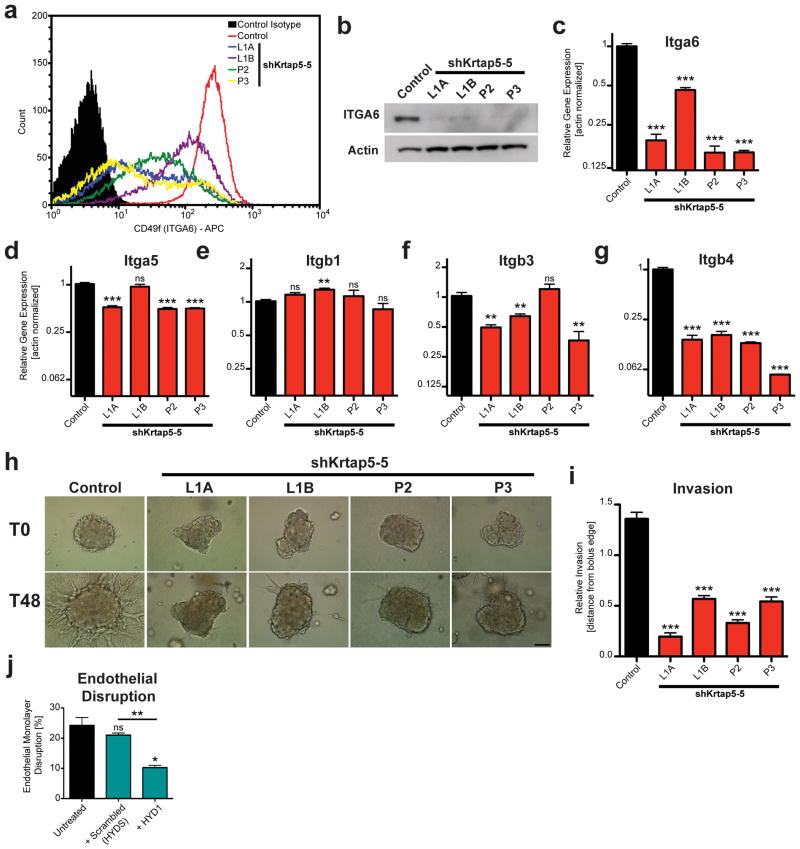
Because keratins are known to interact with hemidesmosomal integrins that link epithelial cells with their basement membrane29, we hypothesized that, by modulating keratin assembly, Krtap5-5 might also affect integrin function at the cell surface. The α6-integrin (Itga6, CD49f) was analyzed because it dimerizes with β4-integrin (Itgb4, CD104) to form a receptor for laminin and is therefore a component of the hemidesmosome. Flow cytometric analysis showed a reduction of Itga6 in Krtap5-5 knockdown cell lines (Figure 4a) that correlated with the magnitude of the knockdown (see Figures 1e–g). Western blot analysis then revealed that the level of total Itga6 protein was reduced after Krtap5-5 knockdown (Figure 4b; Supplementary Figure S4A), indicating that the decrease in cell surface Itga6 presentation was not caused by a defect in cellular transport of the protein. Furthermore, the loss of Itga6 in Krtap5-5 knockdown cells was paralleled at the mRNA level (Figure 4c). The expression levels of other integrins known for their roles in attachment and motility30 also showed distinct alterations after Krtap5-5 knockdown (Figures 4d–g; Supplementary Figure S4B). Itgb4 had the largest decrease in expression (Figure 4g); Itga5 and Itgb3 showed a smaller downregulation in three of the four knockdown cell lines and Itgb1 was not altered. Given the particular effect on α6- and β4-integrins, these observations support the concept that depletion of Krtap5-5 negatively impacts hemidesmosome stability.
Figure 4.
Krtap5-5 stabilizes hemidesmosomal integrins and permits cell invasion. (a–c) α6-integrin (ITGA6, CD49f) expression in control and Krtap5-5 knockdown cells. Flow cytometry for cell surface protein (a), total protein by Western blot (b), mRNA expression (c); ** p<0.01, *** p<0.001, mean ± SEM. (d–g) mRNA expression of α5-integrin (d), β1-integrin (e), β3-integrin (f), and β4-integrin (g); ** p<0.01, *** p<0.001, mean ± SEM. (h, i) Cell invasion of control and Krtap5-5 knockdown cells from a spheroid into a 3D matrix consisting of Matrigel and type I collagen. (h) Representative images at 0 and 48 hours. Scale bar, 100 μm. (i). Quantitation of cell invasion. *** p<0.001, versus control; mean ± SEM; boluses quantitated: n= Control (49), L1A (41), L1B (47), P2 (34), P3 (38). (j) Endothelial monolayer disruption by E0771 cells treated with a α6/β4-integrin peptide blocker (HYD1) vs scrambled peptide (HYDS) after 15 hours. ns, not significant vs control; * p<0.05, ** p<0.01, mean ± SEM.
To evaluate hemidesmosomal function, cells were embedded into a 3D matrix consisting of Matrigel plus type I collagen and cell invasion into the matrix was monitored over time31. There was a significant reduction in invasive ability after Krtap5-5 knockdown (Figures 4h and i; Supplementary Figures S4C and D), consistent with the reduced cell motility (see Figure 2e). A similar observation was made during cell growth in 3D (Supplementary Figure S4E). To determine if hemidesmosomal integrins were required for vascular invasion, we next treated E0771 cells with the hemidesmosomal integrin blocker HYD132,33 and then tested their ability to disrupt an endothelial monolayer. When compared to treatment with a scrambled peptide control, we discovered that blocking hemidesmosomal integrins with HYD1 was sufficient to reduce endothelial invasion (Figure 4j), suggesting that the reduction in α6/β4-integrins following the depletion of Krtap5-5 is relevant for not only extracellular matrix invasion but also vascular invasion. In conclusion, these data demonstrate that Krtap5-5 impacts integrin expression and function, thereby resulting in an attenuated invasive state.
Knockdown of keratin 18 phenocopies depletion of Krtap5-5
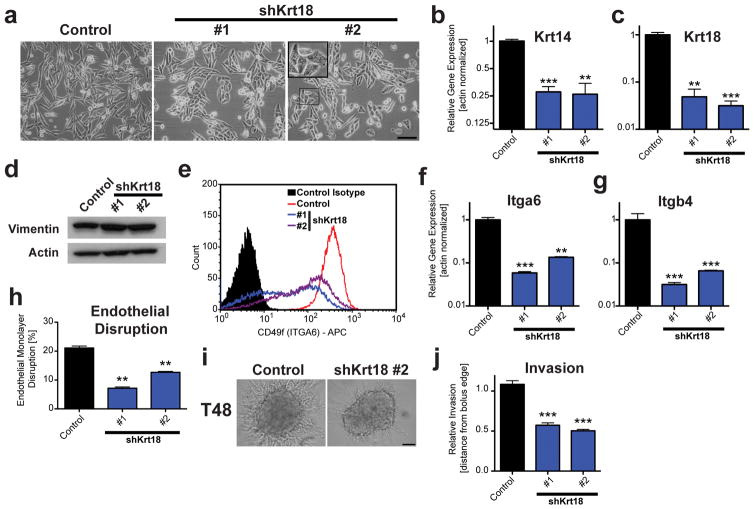
Our characterization revealed that depletion of Krtap5-5 led to a reduction in gene expression for both keratins and integrins. The most dramatic change observed was the >110-fold downregulation of Krt18 mRNA (see Figure 3f), which was prevented by an shRNA-resistant Krtap5-5 expression vector (see Figure 3k). These findings suggested that keratin 18 could be functionally linked to Krtap5-5. Indeed, lentiviral knockdown of Krt18 with two independent shRNAs in E0771 cells phenotypically recapitulated the effects of Krtap5-5 depletion. Akin to the depletion of Krtap5-5, E0771 cells transduced with two independent Krt18 shRNAs presented with an epithelial-like morphology and cell blebbing in vitro (Figure 5a), which correlated with reduced Krt14 and Krt18 expression (Figures 5b and c) in addition to a modest increase in vimentin expression (Figure 5d, Supplementary Figures S5A and B). Knockdown of Krt18 also reduced cell surface localization of Itga6 (Figure 5e) and decreased the mRNA expression of Itga6 and Itgb4 (Figure 5f and g). Functional assessment of Krt18 knockdown cells revealed diminished abilities to disrupt an endothelial monolayer (Figure 5h) and invade from a cell spheroid into surrounding extracellular matrix (Figures 5i and j). Taken together, the observation that the knockdown of Krt18 phenocopies the depletion of Krtap5-5 supports the notion of keratin 18 as the major effector of the cellular phenotype.
Figure 5.
Silencing of Krt18 extensively phenocopies depletion of Krtap5-5. (a) Phase contrast images of control and Krt18 knockdown cells. Inset: Blebbing cell. Scale bar, 100 μm. (b, c) Keratin 14 (b) and Keratin 18 (c) mRNA expression in Krt18 knockdown lines. ** p<0.01, *** p<0.001, vs control; mean ± SEM. (d) Western blot for vimentin. (e) Flow cytometry for α6-integrin (ITGA6, CD49f) surface protein in control and Krt18 knockdown cells. (f, g) Itga6 (f) and Itgb4 (g) mRNA expression in Krt18 knockdown lines. ** p<0.01, *** p<0.001, vs control; mean ± SEM. (h) Endothelial monolayer disruption by E0771 cells at 15 hrs after silencing Krt18. ** p<0.01, mean ± SEM. (i, j) Cell invasion of control and Krt18 knockdown cells from a spheroid into a 3D matrix consisting of Matrigel and type I collagen. (i) Representative images at 48 hours. Scale bar, 100 μm. (j) Quantitation of cell invasion. *** p<0.001, versus control; mean ± SEM; n= 21 (control), n= 32 (shKrt18 #1) and n= 68 (shKrt18 #2) spheroids per cell line.
Disruption of the keratin cytoskeleton broadly alters cytoskeleton regulators and cell surface markers
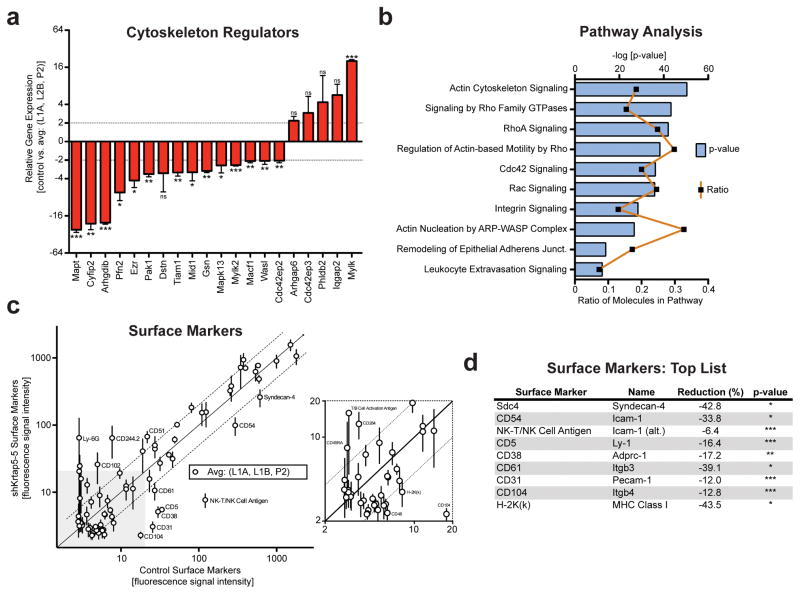
We also assessed the expression of cytoskeleton regulators following the depletion of Krtap5-5, hypothesizing that important regulators would be affected because the switching from keratin to vimentin intermediate filaments led to the disorganization of the actin cytoskeleton (see Figure 3a–c). We used a PCR array to evaluate the expression of 84 cytoskeleton regulators in E0771 cells in response to silencing Krtap5-5 and found that the expression of 15 genes were significantly altered by >2-fold (Figure 6a; Supplementary Table S2). A subsequent pathway analysis indicated a loss of signaling in pathways centering on the actin cytoskeleton, integrin signaling, and leukocyte extravasation signaling in corroboration of our earlier findings (Figure 6b; Supplementary Table S3). We next determined if other cell surface proteins were affected after the loss of the keratin cytoskeleton via depletion of Krtap5-5, building on the effect observed with integrins (see Figures 3a–g). To accomplish this, we conducted a flow cytometric screen of 176 unique cell surface antigens and detected 72 of these in E0771 cells (Supplementary Table S4). A comparison of cell surface antigen expression demonstrated that multiple surface proteins changed in response to Krtap5-5 depletion (Figure 6c). An analysis of the top hits revealed that knockdown of Krtap5-5 reduced the cell surface presence of various markers associated the immune system and cell adhesion (Figure 6d). In addition, the observed flow cytometric reduction in CD104 further confirmed our discovery that β4-integrin is reduced after the depletion of Krtap5-5 (see Figure 4g). These findings suggest that the expression of a keratin-associated protein like Krtap5-5 not only impacts the keratin cytoskeleton but also affects the expression of cytoskeleton regulators and cell surface proteins.
Figure 6.
Manipulating the level of Krtap5-5 broadly impacts on cytoskeleton regulators and cell surface proteins. (a) mRNA profile of cytoskeleton regulators exhibiting >2-fold change. Average of Krtap5-5 knockdown lines (L1A, L1B, P2) vs control. * p <0.5, ** p<0.01, *** p<0.001, mean ± SEM. (b) Pathway analysis (Ingenuity) using the entire cytoskeleton regulator panel after knockdown of Krtap5-5. The most impacted pathways are shown arranged by the p-value (log 10). The ratio depicts the number of molecules affected in the respective pathway. (c, d) Flow cytometry surface protein screen of 72 surface proteins in Krtap5-5 knockdown lines (L1A, L1B, P2) vs control. Averaged data (c) and top hits after statistical analysis (d) are shown. A 2-fold cutoff is denoted by dashed lines; surface proteins residing outside the cutoff are labeled. * p<0.05, ** p<0.01, *** P<0.001 versus control.
Krtap5-5 is required for cancer cell extravasation in vivo
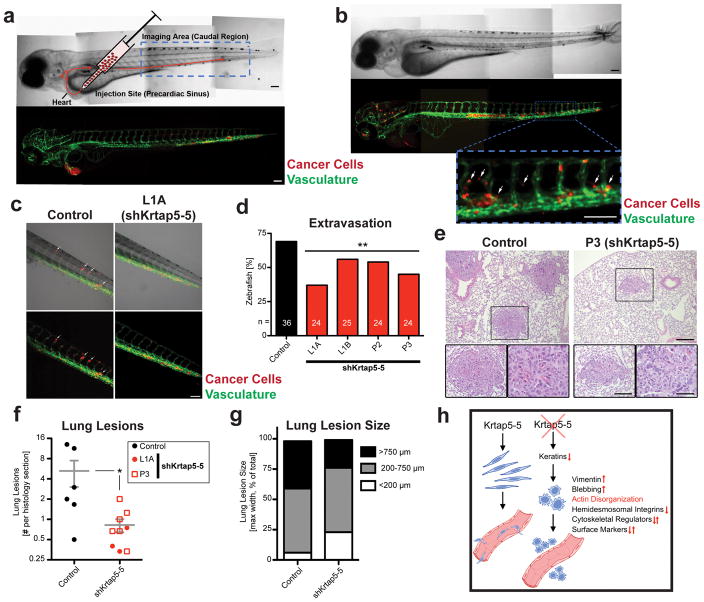
Since Krtap5-5 knockdown altered endothelial monolayer interaction and decreased cancer cell motility and matrix invasion in vitro, we further evaluated the in vivo effect in a recently described zebrafish model10. For this purpose, cancer cells were labeled with a red fluorescent dye and injected into the precardiac sinus of 2-day-old zebrafish embryos (Figure 7a). The ability of cancer cells to travel through the vasculature and then extravasate into the caudal region was visualized (Figure 7b), taking advantage of the embryo’s transparency and GFP expression in the endothelia34. Imaging 24 hours after injection revealed that the knockdown of Krtap5-5 reduced the ability of cancer cells to extravasate from the vasculature and into the surrounding tissues (Figures 7c and d; Supplementary Figure S6A). We next sought to further validate these findings within a syngeneic host for the cancer cells. To accomplish this, either control or Krtap5-5 knockdown E0771 cells were injected into the tail vein of wild-type C57BL/6 mice for an extravasation and lung colonization assay. Histological analysis of the lungs 3 weeks after injecting the cancer cells revealed a significant overall reduction in lung metastic seedings from cells with silenced Krtap5-5 (p=0.02; Figures 7e and f). In addition, there was a trend for smaller lung lesions in mice receiving Krtap5-5 knockdown cells versus the control (Figure 7g). These zebrafish and mice data therefore confirm that the altered cytoskeletal function after the knockdown of Krtap5-5 causes a reduction in vascular invasion. These findings are summarized in the model shown in Figure 7h.
Figure 7.
Krtap5-5 impacts cancer cell extravasation in zebrafish10 and mice. (a) Cancer cells were fluorescently labeled, injected into the precardiac sinus of transparent zebrafish embryos at 2 days post fertilization, and imaged 24 hours later. The endothelia are visualized through coral reef GFP expression under the control of a VEGFR2 promoter. Blue box=imaging area. Scale bar, 0.1 mm. (b) Representative image with cancer cells that have extravasated from the vasculature (arrows). Scale bar, 0.1 mm. (c) Representative images of caudal region, showing extravasation of control and of Krtap5-5 knockdown cells (arrows). (d) Quantitation of extravasation. Zebrafish with >5 extravasating cancer cells were scored as positive events. The number of fish is shown on the bars; ** p<0.01 versus control. (e) Histological example of lung lesions in mice injected with either control or P3 E0771 cells. Representative lesions are shown at different magnifications: Scale bars, 200 μm, 100 μm, 50 μm respectively. (f) Number of lung lesions detectable per lung section in mice injected with either control (n= 6 mice) or shKrtap5-5 cells (n=9 mice). Nonparametric Mann-Whitney U test, p = 0.0282 controls versus shKrtap5-5. (g) Comparison of lung lesion sizes. Data are depicted as percent of total. (h) Graphical summary of the findings in this paper.
Exploring the function of Krtap5-5 in other cell lines and human cancer
We sought to generalize our findings by examining the effect of Krtap5-5 silencing in other murine cell lines. Similar to E0771 cells, the knockdown of Krtap5-5 impacted the expression of both Krt14 and Krt18 in mammary carcinoma 4T1 cells35 and in D10 cells, a primary clonal cell line generated from a pancreatic ductal adenocarcinoma in our lab as described recently36 (Supplementary Table S5). In contrast to E0771 cells, the depletion of Krtap5-5 in these cell lines disparately affected the expression of these keratins, even increasing them in some instances, which ultimately enhanced their ability to invade an endothelial monolayer. These findings imply that Krtap5-5 governs cytoskeletal function and vascular invasion in other cell lines, although its function is likely context-dependent.
Extrapolation of our findings into human cancer is challenging because KRTAP5-5, and keratin-associated proteins in general, are expressed at low levels and rarely exceed the threshold required for detection by gene expression surveys of human cancers deposited in databases. Nevertheless, since our study showed that the expression of Krtap5-5 in murine cancer cells has significant phenotypic effects in vitro and in vivo that were functionally connected to Krt18, we explored the relevance of keratin 18 within the context of the human disease. Although others have demonstrated the KRT18 protein level can be a favorable prognostic factor in breast cancer37, a Kaplan-Meier analysis38 conducted on mRNA expression showed that KRT8 and KRT18 levels correlate with lower relapse-free survival, lower distant metastasis-free survival, as well as lower overall survival (Supplementary Figures S7A–F). We next interrogated the pan-cancer TCGA dataset39 and found that despite KRTAP5-5 expression being below detection for most samples in the dataset, a positive correlation exists between mRNA levels for KRTAP5-5 and KRT18 (Supplementary Figure S7G). This correlation provides additional support for a functional link between KRTAP5-5 and KRT18 in human cancer. Finally, comparing KRTAP5-5 expression across distinct cancers in the dataset indicates that KRTAP5-5 is differentially expressed between cancer types with the highest expression levels occurring in rectal, colon, and pancreatic cancers (Supplementary Figure S7H). These findings from different databases and other cell lines support the significance of keratin intermediate filaments in malignant progression and suggest that modulating the levels of KRTAPs in human cancer could impart significant changes upon keratins to affect patient survival.
DISCUSSION
Krtap5-5 emerged as a surprising candidate from an unbiased, genome-wide RNAi screen to identify genes involved in cancer cell interaction with the endothelium, vascular invasion, and extravasation. In the E0771 mammary cancer cells, its knockdown then exposed a unique vulnerability in their vascular invasive phenotype that consisted of cytoskeletal switching from keratin to vimentin intermediate filaments and disruption of hemidesmosomal integrins. Moreover, the mRNA expression level of KRTAP5-5 positively correlates with KRT18 in human cancers, where KRT18 mRNA expression is further associated with a significantly worse prognosis as measured by time to disease relapse, distant metastasis, and overall survival of patients with breast cancers. In summary, these findings indicate that this pathway is relevant for progression towards a more malignant disease.
Between rounds of panning for endothelial monolayer attachment, the selected cancer cell subpopulations are propagated in tissue culture dishes. This functional approach avoids the selection for shRNA-targeted genes that negatively influence cell growth and survival. Indeed, here we identified a pathway gene, Krtap5-5, that can exert control of cytoskeletal function without resulting in defective growth of cells. The E0771 cells use both keratin and vimentin intermediate filaments to comprise their cytoskeleton, perhaps the result of their epithelial-mesenchymal transition40. The coexpression of vimentin and keratin intermediate filaments has been previously documented in breast cancer cell lines and is thought to enhance their invasive ability41. Breast cancers coexpressing both keratins and vimentin could therefore be more susceptible to the loss of one of the components, e.g. their keratin cytoskeleton, which may in turn have consequences on cell invasion. The outcome of losing the keratin cytoskeleton has been documented by others in keratinocyte models where knockout of keratins did not affect the actin cytoskeleton42 or increased cell invasion43. Our findings deviate from this paradigm in the E0771 mammary cancer model, as depletion of Krtap5-5 not only caused loss of the keratin cytoskeleton but also led to disorganization of the actin cytoskeleton and reduced cell invasion. Our discovery that knockdown of Krtap5-5 or Krt18 affects α6/β4-integrins is supported by others who reported that keratins stabilize hemidesmosomes via their interplay with β4-integrin44.
The observation that knockdown of keratin 18 extensively phenocopied depletion of Krtap5-5 indicates that Krt18 is the major effector of the cellular phenotype. When compared to the knockdown of Krt18, the silencing of Krtap5-5 appears to be a more potent inducer of the intermediate filament subtype switching seen between keratins and vimentin. The depletion of Krtap5-5 dramatically decreased the expression of Krt18, an effect that was prevented by transfection with an shRNA-resistant Krtap5-5 expression vector. Taken together, these experiments suggest that Krtap5-5 is likely a chief regulator of the keratin cytoskeleton in E0771 cells, although it is difficult to rule out that some aspects of the phenotype may result from effects on other highly homologous Krtap genes. Nonetheless, our study demonstrates that the broad influence of Krtap5-5 reaches well beyond keratins and integrins to impact cytoskeleton regulators and other cell surface receptors. From this finding we propose that manipulating the levels of keratin-associated proteins in human cancer might represent a new method to influence the status of cell surface receptors, where altering the exterior of a cancer cell might make it more sensitive to therapy or attack by the immune system.
A significant question is how does the knockdown of low levels of endogenous Krtap5-5 mRNA lead to the observed phenotype? First, it is possible that keratin-associated proteins are intermediate filament regulators that are kept in low abundance in all epithelial cells outside of the hair follicle, and downstream signaling molecules amplify small changes in their expression level. The second possibility raised by our study is that cancer cells may become dependent on a critical pathway for the maintenance of the cytoskeleton that is not normally utilized by cells outside the hair follicle. Regardless of which possibility is true, our data suggest a significant role for keratin-associated proteins in cancer biology.
Krtap5-5 belongs to a large family of related Krtaps, which is then part of an even larger superfamily. Although this study provides a framework for the function of Krtap5-5, there is an intriguing possibility that other cancer types may utilize Krtap5-5 or additional family members in the context of malignant progression. Our data indicate that silencing Krtap5-5 alters the cytoskeleton and modulates vascular invasion in multiple cell lines, yet an increase or decrease in vascular invasion can occur depending on the specific line. We propose that this influence is chiefly inhibitory for the vascular invasive phenotype of E0771 cells because the expression of both Krt14 and Krt18 is decreased in the line. The function of Krtap5-5 in other cell lines may depend on the cytoskeletal context and could explain why silencing Krtap5-5 leads to a gain in keratin expression and the enhancement of vascular invasion. Clearly, more work is needed to pinpoint and describe the functionally relevant keratin-associated proteins in other cell systems. Moving forward, a major technical challenge is the sequence homology shared by keratin-associated proteins, which makes it difficult to distinguish between the different members. Still, their influence on downstream targets, like keratins as well as other effectors, and their phenotypic impact was quite striking in E0771 cells. In summary, this work suggests that Krtap5-5 is a previously unknown regulator of cytoskeletal function in cancer cells and can thus modulate motility and vascular invasion.
METHODS
Cell Culture
E0771 cells (sometimes spelled EO771) were a gift from Dr. Peter Goedegeburre, Washington University in St. Louis, MO and maintained in Dulbecco’s Modified Essential Medium (DMEM; Life Technologies, Carlsbad, CA, USA) that was supplemented with 10% fetal bovine serum. Primary human umbilical vein endothelial cells (HUVECs) were cultured according to supplier’s recommendations (Lonza, Walkersville, MD, USA).
RNAi Screen and shKrtap5-5 Line Generation
E0771 cells were lentivirally transduced with a mouse genome-wide RNAi library (System Biosciences, Mountain View, CA, USA) using a multiplicity of infection of 3.75, which resulted in a low likelihood of there being more than one shRNA per cell. Cells were selected in puromycin (Sigma-Aldrich, St. Louis, MO, USA) for 1 day and then cultured for an additional 8 days prior to the endothelial attachment assay. In the shRNA screen, the transduced E0771 cells were transferred onto confluent monolayer of human umbilical vein endothelial cells (HUVECs). Unattached E0771 cells were collected, propagated on plastic, and then subjected to a total of 4 rounds of selection. Following the 4th round, clonal lines were generated and those retaining an attenuated ability to interact with endothelium had their shRNAs sequenced. Krtap5-5 was identified by first amplifying shRNA from total cellular RNA using proprietary primers (System Biosciences, Mountain View, CA, USA). The resulting cDNAs were then cloned into a p-GEM-T Easy Vector (Promega, Madison, WI, USA), and ligated plasmids were next sent out for sequencing (MCLAB, South San Francisco, CA, USA). This process identified 2 clonal E0771 cell lines (L1A and L1B) that contained an shRNA targeting Krtap5-5. Additional stable Krtap5-5 shRNA knockdown lines (P2 and P3) were generated independently of the screen using unique shRNA sequences inserted into GIPZ lentiviral vectors (Dharmacon, Lafayette, CO USA). Krt18 knockdown lines were generated in the same manner. All stable knockdown lines were selected in puromycin (Sigma-Aldrich, St. Louis, MO, USA).
Motility, Proliferation, and Invasion Assays
Motility “scratch-wound” assays were conducted on confluent cultures of E0771 cells, onto which scratches were made using a 200-μl pipet tip. Wounds were washed with sterile PBS (Life Technologies, Carlsbad, CA, USA) twice, and then fresh culture media containing 5 μg/ml mitomycin C (Sigma-Aldrich, St. Louis, MO, USA) was added to the wounded cultures to inhibit cell proliferation. Microscopy images were taken at the designated timepoints and subjected to color segmentation using the Scratch Assay Analyzer plugin (MiToBo Suite) for ImageJ (NIH, Bethesda, MD, USA). The color segmentation procedure produced black pixels for area covered by cells, and pixel coverage was used to generate the graphs.
Proliferation assays were carried out using the crystal violet stain (Sigma-Aldrich, St. Louis, MO, USA). Cells were seeded into a 96-well plate at a density of 1,000 cells per well. At the designated timepoints, cells were fixed in 4% paraformaldehyde and stained in 0.1% crystal violet. The dye was then solubilized using 10% acetic acid and diluted 1:4 in water before absorbance measurement at 570 nm.
The xCelligence Electric Cell-substrate Impedance Sensing (ECIS) system was used for the endothelial monolayer disruption assay (ACEA Biosciences, San Diego, CA, USA). E-Plate View 16 Arrays (ACEA Biosciences, San Diego, CA, USA) were coated with 0.1% gelatin, and 40,000 HUVECs were added per well and given 24–28 hours to form a stable monolayer. Next, 20,000 cancer cells were added per well and disruption of the endothelial monolayer was then monitored in real time. The change in electrical impedance was measured at 15-minute intervals for the duration of the assay. A monolayer without exposure to cancer cells was used as a control. The HYD1 α6/β4-integrin peptide blocker and its corresponding control32,33 were obtained by gift of Dr. Anne Cress. For the hemidesmosomal integrin blocking experiments, E0771 cells were treated with 30 μg of the peptides in 300 μl of serum-free media for 15 minutes prior to seeding the cancer cells into the endothelial monolayer disruption assay.
The 3D invasion assay was conducted as previously described31. Briefly, 1,000 cancer cells were grown in drops that hung from the lid of a cell culture dish for 72 hours until a cell aggregate had formed. Cellular spheroids were then embedded into an 1:1 admixture of Matrigel (Corning, New York, NY, USA) and type I rat-tail collagen (Millipore, Billerica, MA, USA). Cell invasion out of the aggregates was then monitored over time and microscopy photographs were taken. The invasive distance was a pixel measurement acquired in ImageJ, defined as the pixel length from the longest invasive protrusion to the center of a spheroid minus the pixel length for the radius of a given spheroid.
Krt18 and Krtap5-5 Transfection Assay
The human KRT18-YFP expression vector28 was a generous gift from Dr. Rudolf Leube and Dr. Nicole Schwarz. E0771 cells were transiently transfected with the plasmid or a YFP-only transfection control, and the resulting cellular fluorescence was monitored using a 20x objective lens on an IncuCyte Zoom Imaging System (Essen Bioscience, Ann Arbor, MI, USA). A processing definition was created in the IncuCyte Zoom software to quantitate the number of fluorescent cells in a given microscopy field. Data were graphed as the number of fluorescent cells per microscopy field after transfection with the KRT18-YFP vector minus the number of fluorescent cells visible after transfection with the YFP-only control.
The Krtap5-5 shRNA-resistant vector (Genscript, Piscataway, NJ, USA) used in the shRNA targeting experiment was designed to have the putative P3 shRNA binding site mutated and was Myc-tagged, where the sequence 153-CTCCAGCTGCTGTTGC-168 in wild-type Krtap5-5 was altered to 153-AAGCTCCTGTTGCTGT-168 in the shRNA resistant vector. After transfecting this construct into E0771 cells, stable transfectants were generated after 7-day selection in G418. Transduction with the P3 shRNA proceeded as described above. For experiments involving the shRNA-resistant line, Krtap5-5 gene expression was assessed using a primer pair designed against the gene’s open reading frame.
Gene Expression and Cytoskeleton Regulators Panel
Primers were purchased from Integrated DNA Technologies, Coralville, IA, USA. Cellular RNA was extracted using an RNeasy Kit (QIAGEN, Valencia, CA, USA), cDNA was generated using an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA), and gene expression assays were performed with the iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA). All quantitative PCR reactions were conducted on a Realplex2 PCR Cycler (Eppendorf, Hamburg, Germany). The primer sequences for mouse genes are as follows:
Actb (FWD 5′-GGCGCTTTTGACTCAGGATTTAA-3′,
REV 5′-CCTCAGCCACATTTGTAGAACTTT-3′),
Itga5 (FWD 5′-CTTCTCCGTGGAGTTTTACCG-3′,
REV 5′-GCTGTCAAATTGAATGGTGGTG-3′),
Itga6 (FWD 5′-TGCAGAGGGCGAACAGAAC-3′,
REV 5′-GCACACGTCACCACTTTGC-3′),
Itgb1 (FWD 5′-ACTGTGATGCCGTATATTAGCAC-3′,
REV 5′-GATATGCGTTGCTGACCAACA-3′),
Itgb3 (FWD 5′-CCACACGAGGCGTGAACTC-3′,
REV 5′-CTTCAGGTTACATCGGGGTGA-3′),
Itgb4 (FWD 5′-GCAGACGAAGTTCCGACAG-3′,
REV 5′-GGCCACCTTCAGTTCATGGA-3′),
Krtap5-5 (FWD 5′-GGTTCTCTCCTGGGTCTTTATTC-3′,
REV 5′-GCTTCCAGCAAGAGGAGTTT-3′),
Krtap5-5, Open Reading Frame (FWD-GGGGGCTGCAAGGGAAG-3′,
REV 5′-ACTGGCAACAAGAAGACCCA-3′),
Krt5 (FWD 5′-TCTGCCATCACCCCATCTGT-3′,
REV 5′-CCTCCGCCAGAACTGTAGGA-3′),
Krt8 (FWD 5′-TGTCTACTCGGTCGGACTTCT-3′,
REV 5′-GCTGCTACCTAGCTGACATGC-3′),
Krt14 (FWD 5′-AGCGGCAAGAGTGAGATTTCT-3′,
REV 5′-CCTCCAGGTTATTCTCCAGGG-3′),
Krt18 (FWD 5′-CAGCCAGCGTCTATGCAGG-3′,
REV 5′-CTTTCTCGGTCTGGATTCCAC-3′),
Vim (FWD 5′-CGTCCACACGCACCTACAG-3′,
REV 5′-GGGGGATGAGGAATAGAGGCT-3′)
The expression of cytoskeleton regulators was assessed with the RT2 Profiler qPCR Array (QIAGEN, Valencia, CA, USA). Pathway analysis was conducted on the entire panel using Ingenuity IPA (QIAGEN, Valencia, CA, USA). The RT-qPCR cycle values for all regulators tested are located in Supplementary Table 2.
Western Blotting
Cellular lysates in this study were generally prepared with an NP40-based lysis buffer. The exception is the sample used for the pan-keratin blot, which required a triton-based lysis procedure followed by cytoskeleton protein extraction and urea treatment. Extracted proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and subjected to dry transfer with an iBlot System (Life Technologies, Carlsbad, CA, USA). Antibodies for immunoblotting included the following: anti-β-actin (Millipore, Billerica, MA, USA, product: MAB1501), anti-α6-integrin (Cell Signaling Technology, Boston, MA, USA, product: 3750), anti-pan-cytokeratin (Thermo Scientific, Waltham, MA, USA, product: MA5-13203), anti-slug (Cell Signaling Technology, Boston, MA, USA, product: 3750), anti-tubulin (Cell Signaling Technology, USA, Boston, MA, product: 2148), anti-vimentin (Cell Signaling Technology, Boston, MA, USA, product: 9585), anti-mouse HRP ECL secondary (GE Healthcare, Little Chalfont, UK, product: NA931V), anti-rabbit HRP ECL secondary (GE Healthcare, Little Chalfont, UK, product: NA934V).
Immunofluorescence
General fluorescent imaging was conducted on an Olympus IX-71 inverted microscope (Olympus, Toyko, Japan) and confocal imaging on a Zeiss LSM 510 Meta confocal microscope (Carl Zeiss AG, Jena, Germany). Samples were prepared by growing cells on sterile glass coverslips and then fixing them with either 3.7% paraformaldehyde (F-actin, tubulin) or 100% −20°C methanol (vimentin, pan-keratin). Fixed cells were then permeabilized with 0.2% Triton X-100 and incubated with the following antibodies or probes: Alexa Fluor 647 Phalloidin (Life Technologies, Carlsbad, CA, USA, product: A22287), anti-pan-cytokeratin (Thermo Scientific, Waltham, MA, USA, product: MA5-13203), anti-tubulin (Cell Signaling Technology, Boston, MA, USA, product: 2148), anti-vimentin (Cell Signaling Technology, Boston, MA, USA, product: 9585), anti-rabbit Alexa Fluor 488 secondary (Life Technologies, Carlsbad, CA, USA, product: A21206), anti-mouse Alexa Fluor 488 secondary (Life Technologies, Carlsbad, CA, USA, product: A21202).
In Silico Analyses
The relationship of keratins to breast cancer patient survival was obtained in KM-plot38. Samples from systemically untreated cancer patients were analyzed and the program was allowed to auto select the best cutoff. The TCGA pan-cancer dataset was downloaded using the UCSC Cancer Genome Browser39 and correlations between KRTAP5-5 and KRT18 were examined in human cancer, where samples for which KRTAP5-5 had no expression value reported were trimmed from the dataset for being non-informative. The entire dataset was used to depict KRTAP5-5 expression across various cancer types.
Flow Cytometry and Surface Marker Screen
All cell cultures destined for flow cytometry were dissociated with Accutase (BD Biosciences, USA) to limit cleavage of extracellular receptors. Cells were labeled with the APC-conjugated anti-human/mouse CD49f/ITGA6 antibody (BioLegend, Bethesda, MD, USA, product: 313615) or the APC-conjugated Rat IgG2a,k isotype control (BioLegend, Bethesda, MD, USA, product: 400511) according to manufacturer’s recommendations. The flow cytometry cell surface marker screen was conducted on E0771 cells using the BD Lyoplate Mouse Cell Surface Marker Screening Panel (BD, Franklin Lakes, NJ, USA). The geometric mean of fluorescent values <4 was chosen as the cutoff in accordance with assay negative controls. Values for the entire dataset are contained in Supplementary Table 3.
Zebrafish Extravasation Assay
The overall approach was described recently10. In brief, cancer cells were labeled with DiI stain according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA, USA, product: D3911). The cells were then injected into the precardiac sinus of zebrafish embryos (Tg(kdrl:GRCFP)zn1;34) that were 48-hours post fertilization. Fifty zebrafish larvae were injected with each cell line to account for possible losses due to the injection. We expected to see differences in extravasation of ≥50% at an error rate of 20% with an n = 15 in each group and a p-value of 0.01. Vascular endothelial cells of the embryos were labeled with green reef coral fluorescent protein (GRCFP) that was expressed under a VEGFR2 promoter. Cancer cell extravasation in the caudal region of the embryo was evaluated 24 hours after the injection. The investigator scoring extravasation was blinded to the cell line that was being evaluated. Embryos that exhibited >5 extravasating cancer cells in the tail region were scored positively, while those exhibiting <5 extravasating cancer cells were scored as negative events. The zebrafish embryo experiments were carried out in compliance with the GUACUC (= Georgetown University Animal Care and Use Committee) recommendations: Embryos are generated according to an approved IACUC protocol. The experiments shown here are exempt per federal guidelines since no embryos are allowed to develop beyond 4 days.
Mouse Lung Seeding Assay
The mouse experiments were carried out in compliance with the GUACUC recommendations and conducted according to an approved IACUC animal protocol in male C57BL/6 mice 8–16 weeks of age. One million control, L1A or P3 E0771 cancer cells were injected via the tail vein. We expected to see differences in lung colonization of ≥50% at an error rate of 20% with an n = 6 in each group and a p-value of 0.01. Mice were euthanized 3 weeks after the injection and the lungs were collected for histological analysis after formalin fixation and paraffin embedding. The number of lesions was assessed in at least three separate tissue sections per mouse that were stained for hematoxylin and eosin and is provided as lesions per section (see Fig. 7f). A non-parametric Mann-Whitney U test was used to compare the number of lesions across groups. The size of lung lesions was determined by measuring their maximum width in ImageJ.
Statistics
Statistical comparison of means between two groups was conducted with a two-sided Student’s t-test. For experiments occurring along a time course, a one-way ANOVA test was employed to statistically compare curves in the endothelial monolayer disruption assay, whereas a two-way ANOVA with a Bonferonni post-test evaluated data from the cell motility and keratin stability assays. Prism 5 for Mac (Graphpad Software Inc.) was used to generate figures and carry out statistical analyses, where violations of normal distribution are checked by the analysis program and were not identified. Linear regression was performed to assess correlations between KRTAP5-5 and KRT18 in the human pan-cancer TCGA dataset. In all instances, center values represent the mean and the error bar type is defined in the figure legends, as is the sample size (n) for each group. For all figures, asterisks denote the following p-values: * p <0.5, ** p<0.01, *** p<0.001. Data depicted in the manuscript are from technical replicates that were supported by at least one other independent experiment.
Supplementary Material
Acknowledgments
We thank Peter Johnson of the Georgetown University Microscopy Core for assistance with immunofluorescence, and Dr. Karen Creswell and Alex Joseph of the Georgetown University Flow Cytometry Core for running our FACS samples. We would also like to extend our gratitude to Dr. Rudolf Leube and Dr. Nicole Schwarz for generously providing the KRT18 expression vector used in this study, as well as to Dr. Anne Cress for the HYD1 hemidesmosomal integrin blocker. Finally, we thank Dr. Ian Gallicano for critical evaluation of the manuscript and for providing constructive feedback. This study was supported by NIH/NCI grants CA71508, CA51008, CA177466 (A.W.) and CA113477 (A.T.R.).
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
EB, GS, and AW characterized the gene. EB and AW wrote the paper. CS prepared the RNAi library cells and GS conducted the RNAi screen. EG performed the zebrafish injections and scored extravasation. All authors including GY, LMW, and ATR contributed to experimental design, discussed results, and commented on the manuscript.
References
- 1.Cummings MC, Simpson PT, Reid LE, Jayanthan J, Skerman J, Song S, et al. Metastatic progression of breast cancer: insights from 50 years of autopsies. Am J Pathol. 2014;232:23–31. doi: 10.1002/path.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machesky LM. Lamellipodia and filopodia in metastasis and invasion. FEBS Lett. 2008;582:2102–2111. doi: 10.1016/j.febslet.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 3.Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia. 2007;12:143–152. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]
- 4.Liotta LA, Kohn EC. The microenvironment of the tumour–host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 5.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 8.Reymond N, d’Água BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 9.Chambers AF, Groom AC, MacDonald IC. Metastasis: Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 10.Sharif GM, Schmidt MO, Yi C, Hu Z, Haddad BR, Glasgow E, Riegel AT, Wellstein A. Cell growth density modulates cancer cell vascular invasion via Hippo pathway activity and CXCR2 signaling. Oncogene. 2015;34:5879–5889. doi: 10.1038/onc.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimomura Y, Ito M. Human hair keratin-associated proteins. J Investig Dermatol Symp Proc. 2015;10:230–233. doi: 10.1111/j.1087-0024.2005.10112.x. [DOI] [PubMed] [Google Scholar]
- 12.Wu D-D, Irwin DM, Zhang Y-P. Molecular evolution of the keratin associated protein gene family in mammals, role in the evolution of mammalian hair. BMC Evol Biol. 2008;8:241. doi: 10.1186/1471-2148-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan I, Moldonado E, Vasconcelos V, O’Brien SJ, Johnson WE, Antunes A. Mammalian keratin associated proteins (KRTAPs) subgenomes: disentangling hair diversity and adaptation to terrestrial and aquatic environments. BMC Genomics. 2014;15:779. doi: 10.1186/1471-2164-15-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefkowitz GK, Mukhopadhyay A, Cowing-Zitron C, Yu BD. The post-apoptotic fate of RNAs identified through high-throughput sequencing of human hair. PLoS ONE. 2011;6:e27603. doi: 10.1371/journal.pone.0027603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers MA, Langbein L, Praetzel Wunder S, Winter H, Schweizer J. Human hair keratin associated proteins (KAPs) Int Rev Cytol. 2006;251:209–263. doi: 10.1016/S0074-7696(06)51006-X. [DOI] [PubMed] [Google Scholar]
- 16.Deb-Choudhury S, Plowman JE, Rao K, Lee E, van Koten C, Clerens S, et al. Mapping the accessibility of the disulfide crosslink network in the wool fiber cortex. Proteins. 2014;83:224–234. doi: 10.1002/prot.24727. [DOI] [PubMed] [Google Scholar]
- 17.Fujikawa H, Fujimoto A, Farooq M, Ito M, Shimomura Y. Characterization of the human hair shaft cuticle-specific keratin-associated protein 10 family. J Invest Dermatol. 2013;133:2780–2782. doi: 10.1038/jid.2013.233. [DOI] [PubMed] [Google Scholar]
- 18.Bornschlogl T, Bildstein L, Thibaut S, Santoprete R, Fiat F, Luengo GS, et al. Keratin network modifications lead to the mechanical stifeening of the hair follicle fiber. PNAS. 2016;113:5940–5945. doi: 10.1073/pnas.1520302113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Y, Xie M, Jiang Y, Xiao N, Du X, Zhang W, et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus) Nat Biotechnol. 2013;31:135–141. doi: 10.1038/nbt.2478. [DOI] [PubMed] [Google Scholar]
- 20.Yahagi S, Shibuya K, Obayashi I, Masaki H, Kurata Y, Kudoh J, et al. Identification of two novel clusters of ultrahigh-sulfur keratin-associated protein genes on human chromosome 11. Biochem Biophys Res Co. 2004;318:655–664. doi: 10.1016/j.bbrc.2004.04.074. [DOI] [PubMed] [Google Scholar]
- 21.Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- 22.Jamora C, Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–8. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- 23.Ewens A, Mihich E, Ehrke MJ. Distant metastasis from subcutaneously grown E0771 medullary breast adenocarcinoma. Anticancer Res. 2005;25:3905–3915. [PubMed] [Google Scholar]
- 24.Cobb JP, Hotchkiss RS, Karl IE, Buchman TG. Mechanisms of cell injury and death. Br J Anaesth. 1996;77:3–10. doi: 10.1093/bja/77.1.3. [DOI] [PubMed] [Google Scholar]
- 25.Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 26.Taylor-Papadimitriou J, Stampfer M, Bartek J, Lewis A, Boshell M, Lane EB, et al. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation to in vivo phenotypes and influence of medium. J Cell Sci. 1989;94:403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- 27.Karantza V. Keratins in health and cancer: more than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kölsch A, Windoffer R, Würflinger T, Aach T, Leube RE. The keratin-filament cycle of assembly and disassembly. J Cell Sci. 2010;123:2266–2272. doi: 10.1242/jcs.068080. [DOI] [PubMed] [Google Scholar]
- 29.de Pereda JM, Ortega E, Alonso-García N, Gómez-Hernández M, Sonnenberg A. Advances and perspectives of the architecture of hemidesmosomes: lessons from structural biology. Cell Adh Migr. 2009;3:361–364. doi: 10.4161/cam.3.4.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 31.Berens EB, Holy JM, Riegel AT, Wellstein A. A cancer cell spheroid assay to assess invasion in a 3D setting. J Vis Exp. 2015:e53409. doi: 10.3791/53409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeRoock IB, Pennington ME, Sroka TC, Lam KS, Bowden GT, Bair EL, Cress AE. Synthetic peptides inhibit adhesion of human tumor cells to extracellular matrix proteins. Cancer Res. 2001;61:3308–3313. [PubMed] [Google Scholar]
- 33.Sroka TC, Pennington ME, Cress AE. Synthetic D-amino acid peptide inhibits tumor cell motility on laminin-5. Carcinogenesis. 2006;27:1748–1757. doi: 10.1093/carcin/bgl005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cross LM, Cook MA, Lin S, Chen J-N, Rubinstein AL. Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol. 2003;23:911–912. doi: 10.1161/01.ATV.0000068685.72914.7E. [DOI] [PubMed] [Google Scholar]
- 35.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopluations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 36.Zhang W, Nandakumar N, Shi Y, Manzano Smith A, Graham G, et al. Downstream of mutant KRAS, the transcriptional regulator YAP is essential for neoplastic progression in pancreatic ductal adenocarcinoma. Sci Signal. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaller G, Fuchs I, Pritze W, Ebert A, Herbst H, Pantel K, et al. Elevated keratin 18 protein expression indicates a favorable prognosis in patients with breast cancer. Clin Cancer Res. 1996;2:1879–1885. [PubMed] [Google Scholar]
- 38.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 39.Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, et al. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Weinberg RA. Epithelial-Mesenchymal Transition: At the Crossroads of Development and Tumor Metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol. 1997;150:483–495. [PMC free article] [PubMed] [Google Scholar]
- 42.Ramms L, Fabris G, Windoffer R, Schwarz N, Springer R, Zhou C, et al. Keratins as the main component for the mechanical integrity of keratinocytes. PNAS. 2013;110:18513–18518. doi: 10.1073/pnas.1313491110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seltmann K, Fritsch AW, Kas JA, Magin TM. Keratins significantly contribute to cell stiffness and impact invasive behavior. PNAS. 2013;110:18507–18512. doi: 10.1073/pnas.1310493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seltmann K, Cheng F, Wiche G, Eriksson JE, Magin TM. Keratins Stabilize Hemidesmosomes through Regulation of β4-Integrin Turnover. J Investig Dermatol. 2015;135:1609–1620. doi: 10.1038/jid.2015.46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.