Abstract
BACKGROUND
Severe combined immunodeficiency (SCID) is characterized by arrested T-lymphocyte production and by B-lymphocyte dysfunction, which result in life-threatening infections. Early diagnosis of SCID through population-based screening of newborns can aid clinical management and help improve outcomes; it also permits the identification of previously unknown factors that are essential for lymphocyte development in humans.
METHODS
SCID was detected in a newborn before the onset of infections by means of screening of T-cell–receptor excision circles, a biomarker for thymic output. On confirmation of the condition, the affected infant was treated with allogeneic hematopoietic stem-cell transplantation. Exome sequencing in the patient and parents was followed by functional analysis of a prioritized candidate gene with the use of human hematopoietic stem cells and zebrafish embryos.
RESULTS
The infant had “leaky” SCID (i.e., a form of SCID in which a minimal degree of immune function is preserved), as well as craniofacial and dermal abnormalities and the absence of a corpus callosum; his immune deficit was fully corrected by hematopoietic stem-cell transplantation. Exome sequencing revealed a heterozygous de novo missense mutation, p.N441K, in BCL11B. The resulting BCL11B protein had dominant negative activity, which abrogated the ability of wild-type BCL11B to bind DNA, thereby arresting development of the T-cell lineage and disrupting hematopoietic stem-cell migration; this revealed a previously unknown function of BCL11B. The patient’s abnormalities, when recapitulated in bcl11ba-deficient zebrafish, were reversed by ectopic expression of functionally intact human BCL11B but not mutant human BCL11B.
CONCLUSIONS
Newborn screening facilitated the identification and treatment of a previously unknown cause of human SCID. Coupling exome sequencing with an evaluation of candidate genes in human hematopoietic stem cells and in zebrafish revealed that a constitutional BCL11B mutation caused human multisystem anomalies with SCID and also revealed a prethymic role for BCL11B in hematopoietic progenitors. (Funded by the National Institutes of Health and others.)
Population-based screening of new-borns for severe combined immunodeficiency (SCID) involves the quantification of blood levels of T-cell–receptor excision circles (TRECs), which are DNA by-products of T-cell– receptor rearrangement that indicate thymic production of naive T cells.1 Inadequate TREC levels prompt immunologic investigation to diagnose SCID before infections occur, which permits the timely initiation of therapy; therapy usually involves allogeneic hematopoietic stem-cell transplantation from a healthy donor.2 In addition to enhancing the efficacy of treatment,2,3 newborn screening can reveal previously unknown causes of T-cell lymphopenia.1,4–7
Whole-exome sequencing in persons with rare disorders of immunity has led to the identification of genes that had not previously been associated with SCID.5 However, definitively identifying a causative variant among candidate variants can be challenging. Accordingly, efficient functional tests to study mechanisms of pathogenesis are essential. Zebrafish are useful for understanding human genetics and immunity8–10 because of both the ease with which they can be genetically manipulated and their fidelity in modeling human diseases.11–13 We used whole-exome sequencing in conjunction with functional analysis of a candidate gene in human hematopoietic stem cells and in zebrafish to determine the cause of a unique case of SCID that was found through newborn screening. The patient had “leaky” SCID (i.e., a form of SCID in which a minimal degree of immune function is preserved) and developmental abnormalities, which we traced to a BCL11B variant.
METHODS
PARTICIPANTS AND GENETIC ANALYSIS
Samples from the patient, a male infant who was identified through screening of TRECs at birth, and from his parents were submitted for study after written informed consent had been obtained. The protocol was approved by the institutional review board at the University of California, San Francisco. Genomic DNA from blood was subjected to whole-exome sequencing and analysis7 (see the Methods section in the Supplementary Appendix, available with the full text of this article at NEJM.org). The patient’s BCL11B variant was confirmed by Sanger sequencing of DNA from blood and buccal brushings and was found to be a de novo mutation (it was not present in the DNA of either parent). RNA from blood was used to analyze the diversity of T-cell–receptor Vβ gene families.14
PLASMID CONSTRUCTS, PROTEIN DETECTION, AND FUNCTIONAL ASSESSMENT
Wild-type and p.N441K BCL11B were cloned into pENTR4 vector and expressed in Jurkat cells, followed by activation and flow-cytometric measurement of intracellular interleukin-2.15 To detect protein–protein and protein–DNA interactions, immunoprecipitation, immunoblotting, and chromatin immunoprecipitation sequencing (ChIP-seq) were performed (see the Methods section in the Supplementary Appendix).15,16
LENTIVIRAL TRANSDUCTION AND IN VITRO DIFFERENTIATION OF HUMAN HEMATOPOIETIC PROGENITORS
Wild-type and mutant BCL11B complementary DNAs were subcloned into pLenti CMV/TO GFP-Zeo DEST vector (Addgene)16; lentiviruses expressing epitope-tagged wild-type or mutant BCL11B and green fluorescent protein (GFP) were prepared (see the Methods section in the Supplementary Appendix). BCL11B small interfering RNAs (siRNAs) in lentiviral vectors were obtained from Applied Biological Materials. Human hematopoietic stem cells were selected from normal cord blood or adult peripheral blood with the use of CD34 microbeads (Miltenyi Biotech) after mobilization of stem cells with granulocyte colony-stimulating factor. Stem cells were transduced and differentiated on OP9 and OP9-DL1 monolayers.17 Expression of BCL11B, CCR7, and CCR9 messenger RNA (mRNA) was determined by means of quantitative polymerase chain reaction (qPCR) (primer sequences are provided in the Supplementary Appendix).
ZEBRAFISH EXPERIMENTS
Ectopic expression of wild-type and mutant human BCL11B was achieved by injection of the heat-inducible pSGH2 vector18 into one-cell– stage embryos, which were heated to 37°C for 1 hour at 30 hours after fertilization. The development of T-lymphoid progenitors was assessed by whole-mount in situ hybridization (WISH) for lck at 5 days after fertilization, as described by Zhang et al.19 The zebrafish BCL11B orthologue (bcl11ba, National Center for Biotechnology Information [NCBI] reference sequence NM_001328406) was identified on the basis of homology and synteny.19 Antisense morpholino oligonucleotides to block translation or splicing of zebrafish bcl11ba and ccr9b mRNA were produced and injected into one-cell or two-cell embryos. The efficacy of the bcl11ba-ATG and ccr9b-ATG morpholino oligonucleotides for the silencing of translation was tested by coinjection with mRNA that encoded a fluorescent biosensor linking the target site to mCherry.20 The effects on bcl11ba and ccr9b splicing were assessed by reverse-transcriptase (RT)– PCR.21 For microinjection, mRNAs were synthesized by in vitro transcription. The effects of the mutant bcl11ba on interorbital distance and craniofacial structure in embryos 5 days after fertilization were assessed with the use of Image J software22 and Alcian blue staining,23 respectively. The migration of hematopoietic progenitor cells was examined at 36 hours after fertilization by means of WISH for the detection of c-myb expression. Alterations in expression of the chemotactic receptor ccr9b on bcl11ba knockdown were quantified by quantitative PCR (SYBR Green, Life Technologies).
IN VITRO MIGRATION ASSAYS
The movement of BCL11B lentivirus-transduced human CD34 cells from upper to lower transwell chambers (Life Technologies) in response to CCL19 or CCL25 was quantified by means of flow cytometry after 4 hours of exposure.
RESULTS
PATIENT HISTORY AND IDENTIFICATION OF DISEASE-CAUSING VARIANT
The male infant was born at term to nonconsanguineous, healthy parents who had no known family history of immune deficiency. Screening revealed profound T-cell lymphopenia and no detectable TRECs (Table 1). Abnormal features (Fig. 1A) were accompanied by neonatal teeth, hypotonia, an umbilical hernia, and erythematous psoriaform dermatitis that was responsive to treatment with topical flucinolone. Imaging (not shown) revealed wormian skull bones, mild pulmonary artery stenosis, and the absence of a corpus callosum.
Table 1.
Patient Laboratory Data.*
| Immunologic Finding | Normal Range for Infants |
Value in Patient at 20 Days of Age |
Value in Patient at 2 Yr of Age† |
|---|---|---|---|
| TRECs per microliter/β-actin copies per microliter | >25/>10,000 | 0/68,900‡ | |
| White-cell count (cells/µl) | 9000–30,000 | 9500 | 12,700 |
| Platelet count (platelets/µl) | >150,000 | 333,000 | 337,000 |
| Neutrophil count (cells/µl) | >1500 | 4900 | 6630 |
| Total lymphocyte count (cells/µl) | >2600 | 2600 | 4590 |
| Eosinophil count (cells/µl) | <800 | 600 | 660 |
| CD3+ T-cell count (cells/µl) | 2250–5500 | 182‡ | 2157 |
| CD4+ helper T-cell count (cells/µl) | >1600 | 130‡ | 1239 |
| CD8+ cytotoxic T-cell count (cells/µl) | >560 | 26‡ | 826 |
| CD19+ B-cell count (cells/µl) | 300–2000 | 1326 | 1652 |
| CD16+/CD56+ NK cell count (cells/µl) | 170–1100 | 468 | 643 |
| CD3+/CD4+/CD45RA+ naive cells (% of CD4+) | >50 | Undetectable‡ | 44 |
| CD3+/CD8+/CD45RA+ naive cells (% of CD8+) | >30 | 19‡ | 40 |
| CD45+ lymphocytes proliferating in response to PHA (%)‡ | >50 | 18‡ | 100 |
| CD3+ T cells proliferating in response to PHA (%) | >58 | 43‡ | 100 |
| IgG concentration (mg/dl) | 345–1213 | 1050 | |
| IgA concentration (mg/dl) | 14–159 | 52 | |
| IgM concentration (mg/dl) | 43–200 | 100 | |
| IgE concentration (IU/ml) | 370 | ||
| Antipneumococcal antibodies, 14 serotypes | 12/14 protective | ||
| Antitetanus antibody (IU/ml) | >0.5 | 2.73 |
NK denotes natural killer, PHA phytohemagglutinin, and TREC T-cell–receptor excision circle.
The patient had undergone hematopoietic cell transplantation and vaccination and was receiving no medications or immune globulin.
Value is abnormal.
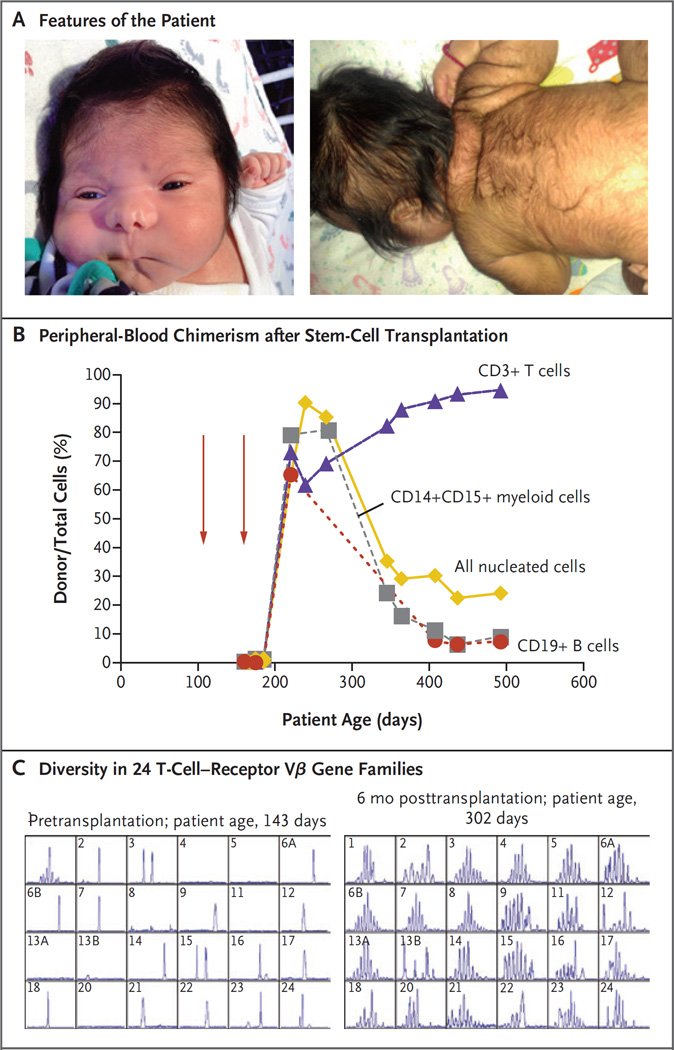
Figure 1. Clinical Findings.
Panel A shows the features of the patient (shown with informed consent). The image on the left shows increased intraorbital distance, short palpebral fissures, abnormal nasal creases, and micrognathia; the image on the right shows ear tag, loose skin folds, and hirsutism. Panel B shows the percentage of peripheral-blood chimerism after infusions of hematopoietic stem cells (red arrows) from a matched unrelated donor. Chimerism has been stable for more than 2 years. Panel C shows T-cell–receptor diversity measured by spectratyping before and 6 months after hematopoietic stem-cell transplantation. Numbers in the upper left of each panel indicate the specific T-cell–receptor Vβ gene family analyzed.
Immunologic studies revealed a T-B+NK+ phenotype, with no naive CD4+ helper T cells, an impaired proliferative response to phytohe-magglutinin (Table 1), and no maternal engraftment. The infant received antiinfective prophylaxis and intravenous immune globulin. Because of persistent T-cell lymphopenia and poor T-cell function, he received anti-CD52 serotherapy and a hematopoietic stem cell transplant at 147 days of age from an unrelated donor who was a match at 10 of 10 HLA markers (Fig. 1B). Engraftment failed, and the patient received a second transplant from the same donor at 204 days of age after treatment with busulfan (targeted steady-state concentration, 300 ng per milliliter), fludarabine, and rabbit antithymocyte globulin. Cyclosporine and methotrexate were given as prophylaxis against graft-versus-host disease. An assessment of post-transplantation chimerism showed full T-cell engraftment (Fig. 1B and Table 1), with normal proliferation to phytohemagglutinin, as well as normal diversity of the T-cell-receptor repertoire (Fig. 1C). By 2 years of age, the patient no longer required any medication and had protective vaccine responses; engraftment has remained stable. However, he has intellectual impairment, with spastic quadriplegia and seizures. He smiles and tracks faces and objects but lacks receptive or expressive language, and he does not sit or hold a bottle. Magnetic resonance imaging (MRI) of the brain at 2.2 years of age revealed, in addition to callosal agenesis, gyri radiating from the third ventricle, parallel configuration of the lateral ventricles, prominent occipital horns, malrotated hippocampi, volume loss of white matter and lentiform nuclei, and T2-weighted hyperintensities in the caudate nuclei.
Chromosome-breakage studies (for the assessment of DNA-repair defects), a copy number array, and sequences of 10 genes that are known to cause B+ SCID failed to pinpoint an anomaly. An analysis of whole-exome sequences from the patient and his parents ruled out nonpaternity; the highest-priority variant found by exome sequencing analysis in the patient (Fig. S1 in the Supplementary Appendix) was a heterozygous de novo missense mutation in BCL11B, p.N441K (c.1323T→G in NCBI reference sequence NM_138576.3; g.99641850A→C in NCBI reference sequence NC_000014.8; ClinVar accession number, SCV000297993). This site, located in a region of BCL11B that encodes the DNA-binding zinc-finger domain of BCL11B (Figs. S2 and S3 in the Supplementary Appendix), is conserved in mammals, zebrafish (bcl11ba), drosophila (DmeI), and yeast (AZF1). BCL11B is a known regulator of early thymocyte development, and somatic BCL11B variants promote human lymphoid and other cancers24; however, its function in the development of human prethymic progenitors had not been investigated, nor had it previously been implicated in a human constitutional genetic syndrome.
ROLE OF BCL11B IN DIFFERENTIATION OF HUMAN AND ZEBRAFISH HEMATOPOIETIC STEM CELLS
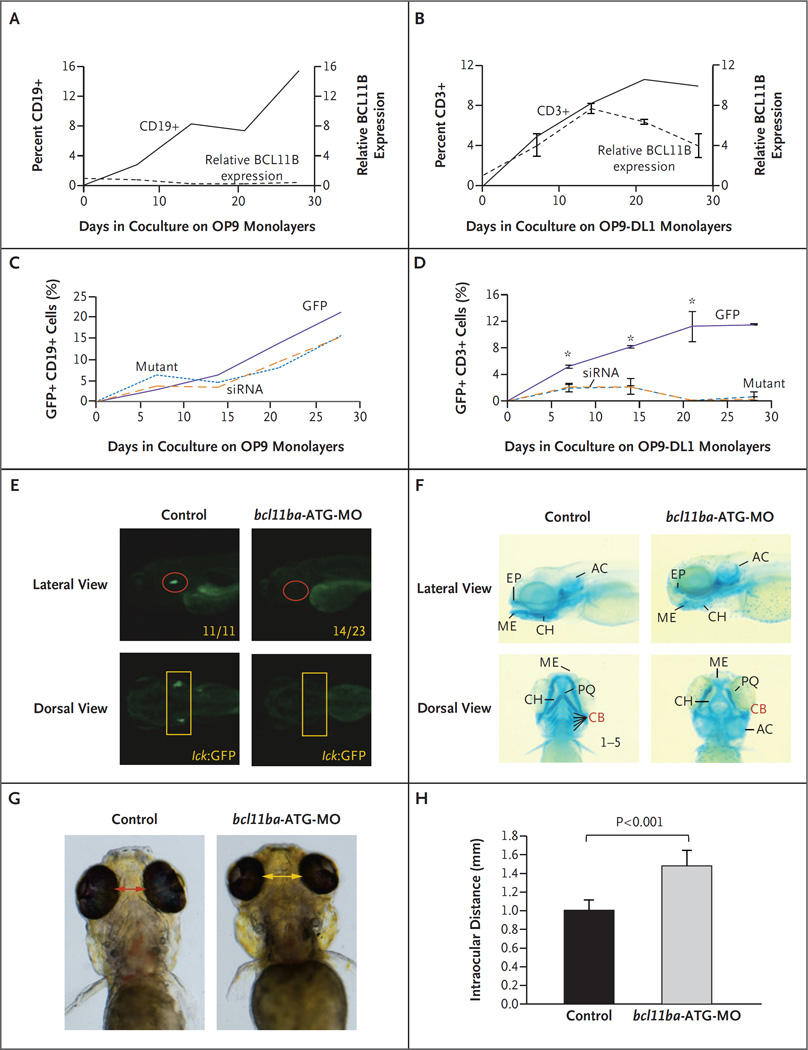
Human hematopoietic stem cells were differentiated into B cells and T cells in vitro on OP9 and OP9-DL1 monolayers, respectively. Although developing B-lineage cells expressed minimal BCL11B mRNA, Notch signaling on OP9-DL1 monolayers induced stem cells to express BCL11B and commit to the T lineage (Fig. 2A and 2B). Moreover, BCL11B played a critical role in the development of human T-cell progenitors, in that their development, but not that of B cells, was impaired by knockdown of BCL11B with the use of siRNA (Fig. 2C and 2D); moreover, ectopic expression of the patient’s mutant construct also abrogated T-cell development, which suggested a dominant negative effect.
Figure 2 (facing page). Role of BCL11B in Human Hematopoietic Stem Cells and Zebrafish Development.
Panels A and B show lymphocyte frequency (left axis, solid lines) and BCL11B messenger RNA (mRNA) expression relative to that of housekeeping gene GAPDH (right axis, dashed lines) during in vitro differentiation of CD34+ human cord-blood cells cultured on the indicated OP9 monolayers. I bars indicate standard deviations. Panels C and D show differentiation of CD34+ human cord-blood cells transduced with green fluorescent protein (GFP) alone, mutant BCL11B–GFP, or BCL11B small interfering RNA (siRNA)–GFP lentivirus. Panel C shows the percentage of CD19+ B cells developing on OP9 monolayers, and Panel D shows the percentage of CD3+ T cells on OP9-DL1 monolayers. An asterisk indicates P<0.05. Panel E shows the effect of morpholino (bcl11ba-ATG-MO; see Fig. S4 in the Supplementary Appendix) knockdown of bcl11ba on T cell development at 5 days after fertilization in transgenic lck:GFP zebrafish embryos. Numbers in the lateral view are the fractions of embryos that have the depicted phenotype. Panel F shows the craniofacial cartilage of control versus bcl11ba morphant zebrafish embryos that were assessed at 5 days after fertilization by Alcian blue staining of craniofacial cartilage structures: AC denotes auditory capsular, CB ceratobranchial, CH ceratohyal, EP ethmoid plate, ME Meckel’s cartilage, and PQ palatoquadrate. Dorsal and lateral views are depicted. Panel G shows measurements of the distance between the eyes, and Panel H shows the distances depicted graphically as the mean for five embryos of each type; T bars indicate standard deviations.
To investigate the role of BCL11B protein in supporting T-cell development in vivo, we used the zebrafish, in which the molecular machinery controlling lymphopoiesis closely resembles that in humans.11 The unique, highly conserved bcl-11ba zebrafish orthologue was identified (Fig. S3 in the Supplementary Appendix), and morpholino oligonucleotides that blocked bcl11ba expression by interfering with the translation of its mRNA were injected into zebrafish embryos; this silenced the expression of a biosensor that contained the bcl11ba morpholino oligonucleotide target site linked to mCherry but not one that lacked the target sequence (Fig. S4 in the Supplementary Appendix). To determine how bcl11ba knockdown affected T-cell development, we injected bcl11ba morpholino oligonucleotides into lck:GFP zebrafish embryos in which developing T cells were transgenically marked with GFP. Knockdown of bcl11ba expression with the use of this bcl11ba antisense oligonucleotide, as well as another antisense oligonucleotide that disrupts bcl11ba pre-mRNA splicing (Fig. S5 in the Supplementary Appendix), blocked the development of lck:GFP–marked T-cell progenitors (Fig. 2E).
Knockdown of bcl11ba in zebrafish also reproduced other developmental abnormalities that were present in the patient, including craniofacial abnormalities and perturbation of the cartilagenous structures in the head (Fig. 2F).25 The interorbital distance was substantially increased in bcl11ba morphant zebrafish, as in the patient (Fig. 2G and 2H). These results strongly suggested that the observed BCL11B mutation was responsible for the patient’s phenotype.
MODE OF ACTION OF THE BCL11B MUTATION
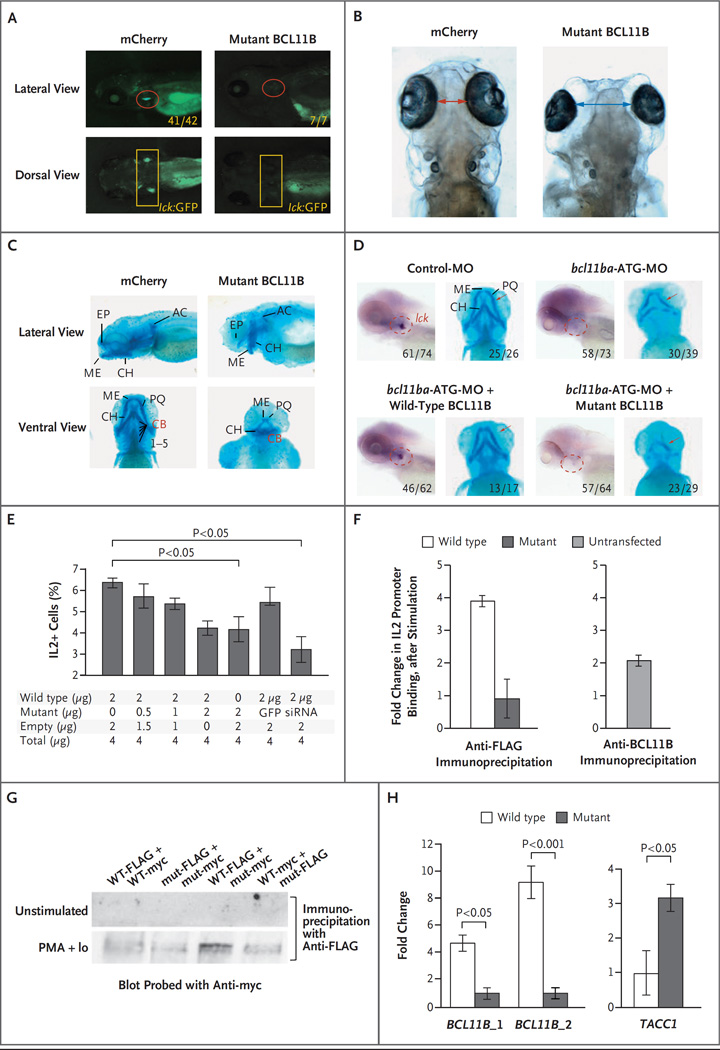
The patient’s heterozygous mutation may produce the observed developmental anomalies by functioning as a dominant negative allele encoding a protein that blocks the function of the remaining wild-type BCL11B protein encoded by the nonmutated BCL11B allele. To test this possibility, we expressed human p.N441K BCL11B in zebrafish and assessed its effects on development. Ectopic expression of the p.N441K BCL11B human protein indeed reproduced the abnormalities that were observed on knockdown of bcl11ba; development of T cells marked by lck:GFP was abrogated (Fig. 3A), as was the development of human T-cell progenitors in vitro (Fig. 2D). Furthermore, embryos expressing the patient’s p.N441K BCL11B had increased intraocular distance (Fig. 3B) and disrupted formation of the cartilagenous cranial structures (Fig. 3C). To determine definitively whether the p.N441K mutation abrogated the function of the protein, we performed rescue experiments in zebrafish embryos in which endogenous bcl11ba expression was knocked down by antisense oligonucleotide treatment. Indeed, although re-expression of wild-type human BCL11B protein restored T-cell development and corrected the craniofacial abnormalities associated with its loss, human p.N441K BCL11B failed to do so (Fig. 3D). These findings, together with our finding that the variant occurred as a de novo mutation that was not found in either genetically confirmed parent, definitively showed that the BCL11B mutation identified in the patient accounted for his disease. Further supporting our claim of causality is the dearth of nonsynonymous variants (in the 1000 Genomes, Exome Sequencing Project, and Exome Aggregation Consortium databases) in the near vicinity of the relevant zinc-finger–encoding region in BCL11B.
Figure 3 (facing page). Dominant Negative Effect of Mutant BCL11B in Zebrafish and Jurkat T Cells.
Panel A shows T-cell development in the zebrafish thymus at 5 days after fertilization after either human mutant BCL11B or mCherry as a control was overexpressed by injection of mRNA (25 pg) into transgenic lck:GFP embryos. Panels B and C show increased intraocular distance (arrows) and disruption of cartilaginous cranial structures 5 days after fertilization in zebrafish overexpressing human p.N441K BCL11B (as in Panel A). The intraocular distance increased from a mean of 1.0 mm to 2.5 mm (five embryos, P<0.001). Panel D shows the ability of intact but not mutant human BCL11B to rescue the arrest in T-cell development and correct the craniofacial abnormalities caused by bcl11ba knockdown. Rescue of T-cell development was evaluated after heat-inducible reexpression (30 hours after fertilization) and whole-mount in situ hybridization (WISH) analysis with an lck probe at 5 days after fertilization to identify thymocytes (within dashed red outlines in the lateral view). The restoration of craniofacial cartilage structure (red arrows) was assessed at 5 days after fertilization with the use of Alcian blue staining. Numbers indicate the proportions of embryos that had the depicted phenotypes. Panel E shows the production of interleukin-2 by Jurkat cells that were stimulated with phorbol myristate acetate (PMA) and ionomycin. The cells were transfected with a constant amount of vector DNA, with increasing proportions of p.N441K BCL11B, GFP vector, or BCL11B siRNA. I bars indicate standard deviations. Panel F shows chromatin immunoprecipitation (ChIP) after stimulation, with PMA and ionomycin, of Jurkat cells (left panel) transfected with FLAG-tagged wild-type or p.N441K BCL11B, and untransfected Jurkat cells (right panel). The fold change in DNA was calculated as the ratio of output DNA to input DNA in cells stimulated with PMA and ionomycin relative to the ratio in unstimulated cells. ChIP in untransfected cells was performed with anti-BCL11B antibody, as a control. Panel G shows the ability of BCL11B to heterodimerize, assessed by transfection of distinctly tagged wild-type (WT) and mutant (mut) BCL11B into Jurkat cells and coprecipitation with the indicated anti-tag antibodies. Extracts were produced from unstimulated Jurkat cells and from cells stimulated with PMA and ionomycin (PMA+Io). Panel H shows verification, by quantitative polymerase chain reaction (PCR), of BCL11B binding sites in genomic loci detected by ChIP-seq analysis of human cord-blood progenitors that were expanded in vitro. DNA bound by FLAG-tagged wild-type or mutant BCL11B was isolated by coprecipitation with anti-FLAG antibody. Fold changes were calculated as the relative quantity of the DNA binding site of interest (BCL11B_1, BCL11B_2, or TACC1), determined by quantitative PCR in ChIP-seq samples from human cord-blood progenitors transduced with FLAG-tagged wild-type BCL11B relative to that in samples transduced with FLAG-tagged mutant BCL11B. Two canonical BCL11B binding sites within the BCL11B locus were bound more extensively (one site 5 times more extensively and one site 10 times more extensively) than were the sites in the p.N441K mutant (left panel). Conversely, a novel TACC1 site (isoform 1, NM_006283.2) was found to have greater binding by the p.N441K mutant than by wild-type BCL11B.
To determine how the p.N441K substitution in the zinc-finger DNA-binding domain affected the ability of BCL11B to transactivate gene expression, we examined a known BCL11B target gene, IL2.15 Variant BCL11B inhibited interleukin-2 production by Jurkat T cells in a dose-dependent manner (Fig. 3E). To assess whether the p.N441K substitution disabled BCL11B by preventing its association with the p300 cofactor,15 we performed co-immunoprecipitation, which revealed that p.N441K BCL11B retained the ability to associate with p300 (Fig. S6 in the Supplementary Appendix). Since the p.N441K mutation is predicted to affect the DNA-binding domain of BCL11B, we used chromatin immunoprecipitation (ChIP) to evaluate BCL11B binding to the consensus site in the IL2 promoter DNA.15 We found that stimulation of Jurkat cells with phorbol myristate acetate and ionomycin markedly increased the binding of wild-type BCL11B but not of p.N441K BCL11B (Fig. 3F).
Nevertheless, this did not explain the dominant negative function of the mutant form of BCL11B. To determine whether p.N441K BCL11B blocked the function of wild-type BCL11B by forming nonfunctional heterodimers, we coexpressed both in Jurkat cells and performed coprecipitation analysis (Fig. 3G). We found that mitogenic stimulation induced the formation of wild-type– mutant BCL11B heterodimers, which suggested that the p.N441K BCL11B in the patient caused the disease by heterodimerization with wild-type BCL11B, producing complexes that were unable to bind to promoter targets in DNA. To determine whether the p.N441K substitution generally disrupted BCL11B binding to DNA — or, alternatively, redirected binding to distinct sites — ChIP-seq analysis was performed after ectopic expression of wild-type or mutant BCL11B in human hematopoietic stem cells. These analyses revealed that binding of the p.N441K BCL11B protein to canonical sites within the coding exons of the BCL11B locus was reduced relative to binding by wild-type BCL11B (Fig. 3H, and the Methods section and Fig. S7 in the Supplementary Appendix), which confirmed that the p.N441K substitution indeed impaired BCL11B binding to already known target DNA sites. Surprisingly, the p.N441K substitution also induced BCL11B binding to a unique site in exon 1 of the TACC1 (transforming acidic coiled-coil containing protein 1) locus that encodes the 5′ untranslated region of transcript variants 1 and 2 (NM_006283.2 and NM_001122824.1). This site was not recognized by wild-type BCL11B (Fig. 3H, and Fig. S7 in the Supplementary Appendix). Thus, the p.N441K mutation altered BCL11B function in two ways, by blocking binding to canonical sites while also redirecting binding to novel sites.
BCL11B AND MIGRATION OF EARLY HEMATOPOIETIC PROGENITORS IN ZEBRAFISH
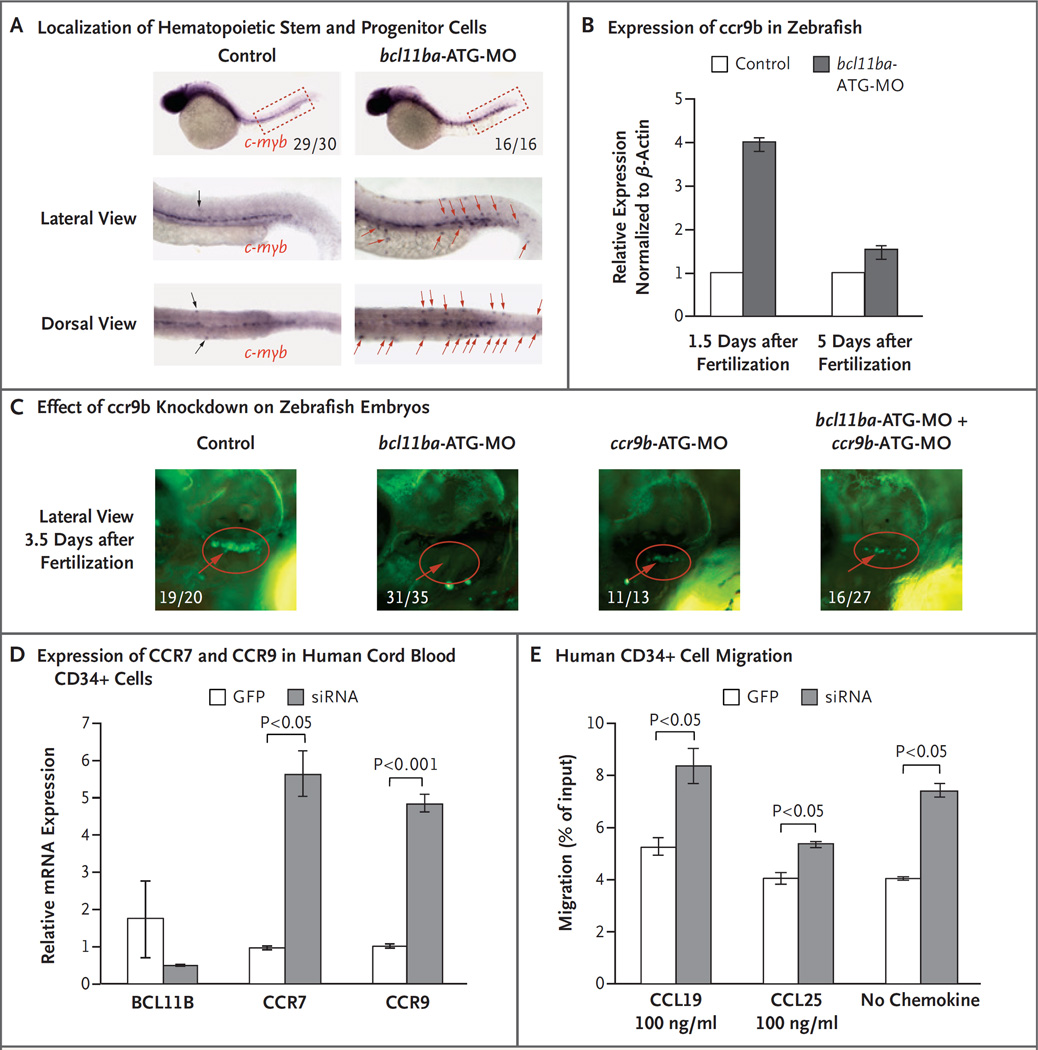
Because zebrafish embryos are transparent, it is possible to observe abnormalities in these embryos that are not obvious in mammalian animal models. The role of Bcl11b in supporting thymocyte development had previously been noted in mice; however, its role in the development of prethymic hematopoietic progenitors was unexplored. When the localization of early hematopoietic progenitors marked by c-myb was investigated in bcl11ba morphant embryos, we found substantial lateral as well as dorsoventral displacement of progenitors, which suggests that bcl11ba contributes to the control of progenitor migration (Fig. 4A, and Fig. S8 in the Supplementary Appendix) but not stem cell emergence (Fig. S9 in the Supplementary Appendix). The migration of progenitors to the thymus is controlled by the chemokine receptors CCR7 and CCR9.26 Accordingly, we assessed the expression of ccr7 and ccr9b (the zebrafish orthologues of human CCR7 and CCR9) in zebrafish by means of quantitative PCR and found that ccr9b mRNA levels were induced at 1.5 days after fertilization, when hematopoietic progenitors emerge, but not later in development, after completion of this process (5 days after fertilization) (Fig. 4B). To determine whether this overexpression of ccr9b impaired thymic seeding, we knocked down ccr9b, using a ccr9b-specific antisense oligonucleotide (Fig. S10 in the Supplementary Appendix), in cd41:GFP zebrafish. Although the knockdown of ccr9b alone did not prevent thymic seeding, when ccr9b was knocked down in combination with bcl11ba, it did produce a partial rescue of thymic seeding in the bcl11ba morphants at 3.5 and 5 days after fertilization (Fig. 4C, and Fig. S10D in the Supplementary Appendix). In human CD34 progenitors, siRNA-mediated repression of BCL11B also increased the expression of CCR9 as well as CCR7 (Fig. 4D) and increased migration of these progenitors both in response to CCR7 and CCR9 ligands (CCL19 and CCL25, respectively) (Fig. 4E) and in the absence of exogenous chemokines. These results suggest that additional molecular effectors controlling migration may also be affected by the absence of BCL11B.
Figure 4. Role of BCL11B in Migration of Hematopoietic Progenitor Cells in Zebrafish and Humans.
Panel A shows c-myb WISH staining at 36 hours (1.5 days) after fertilization to examine the localization of hematopoietic stem and progenitor cells. The fractions indicate the numbers of zebrafish that had the depicted phenotype out of the total number examined. Displaced hematopoietic stem and progenitor cells are marked by black (control morpholino oligonucleotide [MO] injected) and red (bcl11ba-MO injected) arrows. Panel B shows the relative ccr9b expression, measured by quantitative PCR in control embryos and bcl11ba morphants 1.5 and 5 days after fertilization. Expression in triplicate samples was quantified, normalized to that of β-actin and depicted as mean fold change (the expression in experimental embryos relative to the expression in control-injected embryos, which was defined as 1); I bars indicate standard deviations. Panel C shows the effect of morpholino (ccr9b-ATG-MO) knockdown of ccr9b, either alone or in combination with bcl11ba-ATG-MO, on thymic seeding at 3.5 days after fertilization in transgenic cd41:GFP zebrafish embryos (red circles). Numbers are the fraction of embryos that had the depicted phenotype. Panel D shows the expression, normalized to that of housekeeping gene GAPDH, of BCL11B, CCR7, and CCR9 mRNA in human cord-blood CD34+ cells transduced with lentivirus encoding GFP alone or BCL11B siRNA–GFP. Panel E shows the in vitro transwell migration of human CD34+ cells, transduced as in Panel C, in response to CCL19 or CCL25 (100 ng per milliliter) or no chemokine.
DISCUSSION
We identified a de novo constitutional missense mutation in BCL11B as a previously unknown cause of SCID, which illustrates the importance of newborn screening from the perspectives of both public health and scientific discovery. Since 2010, newborn screening in California has increased the survival rate for patients with SCID to 94%1 by enabling the detection and treatment of infants with immunodeficiency before the development of life-threatening infections. This was critical for our patient, who, despite having substantial developmental abnormalities outside the hematopoietic system, had full hematopoietic reconstitution after stem-cell transplantation.
Although at present more than 16 genes, when functionally disrupted, are known to cause SCID,3 our strategy of whole-exome sequencing and functional interrogation of gene candidates in zebrafish has led to the identification of a previously unrecognized heterozygous dominant negative BCL11B variant as a genotype causing SCID. Other features of this previously undescribed syndrome are also attributable to BCL11B inactivation, including craniofacial abnormalities, the absence of corpus callosum, intellectual impairment, abnormal skin, and pulmonary artery stenosis. A number of these defects have been observed in Bcl11b-heterozygous or Bcl11b-deficient mice (reviewed extensively27,28). BCL11B is a Cys2His2 zinc finger transcription factor, and therefore the observed defects may result from perturbed expression of BCL11B target genes.16,29 Nevertheless, the critical target genes required for BCL11B to facilitate morphogenesis and hematopoiesis remain to be identified.
SCID is recognized as being inherited in either an X-linked or an autosomal recessive form, rather than as a de novo autosomal dominant form, as seen in this case. Because some of the abnormalities in our patient also occur in Bcl11b-haploinsufficient mice, either haploinsufficiency or dominant negative function were possible mechanisms to account for the patient’s phenotypes. Our data favor the latter: ectopic expression of the p.N441K allele in BCL11B-sufficient human hematopoietic progenitors and zebrafish embryos resulted in a phenocopy of the patient’s abnormalities. Moreover, p.N441K BCL11B interfered with the function of intact BCL11B protein by forming heterodimers that altered DNA binding. Our chromatin immunoprecipitation data confirmed the decreased binding to canonical target sites and suggested that the p.N441K mutation might also confer new DNA binding specificity on BCL11B, which could contribute to the global anomalies observed in the patient and in zebrafish embryos. The identification of a unique p.N441K BCL11B binding site in TACC1, which encodes a regulator of the centrosome and microtubules that has been implicated in oncogenesis30 and neurogenesis,31 supports this hypothesis. Impairment of BCL11B activity by somatic variants has also been implicated in a broad spectrum of malignant transformations, including in T-cell acute lymphoblastic leukemia.32,33 These variants, as well as the pathogenic mutation we describe here, cluster in regions of BCL11B that encode the zinc finger motifs, including zinc finger 2 (Fig. S2 in the Supplementary Appendix). Structural modeling of the BCL11B variants in T-cell acute lymphoblastic leukemia predicts zinc finger disruption that impairs DNA binding.32 Our functional studies support this interpretation.
Although studies in mice have been useful in elucidating the roles that numerous proteins, including Bcl11b, play in lymphoid development, zebrafish emerge as a useful model with which to rapidly understand the genetic basis for human disease and to gain new biologic insights.13 Indeed, we have established a pipeline for the rapid screening, in zebrafish, of SCID candidate genes that are identified by exome sequencing. Using this approach, we identified the p.N441K BCL11B allele as the cause not only of SCID in this patient but also of craniofacial and other developmental abnormalities. An unresolved question in our study is the mechanistic relationship of our patient’s dominant negative BCL11B mutation to his callosal agenesis. Increases in Bcl11b have been found to disturb callosal neuron specification and cause callosal agenesis in the developing mouse brain.34 The mutation in our patient may mimic the derepression observed in the mouse or, more interestingly, may highlight a critical divergence in callosal neuronal specification between mice and humans.
Finally, the zebrafish model has revealed a previously unknown function of BCL11B: the regulation of hematopoietic progenitor positioning or movement. The role of BCL11B in supporting the commitment of thymic progenitors to the T cell lineage has been studied in other models, including the mouse; however, a requirement for BCL11B in the development of prethymic progenitors was not previously known. We found BCL11B to be essential in controlling the responsiveness of hematopoietic stem cells to chemotactic signals by modulating the expression of receptors CCR7 and CCR9, which direct the movement of progenitors in mammals from the bone marrow to the thymus. This would not have been obvious in mammalian models, but because zebrafish embryos are transparent, this role for BCL11B was recognized.
Supplementary Material
Acknowledgments
Supported by the National Institutes of Health (NIH) (core grant P30 CA006927 to Dr. Wiest; grant R01 AI105776 to Drs. Puck and Brenner; grant R01 AI078248 to Dr. Puck; and R21 AI111208 to Drs. Wiest and Puck; and grant U54 AI082973 to Drs. Puck and Cowan for the Primary Immune Deficiency Treatment Consortium, a part of the Rare Diseases Clinical Research Network, an initiative of the NIH Office of Rare Diseases Research, funded through collaboration between the National Center for Advancing Translational Sciences and the National Institute of Allergy and Infectious Diseases [NIAID]); an appropriation from the Commonwealth of Pennsylvania (to Dr. Wiest); the M.D. Anderson Cancer Center (Leukemia SPORE grant CA100632 to Dr. Zhang); the Fox Chase Cancer Center (William J. Avery fellowship to Dr. Zhang); Tata Consultancy Services (support to Dr. Brenner); NIAID (grant R01 AI105776 to Dr. Brenner); and the Jeffrey Modell Foundation, the Lisa and Douglas Goldman Fund, and the Michelle Platt-Ross Foundation (support to Dr. Puck).
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank the patient’s family for their participation; Christopher Dvorak, Biljana Horn, and Mica Muskat for providing clinical care; Uma Sunderam for bioinformatic and genomic analysis; Yanning Wang and Karly Kondratowicz for technical assistance; and the staff of the DNA Sequencing, Imaging, and Laboratory Animal core facilities of the Fox Chase Cancer Center.
REFERENCES
- 1.Kwan A, Abraham RS, Currier R, et al. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. JAMA. 2014;312:729–738. doi: 10.1001/jama.2014.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shearer WT, Dunn E, Notarangelo LD, et al. Establishing diagnostic criteria for severe combined immunodeficiency disease (SCID), leaky SCID, and Omenn syndrome: the Primary Immune Deficiency Treatment Consortium experience. J Allergy Clin Immunol. 2014;133:1092–1098. doi: 10.1016/j.jaci.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Herz W, Bousfiha A, Casanova JL, et al. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency. Front Immunol. 2014;5:162. doi: 10.3389/fimmu.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verbsky JW, Baker MW, Grossman WJ, et al. Newborn screening for severe combined immunodeficiency: the Wisconsin experience (2008–2011) J Clin Immunol. 2012;32:82–88. doi: 10.1007/s10875-011-9609-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen R, Giliani S, Lanzi G, et al. Whole-exome sequencing identifies tetra-tricopeptide repeat domain 7A (TTC7A) mutations for combined immunodeficiency with intestinal atresias. J Allergy Clin Immunol. 2013;132(3):656–664.e17. doi: 10.1016/j.jaci.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallott J, Kwan A, Church J, et al. Newborn screening for SCID identifies patients with ataxia telangiectasia. J Clin Immunol. 2013;33:540–549. doi: 10.1007/s10875-012-9846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel JP, Puck JM, Srinivasan R, et al. Nijmegen breakage syndrome detected by newborn screening for T cell receptor excision circles (TRECs) J Clin Immunol. 2015;35:227–233. doi: 10.1007/s10875-015-0136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- 9.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 10.Vilboux T, Lev A, Malicdan MCV, et al. A congenital neutrophil defect syndrome associated with mutations in VPS45 . N Engl J Med. 2013;369:54–65. doi: 10.1056/NEJMoa1301296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langenau DM, Zon LI. The zebrafish: a new model of T-cell and thymic development. Nat Rev Immunol. 2005;5:307–317. doi: 10.1038/nri1590. [DOI] [PubMed] [Google Scholar]
- 12.Pannicke U, Hönig M, Hess I, et al. Reticular dysgenesis (aleukocytosis) is caused by mutations in the gene encoding mitochondrial adenylate kinase 2. Nat Genet. 2009;41:101–105. doi: 10.1038/ng.265. [DOI] [PubMed] [Google Scholar]
- 13.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarzotti M, Patel DD, Li X, et al. T cell repertoire development in humans with SCID after nonablative allogeneic marrow transplantation. J Immunol. 2003;170:2711–2718. doi: 10.4049/jimmunol.170.5.2711. [DOI] [PubMed] [Google Scholar]
- 15.Cismasiu VB, Ghanta S, Duque J, et al. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cismasiu VB, Duque J, Paskaleva E, et al. BCL11B enhances TCR/CD28-triggered NF-kappaB activation through up-regulation of Cot kinase gene expression in T-lymphocytes. Biochem J. 2009;417:457–466. doi: 10.1042/BJ20080925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Motte-Mohs RN, Herer E, Zúñiga-Pflücker JC. Induction of T-cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood. 2005;105:1431–1439. doi: 10.1182/blood-2004-04-1293. [DOI] [PubMed] [Google Scholar]
- 18.Bajoghli B, Aghaallaei N, Heimbucher T, Czerny T. An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev Biol. 2004;271:416–430. doi: 10.1016/j.ydbio.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Duc AC, Rao S, et al. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell. 2013;24:411–425. doi: 10.1016/j.devcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 21.Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 22.Bernier R, Golzio C, Xiong B, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robu ME, Larson JD, Nasevicius A, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3(5):e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avram D, Califano D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: impact on immune diseases. J Immunol. 2014;193:2059–2065. doi: 10.4049/jimmunol.1400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes G, van Bakel H, Zhou X, Losic B, Jabs EW. BCL11B expression in intra-membranous osteogenesis during murine craniofacial suture development. Gene Expr Patterns. 2015;17:16–25. doi: 10.1016/j.gep.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zlotoff DA, Bhandoola A. Hematopoietic progenitor migration to the adult thy-mus. Ann N Y Acad Sci. 2011;1217:122–138. doi: 10.1111/j.1749-6632.2010.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kominami R. Role of the transcription factor Bcl11b in development and lymphomagenesis. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:72–87. doi: 10.2183/pjab.88.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- 29.Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- 30.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, Tsai LH. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez A, Kentsis A, Sanda T, et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood. 2011;118:4169–4173. doi: 10.1182/blood-2010-11-318873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumann M, Vosberg S, Schlee C, et al. Mutational spectrum of adult T-ALL. Oncotarget. 2015;6:2754–2766. doi: 10.18632/oncotarget.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranek C, Dittrich M, Parthasarathy S, et al. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc Natl Acad Sci U S A. 2012;109:3546–3551. doi: 10.1073/pnas.1108718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.