Abstract
Objective
Genome-wide association studies (GWAS) have identified multiple obesity susceptibility loci, yet the mechanisms by which these loci influence obesity remain unclear. We hypothesized that alternative splicing could contribute to obesity by regulating the transcriptomic and proteomic diversity of genes in these loci.
Methods
Seventy-two of the 136 genes at the 13 obesity loci encoded multiple protein isoforms based on database search. Thus, we analyzed alternative splicing of these genes in adipose tissue samples from the Metabolic Syndrome in Men (METSIM) population-based study, and from two weight loss intervention studies (surgical and very low calorie diet).
Results
Alternative splicing was confirmed in 11 genes with PCR capillary electrophoresis in human subcutaneous adipose tissue. Interestingly, we observed differential splicing of TRA2B, BAG6 and MSH5 between lean normoglycemic and overweight type 2 diabetic individuals. Of these genes, we detected fat depot dependent splicing of TRA2B and BAG6, and weight loss induced regulation of MSH5 splicing in the intervention studies. Finally, BMI was a major determinant of TRA2B, BAG6 and MSH5 splicing in the combined data.
Conclusions
Our study provides evidence for alternative splicing in obesity loci, suggesting that alternative splicing at least in part mediates the obesity-associated risk in these loci.
Clinical trial number
ISRCTN67529475
Keywords: Adipose tissue, alternative splicing, weight loss
INTRODUCTION
Genome wide association studies (GWAS) increased the number of loci associated with susceptibility to obesity. Polymorphisms at the obesity loci can possibly affect alternative splicing resulting in functionally different protein isoforms. Additionally, expression of several RNA processing genes is reduced in individuals with obesity (1), suggesting link between dysregulation of alternative splicing and obesity.
Alternative splicing of genes is an essential regulatory mechanism responsible for the generation of transcript and protein diversity. Aberrant splicing is increasingly recognized in many forms of diseases and has been linked to obesity, insulin resistance and cholesterol homeostasis (2, 3). Interestingly, rare gene variants in subjects with obesity are enriched in splicing regulatory regions indicating that genetic regulation of splicing may associate with obesity (3).
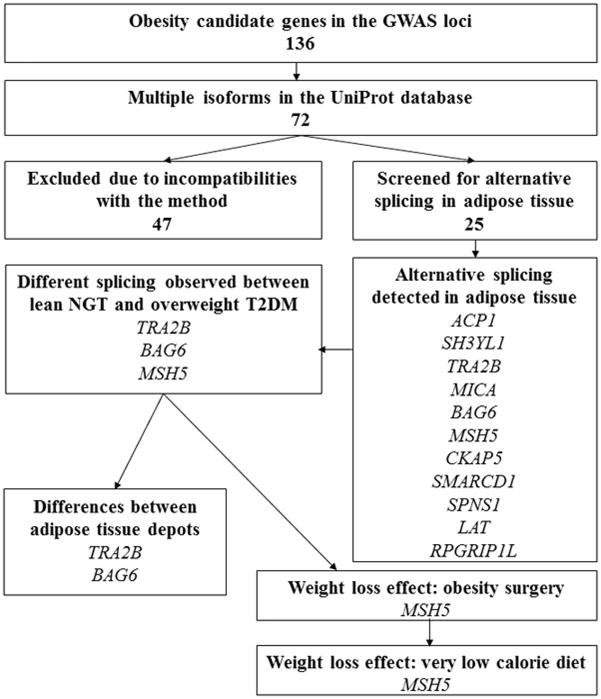
We hypothesized that alternative splicing of genes at obesity loci may contribute to obesity. To investigate this hypothesis, we investigated 136 genes at 13 of the 97 known obesity loci (4, 5, 6). Of these genes, 72 had multiple protein isoforms, and 25 of these resulted from alternative splicing of cassette exons. These genes were screened for alternative splicing in adipose tissue samples in three different study cohorts: a population-based Metabolic Syndrome in Men (METSIM) study and two weight loss intervention studies: Kuopio Obesity Surgery Study (KOBS) and very low calorie diet study (VLCD) (Fig. 1).
Fig. 1.
Analysis pipeline to identify alternatively spliced variants in the obesity loci. The genes were selected based on the known splicing events. Splice variants including 5′ and 3′ splicing events as well as cassette exons bigger that 600bp were excluded from the analysis due to the limitation of the method used.
METHODS
Study subjects
Subcutaneous adipose tissue samples from 46 men were obtained from the METSIM study (7). Two independent follow-up weight loss study groups were used to examine the effect of weight loss on alternative splicing. First, subcutaneous and visceral adipose tissue samples were taken during the Rouxen-Y gastric bypass surgery from a total of 108 participants with severe obesity of the KOBS study. Additionally, subcutaneous fat biopsies were collected a year after the bariatric surgery (7). Second, subcutaneous adipose tissue samples were obtained from 32 non-diabetic participants of a 7-week VLCD intervention study followed by a 24-week weight maintenance period. The tissue biopsies were collected at all visits (baseline, 7 weeks and 24 weeks) (8). Basic characteristics of the study participants are given in Table 1. The study protocols were approved by the Ethics Committee of Northern Savo Hospital District and carried out in the accordance with Helsinki Declaration.
Table 1.
Characteristics of the study groups.
| METSIM | Kuopio Obesity Surgery Study | Very low calorie diet | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
NGT n= 26 |
T2DM n= 20 |
p value |
Baseline n=108 |
1 year Follow-up n=81 |
p value |
Baseline n=32 |
7 wks VLCD (1) n=32 |
24 wks WM (2) n=32 |
p value (1) | p value (2) | |
|
| |||||||||||
| Sex male/female | 26/0 | 20/0 | 34/74 | 26/55 | 9/23 | 9/23 | 9/23 | ||||
|
| |||||||||||
| Age years | 56.5 ± 5.5 | 56.7 ± 5.3 | 0.902 | 47.1±9.1 | 47.6±9.0 | 48.8 ± 8.7 | 49.0 ± 8.8 | 49.5 ± 8.8 | |||
|
| |||||||||||
| Body mass index kg/m2 | 23.1 ± 1.4 | 26.1 ± 3.9 | 0.003 | 45.0±6.3 | 34.6±5.8 | 2×10−37 | 34.7 ± 2.7 | 29.9 ± 2.4 | 30.3 ± 2.8 | 3×10−24 | 2×10−16 |
|
| |||||||||||
| Fasting plasma glucose mmol/l | 5.5 ± 0.3 | 7.1 ± 1.0 | 1×10−6 | 6.7±1.7 | 5.6±0.9 | 2×10−8 | 6.1 ± 0.8 | 5.6 ± 0.6 | 5.7 ± 0.6 | 1×10−4 | 6×10−5 |
|
| |||||||||||
| Fasting insulin pmol/l | 27.6 ± 8.8 | 62.9 ± 47.3 | 0.004 | 135.7±135.5 | 71.4±67.4 | 3×10−10 | 82.8±46.2 | 40.8 ± 19.2 | 51.0 ± 27.0 | 3×10−6 | 4×10−6 |
|
| |||||||||||
| Total cholesterol mmol/l | 5.3 ± 1.0 | 5.7 ± 1.1 | 0.215 | 4.4 ± 1.1 | 4.4 ± 0.9 | 0.291 | 5.1 ± 0.7 | 4.2 ± 0.6 | 5.0 ± 0.6 | 7×10−10 | 0.481 |
|
| |||||||||||
| HDL cholesterol mmol/l | 1.5 ± 0.4 | 1.7 ± 0.6 | 0.217 | 1.1 ± 0.3 | 1.4 ± 0.4 | 3×10−19 | 1.4 ± 0.3 | 1.3 ± 0.3 | 1.6 ± 0.4 | 4×10−3 | 1×10−4 |
|
| |||||||||||
| Triglycerides mmol/l | 1.0 ± 0.3 | 1.8 ± 1.7 | 0.045 | 1.7 ± 0.8 | 1.1 ± 0.5 | 6×10−11 | 1.4 ± 0.6 | 0.9 ± 0.3 | 1.1 ± 0.4 | 4×10−5 | 5×10−4 |
|
| |||||||||||
| Fasting free fatty acids mmol/l | 0.33 ± 0.18 | 0.51 ± 0.23 | 0.007 | 0.64 ± 0.23 | 0.51 ± 0.18 | 1×10−4 | 0.48 ± 0.13 | 0.72 ± 0.23 | 0.45 ±0.20 | 1×10−6 | 0.304 |
Data are expressed as mean ± SD. Data of the same individuals at different time points were compared using paired t test. p values <0.05 are shown in boldface. VLCD, very low calorie diet; WM, weight maintenance
Splicing analysis
PCR-capillary electrophoresis was used to examine the relative ratio of splice variants (3100 DNA Genetic Analyzer, ABI). Quantification of peak areas was performed with Peak Scanner Software v1.0 (ABI). Primers, shown in Table S1, spanning the regions around alternatively spliced regions allow for detection of cassette exons but not alternative 3′/5′ exons.
Statistical analysis
Data are presented as mean±SD. Data from the same individuals at different time points were compared with the two-tailed paired samples t-test. Non-parametric Spearman’s correlation test was used to assess correlations with metabolic traits. Body mass index (BMI), insulin, glucose and free fatty acid (FFA) levels were logarithmically transformed to obtain a normal distribution. The main effects of BMI, fasting insulin, fasting glucose, fasting FFA and age on TRA2B, BAG6 and MSH5 splicing were evaluated by linear regression model.
RESULTS
Selection of candidate genes in obesity loci
One-hundred and thirty-six genes in the 13 known obesity loci were selected (4, 5). All genes in the same haplotype block with the most associated (lowest p-value) obesity single nucleotide polymorphism (SNP) were considered as potentially alternatively spliced genes. Out of these 136 genes, 72 genes had known isoforms derived from alternative splicing, based on the UniProt database (Table S2). From these, we selected 25 genes with known exon cassettes within the genes for PCR capillary electrophoresis analysis. Eleven alternatively spliced genes were detected in adipose tissue. Our analysis pipeline is shown in Fig. 1.
Alternative splicing in lean normoglycemic and overweight type 2 diabetic subjects
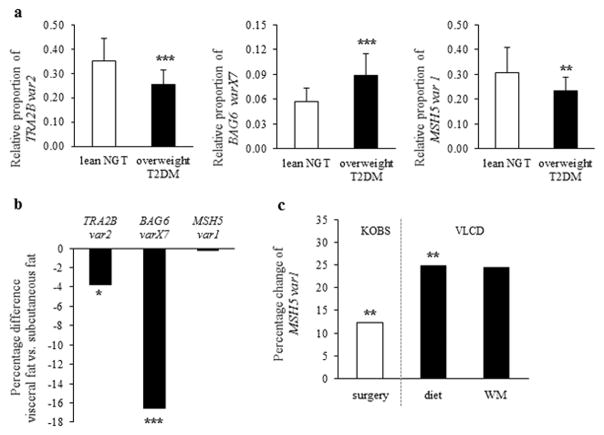
We first investigated the subcutaneous fat splicing profile of these 11 genes, using samples from the METSIM study. Subcutaneous fat distribution of TRA2B var2 was 28.2% lower (p=3×10−5), BAG6 varX7 was 54.1% higher (p=2×10−5) and MSH5 var1 was 24.4% lower (p=0.004) in overweight individuals with type 2 diabetes compared to lean normoglycemic individuals (Fig. 2a). These 3 genes were selected for further analysis (Fig. 1).
Fig. 2.
(a) The relative proportion of TRA2B var2, BAG6 varX7 and MSH5 var1 in the subcutaneous fat of lean normoglycemic individuals (white bars, lean NGT) and overweight type 2 diabetic (black bars, overweight T2DM) individuals in the METSIM study. Mean±SD shown. Comparison made using t- test. (b) Difference in expression levels of TRA2B, BAG6 and MSH5 splice variants between subcutaneous fat and visceral fat in the KOBS study. Comparison made using paired t- test. Total gene expression of TRA2B, BAG6 and MSH5 did not differ between fat depots (data not shown) (c) Change in expression level of MSH5 var1 during the year after gastric bypass surgery (white bar, KOBS study) and during a 7 week very low calorie diet followed by a 24-week weight-maintenance (WM) period (black bar, VLCD study) in subcutaneous fat. Comparison made using paired t- test. *p< 0.05, **p< 0.01, ***p<0.001. No change in total gene expression of TRA2B, BAG6 and MSH5 was detected in response to weight loss (data not shown).
Splicing in subcutaneous and visceral fat
Splicing profiles of TRA2B, BAG6 and MSH5 genes were compared between subcutaneous and visceral fat from 108 participants of the KOBS study. We found a 3.3% lower proportion of TRA2B var2 (p=0.011) and a 32.8% lower proportion of BAG6 varX7 (p=5×10−12) in visceral fat compared to subcutaneous fat. The splicing pattern of MSH5 did not differ between fat depots (Fig. 2b).
Differential alternative splicing in response to weight loss
The splicing profiles of TRA2B, BAG6 and MSH5 were assessed in subcutaneous fat before and after gastric bypass obesity surgery (KOBS study) and dietary weight loss (VLCD study). We detected a 12.4% increase in the proportion of MSH5 var1 after surgery-induced weight loss (p=0.002) and a 24.8% increase in the proportion of MSH5 var1 (p=0.005) after diet induced weight loss (Fig. 2c). We did not detect any difference in splice variant distribution of TRA2B and BAG6 before and after weight loss (data not shown).
Determinants of TRA2B, BAG6 and MSH5 splicing in adipose tissue
To interrogate the determinants of alternative splicing, we pooled the data of the subcutaneous adipose tissue biopsies from the METSIM and KOBS studies (n=154). TRA2B splicing was associated with BMI, fasting insulin and FFA levels in univariate analysis (p=4×10−4, p=0.004 and p=0.001 respectively) whereas forward stepwise multivariate analysis indicated that only BMI was an independent determinant of TRA2B splicing (p=0.001). BMI associated with BAG6 and MSH5 splicing in univariate analysis (p=0.001, p=0.034 respectively) and was an independent predictor of BAG6 and MSH5 splicing profile in forward stepwise multivariate analysis (p=0.002, p=0.015 respectively) (Table S3).
DISCUSSION
The functional relevance of most obesity-associated SNPs remains unknown. SNPs in obesity loci could alter splicing of regional genes, resulting in changes in proteome composition. The current study provides a systematic evaluation for alternative splicing at the 13 GWAS loci.
Our main finding was that 3 genes at two obesity loci were alternatively spliced in type 2 diabetic overweight individuals and at least one of them, MSH5, was also regulated by weight loss and associated with BMI, suggesting functional significance of alternative splicing. The weight-loss-associated effect on alternative splicing of MSH5 demonstrates that obesity risk loci can also be regulated by weight loss, suggesting a potential gene-environment interaction in the regulation of alternative splicing. MSH5 is involved in DNA damage repair (9), and polymorphisms of MSH5 have been associated with cancer, immune diseases and reproductive disorders. The role of MSH5 in obesity remains unknown.
Interestingly, MSH5 and BAG6 genes are localized on chromosome 6p21, suggesting that splicing of genes in the HLA chromosomal region, known to be strongly regulated by splicing, might contribute to obesity.
Additionally, we observed differential splicing of TRA2B and BAG6 between visceral and subcutaneous fat that could contribute to functional differences between adipose tissue depots. TRA2B is a known RNA binding protein that regulates mRNA splicing in a concentration-dependent manner. Importantly, exclusion of exon 2 form TRA2B final mRNA regulates expression of TRA2B itself (10). Our observation that BMI is an independent predictor of TRA2B splicing in subcutaneous fat is in line with a previous report demonstrating reduced expression of TRA2B in individuals with obesity (1). BAG6 plays a role in apoptosis and gene regulation, in addition to regulating protein synthesis and degradation. The function of different BAG6 isoforms remains elusive, but recent studies have demonstrated different sub-cellular expression of BAG6 protein isoforms, with exclusive nuclear expression of the isoform encoded by transcript variant BAG6 varX7 (11). Our analysis indicates BMI as the strongest determinant of BAG6 splicing in subcutaneous fat. A mechanism on how the aberrant splicing of BAG6 contributes to obesity remains to be determined.
It is important to point out the limitations of our study. PCR capillary electrophoresis is not suitable for detection of alternative first and last exons. However, this method allows for efficient screening of subtle changes in splicing of cassette exons and direct comparison of splicing pattern across different tissues. Additionally, the small sample size used in this study does not provide enough statistical power to test an association between the genotypes and splicing of selected candidate genes. However, our study design, which utilized the power of paired tissue samples, allowed us to demonstrate statistically significant tissue-dependent splicing for TRA2B and BAG6, in addition to weight loss-dependent changes in MSH5 splicing in two independent intervention studies. Due to limitations of the method we were not able to study all 97 known obesity loci.
CONCLUSION
Our findings imply that obesity-associated SNPs may act through regulation of splicing and could underlie the pathogenesis of obesity in individuals carrying these risk SNPs. This study revealed the need of utilizing whole transcriptome RNA sequencing to detect all splicing variants in obesity loci in a larger sample size with a longitudinal setting.
Supplementary Material
What is already known about this subject?
There are 97 known loci associated with obesity.
The functional significance remains unknown for the most of the obesity-associated SNPs.
Aberrant splicing has been linked with metabolic diseases.
What does this study add?
Alternative splicing was detected in the 13 GWAS obesity loci.
Differential splicing was observed between overweight type 2 diabetic and lean normoglycemic individuals.
Change in alternative splicing was observed in response to weight loss.
Acknowledgments
Funding: This study was supported by a grant from the Academy of Finland (contracts 124243 to ML and 120979 and 138006 to JP), the Emil Aaltonen Foundation (to JP), the Finnish Diabetes Research Foundation (to ML and JP), the Finnish Cultural Foundation (to JP and DK), the Finnish Heart Foundation (to ML), Tekes–the Finnish Funding Agency for Technology and Innovation (contracts 1510/31/ 06 to ML and 40100/07 to LK) and the NIH grants (HL-095056, HL-28481, and DK093757 to PP).
The authors thank Päivi Turunen, Matti Laitinen, Kristiina Juvonen, Anne Jääskeläinen, Eerika Holma and Tiina Sistonen (University of Eastern Finland) for assistance with tissue biopsies and laboratory analyses.
Abbreviations
- BMI
body mass index
- HOMA-IR
homeostasis model assessment of insulin resistance
- KOBS
Kuopio Obesity Surgery study
- METSIM
Metabolic Syndrome in Men study
- VLCD
very low calorie diet
Footnotes
Disclosure: The authors declare no conflict of interest.
Author contributions: DK acquired, analyzed and interpreted data and wrote the manuscript. PK, SV, II and KHH collected adipose tissue biopsies. EN helped in the analysis of splice variants in the adipose tissue. MK designed the VLCD study. LK is the PI responsible for the VLCD study. HG conducted clinical studies in the KOBS study. PP contributed to the discussion and reviewed the manuscript critically for intellectual content. ML and JK are responsible for the METSIM study, contributed to the discussion and reviewed the manuscript critically for intellectual content. JP is responsible for the KOBS study, designed the study, contributed to the discussion, reviewed and edited the manuscript. JP is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors approved the final version to be published.
References
- 1.Pihlajamaki J, Lerin C, Itkonen P, Boes T, Floss T, Schroeder J, et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011;14:208–218. doi: 10.1016/j.cmet.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sesti G, Marini MA, Tullio AN, Montemurro A, Borboni P, Fusco A, et al. Altered expression of the two naturally occurring human insulin receptor variants in isolated adipocytes of non-insulindependent diabetes mellitus patients. Biochem Biophys Res Commun. 1991;181:1419–1424. doi: 10.1016/0006-291x(91)92097-4. [DOI] [PubMed] [Google Scholar]
- 3.Goren A, Kim E, Amit M, Bochner R, Lev-Maor G, Ahituv N, et al. Alternative approach to a heavy weight problem. Genome Res. 2008;18:214–220. doi: 10.1101/gr.6661308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 6.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pihlajamaki J, Kuulasmaa T, Kaminska D, Simonen M, Karja V, Gronlund S, et al. Serum interleukin 1 receptor antagonist as an independent marker of non-alcoholic steatohepatitis in humans. J Hepatol. 2012;56:663–670. doi: 10.1016/j.jhep.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Karhunen L, Lyly M, Lapvetelainen A, Kolehmainen M, Laaksonen DE, Lahteenmaki L, et al. Psychobehavioural factors are more strongly associated with successful weight management than predetermined satiety effect or other characteristics of diet. J Obes. 2012;2012:274068. doi: 10.1155/2012/274068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tompkins JD, Wu X, Chu YL, Her C. Evidence for a direct involvement of hMSH5 in promoting ionizing radiation induced apoptosis. Exp Cell Res. 2009;315:2420–2432. doi: 10.1016/j.yexcr.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoilov P, Daoud R, Nayler O, Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum Mol Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- 11.Kamper N, Kessler J, Temme S, Wegscheid C, Winkler J, Koch N. A novel BAT3 sequence generated by alternative RNA splicing of exon 11B displays cell type-specific expression and impacts on subcellular localization. PLoS One. 2012;7:e35972. doi: 10.1371/journal.pone.0035972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.