Abstract
Summary
We have implemented a High Performance Computing (HPC) version of ProtTest (Abascal et al., 2007) that can be executed in parallel in multi-core desktops and clusters. This version, called ProtTest 3, includes new features and extended capabilities.
Availability
ProtTest 3 source code and binaries are freely available under GNU license for download from http://darwin.uvigo.es/software/prottest3, linked to a Mercurial repository at Bitbucket (https://bitbucket.org/).
1. Introduction
Recent advances in modern sequencing technologies have resulted in an increasing capability for gathering large data sets. Long sequence alignments with hundred or thousands of sequences are not rare these days, but their analysis imply access to large computing infrastructures and/or the use of simpler and faster methods. In this regard, High Performance Computing (HPC) becomes essential for the feasibility of more sophisticated –and often more accurate– analyses. Indeed, during the last years HPC facilities have become part of the general services provided by many universities and research centers. Besides, multicore desktops are now standard.
The program ProtTest (Abascal et al., 2007) is one of the most popular tools for selecting models of amino acid replacement, a routinary step in phylogenetic analysis. ProtTest is written in Java and uses the program PhyML (Guindon and Gascuel, 2003) for the maximum likelihood (ML) estimation of model parameters and phylogenetic trees and the PAL library (Drummond and Strimmer, 2001) to handle alignments and trees. Statistical model selection can be a very intensive task when the alignments are large and include divergent sequences, highlighting the need for new bionformatic tools capable of exploiting the available computational resources.
Here we describe a new version of ProtTest, ProtTest3, that has been completely redesigned to take advantage of HPC environments and desktop multicore processors, significantly reducing the execution time for model selection in large protein alignments.
2. Prottest 3
The general structure and the Java code of ProtTest has been completely redesigned from a computer engineering point of view. We implemented several parallel strategies as distinct execution modes in order to make an efficient use of the different computer architectures that a user might encounter:
-
(1)
a Java thread-based concurrence for shared memory architectures (e.g., a multi-core desktop computer or a multi-core cluster node). This version also includes a new and richer Graphical User Interface (GUI) to facilitate its use.
-
(2)
an MPJ (Shafi et al., 2009) parallelism for distributed memory architectures (e.g., HPC clusters).
-
(3)
a hybrid implementation MPJ - OpenMP (Dagum and Menon, 1998) to obtain maximum scalability in architectures with both shared and distributed memory (e.g., multicore HPC clusters).
Moreover, ProtTest 3 includes a number of new and more comprehensive features that significantly extend the capabilities of the previous version: (1) more flexible support for different input alignment formats through the use of the ALTER library (Glez-Peña et al., 2010): ALN, FASTA, GDE, MSF, NEXUS, PHYLIP and PIR; (2) up to 120 candidate models of protein evolution; (3) four strategies for the calculation of likelihood scores: fixed BIONJ, BIONJ, ML or user-defined; (4) four information criteria: AIC, BIC, AICc and DT (see Sullivan and Joyce 2005); (5) reconstruction of model-averaged phylogenetic trees (Posada and Buckley, 2004); (6) fault tolerance with checkpointing; and (7) automatic logging of the user activity.
3. Performance Evaluation
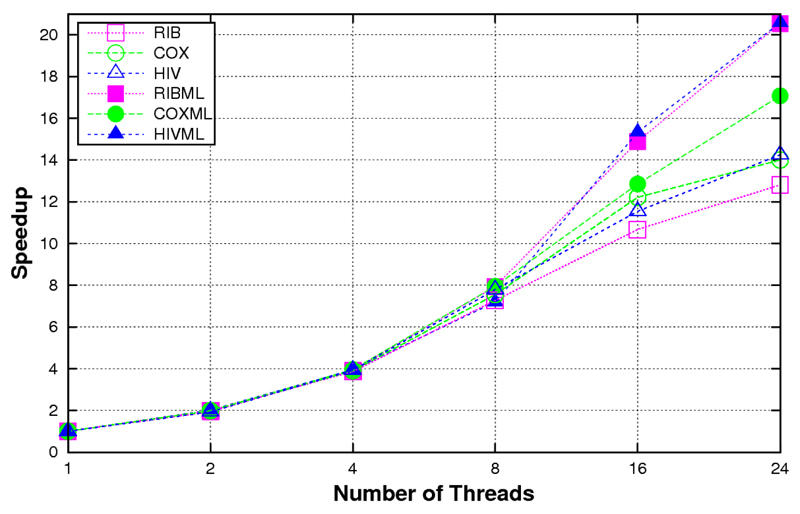
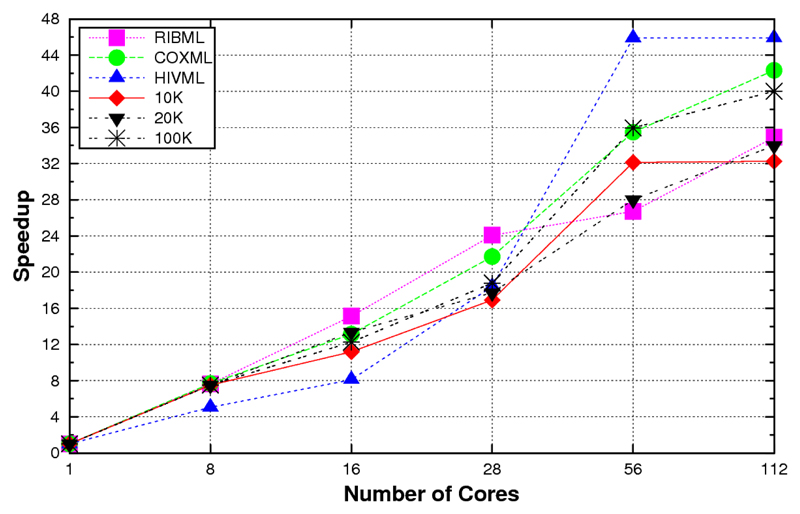
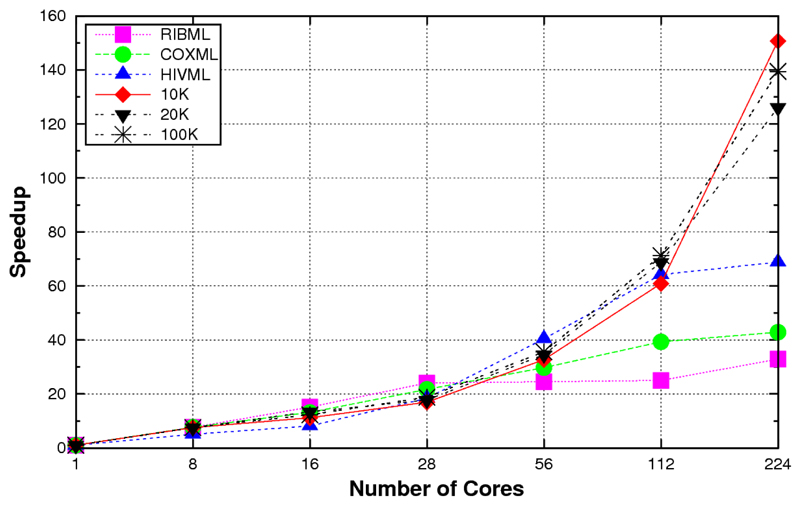
In order to benchmark the performance of ProtTest 3, we computed the running times for the estimation of the likelihood scores of all 120 candidate models from several real and simulated protein alignments (Table 1). When these data were executed in a system with shared memory, e.g., a multicore desktop, the scalability was almost linear as far as there was enough memory to satisfy the requirements. For example, in a shared memory execution in a 24-core node the speedup was almost linear with up to 8 cores, also scaling well with data sets with medium complexity, like HIVML or COXML (Fig. 1). In a system with distributed memory like an cluster, the application scaled well up to 56 processors (Fig. 2). With more processors a theoretical scalability limit exists due to the hetereogeneous nature of the optimization times, from a few seconds for the simplest models to up to several hours for the models that include rate variation among sites (+G). This problem was solved with the hybrid memory approach. In this case, the scalability went beyond the previous limit, reaching up to 150 in the most complex cases with 8-core nodes (Figure 3).
Table 1.
Real and simulated alignments used to benchmark ProtTest 3 performance. In column Size, N indicates the number of sequences and L the length of the alignment. Base tree is the tree used for model likelihood optimization and Seq. exec. time is the time required to calculate the likelihood scores using the sequential version (i.e., a single thread).
| Data set Abbreviation | Protein | Size NxL | Base tree | Seq. exec. time |
|---|---|---|---|---|
| RIB | Ribosomal protein | 21x113 | Fixed BIONJ | 5.5m |
| RIBML | ” | ” | ML tree | 28m |
| COX | Cytochrome C oxidase II | 28x113 | Fixed BIONJ | 9.5m |
| COXML | ” | ” | ML tree | 55m |
| HIV | HIV polimerase | 36x1,034 | Fixed BIONJ | 44m |
| HIVML | ” | ” | ML tree | 160m |
| 10K | Simulated aln | 50x10K | Fixed BIONJ | 9.2h |
| 20K | ” | 50x20K | ” | 24.5h |
| 100K | ” | 50x100K | ” | 80h |
Figure 1.
Speed-ups obtained with the shared memory version of ProtTest 3 according to the numbers of threads used in a 24-core shared memory node (4 hexa-core Intel Xeon E7450 processors) with 12GB memory.
Figure 2.
Speed-ups obtained with the distributed memory version of ProtTest 3 according to the numbers of cores used in a 32-node cluster with 2 quad-core Intel Harpertown processors and 8GB memory per node. Up to 4 processes where executed per node because of the memory requirements of the largest datasets (10K, 20K, 100K).
Figure 3.
Speed-ups obtained with the hybrid memory version of ProtTest 3 according to the numbers of cores used in a 32-node cluster with 2 quad-core Intel Harpertown processors and 8GB memory per node. Up to 4 MPJ Express processes per node and at least 2 OpenMP threads for each ML optimization were executed.
4. Conclusions
ProtTest 3 can be executed in parallel in HPC environments as: (1) a GUI-based desktop version that uses multi-core processors; (2) a cluster-based version that distributes the computational load among nodes; and (3) as a hybrid multi-core cluster version that achieves speed through the distribution of tasks among nodes while taking advantage of multi-core processors within nodes. The new version has been completely redesigned and includes new capabilities like checkpointing, additional amino acid replacement matrices, new model selection criteria and the possibility of computing model-averaged phylogenetic trees. The use of ProtTest 3 results in significant performance gains, with observed speedups of up to 150 on a high performance cluster. For very large alignments this can be equivalent to a reduction of the running time from more than three days to around half an hour. In this way, statistical model selection for large protein alignments becomes feasible, not only for cluster users, but also for the owners of standard multi-core desktop computers. Moreover, the flexible design of ProtTest-HPC will allow developers to extend future functionalities, whereas third-party projects will be able to easily adapt its capabilities to their requirements.
Supplementary Material
Supplementary information: Supplementary data are available at Bioinformatics online.
Acknowledgements
Special thanks to Stephane Guindon for his continuous help with PhyML and to Federico Abascal for his help with the previous version of ProtTest. This work was financially supported by the European Research Council [ERC-2007-Stg 203161-PHYGENOM to D.P.], the Spanish Ministry of Science and Education [BFU200908611 to D.P.] and by the Xunta de Galicia [Galician Thematic Networks RGB 2010/90 to D.P. and GHPC2 2010/53 to R.D.].
References
- Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2007;24(1):1104–1105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Dagum L, Menon R. OpenMP: An industry-standard API for shared-memory programming. IEEE Computational Science and Engineering. 1998;5(1):46–55. [Google Scholar]
- Drummond A, Strimmer K. Pal: an object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics. 2001;17(7):662–663. doi: 10.1093/bioinformatics/17.7.662. [DOI] [PubMed] [Google Scholar]
- Glez-Peña D, Gópromez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D. ALTER: program-oriented conversion of DNA andtein alignments. Nucleic Acids Research. 2010;(38):W14–W18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53(5):793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Shafi A, Carpenter B, Baker M. Nested parallelism for multi-core HPC systems using Java. J Parallel Distr Com. 2009;69(6):532–545. [Google Scholar]
- Sullivan J, Joyce P. Model selection in phylogenetics. Annu Rev Ecol Evol S. 2005;36:445–466. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.