Abstract
SCN8A encephalopathy is a severe, early-onset epilepsy disorder resulting from de novo gain-of-function mutations in the voltage-gated sodium channel Nav1.6. To identify the effects of this disorder on mRNA expression, RNA-seq was performed on brain tissue from a knock-in mouse expressing the patient mutation p.Asn1768Asp (N1768D). RNA was isolated from forebrain, cerebellum, and brainstem both before and after seizure onset, and from age-matched wildtype littermates. Altered transcript profiles were observed only in forebrain and only after seizures. The abundance of 50 transcripts increased more than 3-fold and 15 transcripts decreased more than 3-fold after seizures. The elevated transcripts included two anti-convulsant neuropeptides and more than a dozen genes involved in reactive astrocytosis and response to neuronal damage. There was no change in the level of transcripts encoding other voltage-gated sodium, potassium or calcium channels. Reactive astrocytosis was observed in the hippocampus of mutant mice after seizures. There is considerable overlap between the genes affected in this genetic model of epilepsy and those altered by chemically induced seizures, traumatic brain injury, ischemia, and inflammation. The data support the view that gain-of-function mutations of SCN8A lead to pathogenic alterations in brain function contributing to encephalopathy.
Keywords: RNA-seq, transcriptome, seizure, epileptic encephalopathy, astrocyte, gene expression, sodium channel
1. Introduction
Mutations of the voltage-gated sodium channel gene SCN8A are responsible for the severe disorder Early Infantile Epileptic Encephalopathy type 13 (OMIM #614558), also referred to as SCN8A encephalopathy (Hammer et al., 2016; Meisler et al., 2016; Wagnon and Meisler, 2015). These pathogenic mutations arise de novo in affected individuals and generate gain-of-function missense mutations that increase the activity of sodium channel Nav1.6 (Wagnon et al., 2016). Two commonly observed changes in Nav1.6 are impaired channel inactivation, and a hyperpolarizing shift in voltage dependence of activation that results in premature channel opening (Barker et al., 2016; Blanchard et al., 2015; Estacion et al., 2014; Veeramah et al., 2012; Wagnon et al., 2016). SCN8A encephalopathy is characterized by seizure onset within the first 18 months of life, typically by 4 months of age (Larsen et al., 2015). Co-morbidities include developmental delay and cognitive impairment. Approximately 50% of affected children do not sit or walk, and there is a 10% incidence of SUDEP (sudden unexpected death in epilepsy). Anti-epileptic drugs that target sodium channels provide transient improvement in some patients, but seizure control is usually incomplete (Hammer et al., 2016).
To gain insight into pathogenesis and to identify novel therapeutic targets, we compared gene expression before and after seizure onset in a mouse model of SCN8A encephalopathy. The mutation p.Asn1768Asp (N1768D), identified in an individual with SCN8A encephalopathy and SUDEP (Veeramah et al., 2012), was introduced into the mouse genome by TALEN targeting (Jones and Meisler, 2014). The knock-in mouse model recapitulates the seizure disorder of the patient, with development of severe tonic-clonic seizures between 3 to 4 months of age (Wagnon et al., 2015). The initial seizure initiates an acute downhill progression marked by development of hunched posture, tremor, reduced spontaneous movement, and additional seizures. Many mice succumb to sudden death within 24 hours after the first seizure. This phenotype is incompletely penetrant, and approximately 50% of heterozygous N1768D/+ mice never exhibit visible seizures and survive for more than 18 months.
The effect of seizures on gene expression has previously been studied in non-genetic models of rodents with chemically induced seizures (Ravizza et al, 2001; Tang et al, 2002; Elliott et al, 2003; Wilson et al, 2005; Okamoto et al, 2010), and in human cortical samples obtained after surgery for focal epilepsy (Rakhade et al, 2005; Boer et al, 2010; Beaumont et al, 2012). In this manuscript we describe altered gene expression in a genetic model of sodium channel-dependent epileptic encephalopathy.
2. Materials and Methods
2.1 Animals
The Scn8aN1768D/+ knock-in mouse was previously generated by TALEN targeting (Jones and Meisler, 2014). The initial characterization was carried out on the congenic line B6.Scn8aN1768D (Wagnon et al., 2015). To improve litter size and reproductive performance, we also backcrossed the N1768D mutation to strain C3HeB/FeJ to generate a second congenic line, C3H.Scn8aN1768D. All samples for this study were from the second congenic line, C3H.Scn8aN1768D . The samples for RNA-seq and qRT-PCR included pre-seizure D/+ mice (mean age=6 weeks, n=3) and +/+ littermate controls (n=3), post-seizure D/+ (14 weeks, n=6) and +/+ littermate controls (n=3), and non-seizure D/+(35 weeks, n=2) and +/+ littermate controls (n=2) . As described previously, seizures in these mice have duration of 0.5 to 3 min with onset between 2 and 4 months of age, and occur in 50% of the heterozygous D/+ mice (Wagnon et al, 2015). Death typically occurs within 1 to 3 days after onset. The first seizure is immediately followed by 4 sequelae: tremor, hunched posture, nonresponsiveness to stimuli, and periods of immobility lasting for more than 30 minutes. For this study, D/+ mice were examined every day beginning at 6 weeks of age. For inclusion in the post-seizure group, we required that the mice exhibit none of the four indicators of seizures on the previous day and exhibit seizures as well as all four indicators of seizure activity on the day of sample collection. The samples were thus processed within 24 hours of the first seizure.
2.2 RNA preparation
Total RNA was isolated from dissected forebrain (cerebral cortex plus hippocampus), cerebellum, and brainstem from D/+ mice and +/+ littermate controls using Trizol reagent (Life Technologies). RNA samples were assessed for quality with the High Sensitivity RNA Analysis Kit (Advanced Analytics Fragment Analyzer). RNA concentration was determined using the Quant-iT RiboGreen RNA Assay Kit (Molecular Probes).
2.3 RNA-Seq
Libraries were constructed using a stranded mRNA-Seq Kit (KapaBiosystems TDS KR0960 – v3.15). Average fragment size was assessed with the Fragment Analyzer (Advanced Analytics). Concentration was determined with the Illumina Universal Adaptor-specific qPCR kit (Kapa Biosystems). Equimolar samples were pooled and clustered for sequencing on the HiSeq2500(Illumina). Sequencing was performed using Rapid-Run SBS 2×100bp chemistry (Illumina).
2.4 Sequence analysis
Sample data was demultiplexed, trimmed and quality filtered using Trimmomatic (USADelLab). Fastq files were splice aligned against the NCBIM37 reference genome using Tophat version 2.0.13 running atop Bowtie version 2.2.4. Gene expression counts were obtained using htseq-count version 0.6.1. Both splice alignment and counting were performed with Ensembl Annotation of the NCBIM37 reference genome and raw counts analyzed with DESeq2 version 1.10.1. Results generated by DESeq2 comparing postseizure samples with the age-matched +/+ littermate controls identified 65 transcripts with logarithm of odds (LOD) absolute value of expression change ≥ 3.0, among which 50 were up-regulated and 15 were down-regulated.
2.5 Principal component and heat map analysis
Principal component analysis (PCA) was performed with the plotPCA function using expression counts previously transformed to the log2 scale with the rlogTransformation for the 469 genes with ≥2-fold differential expression between post-seizure D/+ mice (n-4) and wildtype controls (n=3). This plot was also used to check for batch effects and to create a count matrix within DESeq2 producing a PCA plot of the first two principal components. To add information about other non-seizure mice, we projected the coordinates for each individual onto the first two principal components based on variation in expression at the same 469 loci.
Values of the same logarithm transformed expression counts for the 65 genes with ≥3-fold change in transcript abundance were used to generate heatmap plots using the heatmap.2 function from the gplots R package.
2.6 Differential expression analysis
Differential expression was analyzed in DESeq2, version 1.10.1, in R using the workflow described in sections 1.2 and 1.3 of the DESeq2 manual (DESeq2 reference manual: http://www.bioconductor.org/packages/2.13/bioc/vignettes/DESeq2/inst/doc/DESeq2.pdf)
2.7 Pathway analysis
The output of DESeq2 was inputted into QIAGEN’s Ingenuity Pathway Analysis (IPA) software, version 01-01 (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). The 65 genes with ≥3-fold change in transcript abundance were submitted for IPA’s Core Analysis functionality and a regulator network was generated using IPA's Regulator Effects algorithm.
2.8 Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
cDNA was generated from the forebrain RNA samples with the SuperScript III kit (Life Technologies). Taqman probes were obtained from Life Technologies for the genes Ptx3, Gal , Npw, Cdkn1a, Gldn, Serpina3n, Drd4, Mt2, Vim, Gfap, and Hif3a. Probe set IDs are provided in Table S1. Taqman reactions were performed in 15 uL reaction volumes using the Taqman Fast Advance Master Mix. All reactions were run in triplicate on an ABI 7900HT using the SDS 2.4 software (Life Technologies) with ABI384 well Optical PCR plates and AB-1170 Optical PCR film (Fisher Scientific). All samples were run with the endogenous control GAPDH probe set (Life Technologies #Mm99999915_g1). Differential expression analysis was performed using the standard delta-delta CT method (Livak and Schmittgen, 2001). Transcript expression levels were normalized to the average expression of the transcript in wildtype littermate control mice.
2.9 Immunostaining
Brains were fixed overnight at 4°C in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) (8 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, 3 mM KCl pH 7.3), rinsed in PBS and stored at 4°C in 40% sucrose (w/v) in PBS. Frozen sagittal sections (20 µm) were rinsed in Tris buffered saline (TBS) (25 mM Tris, 140 mM NaCl, 3 mM KCl pH 7.5) followed by blocking and permeablization for one hour in 10% normal goat serum in TBS with 0.2% Triton X-100. Sections were immunostained at 4°C overnight with mouse anti-GFAP Cy3-conjugated antibody (Sigma-Aldrich. Clone G A 5) diluted 1:400 in TBS containing 5% normal goat serum and 0.05% Triton X-100. Sections were rinsed with TBS containing 0.05% Triton X-100 and incubated for 30 min in 1:500 DAPI in TBS containing 0.05% Triton X-100. This protocol resulted in very low background staining. Stained sections were imaged with the 10x objective on a Nikon A1R confocal microscope. Tiled images were combined in Adobe Illustrator.
3. Results
3.1 Pre-seizure, post-seizure and non-seizure Scn8aN1768D/+ (D/+) mice
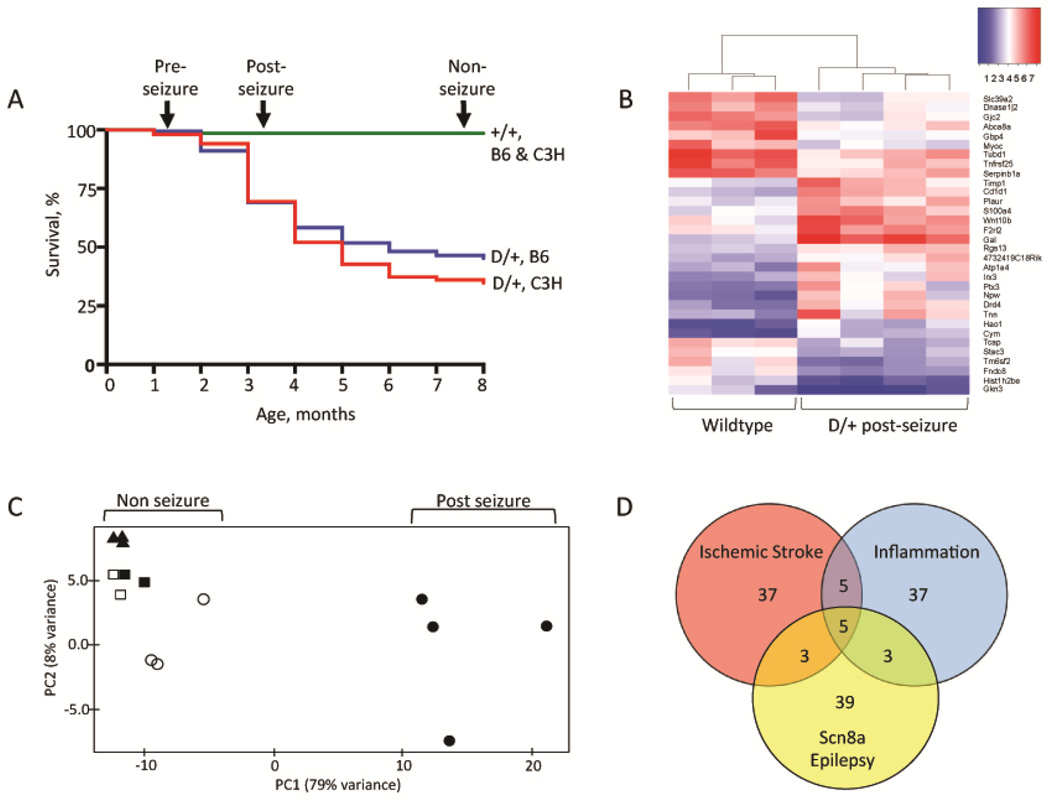
The similar survival curves of D/+ mice in the previously described B6.Scn8aN1768D/+ congenic line (Wagnon et al, 2015) and the newly developed C3H.Scn8aN1768D/+ congenic line are shown in Fig. 1A. Between 2 to 6 months of age, approximately 50% of D/+ mice in both lines exhibit seizure onset followed by sudden death. The acute progression after seizure onset includes hunched posture, tremor, lack of movement, and seizures. For this study, mice between the ages of 3 and 4 months that exhibited all of these phenotypes were sacrificed between 16 and 24 hours after onset and designated "post-seizure" (n=4) (Fig. 1A). The "pre-seizure" group (n=3) was collected at 6 weeks of age. We previously reported that D/+ mice have normal EEG patterns at this age (Wagnon et al, 2015). The "non-seizure" group (n=2) had never demonstrated seizures or any seizure-associated phenotypes at 8 months of age. RNA was prepared from wildtype (+/+) littermates of these three groups of mice at 6 weeks (n=3), 3 to 4 months (n=3), and 8 months (n=2) of age. RNA was prepared from three brain regions for each animal: forebrain (cerebral cortex plus hippocampus), cerebellum and brainstem.
Fig. 1. Transcriptome analysis of Scn8aN1768D/+ mice.
A) Survival curves. RNA was prepared from the three groups of D/+ mice indicated by arrows, and from their +/+ littermates: pre-seizure (6 weeks of age), post-seizure (3 to 4 months of age) and nonseizure (8 months of age). Survival data is based on several hundred animals of genotypes B6.Scn8aN1768D/+ (n = 168); C3H.Scn8aN1768D/+ (n = 150); B6+/+ (n = 297); C3H+/+ (n = 180). B) Heat map of 32 genes with ≥3-fold alteration in D/+ post-seizure mice compared with wildtype littermates. Red, high expression; blue, low expression. (33 highly expressed genes are not included in the display). C) Principal component analysis of the 469 genes with ≥2-fold difference in expression between four post-seizure D/+ mice (solid circles) and three wildtype controls (open circles). Seven additional samples were projected on the plot based on transcript abundance of the same 469 genes: , non-seizure D/+ mice (solid squares), nonseizure littermate controls (open squares); pre-seizure D/+ mice (solid triangles). D) Venn diagram of the 50 most highly elevated transcripts in three models of CNS injury.
3.2. Altered gene expression in forebrain of post-seizure mice
There was excellent concordance between transcriptional profiles of biological replicates. Transcripts from more than 24,000 genes were detected in all RNA samples and included in the differential expression analysis. The two groups of D/+ mice that did not experience seizures, the "pre-seizure" and "non-seizure" groups (Fig. 1A), did not exhibit differences in transcript abundance for any brain region, when compared with their wildtype littermates. Differential expression analysis of forebrain RNA from post-seizure D/+ mice and +/+ littermates, however, identified 50 transcripts whose abundance was increased ≥3-fold and 15 transcripts whose abundance was reduced by ≥3-fold (Table 1). Transcript levels in cerebellum and brainstem of post-seizure mice did not differ from wildtype controls, indicating that forebrain is uniquely sensitive to seizure onset.
Table 1.
Genes with transcript abundance in forebrain that is altered by 3-fold or more after seizures in Scn8aN1768D/+ mutant mice.
| Increase relative to wildtype controls |
Genes |
|---|---|
| 9 x | Galanin |
| 4 – 5 x | Serpina3n, Drd4, Maff, Gldn, Ctla2a, Cd1d1, Atp2c2 |
| 3.5 – 4.0 x | Cdkn1a, Aspg, Col5a3, Serpine1, Apold1, C030019I05Rik, Wnt10b, Fkbp5, Npw |
| 3.2 – 3.5 x | Mt2, Rhoj, Plin4, Map3k6, Blnk, Dpm3, Rxfp2, Arsj, Cd109, F2rl2, A2m, Cym, Tnn, Vim |
| 3 – 3.2 x | Nt5e, Gfap, Plekha2, 4732419C18Rik, Slc2a4, Hif3a, Atp1a4, Ptx3, Angptl7, Hspb6, Irx3, Timp1, Prss23, Rgs13, Ret, Hao1, Plaur, Emp1, S100a4 |
| Decrease relative to wildtype controls |
|
| 3.5–4.0 x | Gjc2, Slc39a2, Abca8a, Tm6sf2 |
| 3.1–3.5 x | Tnfrsf25, Hist1h2be, Tubd1 |
| 3.0 x | Myoc, Dnase1|, Serpin1ba, Fndc8, Gkn3, Tcap, Kcnh4, Stac3 |
The changes in abundance of 65 transcripts in the post-seizure D/+ mice thus represents a response to seizures, rather than a direct effect of the D/+ genotype. A heat map of 32 genes with > 3-fold changes in transcript abundance in post-seizure mice is shown in Fig. 1B. Unsupervised clustering resulted in separate clustering of post-seizure D/+ mice and wildtype littermates. Data from 8-month old non-seizure D/+ mice and their +/+ littermates also clustered with the +/+ mice (not shown).
3.3 Principal Component Analysis
Principal Component Analysis (PCA) was conducted on the 469 forebrain transcripts with ≥2-fold difference in expression between post-seizure D/+ mice (n=4) and wildtype controls.(n=3). The first principal component accounted for 79% of the variance in the expression data (Fig. 1C). We then projected seven additional non-seizure mice onto the plot, based on expression levels of the same 469 genes. These mice included D/+ mice that never exhibited visible seizures and survived for >8 month (n=2), their wildtype littermates (n=2), and 6-week pre-seizure D/+ mice (n=3). Interestingly, all of these mice form a tight cluster close to the original wildtype controls (n=3), indicating that the expression profile for these 469 genes is predominantly explained by seizure status. This is consistent with the lack of altered transcript abundance in the absence of seizures described above.
3.4 Functions of genes with differential expression in post-seizure mice
The molecular functions and physiologic pathways of 65 genes with differential expression of 3-fold or greater were examined by Ingenuity Pathway Analysis. Activation of pathways involved in regeneration of neurites and axons, activation of neuroglia and cell survival, and deactivation of pathways involved in cell death and inflammation were observed (Table 2).
Table 2.
Significantly enriched pathways identified by IPA analysis of 65 genes whose expression is altered ≥3-fold in post-seizure mice
| Function | p-Value | Genes |
|---|---|---|
| A. Predicted increase in activation state | ||
| Inflammatory response | 2.75E-07 | A2m, Blnk, Cd1d, Cdkn1a, Ctla2a, Gal, Gjc2, Npw, Nt5e, Plaur, Ptx3, S100a4, Serpina3, Serpinb1, Serpine1 |
| Regeneration of neurons | 4.99E-07 | A2m, Cdkn1a, Gfap, Mt2, Ret, Vim |
| Outgrowth of neurites | 2.23E-06 | A2m, Cdkn1a, Fkbp5, Gal, Gfap, Ret, S100a4, Serpine1, Tnn, Vim |
| Regeneration of neurites | 3.47E-06 | A2m, Cdkn1a, Gfap, Mt2, Ret |
| Activation of CNS cells | 3.47E-06 | Cdkn1a, Drd4, Gfap, Serpine1, Vim |
| Proliferation of neuronal cells | 7.65E-06 | A2m, Cdkn1a, Fkbp5, Gal, Gfap, Irx3, Ret, S100a4, Serpine1, Tnn, Vim |
| Reactivation of astrocytes | 8.43E-06 | Gfap, Vim |
| Regeneration of axons | 5.57E-05 | A2m, Cdkn1a, Mt2, Ret |
| Activation of neuroglia | 8.41E-04 | Gfap, Gjc2, Serpine1, Vim |
| Mobilization of Ca2+ | 1.55E-03 | A2M, Blnk, F2rl2, Gal, Plaur, Wnt10b |
| Cell survival | 3.65E-03 | Cdkn1a, Emp1, Fkbp5, Hspb6, Mt2, Plaur, Ret, Rhoj, S100a4, Serpine1, Slc2a4, Timp1, Vim |
| B. Predicted decrease in activation state | ||
| Fibrosis | 1.12E-07 | Cd1d, Hspb6, Mt2, Nt5e, Plaur, Ptx3, Ret, S100a4, Serpinb1, Serpine1, Slc2a4, Timp1, Vim |
| Inflammation of body region | 6.58E-07 | A2m, Cd1d, Cdkn1a, Drd4, Gfap, Mt2, Nt5e, Plaur, Plekha2, Ret, S100a4, Serpinb1, Serpine1, Slc2a4, Timp1, Tnfrsf25, Vim |
| Necrosis | 2.38E-05 | A2m, Atp1a4, Blnk, Cd1d, Cdkn1a, Col5a3, Dpm3, Emp1, Fkbp5, Gal, Gfap, Hspb6, Mt2, Myoc, Nt5e, Plaur, Ret, Rjoj, S100a4, Serpina3, Serpine1, Slc2a4, Timp1, Tnfrsf25, Vim |
| Cell death | 2.42E-04 | A2m, Atp1a4, Blnk, Cd1d, Cdkn1a, Col5a3, Dpm3, Emp1, Fkbp5, Gal, Gfap, Hspb6, Mt2, Myoc, Nt5e, Plaur, Ret, Rhoj, Rxfp2, S100a4, Serpina3, Serpine1, Slc2a4, Timp1, Tnfrsf25, Vim, Wnt10b |
| Inflammation of CNS cells | 4.73E-04 | Cd1d, Gfap, Mt2, Nt5e, Serpine1, Timp1, Vim |
We compared these changes with previous studies of chemically and electrically induced seizures and brain injury (see references in Table 3). Of the 50 transcripts that are elevated > 3-fold in D/+ mice after seizures, 11 are elevated in response to induced seizures, and 10 are elevated after brain injury (Table 3). These include the neuropeptide Galanin and the reactive astrocyte markers Gfap and Vimentin (Table 3). Eight of these transcripts were elevated in purified populations of reactive astrocytes (Zamanian et al, 2012). The extent of overlap between elevated expression in response to seizures, traumatic brain injury, and reactive astrocytosis, suggests that a coordinated transcriptional response to brain injury is activated by seizures in Scn8a mutant mice.
Table 3.
Several genes with elevated expression in the current study are also elevated in other models of brain trauma.
| Gene | Induced seizures (rodent) (a–f) |
Traumatic brain Injury (rodent) (g–h) |
Astrocytes, reactive (mouse) (i) |
Astrocytes, epileptic (human) (j) |
Cortical foci (human) (k–m) |
|---|---|---|---|---|---|
| A2m | + | + | + | + | |
| Gfap | + | + | + | + | |
| Vimentin | + | + | + | + | |
| Serpina3n | + | + | + | + | |
| Aspg | + | + | + | ||
| Galanin | + | + | |||
| Mt2 | + | + | |||
| Ptx3 | + | + | |||
| Hif3a | + | ||||
| Atp1a4 | + | ||||
| Serpine1 | + | ||||
| Cdkn1a | + | + | + | ||
| Cd109 | + | + | |||
| Timp1 | + | ||||
| Blnk | + | ||||
| Fkbp5 | + | ||||
| Gldn | + |
Sources: (Ravizza et al., 2001),
3.5 Confirmation of quantitative changes by qRT-PCR
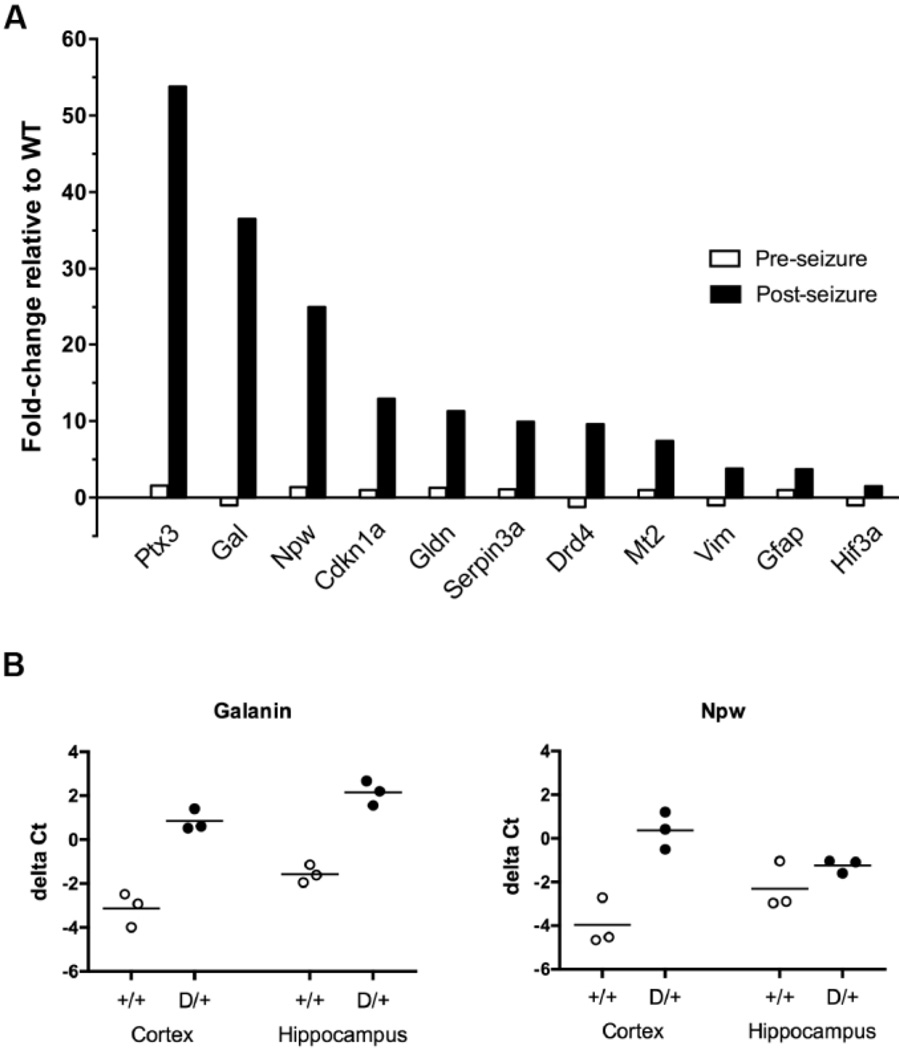
Eleven transcripts with elevated abundance in the RNA-seq data were subjected to confirmation by qRT-PCR, including genes with both high and low constitutive expression: Ptx3, Gal, Npw, Cdkn1a, Gldn, Serpina3n, Drd4, Mt2, Vim, Gfap, and Hif3a. qRT-PCR was carried out in triplicate. The predicted differences between mice with with and without seizures were confirmed for these 11 transcripts (Fig. 2A). There was excellent concordance between RNA-seq and qRT-PCR for 10 of the genes (Pearson correlation coefficient: r=0.764, p=0.006). The 11th gene, Ptx3, appeared to be increased 53-fold by qRT-PCR but only 3-fold by RNA-seq. This discrepancy is likely to be a consequence of the difficulty of accurately measuring the low constitutive expression of this gene by qRT-PCR.
Fig. 2.
Confirmation of selected RNA-seq data by qRT-PCR. Taq-man assays were carried out as described in Methods. A. Analysis of forebrain samples (cortex plus hippocampus). Data represents the ratio of transcript abundance in D/+ mice relative to wildtype littermates. Open bars, pre-seizure; solid bars, post-seizure . Gal, galanin; Gldn, gliomedin. B. Analysis of neuropeptide transcript abundance in cortex versus hippocampus. Each symbol represents an individual mouse of the indicated genotype.
To determine whether the two neuropeptide transcripts are elevated in cortex or in hippocampus, we prepared separate RNA samples from postseizure D/+ mice and littermate controls. qRT-PCR analysis demonstrated that the galanin transcript is elevated in both of these regions, while the NPW transcripts is specifically elevated in cortex (Fig. 2B).
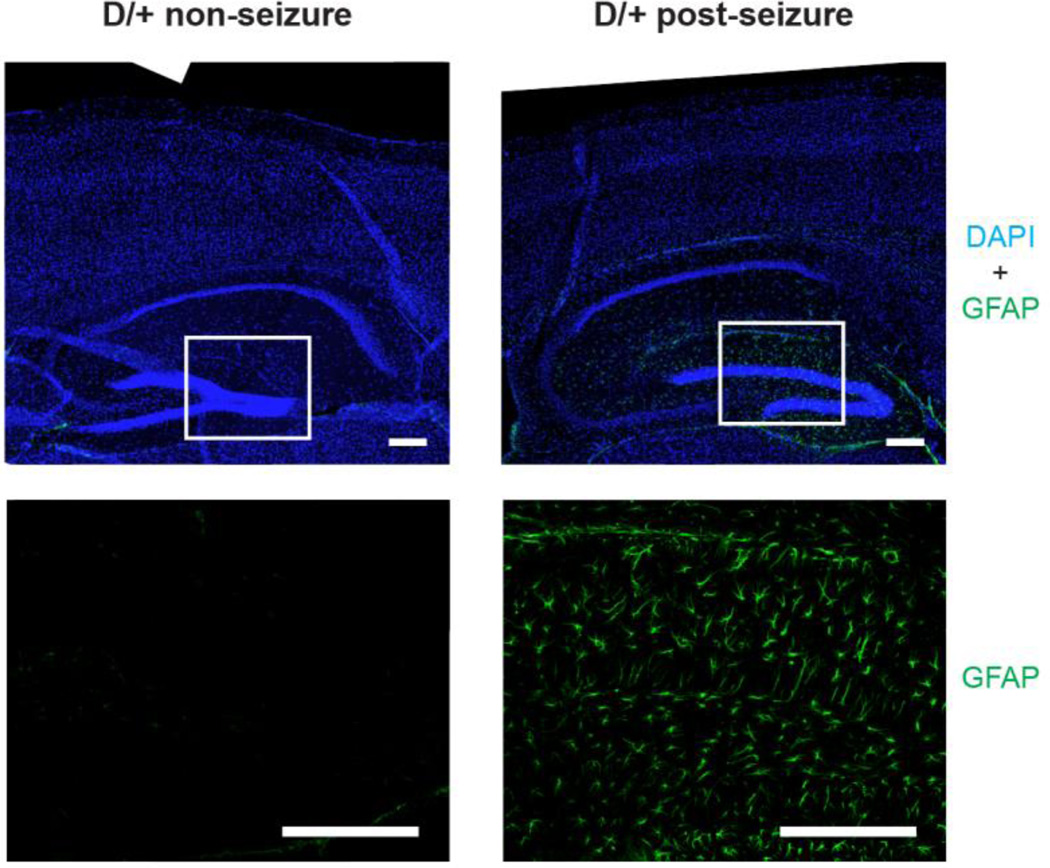
3.6 Hippocampal astrocytosis in post-seizure mice
Expression of GFAP, a marker of reactive astrocytosis, is elevated after seizures. To localize GFAP expression, we carried out immunostaining of sagittal sections of postseizure brain. The major site of GFAP expression is the hippocampus (Fig. 3). This result lends further support to the evidence of astrocytosis from the forebrain RNA expression data.
Fig. 3.
Astrocytosis in the hippocampus of D/+ mice after seizures. Sagittal sections of brain from SCN8A-N1768D/+ non-seizure mice (8 months of age) and post-seizure mice (3 months of age) were immunostained with antibody to GFAP (green) and with DAPI to visualize nuclei (blue). Top panels, composite images from multiple 10x images of forebrain and hippocampus. Bottom panels, higher magnification of boxed areas. Scale bar = 200 µm.
4. Discussion
4.1 Seizures are the cause of altered gene expression in Scn8aN1768D/+ mice
While elevated activity of the mutated sodium channel Nav1.6 is the direct cause of seizures in SCN8A encephalopathy, secondary changes in gene expression may contribute to pathogenicity and provide targets for therapeutic intervention. Using a stringent criterion of ≥3-fold changes in mRNA abundance, we identified 65 transcripts that are differentially expressed in Scn8aN1768D/+ mutant mice after seizures compared with wildtype littermates. These transcriptional changes appear to be a response to seizures, since they are detected after seizure onset but are not present in Scn8aN1768D/+ mice that are too young to experience seizures ("pre-seizure") or in mice with no evidence of seizures at 8 months of age ("non-seizure") (Fig. 1A,C). The data demonstrate a significant transcriptional response to seizures, and an apparent lack of direct transcriptional effect of the Scn8aN1768D/+ mutation. Transcriptional changes were restricted to the forebrain, with no differences observed in cerebellum or brainstem. These data, together with the observed hippocampal astrocytosis, suggest that forebrain circuits are particularly affected. This conclusion is consistent with the observations of elevated persistent current and early afterdepolarization-like action potentials in hippocampal CA1 and CA3 neurons from these mice (Lopez-Santiago et al, 2015), and with the role of the hippocampus in seizure resistance due to Scn8a haploinsufficiency (Makinson et al, 2014). The lack of transcriptional changes in cerebellum is in agreement with the normal activity of cerebellar Purkinje cells from the D/+ mutant mice (MHM and JLW, unpublished observations).
4.2 Genes involved in the transcriptional response
To our knowledge, this is the first study of transcriptome-wide expression changes in an animal model of monogenic human epilepsy. Several of the highly elevated transcripts in post-seizure D/+ mice have also been seen after induced seizures, traumatic brain injury, and neuroinflammation (Table 3). Functional annotation clustering of the 65 genes with ≥3-fold change in transcript abundance after seizures revealed enrichment in functions related to neuronal repair and regeneration (Table 2). These data suggest that the transcriptional changes in post-seizure D/+ mice reflect a generalized response to neuronal damage after seizures.
Two of the most highly elevated transcripts encode the neuropeptides galanin and NPW. In principle, these changes in gene expression may be part of the pathogenic process or part of a protective homeostatic response to seizures. We think that the increased expression of galanin and neuropeptide W (NPW) are likely to be a protective response, because of their well-studied anti-seizure effects of these peptides. For example, overexpression of galanin in several lines of transgenic mice was shown to suppress glutaminergic transmission and reduce seizure susceptibility (Mazarati et al., 2000; Schlifke et al, 2006; Kokaia et al, 2001). Administration of galanin by intra-hippocampal injection or adenovirus mediated delivery was also protective against seizures (Haberman et al., 2003; Lin et al., 2003; Mazarati and Wasterlain, 2002; Mazarati et al., 1998; McCown, 2006). Similar to our observations in D/+ mice, elevated levels of galanin transcripts have been detected after induced seizures (Wilson et al, 2005). Further, the galanin analog NAX 810-2, which penetrates the blood-brain barrier, protects against electrically-induced seizures (Bialer et al., 2013, 2015), as does a similar analog of NPW (Green et al., 2011). IN view of these protective effects, we suggest that additional increases in the levels of galanin and NPW prior to or after seizure onset might be protective in mice with the Scn8a-N1768D mutation.
We also observed a 9.6-fold elevated expression of the dopamine receptor gene Drd4 (Fig. 2), a gene that has not been previously been associated with epilepsy. However, deficiency of Drd4 was reported to increase cortical hyperexcitability (Rubinstein et al., 2001). The increased Drd4 in D/+ mice might counteract their neuronal hyperexcitability, and this might also be accomplished by administration of selective Drd4 agonists. Thus galanin, NPW and Drd4, identified by transcriptome analysis, represent potential therapeutic targets for SCN8A encephalopathy.
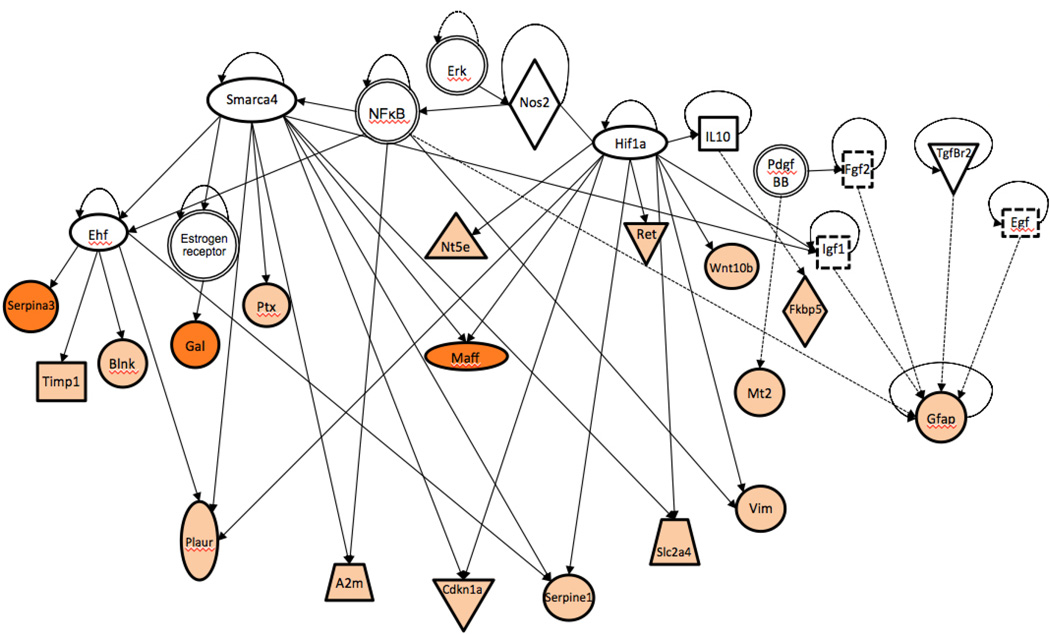
The transcription factors Hif1a and Smarca4 regulate 18 of the 50 induced genes, suggesting the possibility of a coordinated transcriptional response (Fig. 4). HIF-1α is neuroprotective after mild hypoxia, but neurotoxic after more severe hypoxia (Fan et al., 2009). Smarca4 (human gene BRG1) is involved in the developmental switch from neurogenesis to gliogenesis (Deng et al., 2015), and could mediate reactive astrocytosis.
Fig. 4.
Coordinate gene regulation in post-seizure D/+ mice. IPA network analysis of 18 genes with ≥3-fold change in transcript abundance and their shared transcriptional regulators. The key for symbols in the network can be found at: http://ingenuity.force.com/ipa/articles/Feature_Description/Legend.
Neither constitutive expression of the hyperactive Nav1.6 nor the onset of seizures altered the abundance of transcripts encoding pore-forming or accessory subunits of voltage-gated sodium, potassium and calcium channels, with the exception of Kcnh4 which was down regulated in post-seizure D/+ mice (Table S2). Altered "expression" of ion channels has often been invoked as a component of epileptogenesis. Our data suggest that any changes in ion channel function are likely to result from post-transcriptional effects, such as altered subcellular localization, rather than compensatory changes in trancription of ion channel genes.
4.3 Reactive astrocytosis: protective or pathogenic?
Many of the transcripts elevated in D/+ mice are also increased in the course of reactive astrocytosis after traumatic brain injury. The elevated GFAP immunostaining of in hippocampus of D/+ mice confirmed the changes in transcript profiles. Reactive astrocytosis encompasses a spectrum of changes observed after disease and injury to the CNS (Pekny et al., 2016). Reactive astrocytosis is seen in mesial temporal lobe epilepsy and is thought to play a role in epileptogenesis after brain injury and ischemia (Ortinski et al., 2010; Wetherington et al., 2008; Yang et al., 2016). Chronic reactive astrocytosis was sufficient to cause seizures in the mouse (Robel et al., 2015). In our model of monogenic SCN8A epilepsy, reactive astrocytosis occurs in response to seizures and may be neuroprotective via secretion of chemokines, cytokines, growth factors, and extracellular matrix components that promote neuronal survival (Banker, 1980; Bush et al., 1999; Myer et al., 2006; Zhang et al., 2016). Recent studies indicate that reactive astrocytosis comprises a continuum of context-dependent changes that are regulated by specific signaling events (Crunelli et al., 2015; Henneberger, 2016; Sofroniew and Vinters, 2010; Zamanian et al., 2012; Zhang et al., 2016), consistent with either a protective or pathogenic role in Scn8a encephalopathy.
Eleven of the 50 transcripts elevated in post-seizure D/+ mice, or 22%, are also elevated in response to ischemia and/or neuroinflammation (Table 1 and Fig. 1D). Thirty one of the 50 genes are unique to our model, suggesting that an epilepsy-specific subtype of reactive astrocytes may be involved in Scn8a encephalopathy.
Conclusions
We demonstrate alterations in gene expression associated with seizures in a mouse model of SCN8A encephalopathy. The transcriptional changes were not present in mutant mice that had not experienced seizures, ruling out direct effects of Scn8a genotype. Two major effects were elevated expression of neuropeptides and hippocampal astrocytosis. It has been proposed that the term 'epileptic encephalopathy' be restricted to situations in which epileptic seizures lead directly to co-morbidities such as developmental delay and cognitive impairment (Howell et al., 2016). Our data suggest that altered transcription may contribute to development of these co-morbidities. An important question for future research is whether interventions that prevent these transcriptional changes will also reduce co-morbidities. These neuropeptides and genes involved in reactive astrocytosis may provide new targets for prevention of disease progression in intractable childhood epilepsy.
Supplementary Material
Highlights.
-
*
The onset of seizures in a mouse model of SCN8A encephalopathy is accompanied by changes in abundance of a small number of brain transcripts.
-
*
One source of altered transcript abundance is the onset of reactive astrocytosis in the hippocampus.
-
*
In the absense of seizure, there were no alterations in the transcriptome of mice with the Scn8a mutation.
-
*
Elevated levels of the neuropeptides NPW and galanin may play a protective role during the response to seizures.
-
*
There is no compensatory alteration in the abundance of transcripts encoding other voltage-gated ion channels in the Scn8a mutant mice.
Acknowledgments
Support is acknowledged from research grant NIH R01 NS34509 to MHM and a Postdoctoral Fellowship from the Dravet Syndrome Foundation to JLW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida-Suhett CP, et al. Temporal course of changes in gene expression suggests a cytokine-related mechanism for long-term hippocampal alteration after controlled cortical impact. J Neurotrauma. 2014;31:683–690. doi: 10.1089/neu.2013.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker GA. Trophic interactions between astroglial cells and hippocampal neurons in culture. Science. 1980;209:809–810. doi: 10.1126/science.7403847. [DOI] [PubMed] [Google Scholar]
- Barker B, et al. The SCN8A encephalopathy mutation p.Ile1327Val displays elevated sensitivity to the anticonvulsant phenytoin. Epilepsia. 2016 Jul 4; doi: 10.1111/epi.13461. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont TL, et al. Layer-specific CREB target gene induction in human neocortical epilepsy. J Neurosci. 2012;32:14389–14401. doi: 10.1523/JNEUROSCI.3408-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialer M, Johannessen SI, Levy RH, Perucca E, Tomson T, White HS. Progress report on new antiepileptic drugs: a summary of the Eleventh Eilat Conference (EILAT XI) Epilepsy research. 2013;103:2–30. doi: 10.1016/j.eplepsyres.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Bialer M, Johannessen SI, Levy RH, Perucca E, Tomson T, White HS. Progress report on new antiepileptic drugs: A summary of the Twelfth Eilat Conference (EILAT XII) Epilepsy research. 2015;111:85–141. doi: 10.1016/j.eplepsyres.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Blanchard MG, et al. De novo gain-of-function and loss-of-function mutations of SCN8A in patients with intellectual disabilities and epilepsy. J Med Genet. 2015;52:330–337. doi: 10.1136/jmedgenet-2014-102813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer K, et al. Gene expression analysis of tuberous sclerosis complex cortical tubers reveals increased expression of adhesion and inflammatory factors. Brain Pathol. 2010;20:704–719. doi: 10.1111/j.1750-3639.2009.00341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush TG, et al. Leukocyte infiltration, neuronal degeneration, and neurite outgrowth after ablation of scar-forming, reactive astrocytes in adult transgenic mice. Neuron. 1999;23:297–308. doi: 10.1016/s0896-6273(00)80781-3. [DOI] [PubMed] [Google Scholar]
- Crunelli V, et al. Novel astrocyte targets: new avenues for the therapeutic treatment of epilepsy. Neuroscientist. 2015;21:62–83. doi: 10.1177/1073858414523320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, et al. Central nervous system-specific knockout of Brg1 causes growth retardation and neuronal degeneration. Brain Res. 2015;1622:186–195. doi: 10.1016/j.brainres.2015.06.027. [DOI] [PubMed] [Google Scholar]
- Elliott RC, et al. Overlapping microarray profiles of dentate gyrus gene expression during development- and epilepsy-associated neurogenesis and axon outgrowth. J Neurosci. 2003;23:2218–2227. doi: 10.1523/JNEUROSCI.23-06-02218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estacion M, et al. A novel de novo mutation of SCN8A (Nav1.6) with enhanced channel activation in a child with epileptic encephalopathy. Neurobiol Dis. 2014;69:117–123. doi: 10.1016/j.nbd.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, et al. The role and regulation of hypoxia-inducible factor-1alpha expression in brain development and neonatal hypoxic-ischemic brain injury. Brain Res Rev. 2009;62:99–108. doi: 10.1016/j.brainresrev.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Green BR, et al. Analgesic neuropeptide W suppresses seizures in the brain revealed by rational repositioning and peptide engineering. ACS Chem Neurosci. 2011;2:51–56. doi: 10.1021/cn1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman RP, Samulski RJ, McCown TJ. Attenuation of seizures and neuronal death by adeno-associated virus vector galanin expression and secretion. Nature medicine. 2003;9:1076–1080. doi: 10.1038/nm901. [DOI] [PubMed] [Google Scholar]
- Hammer MF, et al. SCN8A Epileptic Encephalopathy. Gene Reviews. 2016 In press. [Google Scholar]
- Henneberger C. Does rapid and physiological astrocyte-neuron signalling amplify epileptic activity? J Physiol. 2016 doi: 10.1113/JP271958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KB, et al. Epileptic encephalopathy: Use and misuse of a clinically and conceptually important concept. Epilepsia. 2016;57:343–347. doi: 10.1111/epi.13306. [DOI] [PubMed] [Google Scholar]
- Jones JM, Meisler MH. Modeling human epilepsy by TALEN targeting of mouse sodium channel Scn8a. Genesis. 2014;52:141–148. doi: 10.1002/dvg.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokaia M, Holmberg K, Nanobashvili A, Xu ZQ, Kokaia Z, Lendahl U, Hilke S, Theodorsson E, Kahl U, Bartfai T, Lindvall O, Hokfelt T. Suppressed kindling epileptogenesis in mice with ectopic overexpression of galanin. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14006–14011. doi: 10.1073/pnas.231496298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, et al. The phenotypic spectrum of SCN8A encephalopathy. Neurology. 2015;84:480–489. doi: 10.1212/WNL.0000000000001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EJ, Richichi C, Young D, Baer K, Vezzani A, During MJ. Recombinant AAV-mediated expression of galanin in rat hippocampus suppresses seizure development. The European journal of neuroscience. 2003;18:2087–2092. doi: 10.1046/j.1460-9568.2003.02926.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez-Santiago LF, Yuan Y, Hull J, Wagnon JL, Frasier CR, Parent JM, Meisler MH, Isom LL. A mouse model of a human SCN8A epileptic encephalopathy mutation exhibits increased persistent sodium current in bipolar and pyramidal hippocampus neurons. Abstract No. 3.142. American Epilepsy Society Annual Meeting. 2015 www.aesnet.org. [Google Scholar]
- Makinson CD, Tanaka BS, Lamar T, Goldin AL, Escayg A. Role of the hippocampus in Nav1.6 (Scn8a) mediated seizure resistance. Neurobiol. Dis. 2014;68:16–25. doi: 10.1016/j.nbd.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarati AM, Liu H, Soomets U, Sankar R, Shin D, Katsumori H, Langel U, Wasterlain CG. Galanin modulation of seizures and seizure modulation of hippocampal galanin in animal models of status epilepticus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:10070–10077. doi: 10.1523/JNEUROSCI.18-23-10070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarati AM, et al. Modulation of hippocampal excitability and seizures by galanin. J Neurosci. 2000;20:6276–6281. doi: 10.1523/JNEUROSCI.20-16-06276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarati A, Wasterlain CG. Anticonvulsant effects of four neuropeptides in the rat hippocampus during self-sustaining status epilepticus. Neuroscience letters. 2002;331:123–127. doi: 10.1016/s0304-3940(02)00847-9. [DOI] [PubMed] [Google Scholar]
- McCown TJ. Adeno-associated virus-mediated expression and constitutive secretion of galanin suppresses limbic seizure activity in vivo. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14:63–68. doi: 10.1016/j.ymthe.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Meisler MH, et al. SCN8A Encephalopathy: Research Progress and Prospects. Epilepsia. 2016 doi: 10.1111/epi.13422. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer DJ, et al. Essential protective roles of reactive astrocytes in traumatic brain injury. Brain. 2006;129:2761–2772. doi: 10.1093/brain/awl165. [DOI] [PubMed] [Google Scholar]
- Okamoto OK, et al. Whole transcriptome analysis of the hippocampus: toward a molecular portrait of epileptogenesis. BMC Genomics. 2010;11:230. doi: 10.1186/1471-2164-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski PI, et al. Selective induction of astrocytic gliosis generates deficits in neuronal inhibition. Nat Neurosci. 2010;13:584–591. doi: 10.1038/nn.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, et al. Astrocytes: a central element in neurological diseases. Acta Neuropathol. 2016;131:323–345. doi: 10.1007/s00401-015-1513-1. [DOI] [PubMed] [Google Scholar]
- Rakhade SN, et al. A common pattern of persistent gene activation in human neocortical epileptic foci. Ann Neurol. 2005;58:736–747. doi: 10.1002/ana.20633. [DOI] [PubMed] [Google Scholar]
- Ravizza T, et al. Dynamic induction of the long pentraxin PTX3 in the CNS after limbic seizures: evidence for a protective role in seizure-induced neurodegeneration. Neuroscience. 2001;105:43–53. doi: 10.1016/s0306-4522(01)00177-4. [DOI] [PubMed] [Google Scholar]
- Robel S, et al. Reactive astrogliosis causes the development of spontaneous seizures. J Neurosci. 2015;35:3330–3345. doi: 10.1523/JNEUROSCI.1574-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M, et al. Dopamine D4 receptor-deficient mice display cortical hyperexcitability. J Neurosci. 2001;21:3756–3763. doi: 10.1523/JNEUROSCI.21-11-03756.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaida M, et al. Electroconvulsive seizure-induced changes in gene expression in the mouse hypothalamic paraventricular nucleus. J Psychopharmacol. 2013;27:1058–1069. doi: 10.1177/0269881113497612. [DOI] [PubMed] [Google Scholar]
- Samal BB, et al. Acute Response of the Hippocampal Transcriptome Following Mild Traumatic Brain Injury After Controlled Cortical Impact in the Rat. J Mol Neurosci. 2015;57:282–303. doi: 10.1007/s12031-015-0626-2. [DOI] [PubMed] [Google Scholar]
- Schlifke I, Kuteeva E, Hokfelt T, Kokaia M. Galanin expressed in the excitatory fibers attenuates synaptic strength and generalized seizures in the piriform cortex of mice. Exp. Neurol. 2006;200:398–406. doi: 10.1016/j.expneurol.2006.02.124. [DOI] [PubMed] [Google Scholar]
- Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, et al. Genomic responses of the brain to ischemic stroke, intracerebral haemorrhage, kainate seizures, hypoglycemia, and hypoxia. Eur J Neurosci. 2002;15:1937–1952. doi: 10.1046/j.1460-9568.2002.02030.x. [DOI] [PubMed] [Google Scholar]
- Veeramah KR, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, et al. Pathogenic mechanism of recurrent mutations of SCN8A in epileptic encephalopathy. Ann Clin Transl Neurol. 2016;3:114–123. doi: 10.1002/acn3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, et al. Convulsive seizures and SUDEP in a mouse model of SCN8A epileptic encephalopathy. Hum Mol Genet. 2015;24:506–515. doi: 10.1093/hmg/ddu470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Meisler MH. Recurrent and Non-Recurrent Mutations of SCN8A in Epileptic Encephalopathy. Front Neurol. 2015;6:104. doi: 10.3389/fneur.2015.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherington J, et al. Astrocytes in the epileptic brain. Neuron. 2008;58:168–178. doi: 10.1016/j.neuron.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, et al. Microarray analysis of postictal transcriptional regulation of neuropeptides. J Mol Neurosci. 2005;25:285–298. doi: 10.1385/JMN:25:3:285. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. Wnt/beta-catenin signaling mediates the seizure-facilitating effect of postischemic reactive astrocytes after pentylenetetrazole-kindling. Glia. 2016;64:1083–1091. doi: 10.1002/glia.22984. [DOI] [PubMed] [Google Scholar]
- Zamanian JL, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.