Abstract
An unprecedented Ebola virus (EBOV) epidemic occurred in 2013–2016 in West Africa. Over this time the epidemic exponentially grew and moved to Europe and North America, with several imported cases and many Health Care Workers (HCW) infected. Better understanding of EBOV infection patterns in different body compartments is mandatory to develop new countermeasures, as well as to fully comprehend the pathways of human-to-human transmission. We have longitudinally explored the persistence of EBOV-specific negative sense genomic RNA (neg-RNA) and the presence of positive sense RNA (pos-RNA), including both replication intermediate (antigenomic-RNA) and messenger RNA (mRNA) molecules, in the upper and lower respiratory tract, as compared to plasma, in a HCW infected with EBOV in Sierra Leone, who was hospitalized in the high isolation facility of the National Institute for Infectious Diseases “Lazzaro Spallanzani” (INMI), Rome, Italy. We observed persistence of pos-RNA and neg-RNAs in longitudinally collected specimens of the lower respiratory tract, even after viral clearance from plasma, suggesting possible local replication. The purpose of the present study is to enhance the knowledge on the biological features of EBOV that can contribute to the human-to-human transmissibility and to develop effective intervention strategies. However, further investigation is needed in order to better understand the clinical meaning of viral replication and shedding in the respiratory tract.
Author Summary
An unprecedented Ebola outbreak occurred in 2013–2016 in West Africa. In order to better understand EBOV infection patterns in different body compartments, we have longitudinally explored the presence of already assessed markers of ongoing EBOV replication (negative sense genomic RNA and positive sense RNA) in the upper and lower respiratory tract, as compared to plasma and other body compartments, in a Health Care Worker infected with EBOV in Sierra Leone, who was hospitalized in the high isolation facility of the National Institute for Infectious Diseases “Lazzaro Spallanzani” (INMI), Rome, Italy. The presence of total EBOV RNA and replication markers was observed in specimens of the lower respiratory tract, even after viral clearance from plasma, suggesting possible local replication. Our results contribute to the knowledge on the biological features of EBOV and shed light on the potential role of respiratory compartment in human-to-human transmissibility.
Introduction
The Ebola virus (EBOV) Makona variant is a negative-sense RNA virus and a member of the Filoviridae. From December 2013 to January 2016, this EBOV Makona caused the largest viral hemorrhagic fever epidemic ever reported in West Africa. [1] Several patients received care in Europe and North America. [2] Among those, two were medically evacuated to the National Institute for Infectious Diseases (INMI) “Lazzaro Spallanzani” of Rome, Italy. The clinical findings of the first imported case to Italy confirmed for Ebola Virus Disease (EVD) have been previously reported [3,4]. Like other patients, who received care outside of Africa, this patient presented a significant lung involvement, consistent with interstitial pneumonia, suggesting that EBOV could cause lung damage, perhaps through replication in these tissues. [4–6] The second imported case to Italy was a Health Care Worker (HCW) working in an Ebola Treatment Center (ETC) in Sierra Leone. He was admitted to INMI for EVD on May 13, 2015 and was discharged in good clinical condition on June 10, 2015. Oral favipiravir (Toyama Chemical Co, Japan), monoclonal antibodies against EBOV (Mil77, Mabworks Beijing China) and sustained hydration were administered, followed by progressive clinical improvement. A longitudinal characterization of cellular immune response has been described in detail elsewhere [7]. At admission, on day three from onset of symptoms, the patient had fever (39.0°C), cough, and a delayed febrile thrombocytopenic syndrome with pericardial effusion; chest X ray showed a basal mild interstitial lung involvement with 89% oxygen saturation. There is no direct evidence suggesting that EBOV can infect lung tissue in humans, although the potential role of EBOV infection of respiratory system has been suggested by clinical in vitro experiments, animal studies and clinical case reports [4,5,8–12]. In addition, viral inclusions can be found inside alveolar macrophages, and free viral particles can be seen within alveolar spaces. In this study, we compared the levels of EBOV negative sense genomic RNA (neg-RNA) and positive sense RNA (pos-RNA), including both mRNAs and anti-genomic RNAs, as indicators of viral replication in clinical specimen of the second EVD patient treated at INMI [13,14], to explore the potential role of different body compartment in EBOV replication.
Results
Plasma and sputum clinical specimens
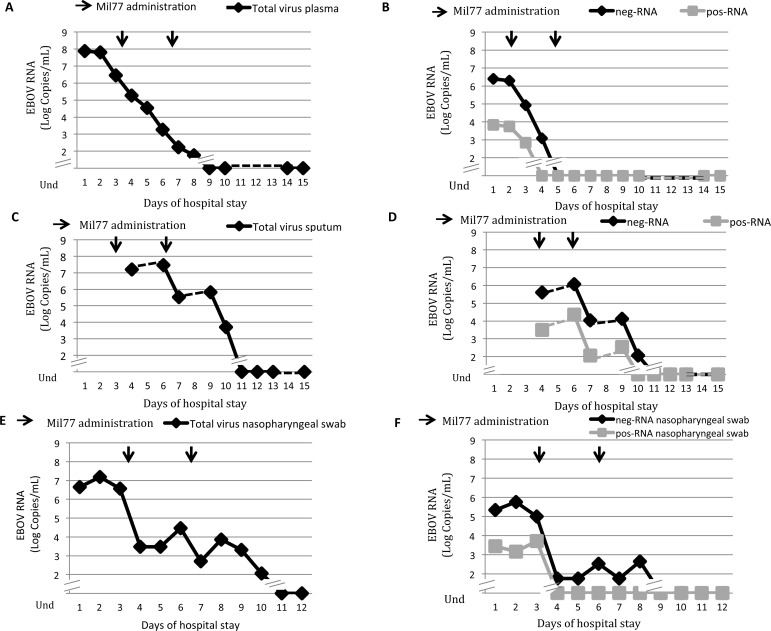
Fig 1 (Panels A and C) shows the trends of total viral RNA in plasma (viremia) and in sputum. High EBOV viremia was detected in the first days of hospital admission (with a peak of 108 copies/mL at day 1) with a marked decrease at day 3, also overlapping with the administration of Mil77. Declining EBOV viremia continued after further administration of MIL77 on day 6, and reached undetectable levels on day 9. In contrast, total EBOV RNA in sputum peaked at day 6 with above 107 copies/mL and remained high at around 106 copies/mL up to day 9 before becoming undetectable at day 11. Fig 1 shows neg-RNA and pos-RNA levels, in plasma (Panel B) and in sputum (Panel D). High levels of neg-RNA and pos-RNA, were detected in plasma up to day 4 and 3, respectively. In the respiratory compartment, neg-RNA was present at a concentration of 106 copies/mL up to day 11, and pos-RNA was present at a concentration of 104 copies/mL up to day 10 (Panel D). The levels of neg-RNA and pos-mRNA in sputum samples were 2 or 3-Log higher than neg-RNA and pos-RNA values in plasma, respectively.
Fig 1. EBOV RNA trends in plasma, sputum and nasopharyngeal specimens during the first 15 days of hospitalization of the second Italian Ebola patient attending the National Institute for Infectious Diseases in Roma (INMI2).
Clinical specimens from the patient were collected daily (plasma and nasopharyngeal swab) and regularly throughout hospitalization (Sputum). Arrows indicate the administration of the experimental drug Mil77 (day 3 and day 6). Dotted lines represent the hypothetical trend of those samples not available for this study. Panel A: trend of total EBOV RNA (viremia), which becomes undetectable at day 9. Panel B: trends of neg-RNA and pos-RNA in plasma, which become undetectable at day 5 and 4, respectively. Panel C: trend of total virus RNA in sputum, which becomes undetectable at day 11. Panel D: trends of neg-RNA and pos-RNA in sputum, which become undetectable at day 11 and 10, respectively. Panel E: trend of total EBOV RNA in nasopharyngeal swab. In nasopharyngeal swab the total EBOV RNA reaches undetectable levels at day 13 of hospitalization. Panel F: trends of neg-RNA and pos-RNA in nasopharyngeal swab, which become undetectable at day 10 and 6, respectively. Symbols are specified in the panels.
Total, neg- and pos-RNA in other clinical specimens
The presence of total, neg- and pos-RNA in nasopharyngeal swab is shown in Fig 1. As depicted, total RNA was present at high levels (106 copies/mL) until day 5, and detected at low levels (103 copies/mL) until day 12 before becoming undetectable at day 13 (Panel E). The prolonged detection of total RNA was paralleled by that of neg-RNA, which was detectable (102−103 copies/mL) until day 10 (Panel B) while pos-RNA became undetectable on day 6 (Panel F). In ocular swabs and urine samples, total RNA remained detectable up to day 15 (103 copies/mL) and day 4 (104 copies/mL), respectively. Interestingly, pos-RNA was always undetectable in all specimens analyzed from these compartments, whereas neg-RNA was detectable around 102 copies/mL both in urine and ocular swab.
Discussion
This study presents novel insights in the potential role of the respiratory compartment in EBOV replication in a human case of EVD. Several clinical findings suggest lung involvement in different cases of EVD who received care in Europe [4] and the USA [5]. The chest radiography performed in patients treated in the USA revealed bilateral pulmonary infiltrates suggestive of pneumonia [5]. Moreover, the first EVD patient treated in Italy developed significant lung injury, consistent with interstitial pneumonia [4]. The role of neg-RNA and pos-RNA levels as surrogate markers of ongoing EBOV replication has already been established in in vitro experiments [13]. We confirmed the specificity of this method using VeroE6 infected cells, where at 24 hours post-infection we observed that the majority of pos-RNA (62%) was cell-associated, while neg-RNA was mostly (84%) shed in the supernatant. In addition, the sensitivity and the linearity of the test were unaffected by sample matrix composition, since plasma and sputum spiked with EBOV viral RNA (Ct 22,12 and Ct 22,41, respectively, for the RNA concentration 105) did not influence tests results as compared to tissue culture medium. Further, no appreciable background noise signal was observed in unspiked plasma and sputum matrices (Ct > 45). Hence, we longitudinally explored the persistence of viral replication markers in clinical samples from the upper and lower respiratory tract of the second EVD patient in Italy, and made comparison with plasma samples. The results indicate the decay of EBOV total RNA, neg-RNA and pos-RNA overtime, which was significantly different in plasma when compared to respiratory samples. In particular, we observed a persistence of total viral RNA and replication markers in sputum, until the second week of hospital stay (up to day 10), when viremia was already undetectable. From these data the possibility that the prolonged detection of EBOV RNA in sputum merely reflects enhanced stability of EBOV RNA in this compartment cannot be ruled out [16]; nevertheless the detection of pos-RNA together with neg-RNA in sputum (until day 9 and 10 of hospital stay, respectively) supports the concept of active viral replication within the respiratory tract, rather than plasma spill-over or prolonged RNA stability. Indeed, the detection of the replication markers in respiratory samples combined with the absence of detectable neg-RNA in the plasma from day 5, while neg-RNA could still be detected for days in respiratory samples, can be explained by active replication of EBOV in lung tissues. Moreover, the absence of detectable levels of pos-RNA in nasopharyngeal swabs (starting day 6 and onward) suggests that the presence of pos-RNA in sputum is likely due to replicating EBOV in the lower rather than in the upper respiratory tract. Additional observations are indicative of a direct role of EBOV replication in lung injury in animals (including nonhuman primates) and in post-mortem examination of human that succumbed with EVD [12]. Moreover, a recent study evaluated the pathogenicity in rhesus macaques of 2 different isolates of the EBOV Makona strains compared to the EBOV Mayinga isolate, showing higher lung pathology in macaques infected with the two Makona strains [17]. In pigs, EBOV can replicate to high titers in the lower respiratory tract and cause severe lung diseases following mucosal exposure [18]. Guinea pigs challenged with aerosolized EBOV can develop lethal interstitial pneumonia [8] while rhesus monkeys demonstrate a significant respiratory involvement [9] with cell-associated EBOV virus antigens present in airway epithelium, alveolar pneumocytes, and macrophages [10,11]. The hypothesis that EBOV can actually replicate into human airway is supported by a recently published review, which collected data from post mortem examinations of 89 fatal cases of filoviral infections (24 of which with EBOV) [12]. This analysis provides evidence of viral replication within the respiratory tract, showing the presence of viral antigens in several lung tissues, with EBOV nucleic acids detected in resident macrophages [12]. Unfortunately, we were only able to recover replication competent virus from plasma collected at presentation and were unable to recover replication competent virus from later plasma samples or other clinical specimens, despite numerous attempts in VeroE6 cell culture (especially from sputum). Since other Authors report difficulties in EBOV isolation from non-blood samples, it is possible that the actual sensitivity of cell culture isolation is not sufficient to rule out virus viability and infectivity [19–20]. To this respect, several factors, among which suboptimal sensitivity of the virus culture system, storage at -80°C and presence of antibodies against EBOV in the clinical samples, may have contributed to unsuccessful virus culture attempts. Despite the lack of supportive data, a key additional question is whether primary pulmonary infection and respiratory transmission of EBOV could actually occur [6]. The presence of virus replication markers here reported adds some support to this possibility, although the evidence is far from being conclusive, due to the absence of recovery of infectious virus in respiratory secretion. Further investigation is needed to better understand the clinical significance of the involvement of the respiratory tract in viral replication and shedding, and possible contribution to respiratory impairment and human–to–human transmission.
Materials and Methods
Patient’s clinical specimens were longitudinally collected for diagnostic purposes. Plasma samples, nasopharyngeal swabs, ocular swabs and urine samples were collected daily (starting from day 1, day 3, day 1 and day 1, respectively) while sputum samples (mucus and phlegm coughed-up from the lower airways) were repeatedly collected (starting from day 4) throughout hospitalization. We made all efforts to uniform the time (morning) and size (4 mL) of sample collection, in order to minimize this as possible source of difference. Inactivation of samples was performed in the BSL4 facility and RNA extraction was carried out in the BSL2 laboratory. RNA extraction was performed with QIAamp Viral RNA Mini Kit (Qiagen) following manufacturer’s instructions. Identical volumes (140 μl) of each sample have been used for RNA extraction. The quality of all the samples was checked using a house-keeping gene (RNaseP) in qRT-PCR according to [15]. Total virus-specific RNA (total RNA) was measured using quantitative Real-time PCR, RealStar Filovirus Screen RT-PCR Kit 1.0 (Altona Diagnostics GmbH), targeting L gene. To measure EBOV-specific negative sense genomic RNA (neg-RNA) and positive sense RNA (pos-RNA) [including both replication intermediate (cRNA) and messenger RNA (mRNA)], L-gene specific reverse or forward primers were used in the reverse transcription step [13,14], using TaqMan Reverse Transcription Reagent kit (Applied Biosystems, Foster City, CA, USA). cDNAs were treated with 1μL of RNase H (2U/ μL) for 20’ at 37°C, to remove remaining viral RNAs, and then amplified using the RealStar Filovirus Screen RT-PCR Kit 1.0, with a modified thermal profile, which omitted the reverse transcription step. The standard curve of the RealStar Filovirus Screen RT-PCR Kit 1.0 has been used to determine the concentration of the virus in the different clinical specimens. In our experimental conditions, the range of the curve is from 109 copies/mL down to 102 copies/mL.
Ethic statement
The Institutional Ethic Board (IEB) of the National Institute for Infectious Diseases “L. Spallanzani” approved the use of residual clinical samples for research purposes. The patient signed a written informed consent for any single procedure or treatment performed, after thoroughly explanation of reasonably anticipated benefits and potential hazards of intervention, and for the publication of the case.
All participants to the study are adults.
Acknowledgments
The contribution of Laboratory of Microbiology and Infectious Disease Biorepository from National Institute for Infectious Diseases “L. Spallanzani”, Rome, Italy, is warmly acknowledged. We gratefully acknowledge the generous assistance and valuable information provided to us by Dr Nikki Shindo, MD, of the Pandemic and Epidemic Diseases Department, World Health Organization, Geneva, Switzerland, specifically for her guidance in the management of the case, and by Dr Michael Jacob, MD, of the Royal Free Hospital, for providing us the experimental drug Mil77 and Heinrich Feldmann and Friederike Feldmann of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, Montana, United States of America for their precious advice in the interpretation of the final results.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants of Italian Ministry of Health, for “Ricerca Corrente” and “Ricerca Finalizzata”, and by European Union Seventh Framework Programme, Grant n°278433-PREDEMICS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dye C. WHO Ebola Response Team. Ebola virus disease in Western Africa–the first 9 months. N Engl J Med. 2015; 372(2):189. [DOI] [PubMed] [Google Scholar]
- 2.Bevilacqua N, Nicastri E, Chinello P, Puro V, Petrosillo N, Di Caro A, Capobianchi MR, Lanini S, Vairo F, Pletschette M, Zumla A, Ippolito G; INMI Ebola Team. Criteria for discharge of patients with Ebola virus diseases in high-income countries. Lancet Glob Health. 2015. December;3(12):e739–40. 10.1016/S2214-109X(15)00205-3 [DOI] [PubMed] [Google Scholar]
- 3.Castilletti C, Carletti F, Gruber CE, Bordi L, Lalle E, Quartu S, Meschi S, et al. Molecular Characterization of the First Ebola Virus Isolated in Italy, from a Health Care Worker Repatriated from Sierra Leone. Genome Announce. 2015; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrosillo N, Nicastri E, Lanini S, Capobianchi MR, Di Caro A, Antonini M, Puro V, et al. Ebola virus disease complicated with viral interstitial pneumonia: a case report. BMC Infect Dis. 2015; 15:432 10.1186/s12879-015-1169-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liddell AM, Davey RT Jr, Mehta AK, Varkey JB, Kraft CS, Tseggay GK, Badidi O, et al. Characteristics and Clinical Management of a Cluster of 3 Patients With Ebola Virus Disease, Including the First Domestically Acquired Cases in the United States. Ann Intern Med. 2015; 163:81–90. 10.7326/M15-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osterholm MT, Moore KA, Kelley NS, Brosseau LM, Wong G, Murphy FA, Peters CJ, et al. Transmission of Ebola viruses: what we know and what we do not know. MBio. 2015; 6:e00137 10.1128/mBio.00137-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrati C, Castilletti C, Casetti R, Sacchi A, Falasca L, Turchi F, Tumino N, et al. Longitudinal characterization of dysfunctional T cell-activation during human acute Ebola infection. Cell Death Dis. 2016. March 31;7:e2164 10.1038/cddis.2016.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twenhafel NA, Shaia CI, Bunton TE, Shamblin JD, Wollen SE, Pitt LM, Sizemore DR, Ogg MM, Johnston SC. Experimental aerosolized guinea pig-adapted Zaire ebolavirus (variant: Mayinga) causes lethal pneumonia in guinea pigs. Vet Pathol. 2015. January;52(1):21–5. 10.1177/0300985814535612 [DOI] [PubMed] [Google Scholar]
- 9.Johnson E, Jaax N, White J, Jahrling P. Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int J Exp Pathol. 1995. August;76(4):227–36. [PMC free article] [PubMed] [Google Scholar]
- 10.Twenhafel NA, Mattix ME, Johnson JC, Robinson CG, Pratt WD, Cashman KA, Wahl-Jensen V, Terry C, Olinger GG, Hensley LE, Honko AN. Pathology of experimental aerosol Zaire ebolavirus infection in rhesus macaques. Vet Pathol. 2013. May;50(3):514–29 10.1177/0300985812469636 [DOI] [PubMed] [Google Scholar]
- 11.Weingartl HM, Embury-Hyatt C, Nfon C, Leung A, Smith G, Kobinger G. Transmission of Ebola virus from pigs to non-human primates. Sci Rep. 2012;2:811 10.1038/srep00811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol. 2015;235(2):153–74 10.1002/path.4456 [DOI] [PubMed] [Google Scholar]
- 13.Hoenen T, Jung S, Herwig A, Groseth A, Becker S. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology. 2010; 403:56–66. 10.1016/j.virol.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 14.Shabman RS, Hoenen T, Groseth A, Jabado O, Binning JM, Amarasinghe GK, Feldmann H, et al. An upstream open reading frame modulates ebola virus polymerase translation and virus replication. PLoS Pathog. 2013; 9:e1003147 10.1371/journal.ppat.1003147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO, 2009. CDC protocol of real-time RT-PCR for influenza A (H1N1). http://www.who.int/csr/resources/publications/swineflu/realtimeptpcr/en/ index.html
- 16.Janvier F, Delaune D, Poyot T, Valade E, Mérens A, Rollin PE, Foissaud V. Ebola Virus RNA Stability in Human Blood and Urine in West Africa's Environmental Conditions. Emerg Infect Dis. 2016; 22:292–4. 10.3201/eid2202.151395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong G, Qiu X, de La Vega MA, Fernando L, Wei H, Bello A, Fausther-Bovendo H, Audet J, Kroeker A, Kozak R, Tran K, He S, Tierney K, Soule G, Moffat E, Günther S, Gao GF, Strong J, Embury-Hyatt C, Kobinger G. Pathogenicity Comparison Between the Kikwit and Makona Ebola Virus Variants in Rhesus Macaques. J Infect Dis. 2016. September 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobinger GP, Leung A, Neufeld J, Richardson JS, Falzarano D, Smith G, Tierney K, Patel A, Weingartl HM. Replication, pathogenicity, shedding, and transmission of Zaire ebolavirus in pigs. J Infect Dis. 2011. July 15;204(2):200–8. 10.1093/infdis/jir077 [DOI] [PubMed] [Google Scholar]
- 19.Fang LQ, Yang Y, Jiang JF, Yao HW, Kargbo D, Li XL, Jiang BG, Kargbo B et al. Transmission dynamics of Ebola virus disease and intervention effectiveness in Sierra Leone PNAS 2016, published ahead of print March 28, 2016 [DOI] [PMC free article] [PubMed]
- 20.Crozier I. Ebola Virus RNA in the Semen of Male Survivors of Ebola Virus Disease: The Uncertain Gravitas of a Privileged Persistence. J Infect Dis. 2016. May 3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.