Abstract
In 1993, lin-4 was discovered as a critical modulator of temporal development in Caenorhabditis elegans and, most notably, as the first in the class of small, single-stranded noncoding RNAs now defined as microRNAs (miRNAs). Another eight years elapsed before miRNA expression was detected in mammalian cells. Since then, explosive advancements in the field of miRNA biology have elucidated the basic mechanism of miRNA biogenesis, regulation, and gene-regulatory function. The discovery of this new class of small RNAs has augmented the complexity of gene-regulatory programs as well as the understanding of developmental and pathological processes in the cardiovascular system. Indeed, the contributions of miRNAs in cardiovascular development and function have been widely explored, revealing the extensive role of these small regulatory RNAs in cardiovascular physiology.
Keywords: miRNA, vascular smooth muscle cells, endothelial cells, cardiac myocytes
INTRODUCTION
Diseases of the cardiovascular system, such as heart attack, stroke, peripheral vascular disease, hypertension, and congenital heart defects, account for many health problems and rank among the leading causes of mortality. Despite tremendous efforts in cardiovascular research and remarkable progress in the understanding of critical cellular and molecular processes during cardiovascular development and pathogenesis, current preventative treatments, early diagnosis, and therapies for cardiovascular disease have not dramatically improved and remain insufficient. Even the wide array of prognostic, diagnostic, and therapeutic tools available for these diseases fails to manage patients suffering from various cardiovascular conditions. Over the past five years, the emergence of small noncoding microRNAs (miRNAs) as integral players in various physiological and patho-physiological processes, including those of the cardiovascular system, has opened new doors for therapeutic developments and interventions (1–8).
MiRNAs, a class of small noncoding RNAs ~22 nucleotides (nt) in length, are involved in diverse biological processes, including developmental timing, differentiation, proliferation, cell death, and metabolism. Although miRNAs were initially recognized as regulators of development specifically in worms and fruit flies, the past decade of research clearly reveals that miRNAs are essential and critical players in mammalian development and the maintenance of homeostasis across a wide array of tissues, including the cardiovascular system. In mice, gene targeting of even a single miRNA may result in developmental abnormalities or aberrant responses to physiological stimuli, signifying the essential function of miRNAs. Individual miRNAs modulate the expression of several dozen or even hundreds of mRNAs that often share related functions and thus govern various biological processes (9). Both the number of miRNAs that are expressed in the cardiovascular system and the diverse functions of individual miRNAs support the idea that the majority of physiological and pathological events in the cardiovascular system may be controlled at least in part by miRNAs. Unlike transcriptional regulation, which involves turning on and off gene expression, miRNAs appear to act by fine-tuning gene expression during development and tissue homeostatic maintenance. During cellular stress or pathophysiological conditions such as hypoxia or oxidative stress, however, cells undergo a state of reduced transcription and translation. In these situations, miRNAs provide a potent, rapid, and efficient means of gene regulation that may allow cells to adapt to or recover from such an abnormal state.
The majority of mRNAs targeted by miRNAs are silenced through direct interaction of the miRNA and mRNA, which causes destabilization of mRNAs or inhibition of protein synthesis. MiRNAs induce gene silencing through association with a large, multiprotein complex termed the RNA-induced silencing complex (RISC). MiRNA-binding sites are commonly found within the 3′ untranslated region (3′UTR) of the target mRNA, but functional miRNA-binding sites can also be found in the 5′ untranslated region (5′UTR) (10) and in the coding region (11). Although the exact nature of mRNA target recognition by miRNAs is still unclear, in general, Watson-Crick base pairing between the miRNA 5′ end, particularly the miRNA seed region (composed of nt 2–7 of miRNA), and the sequence located in the mRNA 3′UTR is an important determinant of functional target sites (12).
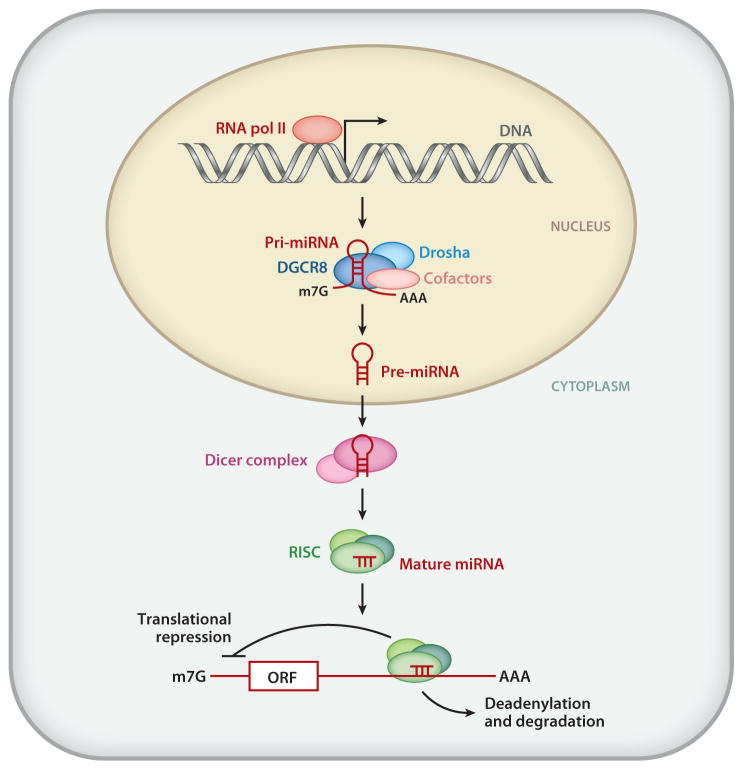
The mechanism that gives rise to mature miRNA involves two rounds of endonucleolytic cleavage performed by the ribonuclease III (RNase III) enzymes Drosha and Dicer (13) (Figure 1). Following transcription by RNA polymerase II, Drosha processes the primary miRNA (pri-miRNA) transcript into an ~70-nt hairpin structure termed the precursor miRNA (pre-miRNA) in the nucleus (13) (Figure 1). Through interaction with exportin-5 and Ran-GTP, the pre-miRNA is transported into the cytoplasm, where it undergoes a second step of processing catalyzed by Dicer (13) (Figure 1). This cleavage event gives rise to a double-stranded ~22-nt product composed of the mature miRNA guide strand and the miRNA’ passenger strand. The mature miRNA is then loaded into the RISC and participates in gene silencing, whereas the passenger strand (miRNA′) is often degraded (13) (Figure 1).
Figure 1.
Schematic representation of miRNA biogenesis and mechanisms of miRNA action. RNA polymerase II (RNA pol II) mediates transcription of miRNA genes. The primary miRNA transcripts (pri-miRNAs) can range from a few hundred nucleotides (nt) to a few kilobases long. In the nucleus, the pri-miRNAs are recognized by ribonuclease III (RNase III) endonuclease Drosha (light blue) together with an essential partner, DiGeorge syndrome critical region gene 8 (DGCR8; also known as Pasha) (dark blue). The Drosha complex also contains other cofactors ( pink) that modulate the catalytic activity of Drosha, such as RNA helicases (p68 and p72), transcription factors [Smad, p53, estrogen receptor α(ERα)], and RNA-binding proteins (KSRP and hnRNP A1). The cleavage product of pri-miRNA, precursor miRNA (pre-miRNA), is ~70 nt long and configures a stem-loop structure. Exportin-5 exports pre-miRNAs from the nucleus to the cytoplasm. In the cytoplasm, another RNase III enzyme, Dicer, cleaves pre-miRNA to generate double-stranded mature miRNA that is ~21 nt long. The mature miRNA duplex is then incorporated into the RNA-induced silencing complex (RISC). In mammals, the RISC is composed of Argonaute proteins 1–4 (Ago1–4) and several cofactors, such as PACT and TARBP1/2. MiRNA loaded onto the RISC undergoes strand separation, interaction with typically the 3′ untranslated regions of mRNA targets through Watson-Crick base pairing between bases 2 and 7 at the 5′ end of the miRNA known as the seed sequence, and repression of mRNA translation through sequestering translational machinery from the miRNAs or mRNA destabilization through the recruitment of poly(A) nuclease, followed by deadenylation. ORF, m7G, and AAA denote open reading frame, 5′ capping, and 3′ poly(A) tail, respectively. Adapted from Reference 111.
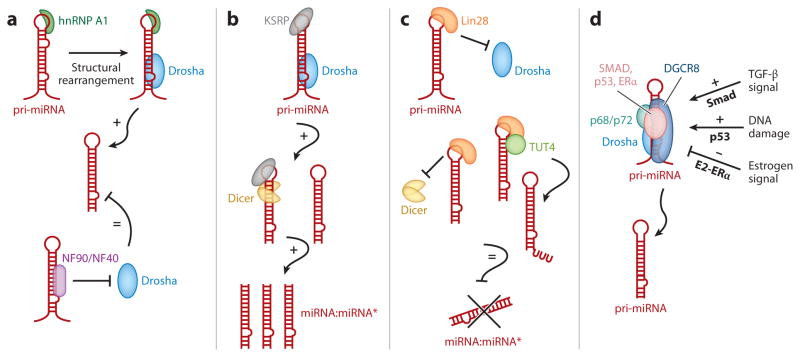
Close to 1,000 miRNAs are found in the human genome, and more than half of these seem to be expressed in a temporally and spatially regulated manner. Although major progress has been made toward understanding the mechanism of miRNA biogenesis, little is known about the mechanisms that regulate miRNA expression. Some miRNAs are encoded in the introns of protein-coding genes, and therefore their expression is coordinated with that of the host genes. There are many examples in which the protein encoded by the host gene and the associated intronic miRNAs participate in regulatory networks that are essential in various biological processes. In cases in which miRNAs are encoded and transcribed as individual genes, levels of miRNAs can be regulated transcriptionally by cis-regulatory elements in the promoter region of the miRNA genes, similar to protein-coding genes. Furthermore, transcription factors (Figure 2d) or RNA-binding proteins (Figure 2a–c) modulate the biogenesis of some miRNAs posttranscriptionally, acting during nuclear cleavage of pri-miRNAs by Drosha (14, 15). Several components of the Drosha-processing complex, such as DiGeorge syndrome critical region gene 8 (DGCR8) and the RNA helicases p68 (DDX5) and p72 (DDX17) (16), interact with transcription factors, including Smads (17), p53 (18), and estrogen receptor α (ERα) (19), which subsequently modulate the catalytic activity of Drosha either positively or negatively (14, 15) (Figure 2d). Several RNA-binding proteins, such as heteronuclear ribonucleoprotein A1 (hnRNP A1) (Figure 2a), the nuclear factor 90/nuclear factor 40 (NF90/NF40) complex (Figure 2a), KH-type splicing regulatory protein (KSRP) (Figure 2b), and Lin28 (Figure 2c), also associate with a subset of pri-miRNAs and regulate processing by Drosha (14, 15). Finally, the stability of miRNAs is regulated under certain environmental conditions, such as cell confluency, developmental stage, and cell cycle status, all of which contribute to dynamic changes in miRNA expression levels (20–23).
Figure 2.
Posttranscriptional mechanisms of regulation of the miRNA biogenesis pathway. MiRNA biosynthesis requires two steps of processing: (1) processing from primary miRNA transcripts (pri-miRNA) to precursor miRNA (pre-miRNA) by the ribonuclease III (RNase III) Drosha in the nucleus and (2) processing from pre-miRNA to a mature miRNA duplex by the RNase III Dicer in the cytoplasm. Both steps can be regulated by various RNA-binding proteins and DNA-binding proteins as summarized in panels a–d. (a) Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) ( green) recognizes the terminal loop of pri-miRNAs (red ) and promotes the structural remodeling of the stem region of pri-miRNAs, which generates a favorable Drosha (light blue)-binding site and enhances processing. The nuclear factor 90/nuclear factor 40 (NF90/NF40) complex ( purple) associates with the stem region of pri-miRNAs, which precludes association with the Drosha-processing complex and inhibits processing. (b) Similar to hnRNP A1 in panel a, KH-type splicing regulatory protein (KSRP) ( gray) binds to the terminal loop of pri-miRNAs and promotes both Drosha (light blue) and Dicer ( yellow) processing. (c) Binding of Lin28 (orange) to the terminal loop of pri-miRNAs prevents the association of both Drosha (light blue) and Dicer ( yellow). Additionally, Lin28 acts as a scaffold to promote the association of terminal RNA uridyltransferase 4 (TUT4; also known as Zcchc11) (light green) with the pre-miRNA. TUT4 promotes the 3′-uridinylation of pre-miRNA, which is then rapidly degraded. (d ) The RNA helicases p68 and p72 (teal ) play a critical role in the posttranscriptional regulation of numerous miRNAs in response to cellular signals, including transforming growth factor β(TGF-β) stimulation, p53-mediated DNA damage response, and estrogen stimulation. The downstream mediators of TGF-βstimulation and DNA damage, Smads and p53 ( pink), promote miRNA processing. Conversely, when bound to E2 ( pink), ERαreduces the processing of a subset of miRNAs. Adapted from Reference 14.
MiRNAS IN CARDIOVASCULAR DEVELOPMENT
The vital role of miRNAs in cardiovascular development and function was revealed by ablation of the general miRNA population in the cardiovascular system via tissue-specific deletion of Drosha, DGCR8, Argonaute 2 (Ago2), or Dicer, essential components in the miRNA biogenesis pathway. Removing these critical genes in mice and fish resulted in lethality during the early gestation period due to severe developmental defects in the cardiovascular system (24). Thus far, however, deletions of individual miRNAs in mice have not exhibited lethality with 100% penetrance, suggesting that a group of miRNAs perform at least partially redundant roles. This finding also highlights the general nature of miRNA-mediated gene regulation, in which an miRNA acts as a tuner rather than as an on/off switch, resulting in the subtle modulation of protein levels on a very modest scale. Although obtaining a complete picture of individual miRNA functions and target mRNAs in the cardiovascular system requires further investigation, the combination of genetic ablation/overexpression and pharmacological knockdown/overexpression of specific miRNAs in whole-animal and cell culture systems has facilitated the understanding of the physiological function of miRNAs both during development and postnatally. Below, the current understanding of the roles of individual miRNAs during cardiac, vascular, and hematopoietic development is summarized.
Function of MiRNAs During Heart Development
The heart is the first organ to function during vertebrate embryogenesis. Cardiac development begins with the fusion of endocardial and myocardial cell layers in the midline to form a bilayered heart tube that already contracts spontaneously and supports the blood supply of the developing embryo (Figure 3). In mammals, the heart later develops into an increasingly complex structure and eventually has four chambers (Figure 3). The importance of miRNA-mediated posttranscrip-tional gene regulation for cardiovascular development has been accessed by generating loss-of-function mutations in genes that encode enzymes essential for miRNA biogenesis, such as Dicer, Drosha, Ago2, and DGCR8. Knockout mice individually lacking these key miRNA-processing genes die during early gestation with severe developmental defects, which prevents further analysis of the role of miRNAs in the cardiovascular system (24). Deletion of Dicer in mice during early heart development [embryonic day 8.5 (E8.5)] by using an Nkx2.5 promoter–driven Cre recom-binase (Nkx2.5-Cre) leads to pericardial edema and defects in the ventricular myocardium (25). Another study using a different Nkx2.5-Cre driver line reported that miRNA-mediated regulation of developmental processes is also essential for cardiac outflow tract morphogenesis and chamber septation (26). Deletion of miRNA signaling in neural crest cells indicates that miRNAs are not required for the survival of these cells but are essential for migration and patterning (27). Deletion of Dicer by using an α-myosin heavy chain (α-MHC/Myh6)-Cre driver line that directs Cre expression in cardiac myocytes but not in cardiac fibroblasts leads to death of mutant mice between postnatal days 0 and 4 (28). Hearts of mutant mice are dilated and exhibit both a dramatic decrease in fractional shortening and a reduced contraction rate (28). These phenotypic results correlate well with cytoskeletal defects and deregulation of proteins important for contractility, cardiac conduction, and calcium handling. Deletion of Dicer in cardiac myocytes 3 weeks after birth results in sudden death and only mild cardiac remodeling, whereas deletion 8 weeks after birth causes massive hypertrophy and induces fibrotic lesions (29). At both ages, strong reexpression of fetal and hypertrophic marker genes was observed (29). Deletion of DGCR8 using a muscle creatine kinase (MCK)-Cre driver line in newborn hearts yields a similar phenotype with dilated cardiomyopathy and heart failure (30). These results confirm that miRNAs produced via the canonical miRNA biogenesis pathway are responsible for the major cardiac phenotype in Dicer and DGCR8 mutants.
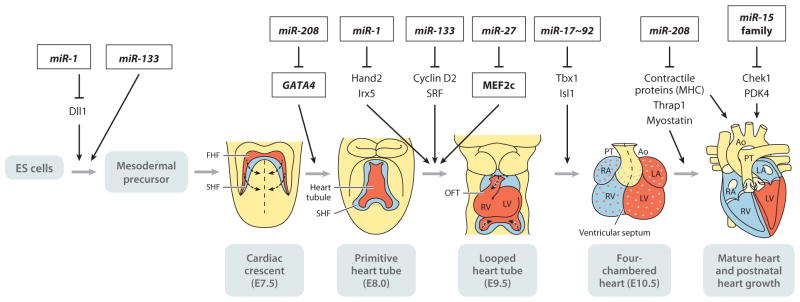
Figure 3.
Schematic representation of mouse heart developmental stages and miRNAs that play a role in the regulation of heart development. Anterior is toward the top. Mouse cardiogenesis begins when two populations of cells termed the first heart field (FHF) (red ) and the second heart field (SHF) (blue) in the mesoderm commit to a cardiogenic fate at embryonic day 7.5 (E7.5). These cardiac cells, localized to the cardiac crescent, migrate ventromedially to form the linear primitive heart tube at E8.0. Shortly after E8.0, rhythmic contractions start. Subsequently, looping morphogenesis, chamber specification, cardiac valve formation, and neural crest cell migration and contribution give rise to the four-chambered heart by E10.5. By E12.5, the four chambers of the heart (RV and LV) are well delineated, and septation (shown as a broken line) of the outflow tract (OFT) is observed. By the late postnatal stages, the OFT is completely septated, and both ventricular septation and atrial septation are complete in preparation for postnatal life. Representative miRNAs and functional targets at the different stages of development are indicated. In the early stages of cardiac development, miR-1 and miR-133 cooperatively promote mesoderm differentiation in embryonic stem (ES) cells and repress ectodermal and endodermal cell fate. At a later stage, miR-1 and miR-133 exhibit opposing effect in cardiomyocyte differentiation. miR-27 is involved in early cardiac development by modulating MEF2c expression. The miR-17~92 cluster promotes myogenic differentiation in the secondary heart field. Deletion of the miR-17~92 cluster causes ventricular-septum defects in mice. The miR-15 family of miRNAs inhibits postnatal proliferation of cardiac myocytes. miR-208a plays a role in cardiomyocyte-specific gene expression, the development of electrophysiological properties of the heart, and hypertrophic growth upon cardiac stress. Abbreviations: Ao, aorta; LA, left atrium; LV, left ventricle; PT, pulmonary tract; RA, right atrium; RV, right ventricle; Dll1, Delta-like-1; Irx5, Iroquois homeobox 5; SRF, serum response factor; Tbx1, T-box 1; Isl1, Islet 1; Thrap1, thyroid hormone receptor–associated protein 1; Chek1, checkpoint kinase 1; PDK4, pyruvate dehydrogenase kinase 4; MHC, myosin heavy chain.
MiRNA expression profiling studies have indicated that more than 90% of cardiac miRNAs are composed of only 18 miRNAs (30), suggesting that either a limited number of abundant miRNAs contribute to cardiac development and function or a large number of low-abundance miRNAs play a role in addition to abundant miRNAs. miR-1 and miR-133, two of the most prolific miRNAs in cardiac myocytes, are derived from common primary transcripts. Transcription of the miR-1/miR-133 bicistron is regulated by well-recognized myogenic transcription factors: MyoD, serum response factor (SRF), and myocyte enhancer factor 2 (MEF2) (31, 32). In the heart, SRF binds the enhancer regions of the miR-1/miR-133 bicistron and regulates their expression in cardiomyocytes (32). In skeletal muscle, MEF2 cooperates with MyoD to bind an intronic enhancer of miR-1/miR-133 to activate transcription (31, 33). miR-1 and miR-133 seem to cooperatively promote mesoderm differentiation in embryonic stem (ES) cells and repress ectodermal and endodermal cell fate (32) (Figure 3). However, miR-1 and miR-133 appear to have opposing roles later in cardiac development, as miR-1 promotes cardiomyocyte differentiation, whereas miR-133 inhibits it (32) (Figure 3). In mice, miR-1 expression is detected at ~E8.5 and increases throughout development. Mice overexpressing miR-1 under the control of a β-myosin heavy chain (β-MHC) promoter show a diminished population of proliferating ventricular myocytes. This state results from the negative regulation of cardiac growth by miR-1, in part via repression of the transcription factor heart and neural crest derivative-2 (Hand-2), which is involved in the expansion of ventricular myocytes (32) (Figure 3). miR-27 displays myocardial expression during early cardiogenesis and plays a role by modulating MEF2c expression (34). Despite the abundance of miR-1 and miR-133 in the heart and evidence for their serving distinct functions, neither of these miRNAs is essential for cardiac cell fate specification, as half of mice with either miR-1 or miR-133 are viable (25, 35). One-half of miR-1-2 homozygous null mice die between E15.5 and just after birth due to ventricular-septum defects and cardiac dysfunction with apparent deregulation of Hand-2 (25). miR-1-2 homozygous null mice that survive until birth often exhibit cardiac arrhythmias and suffer sudden death, indicating the postnatal cardiac function of miR-1 (25). The cardiac arrhythmias in miR-1-2-null mice may be caused by derepression of the transcription factor Iroquois homeobox 5 (Irx5), which regulates the cardiac ventricular repolarization gradient by repressing the level of potassium channel genes, such as the gene encoding the potassium voltage-gated channel subfamily D member 2 (Kcnd2) (36).
miR-208a, miR-208b, and miR-499 are encoded in the introns of Myh6 (α-MHC), Myh7 (β-MHC), and Myh7b, respectively (37). Myh6 expression and Myh7 expression are inversely regulated during cardiac development. In mice, Myh7/miR-208b is expressed in the embryonic heart, and Myh6/miR-208a is expressed predominantly at the postnatal stages. Myh7 is reexpressed in the adult heart only upon cardiac stress. Myh7b/miR-499 is expressed in both the embryonic heart and the adult heart (37). Targeted deletion of miR-208a, however, results in only a mild cardiac phenotype, such as ectopic expression of fast skeletal muscle markers and impaired ability to respond to stress postnatally (38). The mild cardiac phenotype in knockout of miR-208a, miR-208b, or miR-499 in mice suggests nonessential roles of these miRNAs during cardiac development. Potential explanations for the mild phenotype in miRNA knockout mice are (a) multiple miRNAs play partially redundant roles and/or (b) compensatory mechanisms are developed in vivo to adapt to a lack of specific miRNAs. In comparison with cardiac development in mice, cardiac development in zebra fish seems to be more sensitive to inactivation of a single miRNA function, as knockdown of a specific miRNA by using antisense RNA oligonucleotides (ASO) against miRNA (Figure 4) such as miR-143 and miR-138 results in abnormal development of the cardiac chambers and atrioventricular canal in zebra fish (39, 40).
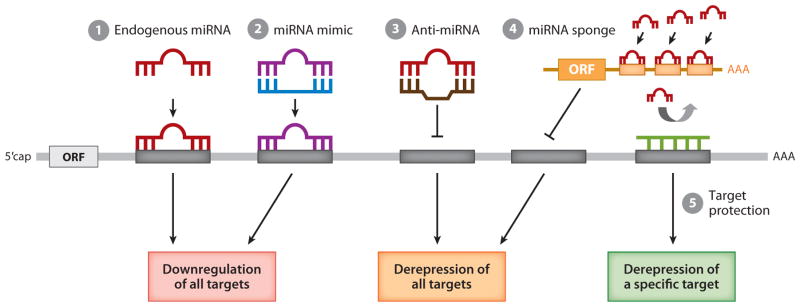
Figure 4.
Schematic representation of approaches to manipulate miRNA activity. The activity of a specific miRNA can be manipulated to modulate the expression level of all miRNA target genes (steps 1–3) or of a specific miRNA target gene (step 4). (1) Under physiological conditions, endogenous miRNA (red ) associates with a partially complementary sequence known as the seed sequence ( gray box) that is often located in the 3′UTR of target gene transcripts. This association results in downregulation of the target. (2) To augment the expression and activity of a specific miRNA, an miRNA mimic ( purple), which is composed of a chemically modified and stabilized ribonucleotide duplex, can be delivered. This structure mimics endogenous miRNA action and further downregulates target gene expression. (3) In contrast, to inhibit the activity of specific endogenous miRNAs, antisense oligonucleotides that are complementary to an endogenous miRNA (anti-miRNA; brown) can be delivered. These antisense oligonucleotides prevent the endogenous miRNA from binding to the seed sequence in the target transcripts, thus derepressing the expression of all target genes. (4) Alternatively, overexpression of exogenous mRNA (orange) that contains multiple copies of the seed sequence of a specific miRNA (an miRNA sponge; orange box) can be used to prevent endogenous miRNA from targeting target gene transcripts. (5) To protect a particular target transcript from silencing by endogenous miRNA (target protection), an antisense oligonucleotide designed to associate with the miRNA target sequence ( green) can be delivered. ORF and m7G denote open reading frame and 5′ capping, respectively. Adapted from Reference 112.
Members of the miR-15 family of miRNAs (i.e., miR-15a/b, miR-16-1/2, miR497/195) exhibit dynamic regulation during postnatal cardiac development and disease (Figure 3). miR-195 and miR-497 are upregulated during early postnatal cardiac development of the mouse. Such upregulation coincides with the exit of cardiac myocytes from cell proliferation and subsequent binucleation (41). Overexpression of miR-195 in the embryonic heart causes premature cell cycle arrest and the repression of mitotic genes (41). The miR-15 family inhibits cardiac myocyte proliferation by repressing the expression of multiple cell cycle regulators, including checkpoint kinase 1 (Chek1) (41). miR-15b is also implicated in the modulation of cellular ATP levels by regulation of the ADP/ATP exchanger in mitochondria in rat neonatal cardiomyocytes (42). Furthermore, inactivation of miR-15 in mice and pigs by using ASO protects against cardiac ischemic injury (43).
Deletion of the miR-17~92 cluster causes severe lung hypoplasia and ventricular-septum defects in mice, leading to early postnatal death (44), whereas loss of paralogous clusters (miR-106a~363 and miR-106b~25) in other organisms does not affect mutant animal viability (Figure 3). Upregulation of the miR-17~92 cluster directly downregulates the cardiac progenitor genes Isl1 and Tbx1, thereby facilitating myocardial differentiation (45).
Function of MiRNAs During Vascular and Blood Development
The formation of the mammalian vascular system is an intricate process including the assembly of mesoderm-derived endothelial precursors (angioblasts) that differentiate into a primitive vascular labyrinth (vasculogenesis), the subsequent sprouting of vessels (angiogenesis) to create a tubular network that remodels into arteries and veins, and the subsequent recruitment of pericytes and vascular smooth muscle cells (VSMCs) to form mature vessels. The initial evidence that miRNAs are essential for angiogenesis is based on the analysis of a hypomorphic Dicer allele in mice that leads to abnormalities in the vascular network in the yolk sac of otherwise normal embryos at E11.5 (46). Endothelial cell (EC)-specific deletion of Dicer using Tie2 promoter–driven Cre (Tie2-Cre) driver mice, however, exhibited no major abnormalities in embryonic angiogenesis and revealed (a) a requirement of miRNAs only for postnatal angiogenesis in tumorigenesis, wound healing, and limb ischemia and (b) reduced responsiveness to vascular endothelial growth factor (VEGF)-induced angiogenesis (46). Although the mild phenotype observed in Dicer deletion by using a Tie2-Cre driver may be due to incomplete recombination resulting in residual activity of Dicer in ECs, miRNAs seem to be less critical for the gene-regulatory program during endothelial development in comparison with other tissue types.
Function of MiRNAs During Endothelial Cell Development
In vitro studies confirm that miRNAs are required for EC function. Reduction or loss of Dicer expression in ECs leads to misexpression of several genes involved in the regulation of EC function, producing abnormal phenotypes that include increased nitric oxide (NO) release, decreased EC growth, and cord formation in matrigel assays. These results suggest that miRNAs are essential in different aspects of EC biology.
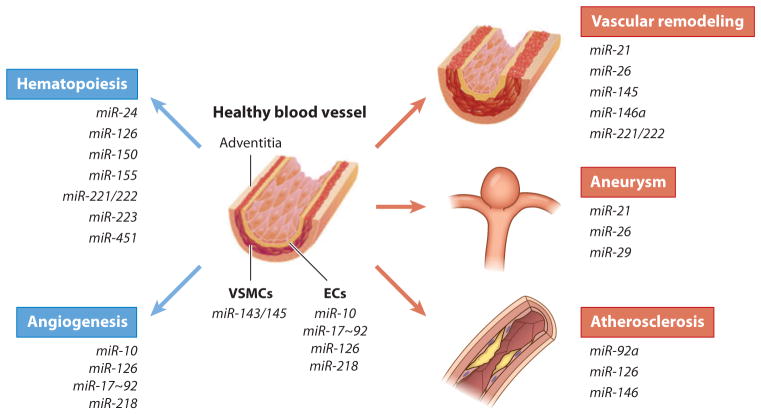
miR-126, an EC-specific miRNA, is encoded by an intron of the epidermal growth factor–like domain 7 (Egfl7) gene (47) (Figure 5). miR-126 expression is induced by blood flow and stimulates angiogenic sprouting of aortic arch vessels (48). Deletion of miR-126 in mice leads to vascular abnormalities with failure of cranial vessel growth and endothelial leakage at reduced penetrance (49, 50). Knockdown of miR-126 in zebra fish by morpholino oligonucleotides results in no gross malformation of vascular patterning of the early embryo but later leads to embryonic lethality as a result of loss of vascular integrity and hemorrhage (51). miR-126 targets the sprouty-related protein Spred1, a regulator of the MAP kinase signaling downstream of VEGF and of fibroblast growth factor (FGF) signaling, as well as phosphoinositide-3-kinase (PI3K)-regulatory subunit 2a (PIK3R2a), a regulator of PI3K signaling (51, 52).
Figure 5.
Role of miRNAs in healthy and diseased vasculature. Various miRNAs, i.e., miR-24, miR-126, miR-150, miR-155, miR-221/222, miR-223, and miR-451, are critical for hematopoiesis. Blood vessels are composed of three layers: the outermost adventitia ( pink), vascular smooth muscle cells (VSMCs; red ), and endothelial cells (ECs; orange). Upon vascular injury or upon pathological stimuli, a healthy blood vessel (left) undergoes vascular remodeling, which is characterized by aberrant proliferation and migration of ECs and/or VSMCs. In some cases an aneurysm or an atherosclerotic lesion forms. Many miRNAs are expressed in a cell type–specific manner. For example, miR-143/145 are specifically expressed in VSMCs and play a role in VSMC development and function. miR-10, miR-17~92, miR-126, and miR-128 are enriched in ECs and play a role in angiogenesis. Aberrant expression of some of these miRNAs is linked to pathological changes in the vasculature, such as vascular remodeling (i.e., miR-21, miR-26, miR-145, miR-146a, and miR-221/222), aortic aneurysm (i.e., miR-21, miR-26, and miR-29), and atherosclerosis (i.e., miR-92a, miR-126, and miR-146). Therefore, these miRNAs are potential therapeutic targets for vascular disorders.
miR-218 is encoded by an intron of the Slit1 and Slit2 genes and controls vascular patterning in mouse retinae by inhibiting the Slit-Robo signaling pathway (52) (Figure 5). Manipulation of miR-218 concentrations in vitro modulates EC migration, and knockdown of miR-218 in the developing retina results in anomalous development of the retinal vascular plexus, suggesting that miR-218 participates in this process (52).
Members of the miR-17~92 cluster are expressed in ECs, and manipulation of these miRNAs in vitro modulates EC properties (53) (Figure 5). In vitro assays in which miR-92a is overexpressed in ECs as well as in zebra fish embryos show impaired angiogenesis and vessel formation, whereas downregulation of miR-92a enhances blood vessel growth and functional recovery after ischemia or myocardial infarction (MI) (54).
miR-10a/10b are encoded in a highly conserved location within the developmental regulator cluster of HOX genes. miR-10 expression is induced after mesoderm specification and is enriched in endothelial cells. miR-10 plays a role in angiogenesis by regulating the level of fms-related tyrosine kinase 1 (FLT1), a cell surface protein that sequesters VEGF and its soluble splice variant, sFLT1. The increase in FLT1/sFLT1 protein levels upon miR-10 knockdown in zebra fish and cultured human umbilical venous endothelial cells inhibits the angiogenic behavior of endothelial cells by antagonizing VEGF receptor-2 signaling (55).
Function of MiRNAs During Vascular Smooth Muscle Cell Development
The developmental origin of VSMCs is diverse, as several independent cell lineages, including the neural crest, somites, the second heart field, and others, contribute to the mature VSMC population. Despite their heterogeneous origin, VSMCs coordinately activate a common set of SMC-specific marker genes, indicating the presence of a common developmental program for VSMC differentiation. Deletion of Dicer in embryonic VSMCs is lethal, resulting in intraperitoneal bleeding and prenatal death of mutant embryos at E16.5–E17.5 (56). This finding confirms the fundamental significance of miRNA-mediated posttranscriptional regulation in embryonic VSMC development.
miR-143 and miR-145, which are encoded by a bicistronic transcript, are highly expressed in VSMCs (57, 58) (Figure 5). The miR-143/miR-145 promoter is positively regulated by a transcription factor complex composed of SRF and members of the myocardin family of coactivators (57, 58). Furthermore, the miR-143/miR-145 promoter contains a Smad response element and is directly activated in response to active transforming growth factor-β (TGF-β) signaling (59). In vitro analysis indicates that miR-143 and miR-145 promote multiple aspects of the VSMC contractile phenotype. In particular, the expression of contractile genes is elevated following overexpression of miR-143 and miR-145. The multiple targets of miR-143 and miR-145 include Krüppel-like factor 4 (KLF4), Krüppel-like factor 5 (KLF5), ELK1, Versican, several actin-remodeling proteins, and angiotensin-converting enzyme (57, 58, 60, 61). Interestingly, all these proteins are antagonistic to VSMC differentiation, and thus repression of these targets by miR-143/miR-145 facilitates VSMC differentiation. In particular, miR-145-mediated repression of KLF4 is required for TGF-β-mediated induction of contractile gene expression (61). Despite the critical role of miR-143 and miR-145 in VSMC differentiation in vitro, mice ablated in both miR-143 and miR-145 are viable, suggesting a possibility that one or more other miRNAs may play a compensatory role in vivo (58, 60, 62). Nonetheless, miR-145-null mice exhibit blood vessels with a reduced medial layer, vascular tone, and blood pressure (58, 60). Furthermore, VSMCs from miR-145-null mice exhibit abnormalities in actin stress fiber formation and phenotype switch in response to vascular injury, indicating a significant role of miR-145 in actin signaling and SRF/myocardin activity and contractility in VSMCs (58).
MiRNAs in Hematopoiesis
The critical role played by miRNAs in hematopoiesis was implicated by miRNA expression pro-filing studies, which indicate dynamic regulation of miRNA levels during the differentiation of hematopoietic stem cells. Mice deleted in Ago2 exhibit abnormalities in erythroid lineage, supporting the essential role of miRNAs during this process. Loss of function of miR-223 and miR-451 in mice suggests the role of these miRNAs in erythroid proliferation and differentiation (Figure 5). miR-24 regulates erythroid differentiation by influencing the expression of human activin type I receptor ALK4 (ACVR1B) (Figure 5). Forced expression of miR-24 leads to a delay of activin-induced maturation of hematopoietic progenitor cells (63). Additionally, the role of miR-155 and miR-150 in the differentiation of B and T lymphocytes, the suppressive role of miR-221 and miR-222 in erythroid differentiation, and the inhibitory effect of miR-181 on hematopoietic differentiation have been reported (64) (Figure 5). Primitive erythropoiesis defines the onset of hematopoiesis in the yolk sac of the early embryo and is initiated by the emergence of progenitors. Analyses of miR-126-null embryos indicate that miR-126 downregulates vascular cell adhesion molecule-1 expression in a mesenchymal cell population, which accelerates the maturation of primitive erythroblasts via a Src family kinase (65). Knockdown of miR-126 promotes erythropoiesis at the expense of thrombopoiesis in zebra fish (66).
MiRNAS IN CARDIOVASCULAR DISEASE
Given the importance of miRNAs in development, it is not surprising that deregulation of miRNA expression is implicated in a variety of human diseases, in particular those of the cardiovascular system. Reexpression of the fetal cardiac gene program in the adult heart or aberrant expression of cardiac-specific miRNAs is associated with the pathogenesis of cardiovascular diseases. Understanding the role of these miRNAs during pathogenesis is critical for the development of novel therapies and diagnostic tools.
Role of MiRNAs in Heart Disease
The mild and nonlethal phenotype of knockout of a single miRNA in mice allows investigators to challenge mice with various cardiovascular insults, such as vascular injury, pressure overload, hypoxia, and pharmacological stimuli. Acute myocardial infarction (AMI) due to coronary artery occlusion is accompanied by a pathological remodeling response that includes hypertrophic cardiac growth and fibrosis, both of which impair cardiac contractility. Therefore, it is critical to elucidate the regulatory mechanisms that are involved in these processes, including the roles of miRNAs. Transgenic overexpression of cardiac-specific miR-208a is sufficient to induce hypertrophic growth in mice (67) (Figure 6). Conversely, deletion of cardiac-specific miR-208a in mice abrogates the reexpression of the fetal β-MHC gene in response to hemodynamic cardiac stress and protects mice from cardiac hypertrophy. Furthermore, systemic delivery of ASO against miR-208a induces potent and sustained silencing of miR-208a in the heart and prevents cardiac remodeling while improving cardiac function and survival during hypertension-induced heart failure in Dahl hypertensive rats (68). This study indicates a potential application of ASO-based therapy in the modulation of cardiac-specific miRNAs, which could lead to the subsequent prevention or amelioration of the cardiac dysfunctions and remodeling that occur during cardiac disease progression. Interestingly, cardiac-specific miR-208a controls systemic energy homeostasis through the regulation of the metabolic gene program by targeting MED13, a subunit of the Mediator complex (69). This intriguing miR-208a–MED13 axis provides a potential link between metabolic disorders, such as obesity and type 2 diabetes, and cardiovascular disease.
Figure 6.
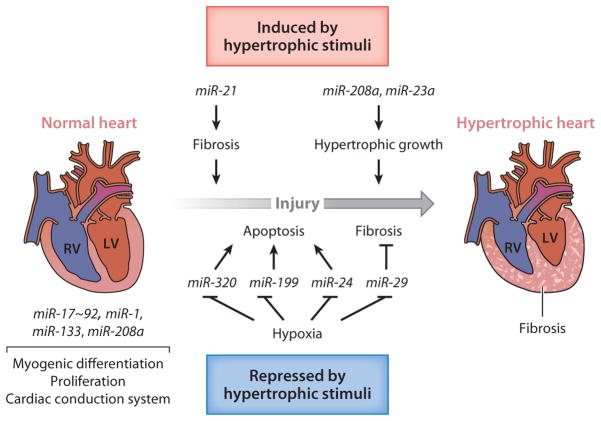
The role of miRNAs in the normal and diseased heart. Upon injury, such as vascular injury, pressure overload, hypoxia, and pharmacological stimuli, a normal heart (left) undergoes pathological remodeling and develops into a hypertrophic heart with fibrosis (right). miRNAs that are expressed in cardiac tissue and contribute to normal function or pathological remodeling along with their corresponding functions are indicated. miR-1 and miR-133 are involved in the development of a normal heart by regulating proliferation, differentiation, and cardiac conduction. Both miR-1 and miR-133 downregulate cell cycle regulators and thereby block proliferation. However, miR-1 and miR-133 exhibit opposing effects in cardiomyocyte differentiation. The miR-17~92 cluster promotes myogenic differentiation in the secondary heart field. miR-208a also contributes to the regulation of the conduction system. After cardiac injury, various miRNAs contribute to pathological remodeling of the heart and the progression to heart failure. miR-29 and miR-21 block and promote cardiac fibrosis, respectively. miR-21 and miR-29 are expressed in cardiac fibroblasts. Whereas miR-29 blocks fibrosis by inhibiting the expression of extracellular matrix (ECM) components, miR-21 promotes fibrosis by stimulating mitogen-activated protein kinase (MAPK) signaling. miR-208 controls this myosin isoform switching, cardiac hypertrophy, and fibrosis. miR-23a is induced in cardiac myocytes by different hypertrophic stimuli and promotes hypertrophic responses. Ischemic injury leads to the downregulation of miR-24, miR-29, and miR-199 in cardiomyocytes; such downregulation promotes inhibition of apoptosis and fibrosis. Downregulation of miR-320 upon ischemic injury contributes to hypertrophic responses as well. RV and LV denote right ventricle and left ventricle, respectively.
miR-23a expression is upregulated in response to various hypertrophic stimuli in cardiomy-ocytes (70, 71). miR-23a is induced by nuclear factor of activated T cells NFATc3, a transcriptional factor that mediates the cardiac stress signaling pathway and promotes pathological hypertrophy (71). Overexpression of miR-23a is sufficient to initiate hypertrophic responses by targeting the anti-hypertrophic protein MuRF1 (muscle-specific ring finger protein 1), whereas downregulation of miR-23a by ASO in mouse abrogates isoproterenol-induced cardiac hypertrophy (71).
Decreased expression of the miR-29 family of miRNAs, miR-24, and miR-320 occurs after MI (72–74) (Figure 6). The miR-29 family targets multiple collagen genes and extracellular matrix (ECM) protein genes, such as fibrillins and elastin, and downregulation of miR-29 upon MI promotes fibrosis and scar formation in the heart (72). miR-24 and miR-320 target the BH3-only domain–containing protein Bim, which positively regulates apoptosis (73), and cardioprotective heat shock protein 20 (74), respectively. Downregulation of miR-24 and miR-320 after MI, therefore, inhibits cardiomyocyte apoptosis. Moreover, miR-199 is downregulated under hypoxic conditions in cardiac myocytes and mediates derepression of sirtuin 1 and hypoxia-inducible factor 1α, a transcription factor essential for the induction of the hypoxia-response gene network (75). Thus, miR-24, miR-29, miR-199, and miR-320 represent potential therapeutic targets for ischemic injury in cardiac myocytes (Figure 6).
miR-21 promotes cardiac hypertrophy and fibrosis in response to pressure overload by facilitating the profibrotic extracellular signal–regulated kinase (ERK)–mitogen-activated protein kinase (MAPK) signaling pathway (76) (Figure 6). Downregulation of miR-21 with cholesterol-modified ASO against miR-21 in mice attenuates cardiac remodeling following thoracic aortic banding (76). Interestingly, neither targeted deletion of miR-21 in mice nor inhibition of miR-21 by differentially modified short ASO improved fibrotic lesions or hypertrophic response after thoracic aortic banding or other cardiac stresses in another study (77), illustrating the complex nature of miR-21 participation in cardiac fibrosis. Further analysis is required to understand these contradictory reports.
Zebra fish regenerate cardiac muscle after severe injuries through the activation and proliferation of spared cardiac myocytes. miR-133, which has a role in cardiac development, shows diminished expression during cardiac regeneration (78). Elevated miR-133 levels after injury inhibit myocardial regeneration, whereas transgenic miR-133 depletion enhances cardiomyocyte proliferation (78), indicating that miR-133 is required to restrict injury-induced cardiomyocyte proliferation.
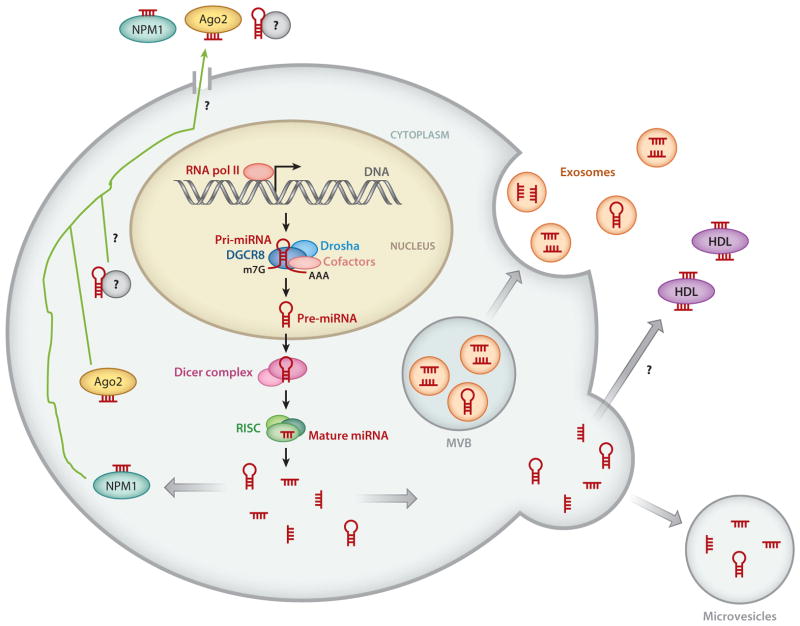
Investigators recently revealed that extracellular miRNAs are stable, circulate in the bloodstream, and may have a crucial function in the cardiovascular system (Figure 7). Both mature miRNAs and pre-miRNAs seem to be packaged into small vesicles termed exosomes, which originate from the endosome and are released from cells when microvesicular bodies fuse with the plasma membrane (79) (Figure 7). MiRNAs can also be released from microvesicles, which are released from the cell through blebbing of the plasma membrane (79) (Figure 7). Once released from the cell, extracellular miRNAs can be found not only in microvesicles and exosomes but also in association with various proteins, including Ago2 (80), nucleophosmin 1 (80), and high-density lipoproteins (81), which protect miRNAs from degradation (Figure 7). Although the precise cellular release mechanisms of miRNAs and the mechanism of regulation remain unknown, these circulating miRNAs may be delivered to recipient cells, where they can regulate levels of target genes (82), thus providing a novel means of distant cell-cell communication. However, whether circulating miRNAs are actively released as a way of cell-cell communication or are passively released as a result of tissue injury remains to be determined.
Figure 7.
Schematic diagram of the mechanisms of release of cellular miRNAs. In the cytoplasm, both precursor miRNAs (pre-miRNAs) and mature miRNAs can be incorporated into small vesicles termed exosomes (orange), which are released from the cell when multivesicular bodies (MVB) are fused to the plasma membrane. Mature miRNAs and pre-miRNAs can also be released by microvesicles. Some miRNAs associate with high-density lipoproteins (HDL; purple) or bind to RNA-binding proteins, such as nucleophosmin 1 (NPM1; teal ) and Argonaute 2 (Ago2; yellow), a component of the RNA-induced silencing complex (RISC) and released through an unknown mechanism ( green arrow). It was initially speculated that dead cells passively release miRNAs. Recent studies suggest that some miRNAs may be actively released into circulation through an unknown mechanism, potentially through a channel. How miRNAs and pre-miRNAs are sorted out to multiple mechanisms of release is unclear. Whether mature miRNA and pre-miRNA are segregated in different compartments is also unclear. RNA Pol II denotes RNA polymerase II.
Various studies point toward the differential release of specific miRNAs during various cardiac injuries. Some reports, for example, indicate that heart-specific miRNAs, such as miR-208a, miR-499, miR-1, and miR-133, are consistently elevated in the plasma of AMI patients within hours after the onset of infarction (83), suggesting that these miRNAs may be useful as stable blood-based biomarkers for AMI. In the early phase (<3 h) of AMI, miR-1, miR-133a, and in particular miR-208a may be even more sensitive than the classic biomarker cardiac Troponin I (cTnI), as these miRNAs achieve their peak before cTnI (83). This earlier miRNA peak suggests leakage of miRNAs that is faster than that of cTnI from damaged cardiac myocytes, most likely due to the fact that cTnI is bound mainly to myofibrils, whereas miRNAs are probably bound to protein complexes in the cytosol, thus allowing for the faster release of miRNAs from damaged cells.
Interestingly, miRNAs in cardiac myocytes are not released into the circulation with identical kinetics, presumably because different miRNAs are bound to different proteins within cells and in the circulation. For example, miR-30c and miR-24, which are highly expressed in the heart, are not detected in the bloodstream after AMI (84). Moreover, studies on circulating miRNAs in patients with heart failure that excluded those with recent cardiac ischemia or infarction found no increase in miRNAs that are detected in AMI patients, but instead showed elevation of a distinct set of miRNAs, including miR-423-5p, miR-18b′, miR-129-5p, miR-1254, miR-675, and miR-622 (85). High abundance of some miRNAs (miR-423-5p, miR-18b′) seems to correlate with the severity of the condition (85). In plasma samples of atherosclerotic coronary artery disease (CAD) patients, miRNAs that are highly expressed in ECs (miR-17, miR-126, miR-92a), VSMCs (miR-145), and inflammatory cells (miR-155) are elevated (86). As distinct circulating miRNAs appear to be detected in different cardiovascular conditions, circulating miRNAs may serve as diagnostic and/or therapeutic markers for various cardiovascular disorders.
Role of MiRNAs in Vascular Disease
Although blood vessels are quiescent in adults and rarely form new branches, the vessel wall is subject to complex regulatory signaling networks that maintain a barrier that does not leak blood but allows for the proper exchange of gas and nutrients between blood and adjacent tissues. Upon vascular injury or environmental changes, the vascular wall undergoes EC migration as well as VSMC phenotypic changes to promote injury repair (87, 88). Abnormalities in such regulatory networks may lead to a variety of vascular diseases. Numerous miRNAs display dramatically altered expression during vascular injury. Aberrant expression of miRNAs has also been correlated with pathological conditions, such as ischemia, tumor angiogenesis, atherosclerosis, pulmonary arterial hypertension (PAH), and the proliferative vessel thickening and obstruction known as restenosis after angioplasty (88).
The TGF-β family of growth factors, including TGF-β and the bone morphogenetic proteins (BMPs), promote VSMC differentiation and prevent the switch of VSMCs to the synthetic phenotype (89). Loss of expression and loss-of-function mutations in the BMP receptor (BMPRII) gene are associated with the pathogenesis of PAH. The Smad proteins, signal transducers of the BMP and TGF-β pathway, induce the expression of miR-21 posttranscriptionally by modulating the Drosha-processing step (17). A target of miR-21 that is critical for this process is programmed cell death protein 4 (PDCD4) (17). Although the precise mechanism is unclear, downregulation of PDCD4 is required for BMP- and TGF-β-mediated VSMC differentiation (17). Similarly, treatment of pulmonary fibroblasts with TGF-β elevated miR-21 and enhanced conversion to myofibroblasts (90), suggesting that miR-21-mediated regulation of contractile genes plays a role in multiple cell types. The relevance of miR-21 in regulating VSMC differentiation in response to BMP signaling is confirmed by the observation that miR-21 levels are reduced in pulmonary artery SMCs from PAH patients (91). In addition to its role in VSMC phenotype regulation, miR-21 can influence the function and migration of angiogenic progenitor cells during CAD (92) (Figure 5). Some studies indicate that miR-21 levels are upregulated after mechanical injury of large vessels (88) and that inactivation of miR-21 with ASO may prevent restenosis (62, 88).
In contrast to the case for miR-21, miR-145, a VSMC-specific miRNA, is downregulated upon mechanical injury (Figure 5). Restoring miR-145 expression to normal levels ameliorates resteno-sis, suggesting that injured vessels undergo dedifferentiation of VSMCs in part through down-regulation of miR-145, thus resulting in vascular remodeling. In addition to decreased neointima formation after vascular injury and hypotension (58, 60, 62), miR-143/miR-145 knockout mice develop spontaneous arteriosclerotic lesions (60). The loss of miR-145 in VSMCs may coincide with loss in responsiveness to external signals, such as angiotensin and adrenergic stimuli. Aberrant expression of miR-145 is reported in lung samples of patients with heritable and idiopathic PAH, which supports a critical role of miR-145 in vascular development, as well as in the maintenance of homeostasis of healthy vasculature (93).
In response to the potent growth simulators platelet-derived growth factors (PDGFs), the VSMC phenotype switches from a highly differentiated contractile phenotype to a dedifferenti-ated synthetic phenotype (87). The induction of miR-221 appears to mediate at least one aspect of the PDGF-mediated VSMC phenotype switch (94, 95). In response to PDGF treatment, pri-miR-221 transcription is rapidly elevated, leading to increased mature miR-221 levels within 3 h (94). Mature miR-221 has multiple effects on the VSMC phenotype, including increased proliferation and migration and reduced expression of contractile genes (94, 95). miR-221 promotes VSMC proliferation through the repression of the cyclin-dependent kinase inhibitor p27Kip1 (94). Inhibition of miR-221, moreover, prevents PDGF-mediated reduction of p27Kip1 and loss of VSMC proliferation (94). Thus, elevated miR-221 levels are required for PDGF-mediated repression of contractile genes (94). However, this effect is independent of the downregulation of p27Kip1, as knockdown of p27Kip1 by small inhibitory RNA (siRNA) has no effect on contractile gene expression (94). These results indicate that the expression of contractile genes and cell growth are not coupled but can instead be regulated by distinct mechanisms. Overexpression of miR-221 is also accompanied by dramatic reduction of myocardin expression (94). Of note, miR-221 and miR-222 are clustered on the X chromosome, are transcribed from a common promoter (96), and share a common seed sequence. The importance of these miRNAs in modulating VSMC phenotype is further supported by the finding that miR-221 and miR-222 are strongly induced in vivo in VSMCs following balloon injury of the vessel (95) (Figure 5). Targeted delivery of ASO against miR-221 and miR-222 at the site of vascular injury reduced VSMC proliferation and intimal thickening in response to vascular injury (95). As does miR-221, miR-26 promotes a synthetic phenotype and proliferation of VSMCs through downregulation of Smad1 and Smad4, signal transducers of the BMP signaling pathway (97). miR-26a is downregulated two- to threefold in mouse models of abdominal aortic aneurysm (AoA) formation (97). Overexpression of miR-21, another miRNA that is critical for VSMC phenotype regulation, protects against AoA progression by targeting phos-phatase and tensin homolog (PTEN) (98). Conversely, inhibition of miR-21 further augments ongoing AoA formation (98). These results suggest that miR-21 and miR-26 may be therapeutic targets for AoA (Figure 5).
As is the case for miR-221 and miR-222, microarray analysis indicates that miR-146a expression is elevated in balloon-injured rat arteries compared with uninjured controls (88). Overexpression of miR-146a increases VSMC proliferation, whereas knockdown of miR-146a attenuates PDGF-mediated increase of VSMC growth (99) (Figure 5). The relevance of miR-146a in vivo is supported by the observation that treatment of balloon-injured rat carotid arteries with ASO against miR-146a resulted in reduced neointima formation and VSMC proliferation (99). Interestingly, a critical target of miR-146a in VSMCs is KLF4 (99), which is a validated target of miR-145 (57, 58). Although a physiological role of miR-146a, miR-221, or miR-222 during VSMC development is yet to be elucidated in the miRNA-null mice, these in vitro results suggest the exciting possibility that inactivation of these miRNAs may be an effective therapy for vascular proliferative disorders such as restenosis or PAH. miR-146a is highly expressed in human atherosclerotic plaques in patients with CAD and modulates Toll-like receptor 4 signaling, which is a critical modulator of inflammatory response, by targeting IRAK1 (IL-1 receptor–associated kinase 1) and TRAF6 (TNF receptor–associated factor 6) (100).
Inhibition of miR-92a enhances functional recovery of ischemic tissues in mice (54), supporting a negative regulatory role of miR-92a in angiogenesis. miR-126-null mice, which successfully complete embryogenesis, have increased mortality after coronary artery ligation (50), indicating that a proper proangiogenic signaling pathway mediated by miR-126 is required after vascular insults such as MI. Furthermore, endothelial progenitor cells (EPCs), which differentiate into ECs, identified in the circulation of CAD patients express lower levels of miR-126 in comparison with EPCs from healthy donors, suggesting a potential role of miR-126 in EPC function and the pathogenesis of CAD (101).
A differential expression profiling study of miRNAs in aortic tissues of young versus aged mice found elevated expression of miR-29 in association with the downregulation of ECM in aged mice (102). Increased expression of miR-29 is also found in human thoracic aneurysms, suggesting that miR-29-mediated downregulation of ECM proteins may promote the formation of aneurysms that is typical in advanced age (102) (Figure 5).
miR-33 regulates cholesterol metabolism in the liver and implicated in the development of atherosclerosis. miR-33 inhibits the expression of the ATP-binding cassette (ABC) transporter ABCA1, thereby attenuating cholesterol efflux to apolipoprotein A1. Silencing of miR-33 with ASO in mice increases hepatic expression of ABCA1 and plasma high-density lipoprotein levels and the regression of atherosclerosis, thus implicating the anti-miR-33 therapy in atherosclerotic vascular disease (103).
Mutations of MiRNAs in Human Diseases
As are protein-coding genes, genomic loci encoding miRNAs are susceptible to mutations that may cause disease or development defects. Furthermore, mutations in the exons encoding the 3′UTR of miRNA target genes, especially within miRNA-binding sites that are critical for miRNA-mediated silencing of targets, can potentially mediate a significant phenotype. Indeed, single-nucleotide polymorphisms (SNPs) are found at a higher frequency at predicted miRNA-binding sites within the 3′UTR. For example, SNPs located in the miR-155-binding site within the 3′UTR of the human angiotensin II type 1 receptor (AGTR1) gene are associated with hypertension (104). Future research may reveal many more SNPs in the 3′UTR that are associated with cardiovascular disease. Whether these SNPs lead to disease as a result of deregulation of target gene expression levels by specific miRNAs remains to be elucidated.
CONCLUSIONS
Cardiovascular development has long been recognized as a complex sequence of processes that requires intricate coordination of cell differentiation, migration, and proliferation, all of which are carefully guided and controlled by temporal and spatial cues. The discovery of miRNAs approximately a decade ago established a completely new paradigm of gene regulation and added another layer of complexity to the understanding of morphogenesis and tissue repair processes in every organ, especially those of the cardiovascular system. In particular, miRNAs provide an efficient mechanism for modulating the expression of a relatively large number of genes during cellular stress, when many transcriptional and translational machines are suppressed. More recent findings that some miRNAs are secreted from cells, possibly affecting gene expression of distant cells, present a new role for miRNAs as a novel mediator of cell-cell communication analogous to cytokines. Although not discussed here, many noncoding RNAs in the mammalian genome are longer in size and are expressed in a spatially and temporally regulated manner similar to that of miRNAs, although most of their functions have not been elucidated (105). Such long noncoding RNAs (lncRNAs) may add another layer of complexity to the mechanism of gene expression regulation in mammalian tissues, including cardiovascular tissues, and may play a significant role in the morphogenesis of the cardiovascular system.
FUTURE ISSUES
Fewer than ten years of active research on miRNAs have elucidated a distinct pattern of expression for both cellular and circulating miRNAs under different pathological conditions, and such research presents the miRNA expression profile as a highly accurate diagnostic tool. Until now, the diagnosis of many cardiovascular diseases, such as hypertrophic cardiomyopathy and pulmonary hypertension, relied heavily on the cardiac imaging technique, which is costly and not quantitative. The detection of circulating miRNAs as a diagnostic or prognostic tool for the treatment of cardiovascular disease or as a set of biomarkers for monitoring the effectiveness of therapies is particularly attractive, as miRNAs are very sensitive, and their detection requires minimal peripheral blood. Furthermore, miRNAs in serum are stable at room temperature and are resistant to freeze-thaw cycles. It is unclear whether circulating miRNAs (a) play an active role under disease conditions and modulate gene expression in recipient cells or (b) are merely released from injured tissue. However, the former possibility is intriguing, as it suggests that circulating miRNAs may further our understanding of human disease pathology.
Overwhelming numbers of studies suggest that the aberrant expression of miRNAs plays a pivotal role in various aspects of cardiovascular development and disease. The supplementation of a single miRNA with beneficial activity by miRNA mimics, the suppression of a single miRNA with pathological activity by ASO or an miRNA sponge, or the protection of a specific target mRNA from a specific miRNA by expressing an oligonucleotide designed to associate with the miRNA recognition sequence could ameliorate or in some cases prevent the progression of the pathological conditions observed in various animal models of cardiovascular disease (68, 76) (Figure 4). Some studies have found that chemically modified short oligonucleotides systemically delivered through tail vein or subcutaneous injection are relatively stable and are delivered to cardiovascular tissues, exhibiting the expected gene-regulatory functions in the recipient tissues and resulting in improved cardiac function (68). Locked nucleic acid (LNA)-containing ASO against the liver-specific miRNA miR-122, which is required for the replication of hepatitis C virus (HCV), have been tested in nonhuman primates and healthy human volunteers, are well tolerated, and have advanced to phase IIa clinical trials as a potential therapy for chronic HCV infection (106–108). However, delivery of these oligonucleotides to the heart currently appears to be less efficient than delivery to the kidney or liver (68). Therefore, using such oligonucleotides in the treatment of cardiovascular disease requires the development of a novel methodology for the more efficient and targeted delivery of oligonucleotides to cardiovascular tissue, e.g., delivery via a stent coated with the oligonucleotides or direct delivery by cardiac catheterization. Further studies on circulating miRNAs and on the mechanism of selective miRNA uptake by target tissues would contribute to the development of more effective delivery of miRNAs or ASO against miRNAs (Figure 7). One of many challenges for future miRNA-based therapies is the fact that a single miRNA modulates more than 100 target genes, and therefore the modulation of a single miRNA level might exhibit both therapeutic and pathological effects. ASO against an miRNA might also be therapeutic in certain tissues and pathological in different or healthy tissues. Currently available miRNA target gene prediction algorithms do not allow us to obtain a comprehensive and accurate view of all target genes of a specific miRNA in a specific tissue type. A single miRNA can target multiple genes that cooperatively function in the common pathway (109). For an enhanced therapeutic effect, administering oligonucleotides against multiple miRNAs could be effective in the same way that a cocktail of small-molecule drugs can more effectively treat a condition than can any of the drugs alone. The other challenge is to determine an effective dose of ASO or miRNA mimic, as the pharmacokinetics of differentially modified short oligonucleotides needs to be carefully evaluated. Injection of LNA-modified anti-miR-122 oligonucleotides in chimpanzees indicated a remarkably long duration of ASO activity of up to 100 days, as judged by the lowering of plasma cholesterol (107), supporting the feasibility of an miRNA-based therapy for the treatment of chronic conditions. Finally, miRNA-based therapies will greatly benefit from the ability to manipulate the recruitment of miRNA to the 3′UTR of specific target mRNA or to modulate the efficiency of silencing by miRNA. To this end, the mechanism of miRNA–target mRNA recognition as well as the mechanism of miRNA/RISC-mediated mRNA silencing must be uncovered.
Last, the recent discovery that the human and other mammalian genomes produce thousands of lncRNAs presents the possibility that, as is the case for miRNAs, the aberrant expression or function of lncRNAs may be responsible for cardiovascular developmental abnormalities and disease. This possibility leads to another avenue of future research (110).
Cardiovascular disease is a leading cause of death that would benefit highly from the development of unconventional therapeutic and diagnostic approaches. The rapid growth of knowledge concerning miRNA biology as well as technical improvements on miRNA detection and manipulation provide the possibility for the application of noncoding RNAs toward novel prognostic, diagnostic, or therapeutic targets in cardiovascular disease in the near future.
SUMMARY POINTS.
MicroRNAs (miRNAs) are encoded in the mammalian genome and negatively regulate the expression of more than 100 genes posttranscriptionally via translational inhibition or destabilization of mRNAs.
Many miRNAs are specifically expressed in cardiovascular tissues or for a specific window of time during cardiovascular development and play a critical role in various aspects of cardiovascular development and disease processes.
Molecules essential for the miRNA biogenesis pathway and for miRNA stability are regulated by various growth factor signaling and environmental cues that control the level of miRNA expression.
Aberrant expression of miRNAs plays a role in developmental defects and the pathogenesis of the cardiovascular system.
MiRNAs are secreted from tissues and are detected in the circulation.
The level of a specific miRNA can be manipulated in cultured cells and whole animals by using chemically modified RNA oligonucleotides with sense or antisense miRNA sequences.
There is a strong potential for the successful development of miRNA-targeted therapies for cardiovascular disease. In addition, the use of miRNAs, especially extracellular miRNAs, as biomarkers for cardiovascular disease is under development.
Further elucidation of basic miRNA biology, including the precise mechanism of gene-silencing action and the mechanism for the regulation of miRNA activity, will allow for clinical applications of miRNAs as therapeutic and diagnostic tools.
Acknowledgments
The author apologizes to all colleagues whose work could not be cited due to space restrictions and thanks Ms. Sheu-Gruttadauria for editorial support of the review. A.H. is supported by grants from the National Heart, Lung, and Blood Institute; the LeDucq Foundation Transatlantic Network for Excellence in Cardiovascular Research Program; and the American Heart Association.
Glossary
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- UTR
untranslated region
- Pri-miRNA
primary miRNA
- Pre-miRNA
precursor miRNA
- DGCR8
DiGeorge syndrome critical region gene 8
- Ago
Argonaute
- ASO
antisense oligonucleotides
- VSMC
vascular smooth muscle cell
- EC
endothelial cell
- ECM
extracellular matrix
- LNA
locked nucleic acid
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–58. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–48. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 5.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–32. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–29. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 7.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–32. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–80. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 9.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 10.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′UTR as in the 3′UTR. Proc Natl Acad Sci USA. 2007;104:9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 14.Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148:381–92. doi: 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–32. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 17.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 19.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–47. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Rissland O, Hong S, Bartel D. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2012;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang HW, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci USA. 2009;106:7016–21. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19:R169–75. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 26.Saxena A, Tabin CJ. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc Natl Acad Sci USA. 2010;107:87–91. doi: 10.1073/pnas.0912870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang ZP, Chen JF, Regan JN, Maguire CT, Tang RH, et al. Loss of microRNAs in neural crest leads to cardiovascular syndromes resembling human congenital heart defects. Arterioscler Thromb Vasc Biol. 2010;30:2575–86. doi: 10.1161/ATVBAHA.110.213306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111–16. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–76. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 30.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res. 2009;105:585–94. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104:20844–49. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 33.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chinchilla A, Lozano E, Daimi H, Esteban FJ, Crist C, et al. MicroRNA profiling during mouse ventricular maturation: a role for miR-27 modulating Mef2c expression. Cardiovasc Res. 2011;89:98–108. doi: 10.1093/cvr/cvq264. [DOI] [PubMed] [Google Scholar]
- 35.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–54. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–58. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–73. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–79. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 39.Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, et al. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development. 2010;137:1887–96. doi: 10.1242/dev.050526. [DOI] [PubMed] [Google Scholar]
- 40.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci USA. 2008;105:17830–35. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109:670–79. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishi H, Ono K, Iwanaga Y, Horie T, Nagao K, et al. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem. 2010;285:4920–30. doi: 10.1074/jbc.M109.082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a microRNA-mediated mechanism. Dev Cell. 2010;19:903–12. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–35. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt M, Paes K, De Maziere A, Smyczek T, Yang S, et al. EGFL7 regulates the collective migration of endothelial cells by restricting their spatial distribution. Development. 2007;134:2913–23. doi: 10.1242/dev.002576. [DOI] [PubMed] [Google Scholar]
- 48.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–93. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 50.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–84. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. 2010;107:1336–44. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, et al. Members of the microRNA-17–92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944–50. doi: 10.1182/blood-2010-01-264812. [DOI] [PubMed] [Google Scholar]
- 54.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, et al. MicroRNA-92a controls an-giogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–13. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 55.Hassel D, Cheng P, White MP, Ivey KN, Kroll J, et al. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ Res. 2012;111:1421–33. doi: 10.1161/CIRCRESAHA.112.279711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–26. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–78. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long X, Miano JM. Transforming growth factor-β1 (TGF-β1) utilizes distinct pathways for the transcriptional activation of microRNA 143/145 in human coronary artery smooth muscle cells. J Biol Chem. 2011;286:30119–29. doi: 10.1074/jbc.M111.258814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Investig. 2009;119:2634–47. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, et al. Down-regulation of Krüppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-β and bone morphogenetic protein 4. J Biol Chem. 2011;286:28097–110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–98. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q, Huang Z, Xue H, Jin C, Ju XL, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–95. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- 64.Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol. 2010;84:1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 65.Sturgeon CM, Chicha L, Ditadi A, Zhou Q, McGrath KE, et al. Primitive erythropoiesis is regulated by miR-126 via nonhematopoietic Vcam-1+ cells. Dev Cell. 2012;23:45–57. doi: 10.1016/j.devcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 66.Grabher C, Payne EM, Johnston AB, Bolli N, Lechman E, et al. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia. 2011;25:506–14. doi: 10.1038/leu.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Investig. 2009;119:2772–86. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–47. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–83. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin Z, Murtaza I, Wang K, Jiao J, Gao J, et al. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci USA. 2009;106:12103–8. doi: 10.1073/pnas.0811371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, et al. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med. 2011;203:549–60. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren XP, Wu J, Wang X, Sartor MA, Qian J, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rane S, He M, Sayed D, Vashistha H, Malhotra A, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1a and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–84. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 77.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Investig. 2010;120:3912–16. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin VP, Lepilina A, Smith A, Poss KD. Regulation of zebrafish heart regeneration by miR-133. Dev Biol. 2012;365:319–27. doi: 10.1016/j.ydbio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–95. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 80.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 83.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 84.D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–39. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 86.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–84. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 87.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 88.Ji R, Cheng Y, Yue J, Yang J, Liu X, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 89.Davis-Dusenbery BN, Wu C, Hata A. Micromanaging vascular smooth muscle cell differentiation and phenotypic modulation. Arterioscler Thromb Vasc Biol. 2011;31:2370–77. doi: 10.1161/ATVBAHA.111.226670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–97. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Caruso P, MacLean MR, Khanin R, McClure J, Soon E, et al. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30:716–23. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- 92.Fleissner F, Jazbutyte V, Fiedler J, Gupta SK, Yin X, et al. Short communication: Asymmetric dimethylarginine impairs angiogenic progenitor cell function in patients with coronary artery disease through a microRNA-21-dependent mechanism. Circ Res. 2010;107:138–43. doi: 10.1161/CIRCRESAHA.110.216770. [DOI] [PubMed] [Google Scholar]
- 93.Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, et al. A role for miR-145 in pulmonary arterial hypertension: evidence from mouse models and patient samples. Circ Res. 2012;111:290–300. doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- 94.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–38. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–87. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, et al. Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–43. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci Transl Med. 2012;4:122ra122. doi: 10.1126/scitranslmed.3003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun SG, Zheng B, Han M, Fang XM, Li HX, et al. miR-146a and Krüppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12:56–62. doi: 10.1038/embor.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, et al. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci. 2010;119:395–405. doi: 10.1042/CS20100003. [DOI] [PubMed] [Google Scholar]
- 101.Zhang Q, Kandic I, Kutryk MJ. Dysregulation of angiogenesis-related microRNAs in endothelial progenitor cells from patients with coronary artery disease. Biochem Biophys Res Commun. 2011;405:42–46. doi: 10.1016/j.bbrc.2010.12.119. [DOI] [PubMed] [Google Scholar]
- 102.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res. 2011;109:1115–19. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 103.Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–73. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–13. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Derrien T, Guigo R, Johnson R. The long non-coding RNAs: a new (p)layer in the “dark matter. Front Genet. 2011;2:107. doi: 10.3389/fgene.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–99. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 107.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haussecker D, Kay MA. miR-122 continues to blaze the trail for microRNA therapeutics. Mol Ther. 2010;18:240–42. doi: 10.1038/mt.2009.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–25. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kang H, Hata A. MicroRNA regulation of smooth muscle gene expression and phenotype. Curr Opin Hematol. 2012;19:224–31. doi: 10.1097/MOH.0b013e3283523e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–42. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]