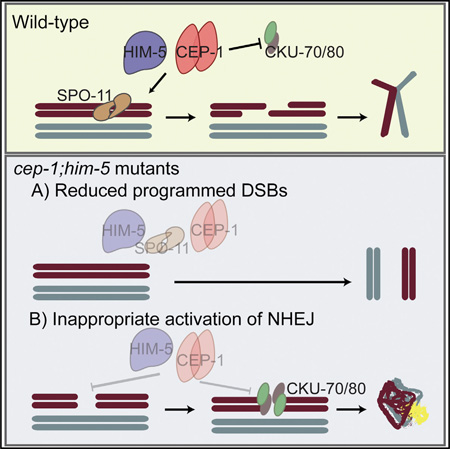
SUMMARY
The passage of genetic information during meiosis requires exceptionally high fidelity to prevent birth defects and infertility. Accurate chromosome segregation during the first meiotic division relies on the formation of crossovers between homologous chromosomes and a series of precisely controlled steps to exchange genetic information. Many studies have hinted at a role for p53 in meiosis, but how it functions in this process is poorly understood. Here, we have identified a cooperative role for the p53-like protein CEP-1 and the meiotic protein HIM-5 in maintaining genome stability in the C. elegans germline. Loss of cep-1 and him-5 results in synthetic lethality that is dependent on the upstream DNA damage checkpoint but independent of the downstream core apoptotic pathway. We show that this synthetic lethality is the result of defective crossover formation due to reduced SPO-11-dependent double-strand breaks. Using cep-1 separation-of-function alleles, we show that cep-1 and him-5 also suppress inappropriate activation of the nonhomologous end joining (NHEJ) pathway. This work reveals an ancestral function for the p53 family in ensuring the fidelity of meiosis and establishes CEP-1 as a critical determinant of repair pathway choice.
Graphical abstract
INTRODUCTION
Faithful segregation of chromosomes during the first meiotic division relies on recombination between homologous chromosomes, resulting in the generation of crossovers (COs). Crucial to this process is the induction and repair of programmed DNA double-strand breaks (DSBs), as well as the proper pairing and synapsis between the homologs. Therefore, in addition to generating genetic diversity, meiotic recombination is important for the maintenance of genome stability during gametogenesis.
Tumor suppressor p53 promotes genome stability through its well-established roles in the DNA damage response, such as transcriptional control of cell-cycle progression, apoptosis, and DNA repair [1]. In addition to protecting the genome from exogenous damage, multiple studies have demonstrated roles for the three mammalian p53 family members (p53, p63, and p73) in regulating reproductive fitness [2]. For example, successful implantation of fertilized eggs in mice is regulated by p53 through the transactivation of leukemia inhibitory factor (Lif) [3]. On the other hand, p63 protects the female germline by eliminating damaged oocytes [4], whereas p73 is essential for sperm cell adhesion and maturation of testes [5]. Furthermore, p73 prevents abnormal chromosome segregation by regulating the spindle assembly checkpoint, which senses improper mitotic spindle attachments [6]. Earlier studies on p53 have also suggested an important role in meiosis due to its interactions with meiotic recombination proteins and intermediates [7–11]. Recently, it was reported that Spo11 activity stimulates p53 activation in the reproductive tissues of Drosophila and mice [12]. Consistent with this, ablation of the sole Caenorhabditis elegans p53 family member CEP-1 causes a high incidence of males (Him) phenotype [13], a consequence of X chromosome nondisjunction. Taken together, these results point to an active role for the p53 family in meiosis, but how this function is accomplished remains undefined.
In this study, we took advantage of the powerful genetics and cell biology tools of C. elegans to understand how CEP-1 promotes meiotic fidelity. We utilized two deletion alleles of cep-1, gk138, and lg12501, both of which cause resistance to DNA-damage-induced germ cell apoptosis but have distinct chromosome nondisjunction phenotypes. The lg12501 allele is ~2% Him compared with cep-1(gk138) mutants, which do not exhibit a Him phenotype. We combined these alleles with mutations in the high incidence of males-5 gene (him-5), which encodes a novel protein required for DSB induction. We show that CEP-1 cooperates with HIM-5 to ensure CO generation by promoting the formation of programmed DSBs. The gk138 allele reveals additional roles for CEP-1 and HIM-5 in committing DNA repair to homologous recombination (HR) by actively inhibiting the error-prone nonhomologous end joining (NHEJ) pathway. This work suggests an ancient role for the p53 family of proteins in ensuring genome stability in the germline, which was later co-opted in the soma of more-complex multicellular organisms to suppress tumor formation.
RESULTS
cep-1 and him-5 Cooperate to Maintain Chromosome Stability during Meiosis
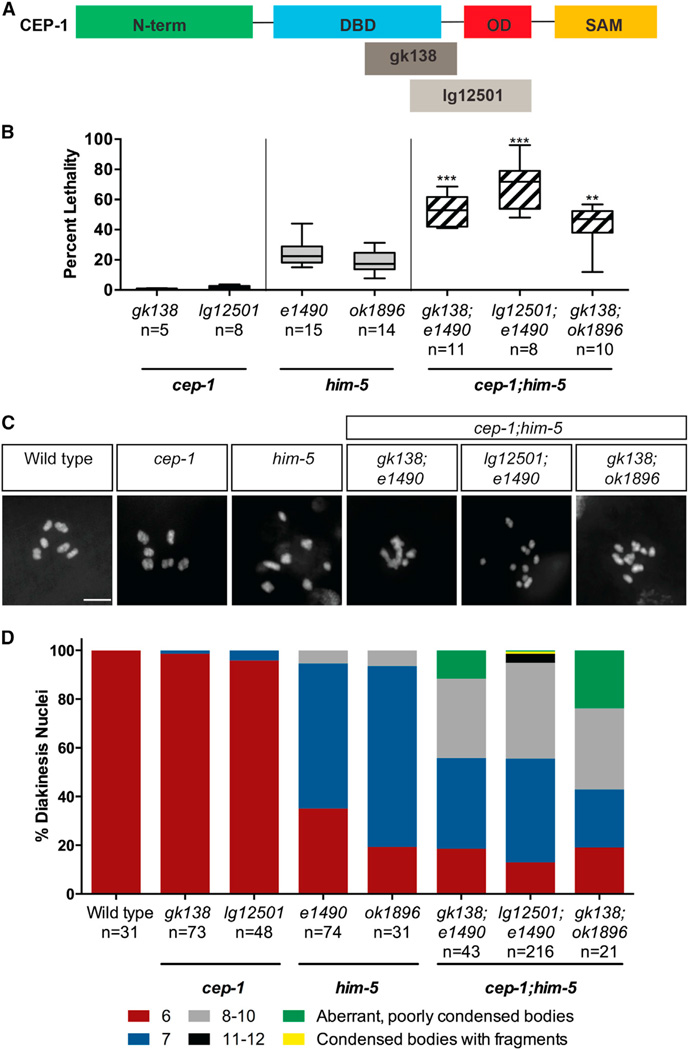
We previously observed a mild Him phenotype when cep-1 was ablated by RNAi [13] that is phenocopied by cep-1(lg12501), but not the cep-1(gk138) allele (Table S1). Because both of these alleles are completely resistant to DNA-damage-induced germline apoptosis [14, 15], this reveals a separation of function for CEP-1 in preventing X chromosome nondisjunction. The lg12501 allele contains a frameshift deletion that removes part of the DNA-binding domain (DBD) and a portion of the C terminus that includes the oligomerization domain (OD), whereas the gk138 allele is an in-frame deletion that removes part of the DBD but retains the OD and sterile alpha motif (SAM) domains (Figure 1A). To determine at which step in the meiotic program CEP-1 functions, we surveyed its genetic interactions with mutants defective in various steps of meiotic development. Our genetic analyses uncovered a synthetic lethal interaction between cep-1 and the C. elegans meiotic gene him-5. cep-1 mutants have negligible levels of embryonic lethality whereas him-5 mutants produce ~25% dead eggs and ~35% male progeny due to nondisjunction of autosomal and sex (X) chromosomes, respectively [16, 17]. Remarkably, cep-1;him-5 double mutants produce ~40%–70% dead eggs, but the frequency of males compared with him-5 single mutants remains unchanged (Figure 1B; Table S1). We wondered whether this was due to an expansion from X-biased to autosomal nondisjunction when both cep-1 and him-5 are ablated. To check this, we stained cep-1;him-5 worms with DAPI and quantified the number of DAPI-stained bodies in the most-mature diakinesis-arrested oocytes. Consistent with high levels of embryonic lethality, cep-1;him-5 mutants display an array of chromosomal abnormalities that include poorly condensed chromosomes and univalents (8–12 DAPI bodies) or a combination of both (Figures 1C, 1D, and S1). Taken together, these results indicate that cep-1 and him-5 cooperate to maintain chromosome stability during meiosis.
Figure 1. cep-1 and him-5 Cooperate to Maintain Chromosome Stability during Meiosis.
(A) Structural features of CEP-1 protein showing the N-terminal domain (N-term), DNA-binding domain (DBD), oligomerization domain (OD), and C-terminal sterile alpha motif (SAM). The gk138 allele is a 1,630-bp in-frame deletion spanning the DBD that is predicted to generate a truncated protein retaining the OD and SAM domains. The lg12501 allele is a 1,231-bp frameshift deletion that removes part of the DBD and OD.
(B) Quantification of lethality among F1 progeny of cep-1 and him-5 single mutants and cep-1;him-5 double mutants. Data are represented as mean ± SEM; n, broods counted. Statistical comparisons between him-5 single mutants and cep-1;him-5 double mutants were performed using the two-tailed Mann-Whitney test; 95% CI. ***p < 0.0001, **p = 0.002. See also Table S1.
(C) Representative confocal images of DAPI-stained diakinesis chromosomes of young adult animals (day 1 or ~24 hr post-L4 stage) of the indicated genotypes. The scale bar represents 5 µm. See also Figure S1.
(D) Quantification of DAPI-stained bodies in diakinesis oocytes of young adult animals (day 1) of the indicated genotypes. n, number of oocytes.
Synthetic Lethality Is Independent of Apoptosis but Involves the Checkpoint
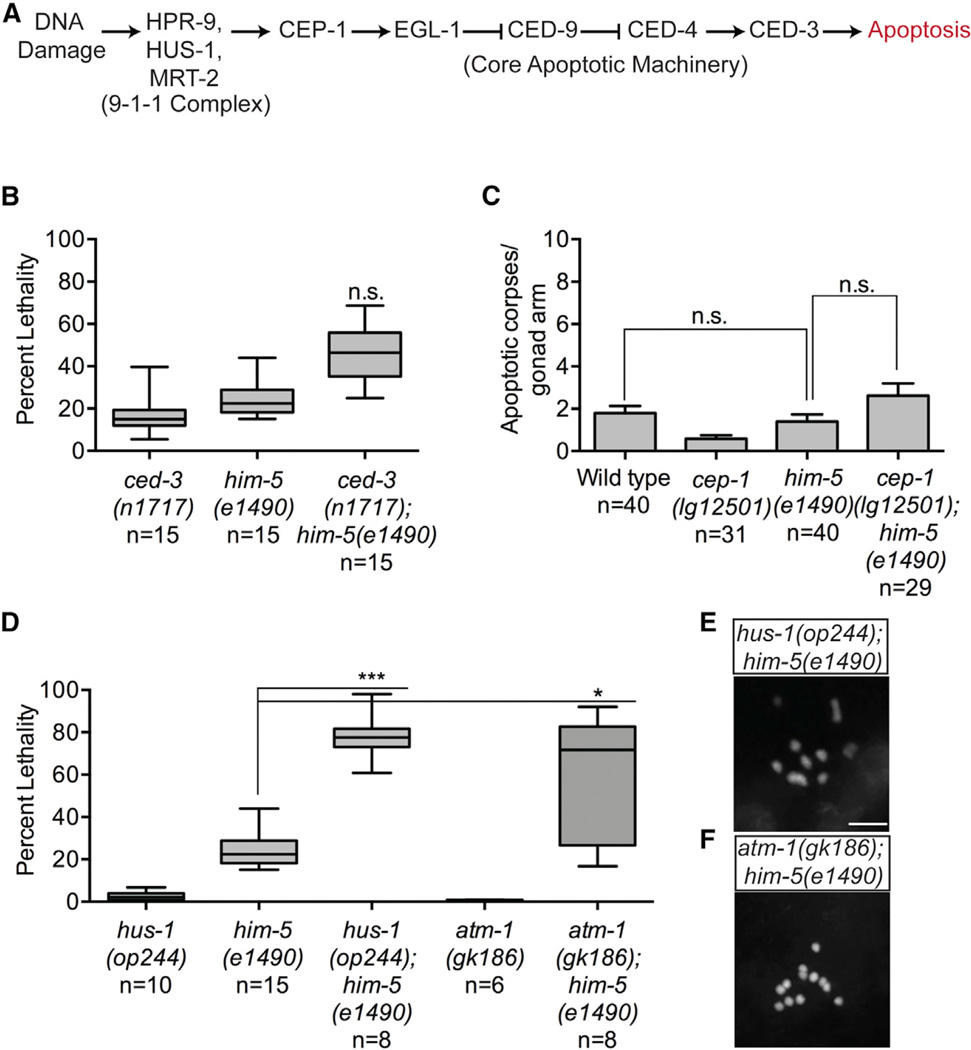
Similar to other members of the p53 family, CEP-1 has a well-characterized role in DNA-damage-induced apoptosis [13, 18]. Therefore, to determine whether the synthetic lethality observed in cep-1;him-5 mutants was due to a failure in culling defective germ cells, we tested genetic interactions between him-5 and the core apoptosis pathway, downstream of cep-1 (Figure 2A). Unlike cep-1, mutations in the caspase homolog ced-3 result in ~15% larval lethality [19]. Progeny from ced-3(n717);him-5(e1490) double mutants exhibited the expected combined levels of lethality for both single mutants (Figure 2B), in contrast to the synthetic lethality observed in cep-1;him-5 double mutants. Consistent with this, diakinesis nuclei of ced-3(n717);him-5(e1490) mutants have the expected number of condensed bivalent chromosomes as him-5 alone (Figure S2A). Furthermore, him-5 mutants do not have abnormal levels of physiological germline apoptosis (Figure 2C). Thus, cep-1 promotes chromosome stability by a mechanism that is independent of its role in apoptosis.
Figure 2. Synthetic Lethality with him-5 Is Independent of Apoptosis but Involves the DDR Checkpoint.
(A) Simplified schematic of the DNA damage checkpoint pathway in the C. elegans germline.
(B) Quantification of total lethality among F1 progeny of ced-3(n717) and him-5(e1490) single mutants and ced-3(n717);him-5(e1490) double mutants. Data are represented as mean ± SEM; n, broods counted. Statistical comparisons between combined lethality of ced-3(n717) and him-5(e1490) single mutants and ced-3(n717);him-5(e1490) double mutants were performed using the two-tailed Mann-Whitney test; 95% CI. p = 0.3508. See also Figure S2A.
(C) Quantification of apoptotic corpses in cep-1(lg12501) and him-5(e1490) single and cep-1(lg12501);him-5(e1490) double mutants under physiological conditions. Data are represented as mean ± SEM; n, germlines counted. Statistical comparisons between wild-type and him-5 single mutants were performed using the two-tailed Mann-Whitney test; 95% CI. p = 0.1704.
(D) Quantification of total lethality among F1 progeny of hus-1(op244), him-5(e1490), and atm-1(gk186) single mutants and hus-1(op244);him-5(e1490) and atm-1(gk186);him-5(e1490) double mutants. Data are represented as mean ± SEM; n, broods counted. Statistical comparisons between genotypes were performed using the two-tailed Mann-Whitney test; 95% CI. him-5(e1490) versus hus-1(op244); him-5(e1490), ***p < 0.0001;him-5(1490) versus atm-1(gk186);him-5(e1490), *p = 0.0159.
(E) Representative confocal image of DAPI-stained diakinesis chromosomes of young adult (Day 1) hus-1(op244);him-5(e1490) mutants. Scale bar represents 5 µm. See also Figure S2B.
(F) Representative confocal image of DAPI-stained diakinesis chromsomes of young adult (Day 1) atm-1(gk186);him-5(e1490) mutants. Scale bar represents 5 µm. See also Figure S2C.
The C. elegans DNA damage response (DDR) initiates with recognition of lesions by the conserved 9-1-1 checkpoint complex, which transduces damage signals through the Ataxia-telangiectasia-mutated kinase ortholog ATM-1 to activate CEP-1 [20] (Figure 2A). Through mechanisms that are poorly understood, activation of the DDR can lead to cell-cycle arrest allowing for DNA repair or, if the damage is severe, apoptotic cell death [21]. To determine whether the DDR checkpoint upstream of cep-1 cooperates with him-5 to promote genome stability, we created a hus-1(op244);him-5(e1490) double mutant strain and quantified survival of F1 progeny. The hus-1 gene is homologous to S. pombe Hus1, a component of the 9-1-1 complex [20]. Similar to cep-1, we observed a dramatic increase in lethality, from ~25% in him-5(e1490) single mutants to ~80% in hus-1(op244);him-5(e1490) double mutants (Figure 2D). In addition, these mutants had diakinesis chromosomes that resembled those in cep-1(lg12501);him-5(e1490) mutants (Figures 2E and S2B). Likewise, a loss-of-function allele in atm-1 also caused synthetic lethality and abnormal diakinesis chromosomes when combined with a him-5 mutation (Figures 2F and S2C). Collectively, these results indicate that the role of CEP-1 in meiosis is dependent on the upstream DDR checkpoint, but not the downstream apoptosis pathway.
cep-1 and him-5 Promote Crossover Formation
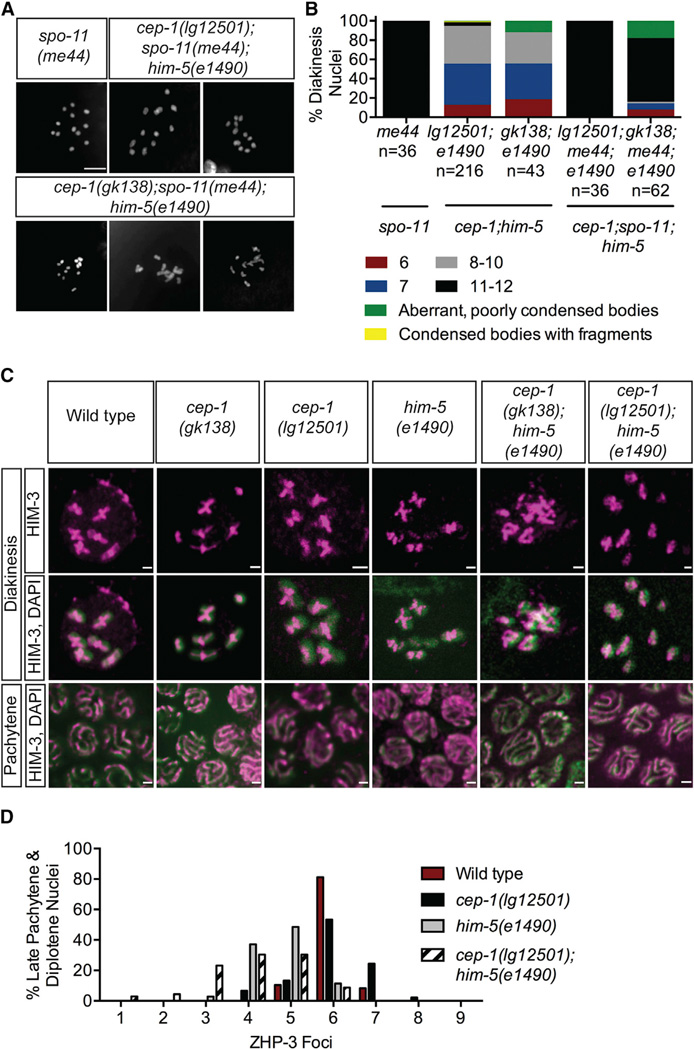
Errors during mitosis or meiosis can result in aneuploid germ cells. To determine whether the chromosomal abnormalities in cep-1;him-5 mutants were acquired during meiosis, we asked whether they were dependent upon the induction of programmed DSBs by the SPO-11 nuclease. spo-11 single mutants contain 12 univalent chromosomes because programmed DSBs are not generated, which prevents formation of chiasmata required for interhomolog association [22]. The diakinesis nuclei of all cep-1(lg12501);spo-11(me44);him-5(e1490) triple mutants examined contained 11 or 12 condensed DAPI bodies (Figures 3A and 3B), indicating that spo-11 is epistatic to cep-1(lg12501);him-5(e1490). Because SPO-11 catalyzes the initial step of meiotic recombination, these results suggest that the chromosomal defects that arise in cep-1(lg12501);him-5(e1490) are due to errors acquired during meiotic recombination. In contrast to lg12051, triple mutants with the cep-1(gk138) allele contained a substantial number of diakinesis nuclei with <11 condensed univalents (8%–16%), condensed but fragmented DAPI bodies (2%), and aberrant, poorly condensed DAPI bodies (12%–18%), in addition to 11 or 12 well-formed DAPI bodies (Figures 3A, 3B, S3A, and S3B). This suggests that a fraction of the chromosomal abnormalities observed in the cep-1(gk138) background may arise independently of SPO-11-induced breaks.
Figure 3. cep-1 and him-5 Promote Crossover Formation.
(A) Representative confocal images of DAPI-stained diakinesis chromosomes of young adult animals of the indicated genotypes. The scale bar represents 5 µm. See also Figures S3A and S3B.
(B) Quantification of DAPI-stained bodies in diakinesis oocytes of young adult animals of the indicated genotypes. n, number of oocytes. Data for cep-1;him-5 double mutants were taken from Figure 1D.
(C) Pachytene and diakinesis nuclei of wild-type (1), cep-1(gk138) (2), cep-1(lg12501) (3), him-5(e1490) (4), cep-1(gk138);him-5(e1490) (5), and cep-1(lg12501);him-5(e1490) (6) stained with antibody against the chromosome axis protein HIM-3. See also Figures S3C–S3G.
(D) Frequency distribution of ZHP-3 foci in late pachytene and diplotene nuclei of the indicated genotypes. Statistical comparisons between genotypes were performed using the two-tailed Mann-Whitney test; 95% CI. Wild-type versus him-5(e1490), p < 0.0001;him-5(e1490) versus cep-1(lg12501);him-5(e1490), p = 0.0070.
During meiotic prophase I, chromosomes reorganize to establish associations between homologous pairs. Critical to the stabilization of homologous pairing is synapsis, the localization of meiosis-specific proteins along the lengths of homologs, which form a scaffold called the synaptonemal complex (SC) [23]. Because synapsis is required for generating COs, mutations in SC components exhibit 12 univalent chromosomes instead of six bivalents [24, 25]. Because diakinesis oocytes of cep-1;him-5 mutants also contain a substantial number of univalents, we asked whether these are due to defective homolog pairing, synapsis, and/or chromosome axis establishment. To test this, germlines were dissected and stained with antibodies against HIM-3, a meiosis-specific axis protein required for synapsis; SYP-1, an integral component of the SC [24, 26]; or FISH probes for the X and V chromosomes. The establishment of pairing, synapsis, and the chromosome axis were indistinguishable from wild-type (Figures 3C, S3C, S3E, and S3F). Nonetheless, the localization of these markers on diakinesis chromosomes was abnormal in cep-1;him-5 compared to wild-type controls (Figures 3C, S3C, and S3G). As expected for HIM-3 staining, wild-type, cep-1, and him-5 single mutants contain discernible cruciform structures, one on each bivalent indicating successful chiasma formation. Strikingly, HIM-3 appears to be fused and aggregated in cep-1(gk138);him-5(e1490) mutants. On the other hand, cep-1(lg12501);him-5(e1490) had fewer HIM-3 cruciforms and more univalents with long single tracks of HIM-3, similar to those observed in spo-11 mutants [27]. Consistent with these results, both cep-1(gk138);him-5(e1490) and cep-1(lg12501);him-5(e1490) mutants displayed defects in reconfiguration of the SC at diplotene; close inspection of early diakinesis nuclei revealed DAPI bodies lacking SYP-1 staining (Figure S3G). Therefore, the chromosomal abnormalities in diakinesis oocytes of cep-1;him-5 mutants are likely due to defects in CO formation rather than synapsis failure.
The abnormal chromosome phenotypes and disorganized chiasmata structures observed in cep-1;him-5 mutants suggest a reduction, but not a complete absence, of COs. To test this, we quantified the number of ZHP-3 foci in late pachytene and diplotene nuclei of double mutants and compared it with wild-type and single mutants. ZHP-3 initially localizes along the lengths of the SC and eventually becomes restricted to foci that mark CO sites [28]. In wild-type worms, six ZHP-3 foci were observed in ~80% late pachytene/diplotene nuclei, indicating the expected single CO event per bivalent. Similarly, the majority of late pachytene/diplotene nuclei examined in cep-1(lg12501) mutants contained six ZHP-3 foci. However, there was a significant reduction in ZHP-3 foci in him-5(e1490) mutants that was reduced further in cep-1(lg12501);him-5(e1490) double mutants. Consistent with defective chiasma formation, approximately 60% of nuclei in cep-1(lg12501);him-5(e1490) mutants contained fewer than five ZHP-3 foci (Figure 3E). Taken together, the altered localization of lateral and axis SC components, and reduced number of ZHP-3 foci, reveal cooperative roles for CEP-1 and HIM-5 in CO generation.
cep-1 and him-5 Regulate Meiotic DSB Formation
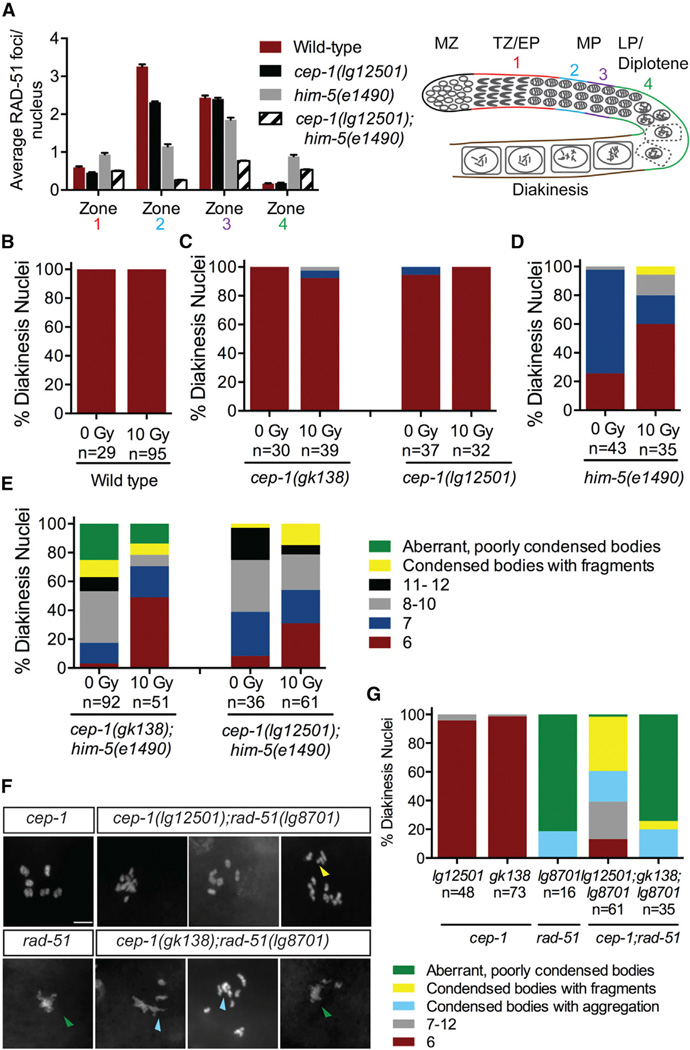
Defects in CO formation can be the result of limited DSBs generated by SPO-11. After DSBs are created by SPO-11, the DNA ends are resected to generate 3′ single-strand overhangs that are subsequently bound by the RecA-like protein RAD-51 [29]. Because HIM-5 has been shown to help promote meiotic DSB formation [30], we wondered whether CEP-1 also cooperates with HIM-5 in this process. We reasoned that the condensed univalents observed in cep-1(lg12501);him-5(e1490) mutants are due to defective programmed DSB induction, which results in a decreased number of COs. To test this, we examined RAD-51 localization and observed a significant reduction in the number of RAD-51 foci in cep-1(lg12501);him-5(e1490) double mutants compared with wild-type and single mutants (Figure 4A; Table S2). Together with the defects in bivalent formation, this indicates that CEP-1 and HIM-5 cooperate in the generation of COs at an early step of meiotic DSB formation.
Figure 4. cep-1 and him-5 Regulate Meiotic DSB Formation.
(A) Quantification of RAD-51 foci in the meiotic germlines of worms of the indicated genotypes and schematic of the C. elegans germline. Each gonad was divided into four meiotic zones (TZ/EP, transition zone/early pachytene; MP, mid-pachytene; LP/diplotene, late pachytene). Error bars represent SD. See also Table S2.
(B) Quantification of DAPI-stained bodies in diakinesis oocytes of wild-type worms 24 hr post-IR treatment. See also Figure S4A.
(C) Quantification of DAPI-stained bodies in diakinesis oocytes of cep-1(gk138) and cep-1(lg12501) single mutants 24 hr post-IR treatment. See also Figure S4B.
(D) Quantification of DAPI-stained bodies in diakinesis oocytes of him-5(e1490) single mutants 24 hr post-IR treatment. See also Figure S4C.
(E) Quantification of DAPI-stained bodies in diakinesis oocytes of cep-1(gk138);him-5(e1490) and cep-1(lg12501);him-5(e1490) double mutants 24 hr post-IR treatment. See also Figure S4D.
(F) Representative confocal images of DAPI-stained diakinesis chromosomes of adult animals of indicated genotypes. The scale bar represents 5 µm. Image for cep-1 single mutant was taken from Figure 1C. Green triangle indicates aberrant, poorly condensed chromosomes. Yellow triangle indicates chromosome fragments. Light blue triangle indicates condensed bodies with aggregations. See also Figure S1.
(G) Quantification of DAPI-stained bodies in diakinesis oocytes of young adult animals of indicated genotypes. n, number of oocytes. Data for cep-1 single mutants were taken from Figure 1C.
As shown previously, ionizing radiation (IR) can generate DSBs that bypass the requirement for SPO-11 and HIM-5 [22, 30]. Because CEP-1 helps HIM-5 promote meiotic DSB formation, we asked whether IR could restore DSB formation in cep-1;him-5 mutants. Young adult worms were exposed to a dose of IR (10 Gy) previously shown to restore chiasmata formation in spo-11 [31, 32] and him-5 single mutants [30]. Examination of diakinesis nuclei 24 hr after IR treatment dramatically increased the percentage of bivalents in cep-1;him-5 double mutants from 3% to 49% in the gk138 background and 8% to 31% in the lg12501 background (Figures 4B–4E), supporting roles for CEP-1 and HIM-5 in break induction. In agreement with this, we also observed a significant increase in survival of progeny from irradiated cep-1;him-5 mutants (Figures S4A–S4D). However, IR failed to rescue the aberrant, poorly condensed chromosomes in cep-1(gk138);him-5(e1490) mutants, suggesting additional roles for CEP-1 and HIM-5 downstream of DSB induction in repair.
Mutations in genes involved in break formation (e.g., spo-11 and rad-50) resolve the diakinesis aggregation phenotype of rad-51 mutants [33, 34]. Because we uncovered a role for CEP-1 in DSB formation, we tested whether a mutation in cep-1 will also suppress the chromosomal aggregations in rad-51 mutants. Strikingly, unlike rad-51 single mutants that have poorly condensed, aggregated chromosomes, cep-1(lg12501);rad-51(lg8701) double mutants contain condensed DAPI bodies (Figures 4F and 4G). In contrast, cep-1(gk138);rad-51(lg8701) double mutants look similar to rad-51(lg8701) single mutants that have mainly aggregated chromosomes. These results provide strong evidence supporting a key role for CEP-1 in assisting in the formation of meiotic breaks. The suppression of chromosomal fusions in rad-51 mutants by the cep-1(lg12501) allele, but not the gk138 allele, suggests an additional defect of the latter in managing repair pathway decisions.
cep-1 and him-5 Prevent Inappropriate NHEJ Activity
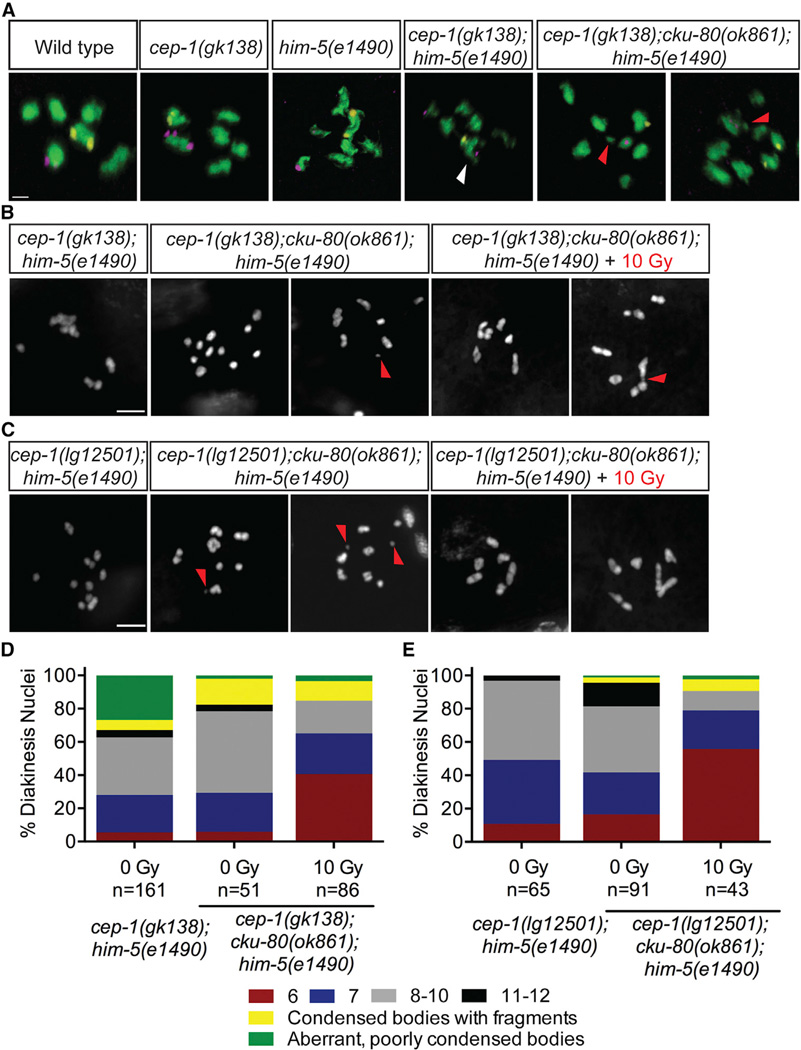
Because IR-induced breaks failed to suppress the aggregation in cep-1(gk138);him-5(e1490) double mutants, we hypothesized that these aberrant, poorly condensed chromosomes might arise from inappropriate associations between nonhomologous chromosomes. To test this, we performed fluorescence in situ hybridization (FISH) using probes labeling two different chromosomes (V and X), as described previously [35]. Normally, these probes label separate DAPI bodies (chromosomes), whereas a single DAPI body containing both probes would suggest a physical association between chromosomes V and X. As expected, we only observed distinct DAPI bodies labeling for each probe in the diakinesis nuclei of wild-type, cep-1, and him-5 single mutants. However, probes for chromosomes V and X were frequently observed in the same DAPI bodies of cep-1(gk138); him-5(e1490) double mutants, indicative of nonhomologous chromosome associations (Figure 5A; Table S3; Movie S1). In agreement with our cytological analyses using DAPI, probes for chromosome V were often located in separate DAPI bodies in the same nucleus, indicative of univalent and/or fragmented chromosomes. Therefore, the combined loss of CEP-1 and HIM-5 causes massive genomic instability in the germline that manifests as condensed univalents, fragmented chromosomes, and inappropriate engagements between nonhomologous chromosomes. Importantly, chromosomal aggregations are only observed in the gk138 allele background, revealing an unexpected role for CEP-1 in preventing nonhomologous chromosome associations.
Figure 5. cep-1 and him-5 Prevent Inappropriate NHEJ Activity.
(A) Diakinesis nuclei of day 4 animals of the indicated genotypes labeled with FISH probes to chromosomes V (yellow) and X (magenta). DNA is shown in green. White arrow indicates a single DAPI body containing both FISH probes, demonstrating a fusion between nonhomologous chromosomes. Red triangle indicates chromosome fragments. White triangle indicates a single DAPI body containing probes for chromosomes V and X. The scale bar represents 1 µm. See also Table S3.
(B) Representative confocal images of DAPI-stained diakinesis chromosomes of cep-1(gk138);cku-80(ok1861);him-5(e1490) triple mutants 24 hr after mock (0 Gy) or IR (10 Gy) treatment. Image of cep-1(gk138);him-5(e1490) mutant is shown for comparison. Scale bar represents 5 µm. See also Figures S5A–S5C.
(C) Representative confocal images of DAPI-stained diakinesis chromosomes of cep-1(lg12501);cku-80(ok1861);him-5(e1490) triple mutants 24 hr after mock (0 Gy) or IR (10 Gy) treatment. Image of cep-1(lg12501);him-5(e1490) mutant is shown for comparison. Scale bar represents 5 µm. See also Figures S5A–S5C.
(D) Quantification of DAPI-stained bodies in diakinesis oocytes of cep-1(gk138);him-5(e1490) double and cep-1(gk138);cku-80(ok861);him-5(e1490) triple mutants 24 hr after mock (0 Gy) or IR (10 Gy) treatment.
(E) Quantification of DAPI-stained bodies in diakinesis oocytes of cep-1(lg12501);him-5(e1490) double and cep-1(lg12501);cku-80(ok861);him-5(e1490) triple mutants 24 hr after mock (0 Gy) or IR (10 Gy) treatment.
In C. elegans, mutations in HR components such as rad-51 (Rad51), brc-2 (Brca2), and com-1 (CtIP) severely compromise the ability of germ cells to form chiasmata. This results in aggregated chromosomes that arise from inappropriate activation of NHEJ, which can also cause chromosome translocations and deletions [36, 37]. Because cep-1(gk138);him-5(e1490) double mutants have aberrant chromosomes that resemble these mutants, we asked whether these were the consequences of deleterious NHEJ activity. To test this, we inhibited NHEJ by introducing a cku-80 (Ku80) mutation into cep-1;him-5 mutants and assessed diakinesis chromosome morphology. Ku80 forms a heterodimer with Ku70 and functions at an early step of NHEJ by binding DNA ends to prevent their resection, which is critical for generating HR intermediates [38, 39]. Introducing a cku-80 mutation resolved the chromosomal aggregations in cep-1(gk138);him-5(e1490) mutants but increased the frequency of condensed univalent and fragmented chromosomes (Figures 5A, 5B, and 5D; Table S3). Because chiasma formation is not restored by removing cku-80, there is only a modest, albeit significant, increase in the fraction of surviving progeny of cep-1(gk138);cku-80(ok861);him-5(e1490) triple mutants compared to cep-1(gk138);him-5(e1490) double mutants (Figure S5A). Similarly, introducing a mutation in the NHEJ ligase lig-4 [38] into cep-1(gk138);him-5(e1490) mutant also rescued chromosome aggregation to univalent morphology (Figure S5B). In contrast to the results with the cep-1(gk138) allele, introducing a cku-80 mutation into cep-1(lg12501);him-5(e1490) mutants failed to increase progeny survival or rescue chromosome defects (Figures S5A, 5C, and 5E). This is expected because the cep-1(lg12501) allele causes predominantly univalent chromosomes in combination with the him-5 allele, rather than aggregated chromosomes observed with the cep-1(gk138) allele. Collectively, these results establish cooperative roles for CEP-1 and HIM-5 in suppressing deleterious NHEJ activity.
cep-1 and him-5 Promote DSB Repair
Because the frequency of chromosomal aggregations of cep-1(gk138);him-5(e1490) double mutants is similar to cep-1(gk138);spo-11(me44);him-5(e1490) triple mutants, we hypothesized that they arise from DSBs repaired by NHEJ (Figure 3B). Therefore, we created a cep-1(gk138);cku-80(ok861);spo-11(me44);him-5(e1490) quadruple mutant and observed a suppression of these defects (Figures S6C and S6G). This indicates that, in the absence of cep-1 and him-5, SPO-11-independent breaks are inappropriately repaired by NHEJ.
Because IR rescued the univalents (but not aggregated chromosomes) in cep-1;him-5 mutants, and introducing a cku-80 (or lig-4) mutation rescued aggregation but resulted in univalents, we next asked whether treating cep-1(gk138);cku-80(ok861); him-5(e1490) triple mutants with IR would rescue the univalents to bivalent morphology. Indeed, IR treatment partially restored bivalent chromosomes in these triple mutants (Figures 5B and 5D). This was also observed in cep-1(gk138);lig-4(tm750);him-5(e1490) triple mutants (Figures S5B and S5C), confirming a cooperative role for CEP-1 and HIM-5 in creating DSBs. We observed that a substantial number of diakinesis nuclei in both triple mutants contained fragmented chromosomes after irradiation (Figures 5B–5E, S5B, and S5C), indicative of unrepaired DSBs. Therefore, introducing exogenous DSBs and inhibiting NHEJ is not sufficient to rescue all the chromosomal defects of cep-1;him-5 mutants. This strongly suggests that CEP-1 and HIM-5 are also required to promote meiotic DSB repair after assisting in the generation of programmed DSBs. In agreement with this, IR also failed to restore bivalents in cep-1(gk138); spo-11;him-5 triple mutants and cep-1(gk138);cku-80(ok861); spo-11(me44);him-5(e1490) quadruple mutants to the same levels observed in spo-11 single and cep-1;spo-11 or spo-11;him-5 double mutants (Figures S6A–S6C and S6E–S6G). Therefore, CEP-1 and HIM-5 function to ensure chromosome stability at multiple steps of the meiotic program.
In summary, taking advantage of cep-1 separation-of-function alleles, we show that cep-1 cooperates with him-5 to ensure CO formation by regulating fundamental and essential steps of meiotic recombination. Specifically, cep-1 and him-5 promote the induction of SPO-11-dependent DSBs and ensure their accurate repair. We show that these roles of CEP-1 require the upstream DDR checkpoint, but not the downstream apoptosis pathway. Furthermore, we uncover additional roles for cep-1 and him-5 in inhibiting erroneous NHEJ-dependent repair of DSBs that arise independent of SPO-11.
DISCUSSION
Numerous studies support the importance of tumor suppressor p53 in the regulation of stress responses, but very little is known about its normal physiological function. Multiple lines of evidence suggest that the tumor-suppressive function of the p53 family has been derived from primordial activities. In support of this, p53-like proteins have been identified in simple organisms such as amoeba and unicellular protists. Therefore, it is unlikely that the original function of this family of proteins is to suppress tumor formation. Furthermore, late-onset diseases, such as cancer, are likely not a source of selective pressure for the evolution of the p53 family because the human lifespan has been fairly short until recently [40].
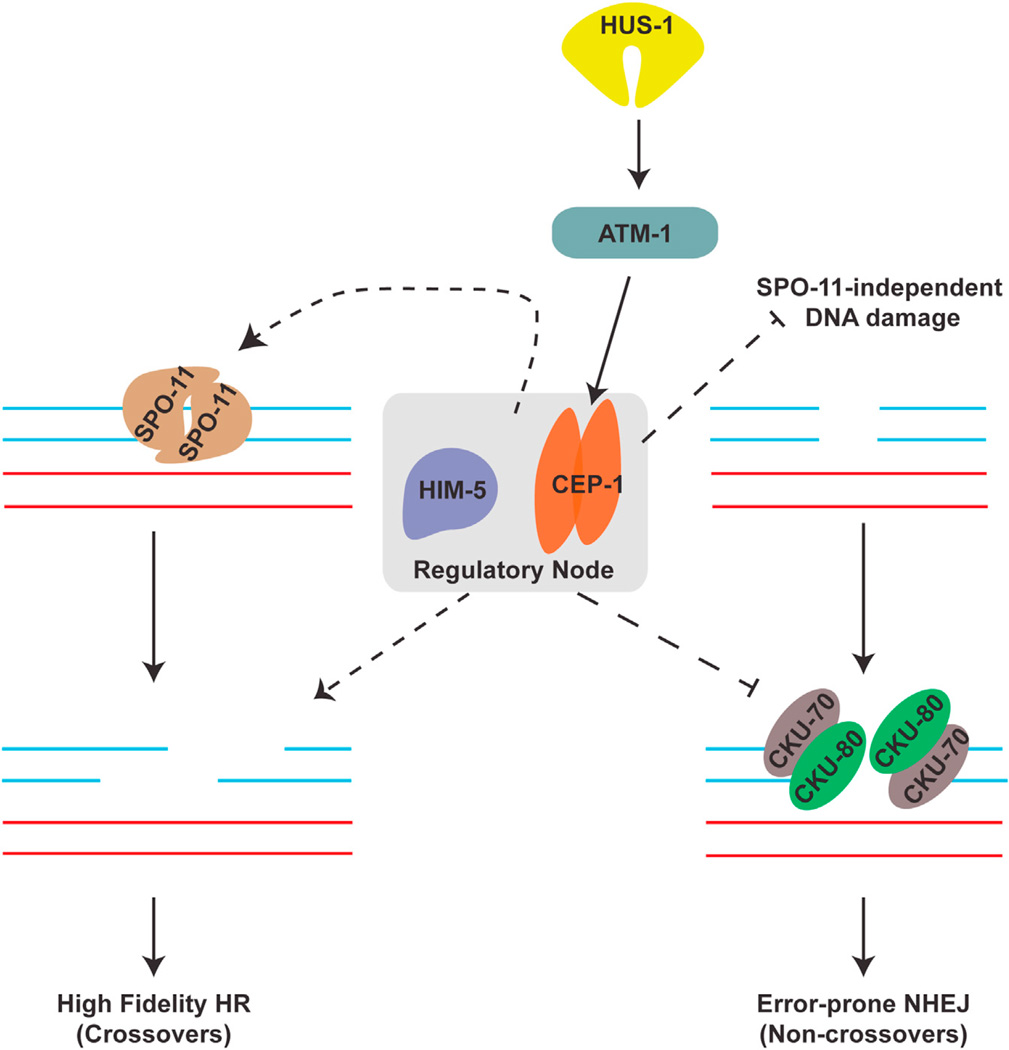
In this study, we describe cooperative roles for the p53-like protein CEP-1 and the meiotic protein HIM-5 in promoting programmed DSBs and suppression of error-prone NHEJ. We propose that CEP-1 and HIM-5 form a critical regulatory node that monitors distinct, but intimately linked, steps of meiotic recombination to maintain genome stability (Figure 6).
Figure 6. cep-1 and him-5 Cooperate to Maintain Genome Stability during Meiosis.
Schematic highlighting the regulatory node consisting of HIM-5 and CEP-1, which regulates SPO-11-dependent break formation and ensures fidelity of repair by suppressing the error-prone NHEJ pathway. The dashed lines represent novel roles described in this study.
CEP-1 Separation of Function
Our discovery of multiple roles for CEP-1 in meiosis was facilitated by the identification of cep-1 separation-of-function alleles. The striking difference between the chromosome morphology in cep-1(lg12501);him-5(e1490) versus cep-1(gk138); him-5(e1490) mutants indicates that specific activities of CEP-1 are perturbed whereas others are retained in an allele-dependent manner. For example, both cep-1 alleles are completely resistant to DNA-damage-induced germ cell apoptosis because the proteins are incapable of transcriptionally activating the pro-apoptotic egl-1 gene [15, 20]. However, only the lg12501 allele exhibited a mild Him phenotype that was similar to cep-1 depletion by RNAi [13]. This hinted at additional roles for CEP-1 in meiotic chromosome segregation, which was greatly enhanced when combined with him-5 mutations. In combination with him-5 mutants, both cep-1 alleles exhibit defects in programmed DSB formation. However, only the gk138 allele, which does not have a Him phenotype alone, resulted in chromosome aggregations that arise as a result of defective repair pathway choice.
CEP-1 Links Break Formation with Repair
It is well appreciated that meiotic DSB formation is a complex and tightly coordinated process involving functional subgroups of proteins. Although the Spo11 nuclease initiates meiotic DSB formation, its recruitment and enzymatic activity requires accessory proteins as well as a number of other factors essential for the proper distribution of DSBs across the chromosomal landscape. For example, in S. cerevisiae, at least nine accessory proteins function as co-factors of Spo11 [41]. In C. elegans, the chromatin factors HIM-5 [30], HIM-17 [42], and XND-1 [35], and the recently identified paralogs DSB-1 [32] and DSB-2 [31], all contribute to meiotic DSB formation. In particular, HIM-5 is crucial for DSB formation on the X chromosome and, to a lesser degree, on the autosomes. On the other hand, CEP-1 has a minor role in preventing X chromosome nondisjunction as indicated by the mild Him phenotype observed in cep-1(lg12501) mutants, or by ablation of cep-1 using RNAi [13]. Strikingly, combined mutations in cep-1 and him-5 result in DSB formation defects on all chromosomes that can be partially rescued by IR, indicating cooperative roles for CEP-1 and HIM-5 in regulating SPO-11-dependent break formation across the chromosomal landscape.
In addition to Spo11 accessory proteins, certain histone modifications also influence meiotic DSB induction. In C. elegans, chromatin factors XND-1 and HIM-17 promote DSB formation, in part through the regulation of chromatin modifications [35, 42–44]. Likewise, work in S. cerevisiae and S. pombe demonstrated that histone modifications are important for meiotic DSB formation, further demonstrating that DSB induction is intimately linked to chromatin organization [44, 45]. Interestingly, in addition to its well-characterized transcription-dependent roles, p53 also indirectly promotes chromatin remodeling during nucleotide excision repair by recruiting the histone acetyltransferase p300 [46]. We considered the possibility that CEP-1 may alter chromatin structure to influence DSB repair because both the gk138 and lg12501 alleles are predicted to produce truncated proteins that are transcriptionally dead. However, after surveying global histone acetylation in cep-1(lg12501);him-5(e1490) mutants, we did not observe any obvious defects. We also performed immunohistochemical analysis of several chromatin marks, including H2AK5Ac, H3K4me2, H3K56Ac, H3K9me3, and H2BK5Ac, but found no differences in these histone modifications (data not shown). Although it is possible that other chromatin modifications that impact break formation and repair pathway choice are altered in cep-1;him-5 mutants, we suspect this occurs by a different mechanism.
The presence of chromosome fragments and the incomplete restoration of bivalents in cep-1;him-5 mutants after introducing DSBs by IR hinted at roles for CEP-1 and HIM-5 in repair, downstream of break formation. Programmed induction of DSBs and meiosis-specific DNA repair are essential for the formation of chiasmata and accurate segregation of chromosomes. One of the challenges during prophase I is to ensure that these steps are properly executed such that homologous chromosomes are linked by at least one CO prior to meiosis I division. Therefore, it is not surprising that proteins, such as CEP-1 and HIM-5, form functional nodes that couple critical steps of the recombination pathway that reinforce fidelity of DNA repair, leading to genome stability. Having proteins that coordinate DSB formation with repair during meiosis ensures that breaks are only induced if they can be accurately repaired through mechanisms that would lead to the generation of COs. In support of this, work in different organisms has demonstrated dual roles for Mre11 in meiotic DSB formation as well as DNA end processing [47].
cep-1 and him-5 Inhibit NHEJ Repair
How do CEP-1 and HIM-5 ensure genome stability? Notably, the cep-1(gk138) separation-of-function allele reveals a crucial role for CEP-1 in antagonizing NHEJ. Unlike the cep-1(lg12501) allele, when cep-1(gk138) is combined with him-5 alleles, chromosomal aggregations are observed. Interestingly, these aggregations are present even in the absence of programmed DSBs (Figures 3A, 3B, and S6). We show that these aberrant, poorly condensed chromosomes are dependent on the NHEJ pathway, indicating crucial roles for CEP-1 and HIM-5 in suppressing erroneous NHEJ activity (Figures S6C and S6G).
There are several ways CEP-1 and HIM-5 could function to prevent inappropriate NHEJ activity in the absence of programmed DSBs. Recent work showed that germ cells with mitotic errors progress into meiosis and are eventually culled in a CEP-1-dependent manner [48]. We considered the possibility that mitotic damage occurring in worms lacking cep-1 and him-5 is propagated into the meiotic stages. Because CEP-1 is required for culling damaged cells by apoptosis, it is likely that the damage persists and becomes inappropriately repaired by the NHEJ machinery. However, because we did not observe similar defects when him-5 was co-ablated with the caspase gene ced-3, it is unlikely that inhibition of cell death alone causes these defects. Alternatively, germ cells harboring mitotic errors may be repaired by the NHEJ machinery before they enter meiosis. Consistent with this, we did not observe a striking difference in the number of RAD-51 foci in the mitotic region of cep-1(gk138);him-5(e1490) double and cep-1(gk138);spo-11(me44);him-5(e1490) triple mutants in comparison to wild-type worms (data not shown). This is not surprising because commitment to NHEJ repair occurs prior to resection and RAD-51 loading.
Given that the gk138 and lg12501 alleles are predicted to generate truncated proteins that are not able to activate transcription of its target genes, CEP-1 may serve as a scaffold to recruit factors that inhibit NHEJ activity. Recent studies in different model systems showed that DNA end resection commits DSB repair to HR; therefore, CEP-1 may antagonize NHEJ by associating with components of the end resection machinery such as MRE-11 and COM-1 [37, 49, 50]. Alternatively, CEP-1 may influence other factors that affect the enrichment of Ku70/Ku80 to break sites. Identification of these CEP-1-interacting proteins should help resolve the biochemical mechanisms of this process.
Ancient Role for the p53 Family in Meiosis
A link between p53 and meiotic recombination has been proposed on the basis of observations made in different organisms. For example, high levels of p53 were observed in mice and rat testes, and its absence results in giant multinucleated testicular cells, presumably due to failed meiotic division [51]. p53 also binds Holliday junctions [11], and recent work in Drosophila and mice showed that p53 is activated in response to Spo11- induced breaks [12]. Although, collectively, these observations suggest that p53 has a role in meiotic development, a functional role has yet to be ascribed. Our work advances these observations by providing a mechanistic framework in which the p53-like protein CEP-1 regulates specific steps of the meiotic program. Whether the meiotic functions we have uncovered for CEP-1 in C. elegans are retained by the p53 family of proteins in other organisms warrants further investigation. Moreover, the presence of p53 paralogs p63 and p73 may confer additional layers of complexity to this regulation in vertebrates [52]. Our findings raise the possibility that maintenance of genome stability during meiosis may have laid the foundations for shaping the p53 tumor suppressor network that ensures genome stability and prevents cancer in dividing somatic and stem cells.
EXPERIMENTAL PROCEDURES
C. elegans Genetics
Strains used in this study are listed in the Supplemental Experimental Procedures. All strains were maintained at 20°C under standard conditions. Experiments were performed at 20°C.
Immunofluorescence and Imaging
Immunofluorescence was performed as described previously [30], with minor modifications. Primary antibodies used were HIM-3 (a kind gift from M. Zetka), RAD-51 (Novus), ZHP-3 (a kind gift from V. Jantsch), and SYP-1 (a kind gift from A. Villeneuve). Images were acquired using the Olympus IX81 Quorum Spinning Disk confocal or the Nikon A1R confocal microscope. z stacks were analyzed using the Volocity Image analysis software (PerkinElmer).
Fluorescence In Situ Hybridization
FISH was performed using probes to XR and 5S as described previously [35].
Highlights.
CEP-1 cooperates with HIM-5 to ensure meiotic crossover formation
CEP-1 and HIM-5 promote induction of programmed double-strand breaks
CEP-1 and HIM-5 ensure DSB repair fidelity by antagonizing NHEJ activity
In Brief.
Mateo et al. report a key role for the C. elegans p53 family member CEP-1 in coordinating meiotic double-strand break formation with suppression of error-prone repair by the NHEJ pathway. This ensures high fidelity of repair during meiosis and may represent the ancestral function from which p53 evolved its tumor-suppressive properties.
Acknowledgments
W.B.D. was supported by the Canadian Institutes for Health Research (MOP-137089) and a Pitblado small grant from the Garron Family Cancer Centre, Hospital for Sick Children. J.L.Y. was supported by NIH grant R01 GM104007 and Magee-Womens Research Institute startup funds. A.-R.F.M. was supported by a doctoral research award from the Canadian Institutes for Health Research and Fanconi Canada. Strains were provided by the National Bioresource Project (Japan) and the Caenorhabditis Genetics Center, funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Antibodies were kindly provided by Dr. Monique Zetka (HIM-3), Dr. Verena Jantsch (ZHP-3), and Dr. Anne Villeneuve (SYP-1).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, three tables, Supplemental Experimental Procedures, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2016.03.036.
AUTHOR CONTRIBUTIONS
A.-R.F.M. performed most of the experiments. W.B.D., J.L.Y., A.K.J., Z.K., O.M., B.Y., and A.N. performed experiments. A.-R.F.M., J.L.Y., and W.B.D. wrote the manuscript.
REFERENCES
- 1.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat. Rev. Mol. Cell Biol. 2011;12:259–265. doi: 10.1038/nrm3086. [DOI] [PubMed] [Google Scholar]
- 3.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 4.Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- 5.Holembowski L, Kramer D, Riedel D, Sordella R, Nemajerova A, Dobbelstein M, Moll UM. TAp73 is essential for germ cell adhesion and maturation in testis. J. Cell Biol. 2014;204:1173–1190. doi: 10.1083/jcb.201306066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, Tsao MS, Iovanna JL, Jurisicova A, Melino G, Mak TW. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc. Natl. Acad. Sci. USA. 2009;106:797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stürzbecher HW, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 8.Linke SP, Sengupta S, Khabie N, Jeffries BA, Buchhop S, Miska S, Henning W, Pedeux R, Wang XW, Hofseth LJ, et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003;63:2596–2605. [PubMed] [Google Scholar]
- 9.Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p53-dependent gene expression. Proc. Natl. Acad. Sci. USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl. Acad. Sci. USA. 1998;95:13869–13874. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Cavallo L, Griffith J. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J. Biol. Chem. 1997;272:7532–7539. doi: 10.1074/jbc.272.11.7532. [DOI] [PubMed] [Google Scholar]
- 12.Lu WJ, Chapo J, Roig I, Abrams JM. Meiotic recombination provokes functional activation of the p53 regulatory network. Science. 2010;328:1278–1281. doi: 10.1126/science.1185640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 14.Deng X, Hofmann ER, Villanueva A, Hobert O, Capodieci P, Veach DR, Yin X, Campodonico L, Glekas A, Cordon-Cardo C, et al. Caenorhabditis elegans ABL-1 antagonizes p53-mediated germline apoptosis after ionizing irradiation. Nat. Genet. 2004;36:906–912. doi: 10.1038/ng1396. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode CAENORHABDITIS ELEGANS. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broverman SA, Meneely PM. Meiotic mutants that cause a polar decrease in recombination on the X chromosome in Caenorhabditis elegans. Genetics. 1994;136:119–127. doi: 10.1093/genetics/136.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 19.Xue D, Shaham S, Horvitz HR. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 1996;10:1073–1083. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, Vidal M, Hengartner MO. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 21.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 22.Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 23.Bhalla N, Dernburg AF. Prelude to a division. Annu. Rev. Cell Dev. Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 25.Smolikov S, Eizinger A, Schild-Prufert K, Hurlburt A, McDonald K, Engebrecht J, Villeneuve AM, Colaiácovo MP. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics. 2007;176:2015–2025. doi: 10.1534/genetics.107.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zetka MC, Kawasaki I, Strome S, Müller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabeshima K, Villeneuve AM, Colaiácovo MP. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 2005;168:683–689. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 30.Meneely PM, McGovern OL, Heinis FI, Yanowitz JL. Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics. 2012;190:1251–1266. doi: 10.1534/genetics.111.137463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, Dernburg AF, Villeneuve AM. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 2013;9:e1003674. doi: 10.1371/journal.pgen.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 2013;9:e1003679. doi: 10.1371/journal.pgen.1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, La Volpe A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–479. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner CR, Kuervers L, Baillie DL, Yanowitz JL. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature. 2010;467:839–843. doi: 10.1038/nature09429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemmens BB, Johnson NM, Tijsterman M. COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet. 2013;9:e1003276. doi: 10.1371/journal.pgen.1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clejan I, Boerckel J, Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nat. Rev. Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 41.Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2014;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc. Natl. Acad. Sci. USA. 2004;101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, de La Roche Saint-André C. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–1967. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J. 2003;22:975–986. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 48.Stevens D, Oegema K, Desai A. Meiotic double-strand breaks uncover and protect against mitotic errors in the C. elegans germline. Curr. Biol. 2013;23:2400–2406. doi: 10.1016/j.cub.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin Y, Smolikove S. Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol. Cell. Biol. 2013;33:2732–2747. doi: 10.1128/MCB.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 51.Rotter V, Schwartz D, Almon E, Goldfinger N, Kapon A, Meshorer A, Donehower LA, Levine AJ. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc. Natl. Acad. Sci. USA. 1993;90:9075–9079. doi: 10.1073/pnas.90.19.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gersten KM, Kemp CJ. Normal meiotic recombination in p53-deficient mice. Nat. Genet. 1997;17:378–379. doi: 10.1038/ng1297-378. [DOI] [PubMed] [Google Scholar]