Abstract
Glucocorticoid signaling through the glucocorticoid receptor (GR) plays essential roles in the response to stress and in energy metabolism. This hormonal action is integrated to the transcriptional control of GR-target genes in a cell type-specific and condition-dependent manner. In the present study, we found that the GR regulates the angiopoietin-like 4 gene (ANGPTL4) in a CCCTC-binding factor (CTCF)-mediated chromatin context in the human hepatic HepG2 cells. There are at least four CTCF-enriched sites and two GR-binding sites within the ANGPTL4 locus. Among them, the major CTCF-enriched site is positioned near the ANGPTL4 enhancer that binds GR. We showed that CTCF is required for induction and subsequent silencing of ANGPTL4 expression in response to dexamethasone (Dex) and that transcription is diminished after long-term treatment with Dex. Although the ANGPTL4 locus maintains a stable higher-order chromatin conformation in the presence and absence of Dex, the Dex-bound GR activated transcription of ANGPTL4 but not that of the neighboring three genes through interactions among the ANGPTL4 enhancer, promoter, and CTCF sites. These results reveal that liganded GR spatiotemporally controls ANGPTL4 transcription in a chromosomal context.
Introduction
The glucocorticoid receptor (GR) is a member of a family of transcription factors that regulate biological processes, such as basal and stress-associated homeostasis, energy metabolism, and the immune response in a cell-type and condition-dependent manner [1, 2]. In the absence of ligand, GR is present in the cytoplasm in a complex with chaperons such as heat-shock proteins. Upon ligand-induced activation, GR dissociates from the complex and translocates to the nucleus, typically by binding to the glucocorticoid response elements (GREs) to activate or repress transcription of target genes. After the gene control, GR dissociates from its ligand or is degraded [2].
Although evidence indicates that the GR regulates gene expression through binding to promoter regions [3–5], recent genome-wide studies reveal that the GR mainly binds to distal enhancer regions [6] to regulate target-gene activity through long-range interactions between the promoter and enhancer [7–9]. The GR target Lcn2 encoding the acute-phase protein lipocalin-2 is co-regulated through long-range interactions with Ciz1 located approximately 30-kb upstream from the Lcn2 GRE [7]. Further, genomic interaction profiling revealed that numerous GREs interact with the Lcn2 GRE before GR binding [10]. Moreover, the contact loci were enriched in DNase I-hypersensitive sites, including the consensus motif for the CCCTC-binding factor (CTCF). FKBP51, which encodes the GR chaperon FKBP51, is regulated by GR-responsive enhancers far from the transcription start site [9]. FKBP51 and these enhancers are bordered by CTCF-binding sites, which likely contribute to long-range interactions and loop formation. However, whether CTCF contributes to the regulation of GR target genes remains to be determined.
The angiopoietin-like 4 gene (ANGPTL4) is a primary target of GR and is dominantly expressed in the liver and adipose tissue [11–16]. ANGPTL4 is a secreted protein that inhibits extracellular lipoprotein lipase (LPL), which hydrolyzes triglycerides (TGs) to free fatty acids and glycerol [17–19]. Plasma TG levels are reduced in ANGPTL4-null mice and are increased in mice that overexpress ANGPTL4 in the liver [20]. ANGPTL4-null mice exhibit reductions in glucocorticoid excess-induced changes associated with hypertriglyceridemia, hepatic steatosis, and visceral obesity [15]. These findings establish the role of ANGPTL4 in GR-mediated lipid metabolism. However, the mechanism of the regulation of ANGPTL4 by CTCF as well as GR has not been investigated.
Higher-order chromosome conformations, such as chromatin looping, mediate long-range physical interactions between distal regulatory elements and their target genes [21]. The chromatin insulator is a genomic boundary element that controls enhancer activity and the formation of chromatin loops [22]. CTCF is an insulator-binding protein that cooperates with the cohesin complex and other chromatin proteins [23–27]. Genome-wide studies show that CTCF binds several tens of thousands of sites in the mammalian genome. Approximately 50% of the CTCF-binding sites reside within intergenic regions, and the others are present near promoters and within gene bodies [28, 29]. Moreover, CTCF-binding sites are frequently located on the border between transcriptionally active and repressed genes and between different histone modification domains [30]. Although most CTCF-binding sites are conserved among tissues, some of the cell and tissue type-specific sites overlap with transcriptional enhancers, suggesting that CTCF adjusts the interaction between the enhancer element and promoter [29]. For example, CTCF/cohesin-mediated higher-order chromatin mediates basal and inducible gene expression [31–34].
Here, we report that GR regulates ANGPTL4 in a CTCF-mediated chromatin context in the human hepatic carcinoma cell line HepG2. We identified one GR-binding enhancer site adjacent to a liver-specific CTCF-enriched site. Moreover, in HepG2 cells treated with dexamethasone (Dex), CTCF was required for induction and subsequent silencing of ANGPTL4, likely via an enhancer–promoter interaction. Therefore, liganded GR can target ANGPTL4 but not three neighboring genes. Further, induction of ANGPTL4 transcription was decreased by long-term glucocorticoid treatment. These results indicate that liganded GR spatiotemporally controls the transcription of ANGPTL4 depending on the chromosomal context.
Materials and Methods
Cell Culture
Human hepatocellular carcinoma HepG2 cells were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). HepG2 cells were cultured in DMEM (Sigma-Aldrich) supplemented 10% (v/v) fetal bovine serum (FBS). The cells were treated with 100 nM Dex (Sigma-Aldrich) after the cells were cultured for 24 to 48 h in DMEM (Sigma-Aldrich) containing 10% (v/v) dextran-coated charcoal (DCC)-treated FBS.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was isolated from cultured cells with TRIzol (Invitrogen). For cDNA synthesis, 500 ng of total RNA was reverse-transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s instructions. We used an ABI Prism 7300 (Applied Biosystems) and SYBR Green to perform qRT-PCR. Each experiment was performed at least three times. Expression levels were normalized to those of 36B4 mRNA encoding ribosomal protein, large, P0. Primer sequences are listed in S1 Table.
Gene Knockdown
GL3 (control) and CTCF-specific siRNAs were used as previously reported [31]. The RAB11B-AS-siRNA sequence is as follows: 5´-GACCAAAUAACUAAUGAGA(dT)(dT)-3´and 5´-UCUCAUUAGUUAUUUGGUC(dU)(dG)-3´. Cells were transfected for 48 h with siRNAs in the presence of Lipofectamine RNAiMAX (Invitrogen).
Chromatin Immunoprecipitation Sequencing (ChIP-Seq)
HepG2 cells (2 × 107) were treated with Dex (100 nM) for 3 h and then cross-linked with 1% formaldehyde at room temperature for 10 min. The cells were lysed with the RIPA buffer supplemented with protease inhibitors. The lysates were sonicated using a Branson Bioruptor to yield 200–500 bp DNA fragments. ChIP was performed using anti-glucocorticoid receptor (GR) antibodies (sc-8992; Santa Cruz Biotechnology, Inc.). DNA fragments were collected using Dynabeads Protein A/G (Life Technologies). Purified DNA (20 ng) was used to construct libraries for sequencing using an Ion Fragment Library Kit (Life Technologies). High-throughput sequencing was performed using Proton semiconductor sequencers (Life Technologies) according to the manufacturer's instructions. Proton sequence data were aligned to the human reference genome hg19. Peaks were detected using the MACS algorithm included with Avadis NGS software (Agilent Technologies). GR binding sites were detected according to a cut-off value of P = 10−5 and their enrichment by a factor of 10 vs input. ChIP-seq data for CTCF and histone modifications in HepG2 cells were obtained from the ENCODE/Broad Institute via the UCSC Genome Browser website (http://genome.ucsc.edu/). Gene Expression Omnibus (GEO) accession numbers are as follows: GSM733645, CTCF; GSM733743, H3K27Ac; GSM798321, H3K4me1; and GSM733737, H3K4me3.
ChIP-qPCR Analysis
HepG2 cells were cross-linked with 1% formaldehyde at room temperature for 10 min. Nuclear lysates were sonicated using the Bioruptor to yield DNA fragments of approximately 200–500 bp. ChIP was performed using the antibodies as follows: anti-CTCF (07–729, Millipore), anti-GR (sc-8992, Santa Cruz Biotechnology, Inc.), anti-H3K27ac (ab4729, Abcam), anti-RNA Pol II serine-5 phosphorylation (provided by Dr. Hiroshi Kimura). Rabbit IgG (sc-2027, Santa Cruz Biotechnology, Inc.) served as a control. Purified DNA was used in quantitative PCR analyses performed using an ABI Prism 7300 System (Applied Biosystems) and SYBR Green. The threshold was set to intersect the linear segment of the PCR amplification curve, and the number of cycles (Ct) required to reach the threshold was counted. We generated a standard curve using the input sample and calculated the enrichment by applying the Ct value of each sample. Primer sequences are listed in S1 Table. ChIP analysis of RNA Pol II serine-5 phosphorylation included the addition of a phosphatase inhibitor to the reaction.
Chromosome Conformation Capture (3C) Assay
Formaldehyde cross-linked chromatin from HepG2 cells was digested with DpnII overnight, followed by ligation catalyzed by T4 DNA ligase at 16°C for 1 h. To prepare standard curves, a bacterial artificial chromosome spanning the ANGPTL4 locus, RPCI11.C-978J4, was digested with Sau3AI, which is insensitive to Dam methylation, followed by random ligation. After reversing the cross-links, genomic DNA was purified using phenol extraction and ethanol precipitation. We used an ABI Prism 7300 (Applied Biosystems) and a Thunderbird SYBR qPCR Mix (Toyobo) to assess the ligated products. The 3C-qPCR data were normalized to the value of the loading control, using internal primers located within ANGPTL4. The relative frequencies of interactions between the reference and its physically adjacent site were normalized to 1. We used the Student t test to evaluate the results of more than three independent experiments. Primer sequences are listed in S1 Table.
Statistical Analysis
Differences between two groups were analyzed using the Student t test. To compare three groups, we performed ANOVA followed by Tukey-Kramer post hoc test. P < 0.05 indicates a statistically significant difference.
Data Availability
ChIP-seq datasets of GR are deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE85343.
Results
Distribution of CTCF and GR-enriched Sites within the ANGPTL4 Locus
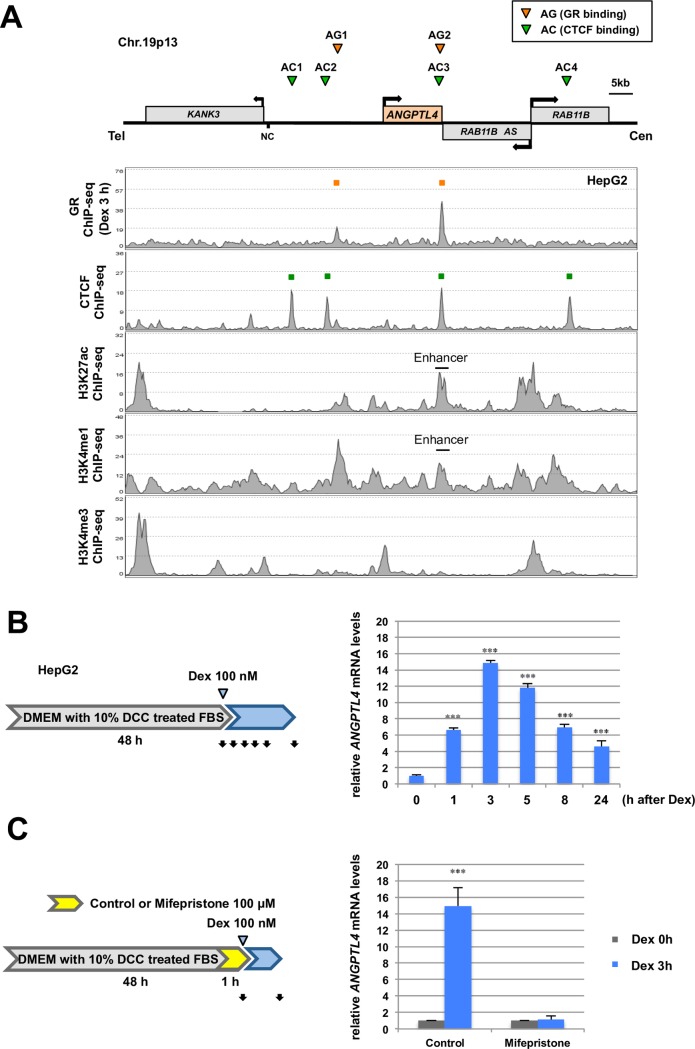
The ANGPTL4 locus is located on human chromosome 19p13 where KANK3, ANGPTL4, RAB11B, and RAB11B AS are clustered within an approximately 80-kb region. Among them, ANGPTL is a direct target of GR signaling [35]. To examine the chromatin context within this locus, we used publicly available ChIP-Seq data to conduct a survey of genome-wide CTCF-enriched sites in cell lines derived from different tissues. There are at least four CTCF-enriched sites in the ANGPTL4 locus in HepG2 cells (Fig 1A), which contain a CTCF-binding consensus motif (S1A Fig). We designated AC1 to AC4. The AC3 site is unique to HepG2 cells (S2 Fig), suggesting that AC3 plays a cell-type-specific role in regulating ANGPTL4 transcription.
Fig 1. Distribution of glucocorticoid receptor and CTCF in human ANGPTL4 gene locus.
(A) Enrichment of the glucocorticoid receptor (GR), CTCF, and modified histone H3 in the ANGPTL4 locus of HepG2 cells. KANK3, ANGPTL4, RAB11B-AS, and RAB11B are located across an approximately 80-kb region. The arrow at the transcription start site of each gene indicates the direction of transcription. According to publically available data and our ChIP-Seq results, two GR-binding sites (designated AG1 and AG2) and four CTCF-enriched sites (designated AC1–AC4) are indicated in orange and green, respectively. NC, negative control. Modifications of histone H3, such as acetylation and methylation, are shown. AG2/AC3 sites are close to each other, and AG2 is an enhancer that has been demonstrated in rat cells [15]. (B) Induction of ANGPTL4 transcription by dexamethasone (Dex). HepG2 cells were grown in DMEM medium supplemented with 10% dextran-coated charcoal (DCC)-treated FBS and were treated with Dex (100 nM). Black arrows show the sampling times of the assays. (C) ANGPTL4 as a direct GR target in HepG2 cells. The GR antagonist mifepristone was added to the medium (100 μM for 1 h) before Dex treatment. The relative expression level is indicated as a value normalized to the level of 36B4 mRNA. Asterisks indicate statistically significance between control (Dex 0 h) and Dex-treated cells at each time point. ***P < 0.005.
We performed ChIP-Seq analysis of HepG2 cells treated with 100 nM Dex for 3 h to identify genome-wide GR binding sites (Fig 1A), and identified the GR-enriched sites (AG1 and AG2) in the ANGPTL4 locus, which possess GREs (S1A Fig). ChIP-Seq data indicated that the AG1 and AG2 sites were marked with acetylated lysine residue 27 and mono-methylated lysine residue 4 of histone H3 (H3K27Ac and H3K4me1, respectively), suggesting that they are potential enhancers. These findings are consistent with the evolutionary conservation of the AG2 sequence among rats, mice, and humans, which exhibits GR-responsive enhancer activity in rat H4IIE cells [15]. Further, AG2 and AC3 reside within an approximately 100-bp contiguous region (S1B Fig), suggesting that these sites cooperate in the GR-dependent expression of ANGPTL4.
Dexamethasone Induces ANGPTL4 Expression
We next tested the expression levels of ANGPTL4 in HepG2 cells treated with Dex. Before Dex addition, the cells were grown in DMEM medium supplemented with 10% DCC-treated FBS for the indicated times to minimize the effects of basal hormone levels. The level of ANGPTL4 mRNA immediately increased by a factor of 15 at 3 h after Dex addition and decreased from 3 to 24 h (Fig 1B). In contrast, Dex treatment did not affect the levels of the three neighboring genes (S3A Fig), indicating that ANGPTL4 is a specific GR target. Western blot analysis showed that ANGPTL4 protein was maximally induced at 3 h after Dex addition and decreased from 3 to 24 h (S3B Fig).
To determine whether ANGPTL4 was directly regulated by liganded GR, we analyzed HepG2 cells treated with the GR antagonist mifepristone (100 μM for 1 h) before Dex treatment. Mifepristone completely inhibited ANGPTL4 expression 3 h after Dex addition (Fig 1C), indicating that ANGPTL is the only GR target in this chromosomal region.
Dexamethasone Treatment Alters Chromatin Status at the ANGPTL4 Locus
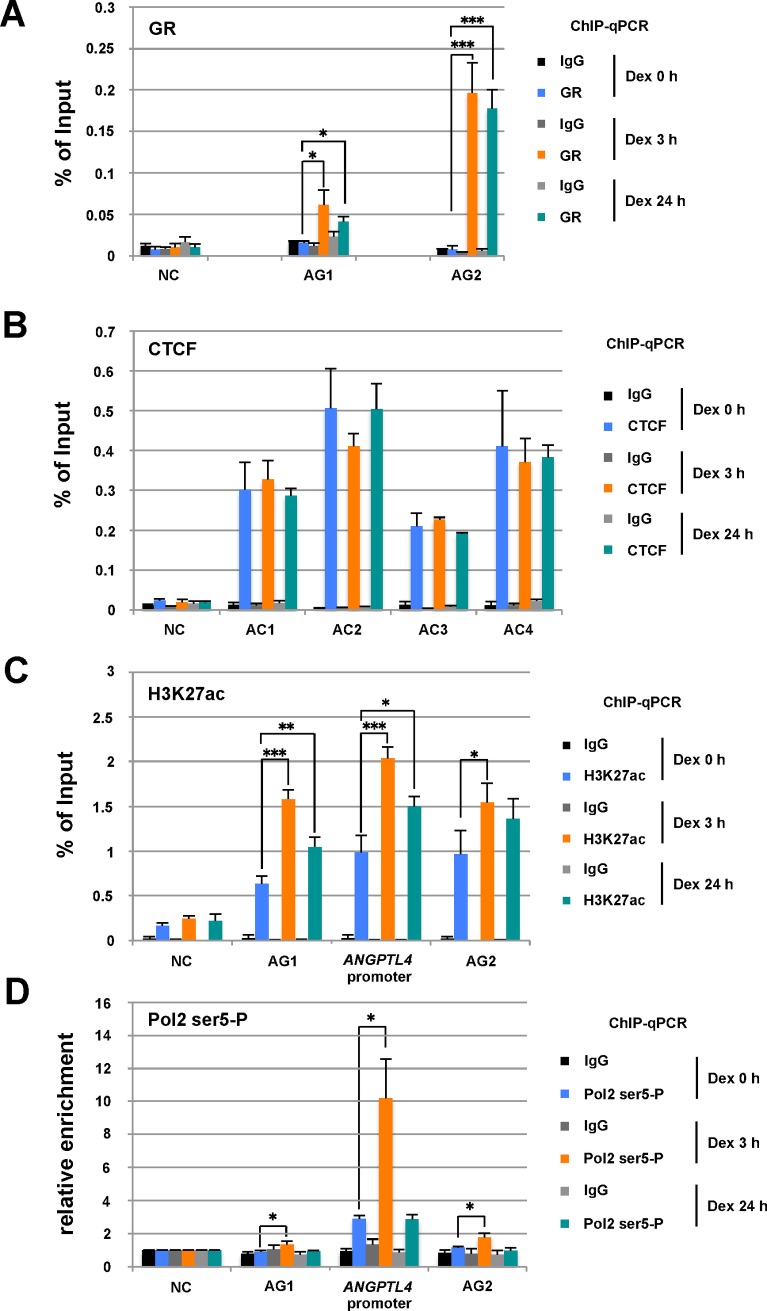
We performed ChIP-qPCR analyses to assess the effect of Dex on the chromatin status of the ANGPTL4 locus (Fig 2). We first examined the levels of enrichment of GR at AG1 and AG2. GR was not detected at either AG site under basal conditions (Dex 0 h), while GR bound these AG sites 3 h after Dex addition (Dex 3 h) (Fig 2A). GR was similarly retained 24 h after Dex treatment (Dex 24 h), although the levels of ANGPTL4 mRNA decreased at this time (Fig 1B). In contrast, CTCF-enrichment did not significantly change at AC1 to AC4 after Dex treatment (Fig 2B), suggesting that GR does not compete with CTCF for DNA binding at the AC3 and AG2 sites, despite their proximity.
Fig 2. Enrichment of GR, CTCF, acetyl-H3K27, and RNA polymerase II at the ANGPTL4 gene locus in cells treated with Dex.
(A) Enrichment of glucocorticoid receptor (GR) in cells treated with Dex. As shown in Fig 1B, HepG2 cells were treated with Dex for 24 h. ChIP-qPCR analysis was performed using an anti-GR antibody and an anti-rabbit IgG (control), followed by quantitative PCR using specific primers for each AG site and the control (NC). (B–D) Enrichment of CTCF, acetyl-H3K27 (H3K27ac), and active RNA polymerase II (Pol2 ser5-P) in cells treated with Dex. ChIP-qPCR analyses were performed using an anti-CTCF antibody and an anti-rabbit IgG (control) (B), anti-H3K27ac (C), and anti-Pol2 ser5-P (D), followed by quantitative PCR using specific primers for each indicated site. Relative enrichment of the control (NC) site was normalized to 1 (D). Asterisks indicate statistically significance between control (Dex 0 h) and Dex-treated cells at each time point. *P < 0.05, **P < 0.01, ***P < 0.005.
We next determined the enrichment of the active chromatin mark H3K27Ac in the ANGPTL4 promoter and enhancer elements AG1 and AG2 (Fig 2C). After Dex treatment for 3 h (Dex 3 h), the H3K27Ac mark increased by a factor of approximately 2 at these sites compared with the basal condition and was detected 24 h after Dex addition. Further, we determined the enrichment of RNA polymerase II (Pol II) at the ANGPTL4 promoter and enhancers (Fig 2D). Active Pol II was enriched at the ANGPTL4 promoter by approximately a factor of 4 at 3 h after Dex treatment and then decreased to the basal level at 24 h. In contrast, enrichment of Pol II enrichment at AG1 and AG2 was moderately detected 3 h after Dex addition, indicating that liganded GR bound to distal enhancers and activated transcription mediated by the ANGPTL4 promoter.
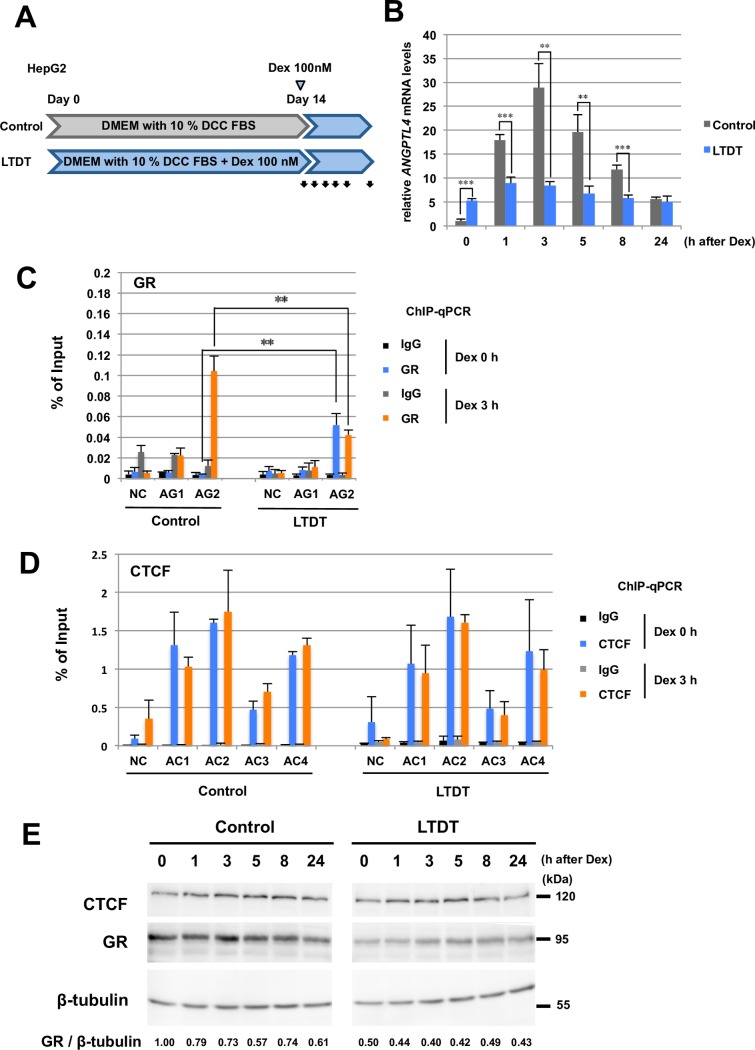
Long-term Dexamethasone Treatment Attenuates ANGPTL4 Induction and Down-regulates GR Expression
An excess of endogenous or exogenous glucocorticoid causes hypertriglyceridemia, insulin resistance, fatty liver, hepatic steatosis, and obesity as well as Cushing’s syndrome [36]. These changes may be associated, in part, with ANGPTL4 levels. Therefore, we examined whether Long-Term Dex Treatment (shortly LTDT; 100 nM for 14 days) altered ANGPTL4 expression (Fig 3A). Control HepG2 cells were cultured in DMEM medium containing 10% DCC-treated FBS. In control cells, ANGPTL4 expression increased by approximately 30-fold 3 h after Dex addition and then gradually decreased at 24 h (Fig 3B). ANGPTL4 induction in response to Dex treatment was significantly diminished in the LTDT cells (maximally 1.7-fold at 1 h). The basal level of ANGPTL4 expression in LTDT cells was higher by a factor of approximately 5, compared with that of the control cells.
Fig 3. Long-term dexamethasone treatment inhibits the induction of ANGPTL4 transcription, together with down-regulation of the GR.
(A) Protocol for Long-Term Dexamethasone Treatment (LTDT). For LTDT, HepG2 cells were initially cultured in DMEM medium supplemented with 10% DCC-treated FBS and 100 nM dexamethasone (Dex) for 14 days. Black arrows show sampling times. (B) Decrease in ANGPTL4 induction in LTDT cells. (C) Decreased enrichment of GR after Dex treatment of LTDT cells. ChIP-qPCR analysis was performed using an anti-GR antibody and an anti-rabbit IgG (control), followed by quantitative PCR using specific primers for each AG site and the control (NC). (D) CTCF enrichment in control and LTDT cells. ChIP-qPCR analysis was performed using an anti-CTCF antibody and anti-rabbit IgG (control), followed by quantitative PCR using specific primers for each AC site. (E) Expression of CTCF and GR after Dex treatment of control and LTDT cells. The amount of GR decreased in most LTDT cells. The relative level of GR normalized to that of β-tubulin is shown below. Uncropped image of western blot analysis is shown in S6 Fig. Asterisks indicate statistically significance between control and LTDT cells at each time point. **P < 0.01, ***P < 0.005.
We next examined GR enrichment at AG sites in the control and LTDT cells (Fig 3C). GR enrichment at AG2 differed significantly between these conditions. GR did not bind AG2 under basal conditions (Dex 0 h) in control cells and was significantly enriched by Dex treatment (Dex 3 h). In contrast, moderate GR binding at AG2 in LTDT cells was detected initially (Dex 0 h), and the levels were approximately 50% of those of the Dex-treated control cells and did not increase after Dex addition (Dex 3 h). Thus, newly added Dex reduced ANGPTL4 induction in LTDT cells and did not increase GR binding to AG2.
We next checked whether CTCF was involved in the reduction of GR enrichment at AG2 in LTDT cells (Fig 3D). ChIP-qPCR analysis using anti-CTCF antibodies revealed that the amounts of CTCF at the AC sites were comparable in the control and LTDT cells. Moreover, we did not detect significant changes in GRα (encoding GR) and CTCF mRNAs in control and LTDT cells (S4A Fig).
To clarify the mechanism of reduced GR binding to AG2 in LTDT cells, we performed western blot analysis of whole-cell lysates and found that the amount of GR significantly decreased in LTDT cells (Fig 3E). Further, we detected a decrease in the level of GR in nuclear extracts prepared from LTDT cells (S4B Fig), suggesting that a negative feedback mechanism regulates GR levels in LTDT cells. Together, these results suggest that GR targeted ANGPTL4 and the interaction was affected by long-term exposure of cells to excess concentrations of Dex.
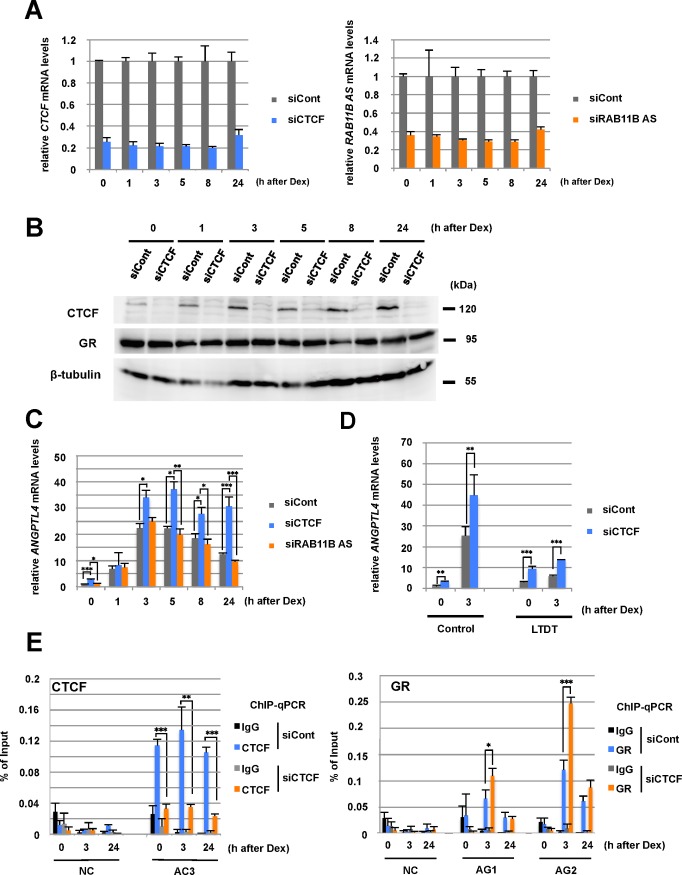
Loss of CTCF Deregulates ANGPTL4 Expression Induced by DEX
To determine the role of CTCF in the regulation of ANGPTL4 transcription, we performed RNA interference-mediated knockdown of CTCF expression in HepG2 cells. We conducted qRT-PCR and western blot analyses to confirm the reduction in the levels of CTCF mRNA and protein, respectively (Fig 4A, left and 4B). In addition, the depletion of CTCF did not affect GR expression at the protein level (Fig 4B and S5A Fig). When the CTCF knockdown cells were treated with Dex, ANGPTL4 mRNA levels significantly increased 3 h after Dex addition, and induction was maintained for 24 h (Fig 4C). In contrast, ANGPTL4 expression increased 3 h after treatment and then decreased in control cells. Similarly, the increased level of ANGPTL4 mRNA in the CTCF-knockdown cells was detected in LTDT cells (Fig 4D). Moreover, we confirmed that the expression of another GR-target gene FKBP5 was hardly affected by CTCF knockdown (S5A Fig).
Fig 4. The role of CTCF in regulating ANGPLT4 transcription.
(A) qRT-PCR analysis of HepG2 cells transfected with the siRNAs (siCont, siCTCF, and siRAB11B AS) for 48 h and then treated with Dex (100 nM). Expression levels were normalized to those of 36B4 transcripts. (B) Western blot analysis of CTCF and GR expression in siRNA-transfected cells. Asterisks indicate statistically significance among siRNA-transfected cells at each time point. (C) qRT-PCR analysis of ANGPTL4 mRNA expression in siRNA-transfected HepG2 cells treated with Dex (see Fig 4A). (D) qRT-PCR analysis of ANGPTL4 mRNA expression in siRNA-transfected LTDT cells treated with Dex. Expression levels were normalized to those of 36B4 transcripts. (E) Enrichment of CTCF at AC3 and GR at AG sites in siRNA-transfected cells. ChIP-qPCR analysis was performed using anti-CTCF, anti-GR, and anti-rabbit IgG (control) antibodies, followed by quantitative PCR using primers specific for each site. Asterisks indicate statistically significance between control and CTCF-knockdown cells at each time point. *P < 0.05, **P < 0.01, ***P < 0.005.
Several lines of evidence suggest that antisense transcripts influence sense-strand transcription [37]. Further, it has been reported that CTCF has large RNA interactions in mammalian cells [38, 39]. In the ANGPTL4 locus, the direction of RAB11B-AS transcription is opposite of that of ANGPTL4 and RAB11B (Fig 1A and S5B Fig) and extends to the AC3/AG2 sites in the 3´-region of ANGPTL4 gene (S5C Fig). To test whether the RAB11B-AS transcripts influenced ANGPTL4 expression, we inhibited the expression of RAB11B-AS in HepG2 cells (Fig 4A, right) and found that Dex-induced ANGPTL4 expression was unchanged (Fig 4C).
We next performed ChIP-qPCR analysis of CTCF-knockdown HepG2 cells to determine whether depletion of CTCF affected GR binding to the AG1 and AG2 sites. (Fig 4E). We detected a significant reduction of CTCF enrichment at AC3, and the loss of CTCF caused an increase in GR binding to AG1 and AG2 by a factor of approximately 2 at 3 h after Dex treatment. These data suggest that CTCF negatively regulated the induction of ANGPTL4 transcription by reducing the amount of liganded GR bound to AG sites. Further, at 24 h after Dex addition, GR enrichment at the AG sites was equivalent in the control and CTCF-knockdown cells, although ANGPTL4 was expressed at a high level when CTCF was depleted (Fig 4C). Moreover, the decrease in the level of ANGPTL4 mRNA coexisted with a high level of GR binding to the AG sites 24 h after Dex addition (Figs 1B and 2A). Together, these results suggest that Dex-induced ANGPTL4 expression was regulated by the CTCF-mediated chromatin context as well as by GR binding to the enhancers. We also observed that the expression level of nearby genes of ANGPTL4 was partly increased by CTCF knockdown (S5D Fig), suggesting that the activity of the ANGPTL4 enhancer may spread to nearby genes in the CTCF-knockdown cells. Thus, CTCF was required for proper induction and subsequent silencing of ANGPTL4 in cells treated with Dex.
Dexamethasone-induced Chromatin Conformation Changes in the ANGPTL4 locus
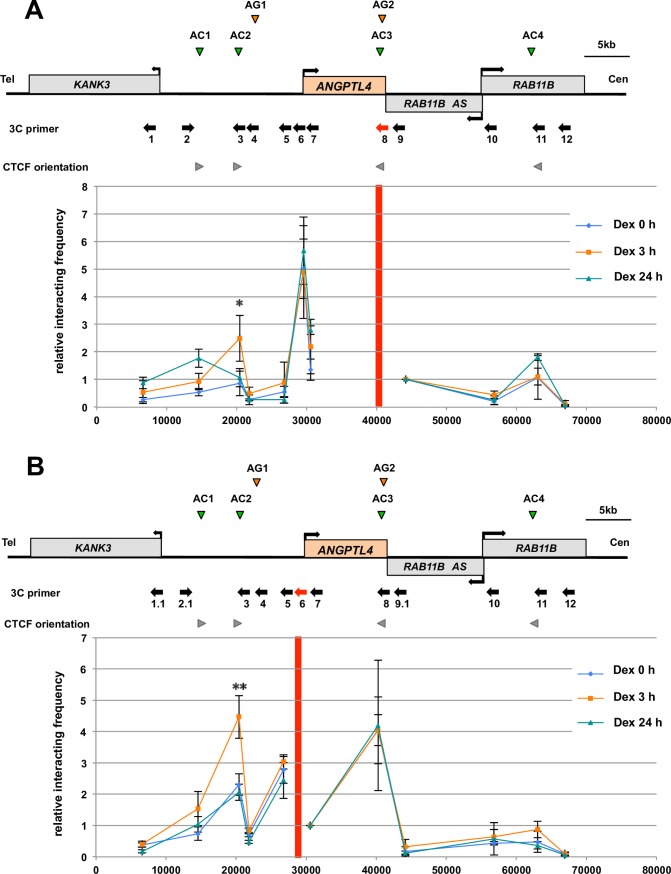
To investigate higher-order chromatin regulation in the ANGPTL4 locus, we performed the chromosome conformation capture (3C) assay of Dex-treated HepG2 cells. The experiments were performed under the conditions as follows: basal condition (Dex 0 h), Dex treatment for 3 h (Dex 3 h) and 24 h (Dex 24 h). There are at least four CTCF-enriched sites (AC1–AC4) and two GR-binding enhancer sites (AG1 and AG2) in the ANGPTL4 locus (Figs 1A and 5). DpnII was used to convert these sites and the ANGPTL4 promoter into distinct fragments, except for the composite AC3/AG2 site. Using quantitative PCR analyses of the intramolecular ligation products, we first tested the interaction frequency of AC3/AG2 as a reference, which is the counterpart of a previously demonstrated enhancer element [15], with the other 11 DpnII fragments. The interaction frequency between AC3/AG2 and the ANGPTL4 promoter was consistently high under basal and Dex conditions (Fig 5A). The interaction between AC3/AG2 and AC2 increased by a factor of 2.5 at 3 h after Dex addition and then decreased to the basal level at 24 h. Dex treatment induced a significant dynamic change at AC2, because the interaction frequencies of AC3/AG2 with the other sites were largely constant.
Fig 5. Specific changes in higher-order chromatin conformation of the ANGPTL4 locus in cells treated with dexamethasone.
(A) Chromosome conformation capture (3C) assays were performed using of DpnII-digested fragments containing each AC/AG site and the ANGPTL4 promoter. AC3/AG2 sites reside in the same fragment, and AG2 is the ANGPTL4 enhancer [15]. The relative interaction frequencies of the reference AC3/AG2 fragment (indicated with red) with other DpnII fragments were determined using qPCR analysis of at least three distinct samples from HepG2 cells treated with Dex. Gray arrowheads indicate the orientation of CTCF-binding sites. (B) The relative interaction frequencies of the reference ANGPTL4 promoter (indicated with red) with other DpnII fragments in Dex-treated cells. PCR amplification using internal primers derived from the ANGPTL4 locus was used as a loading control to normalize the amount of DNA fragments. The efficiencies of DpnII digestions and subsequent ligations were determined at each restriction site. The relative frequencies of interactions between the reference and its closest site in the control state (Dex 0 h) were normalized to 1. Asterisks indicate statistically significance between control (Dex 0 h) and Dex-treated cells (Dex 3h). *P < 0.05, **P < 0.01.
To further evaluate the higher-order chromatin conformation in this locus, we performed the 3C assay using the ANGPTL4 promoter as a reference (Fig 5B). The interaction frequency of the ANGPTL4 promoter with AC2 significantly increased 3 h after Dex addition and then decreased to the basal level at 24 h, similar to case of the reference AC3/AG2 (Fig 5A). The interaction of the ANGPTL4 promoter with other fragments was unchanged during Dex treatment, and the ANGPTL4 promoter possibly interacted with the AC3/AG2 sites. We repeated the 3C assay of CTCF-knockdown and LTDT cells, but the data were not reproducible because of the higher-order chromatin instability of the ANGPTL4 locus under these conditions (data not shown). Together, our results suggest that 1) the ANGPTL4 locus maintains the stable higher-order chromatin conformation under basal conditions and Dex treatment and 2) ANGPTL4 is selectively activated by liganded GR via the associations among the enhancer, promoter, and CTCF sites.
Discussion
In the present study, we show that GR regulated ANGPTL4 in a CTCF-mediated chromatin context in the human hepatic carcinoma cell line HepG2. Liganded GR selectively targeted ANGPTL4 but not the three neighboring genes via interactions among the enhancer, promoter, and CTCF sites. In CTCF-depleted cells, ANGPTL4 transcription was more inducible by Dex and was persistently up-regulated for 24 h, indicating that CTCF was required for proper induction and subsequent silencing of transcription. Further, long-term Dex treatment subsequently diminished the induction of ANGPTL4 transcription. Our results therefore identified the mechanism of the regulation of ANGPTL4 transcription by CTCF as well as GR.
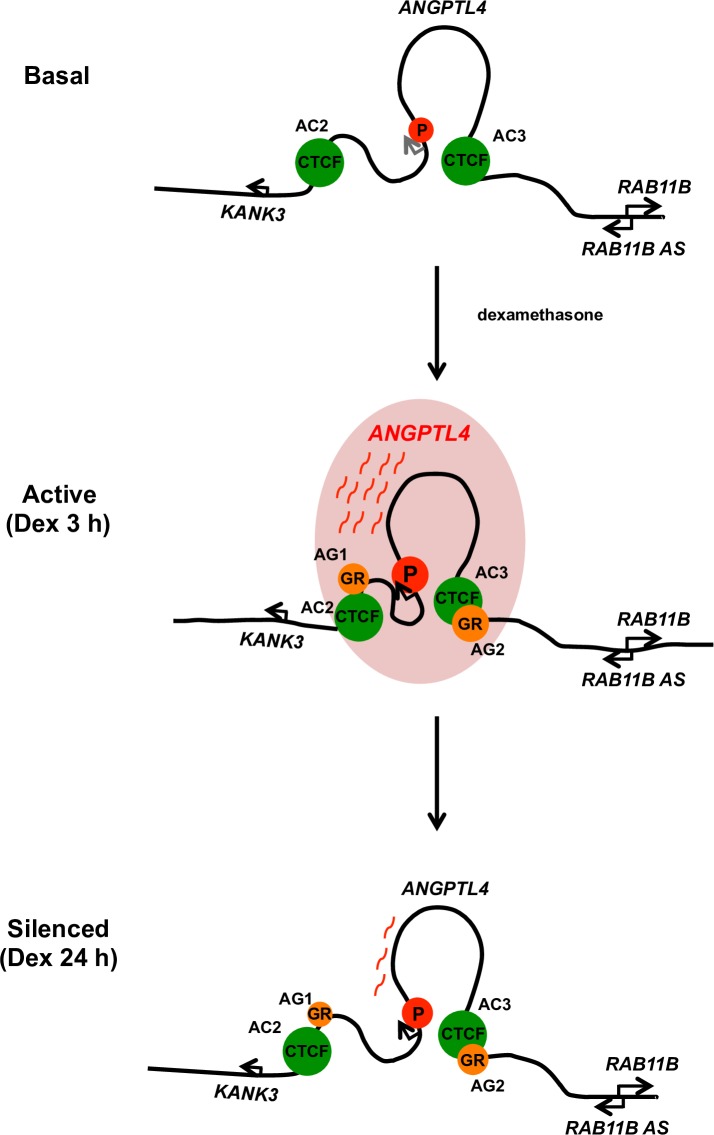
Fig 6 shows a model of the regulation of ANGPTL4 transcription and the conformation of higher-order chromatin in cells treated with Dex. In its basal state, ANGPTL4 is not activated in the absence of glucocorticoid. However, the higher-order chromatin structure of the locus is prearranged. In the transcriptionally active state (Dex 3 h), liganded GR binds to AG sites and increases the affinities of the interactions among enhancer, promoter, and CTCF sites in ANGPTL4, which specifically activate ANGPTL4 but not the neighboring genes KANK3, RAB11B-AS, and RAB11B. Since AC2 and AC3 show convergent orientations to the CTCF-binding sites, these may facilitate the formation of a chromatin loop [40]. In the silenced state (Dex 24 h), the positions of the enhancer, promoter, and CTCF sites are restored to the basal state when ANGPTL4 transcription is down-regulated. During these processes, CTCF is required for both induction and subsequent silencing of the ANGPTL4 gene.
Fig 6. Model of ANGPTL4 gene regulation and higher-order chromatin in cells treated with Dex.
In its basal state, ANGPTL4 is not activated in the absence of Dex-bound GR within the preexisting higher-order chromatin structure. When ANGPTL4 is transcriptionally active (Dex 3 h), liganded GR binds to AG sites and enhances interactions between enhancer, promoter, and CTCF sites to selectively induce ANGPTL4 but not the neighboring KANK3, RAB11B-AS, and RAB11B genes. In the silenced state (Dex 24 h), the positions of enhancer, promoter, and CTCF sites are restored, leading to down-regulation of ANGPTL4 transcription. CTCF is required for proper induction and subsequent silencing of the ANGPTL4 transcription.
Glucocorticoids are essential regulators of lipid homeostasis and energy metabolism, mainly via GR signaling. Dex treatment of HepG2 cells showed that ANGPTL4 was induced after 3 h and was silenced after 24 h. During this reaction, ANGPTL4 locus maintained a higher-order chromatin conformation during basal and induced states, together with specific interactions between the ANGPTL4 enhancer and promoter that were required for selective transcriptional activity. In contrast, under long-term Dex treatment, ANGPTL4 induction was significantly inhibited, at least in part, by the decrease in the level of GR. When the levels of glucocorticoids are up-regulated in vivo [41] under fasting or starvation, lipolysis in adipose tissue releases TGs and free fatty acids to other tissues, including the liver. Glucocorticoids may induce ANGPTL4 transcription to promote the catabolism. In contrast, long-term elevation of endogenous or exogenous glucocorticoids causes hypertriglyceridemia, insulin resistance, fatty liver, and obesity [36]. Such a long-term response is likely to produce cellular memory against glucocorticoid exposure. These changes may partly correlate with the level of ANGPTL4, because we show here that this protein was down-regulated in LTDT cells.
The siRNA-mediated inhibition of CTCF expression in HepG2 cells shows that ANGPTL4 transcription was highly induced by 3 h and was subsequently up-regulated for 24 h after Dex addition. Since there was no difference in liganded GR binding 24 h after Dex addition to control and CTCF-depleted cells, enhancer-promoter interactions in ANGPTL4 may contribute to persistent activation. Moreover, CTCF-binding is known to be affected by DNA methylation in the genome [42, 43], and CTCF protects adjacent sequences against de novo CpG methylation[44, 45]. Indeed, IGF2 imprinting is altered in aged and senescent human epithelial cells [46]. Our previous study shows that CTCF is down-regulated during cellular senescence [33]. Thus, the age-dependent increase of abnormal lipid metabolism may be linked to a reduction in CTCF-mediated regulation of ANGPTL4 transcription, possibly caused by changes in DNA methylation and the down-regulation of CTCF levels. In conclusion, our study of the human ANGPTL4 locus sheds light on the molecular basis of the regulation of GR target genes and its biological significance.
Supporting Information
(A) The consensus motifs of four CTCF-binding sequences (AC1–AC4) and two GR-binding sequences (AG1 and AG2). (B) The close localization of AC3 with AG2 in the 3’-region of ANGPTL4.
(TIF)
ChIP-Seq data reveal four major CTCF-enriched sites in the ANGPTL4 locus. The CTCF-enriched AC3 site is uniquely present in the genome of HepG2 cells.
(TIF)
(A) The expression patterns the KANK3, ANGPTL4, RAB11B, and RAB11B AS genes in cluster. In cells treated with Dex, ANGPTL4 mRNA was only induced by Dex. **P < 0.01. (B) Western blot analysis of ANGPTL4 expression in HepG2 cells under the presence or absence of Dex.
(TIF)
(A) Analysis of the expression of glucocorticoid receptor (GRα) and CTCF mRNAs in LTDT cells. In the presence of Dex, there were no significant differences in the levels GRα and CTCF transcripts in control and LTDT cells. *P < 0.05. (B) Western blot analysis of GR expression in LTDT cells. Uncropped image of western blot analysis is shown in S6 Fig.
(TIF)
(A) qRT-PCR analysis of GRα (left) and FKBP5 (right) in siRNA-transfected HepG2 cells treated with Dex. (B) The transcribed sequence of RAB11B and RAB11B AS in the human ANGPTL4 locus. The arrow indicates the transcriptional start site. (C) Close localization of AC3/AG2 with the 3´-region of RAB11B AS. (D) qRT-PCR analysis of four genes within human ANGPTL4 locus in siRNA-transfected HepG2 cells treated with Dex. The relative expression level is indicated as a value normalized to the level of 36B4 mRNA. Asterisks indicate statistically significance between control and CTCF-knockdown cells at each time point. *P < 0.05, **P < 0.01, ***P < 0.005.
(TIF)
(A) Uncropped images of Fig 3E. (B) Uncropped image of S4B Fig.
(TIF)
(PDF)
Acknowledgments
We thank Dr. H. Kimura (Tokyo Institute of Technology) for the antibody against RNA polymerase II and Dr. Takashi Seki for assistance with high-throughput sequencing, and the members of our laboratory for discussions and technical support.
Data Availability
ChIP-seq datasets of GR are available from the Gene Expression Omnibus (GEO) database (accession number GSE85343).
Funding Statement
This work was supported by the Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research (JSPS KAKENHI) Grant Numbers JP15H04707 and JP15K15068 (https://www.jsps.go.jp/english/e-grants/), by grants from Takeda Science Foundation (http://www.takeda-sci.or.jp/), and the Japan Agency for Medical Research and Development (CREST) (http://www.amed.go.jp/en/) (MiN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baschant U, Tuckermann J. The role of the glucocorticoid receptor in inflammation and immunity. J Steroid Biochem Mol Biol. 2010; 120(2–3): 69–75. 10.1016/j.jsbmb.2010.03.058 [DOI] [PubMed] [Google Scholar]
- 2.Patel R, Williams-Dautovich J, Cummins CL. Minireview: new molecular mediators of glucocorticoid receptor activity in metabolic tissues. Mol Endocrinol. 2014; 28(7): 999–1011. 10.1210/me.2014-1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payvar F, DeFranco D, Firestone GL, Edgar B, Wrange O, Okret S, et al. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell. 1983; 35(2 Pt 1): 381–392. [DOI] [PubMed] [Google Scholar]
- 4.Scheidereit C, Geisse S, Westphal HM, Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983; 304(5928): 749–752. [DOI] [PubMed] [Google Scholar]
- 5.Perlmann T, Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988; 7(10): 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008; 29(5): 611–624. 10.1016/j.molcel.2008.02.010 [DOI] [PubMed] [Google Scholar]
- 7.Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, Hager GL. Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J Biol Chem. 2009; 284(10): 6048–6052. 10.1074/jbc.C800212200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009; 19(12): 2163–2171. 10.1101/gr.097022.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paakinaho V, Makkonen H, Jaaskelainen T, Palvimo JJ. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol Endocrinol. 2010; 24(3): 511–525. 10.1210/me.2009-0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, et al. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011; 21(5): 697–706. 10.1101/gr.111153.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kersten S, Mandard S, Tan NS, Escher P, Metzger D, Chambon P, et al. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J Biol Chem. 2000; 275(37): 28488–28493. 10.1074/jbc.M004029200 [DOI] [PubMed] [Google Scholar]
- 12.Kim I, Kim HG, Kim H, Kim HH, Park SK, Uhm CS, et al. Hepatic expression, synthesis and secretion of a novel fibrinogen/angiopoietin-related protein that prevents endothelial-cell apoptosis. Biochem J. 2000; 346 Pt 3: 603–610. [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon JC, Chickering TW, Rosen ED, Dussault B, Qin Y, Soukas A, et al. Peroxisome proliferator-activated receptor gamma target gene encoding a novel angiopoietin-related protein associated with adipose differentiation. Mol Cell Biol. 2000; 20(14): 5343–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeton EK, Fletcher TM, Baumann CT, Hager GL, Smith CL. Glucocorticoid receptor domain requirements for chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter in different nucleoprotein contexts. J Biol Chem. 2002; 277(31): 28247–28255. 10.1074/jbc.M203898200 [DOI] [PubMed] [Google Scholar]
- 15.Koliwad SK, Kuo T, Shipp LE, Gray NE, Backhed F, So AY, et al. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor) is a direct glucocorticoid receptor target and participates in glucocorticoid-regulated triglyceride metabolism. J Biol Chem. 2009; 284(38): 25593–25601. 10.1074/jbc.M109.025452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koliwad SK, Gray NE, Wang JC. Angiopoietin-like 4 (Angptl4): A glucocorticoid-dependent gatekeeper of fatty acid flux during fasting. Adipocyte. 2012; 1(3): 182–187. 10.4161/adip.20787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hato T, Tabata M, Oike Y. The role of angiopoietin-like proteins in angiogenesis and metabolism. Trends Cardiovasc Med. 2008; 18(1): 6–14. 10.1016/j.tcm.2007.10.003 [DOI] [PubMed] [Google Scholar]
- 18.Mattijssen F, Alex S, Swarts HJ, Groen AK, van Schothorst EM, Kersten S. Angptl4 serves as an endogenous inhibitor of intestinal lipid digestion. Mol Metab. 2014; 3(2): 135–144. 10.1016/j.molmet.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang R. The ANGPTL3-4-8 model, a molecular mechanism for triglyceride trafficking. Open Biol. 2016; 6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koster A, Chao YB, Mosior M, Ford A, Gonzalez-DeWhitt PA, Hale JE, et al. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005; 146(11): 4943–4950. 10.1210/en.2005-0476 [DOI] [PubMed] [Google Scholar]
- 21.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011; 12(4): 283–293. 10.1038/nrg2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014; 15(4): 234–246. 10.1038/nrg3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006; 23(5): 733–742. 10.1016/j.molcel.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 24.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008; 451(7180): 796–801. 10.1038/nature06634 [DOI] [PubMed] [Google Scholar]
- 25.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008; 105(24): 8309–8314. 10.1073/pnas.0801273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008; 132(3): 422–433. 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- 27.Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci. 2009; 122(Pt 9): 1275–1284. 10.1242/jcs.039990 [DOI] [PubMed] [Google Scholar]
- 28.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007; 128(6): 1231–1245. 10.1016/j.cell.2006.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Tian Y, Shu W, Bo X, Wang S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One. 2012; 7(7): e41374 10.1371/journal.pone.0041374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009; 19(1): 24–32. 10.1101/gr.082800.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, et al. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009; 28(9): 1234–1245. 10.1038/emboj.2009.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe T, Ishihara K, Hirosue A, Watanabe S, Hino S, Ojima H, et al. Higher-order chromatin regulation and differential gene expression in the human tumor necrosis factor/lymphotoxin locus in hepatocellular carcinoma cells. Mol Cell Biol. 2012; 32(8): 1529–1541. 10.1128/MCB.06478-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirosue A, Ishihara K, Tokunaga K, Watanabe T, Saitoh N, Nakamoto M, et al. Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell. 2012; 11(3): 553–556. 10.1111/j.1474-9726.2012.00809.x [DOI] [PubMed] [Google Scholar]
- 34.Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347(6225): 1017–1021. 10.1126/science.1262088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA. 2004; 101(44): 15603–15608. 10.1073/pnas.0407008101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibli-Rahhal A, Van Beek M, Schlechte JA. Cushing's syndrome. Clin Dermatol. 2006; 24(4): 260–265. 10.1016/j.clindermatol.2006.04.012 [DOI] [PubMed] [Google Scholar]
- 37.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013; 14(12): 880–893. 10.1038/nrg3594 [DOI] [PubMed] [Google Scholar]
- 38.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013; 153(7): 1537–1551. 10.1016/j.cell.2013.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kung JT, Kesner B, An JY, Ahn JY, Cifuentes-Rojas C, Colognori D, et al. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell. 2015; 57(2): 361–375. 10.1016/j.molcel.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015; 60(4): 676–684. 10.1016/j.molcel.2015.09.023 [DOI] [PubMed] [Google Scholar]
- 41.Rose AJ, Herzig S. Metabolic control through glucocorticoid hormones: an update. Mol Cell Endocrinol. 2013; 380(1–2): 65–78. 10.1016/j.mce.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 42.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000; 405(6785): 482–485. 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- 43.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000; 405(6785): 486–489. 10.1038/35013106 [DOI] [PubMed] [Google Scholar]
- 44.Butcher DT, Mancini-DiNardo DN, Archer TK, Rodenhiser DI. DNA binding sites for putative methylation boundaries in the unmethylated region of the BRCA1 promoter. Int J Cancer. 2004; 111(5): 669–678. 10.1002/ijc.20324 [DOI] [PubMed] [Google Scholar]
- 45.Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005; 8(1): 31–42. [DOI] [PubMed] [Google Scholar]
- 46.Fu VX, Schwarze SR, Kenowski ML, Leblanc S, Svaren J, Jarrard DF. A loss of insulin-like growth factor-2 imprinting is modulated by CCCTC-binding factor down-regulation at senescence in human epithelial cells. J Biol Chem. 2004; 279(50): 52218–52226. 10.1074/jbc.M405015200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The consensus motifs of four CTCF-binding sequences (AC1–AC4) and two GR-binding sequences (AG1 and AG2). (B) The close localization of AC3 with AG2 in the 3’-region of ANGPTL4.
(TIF)
ChIP-Seq data reveal four major CTCF-enriched sites in the ANGPTL4 locus. The CTCF-enriched AC3 site is uniquely present in the genome of HepG2 cells.
(TIF)
(A) The expression patterns the KANK3, ANGPTL4, RAB11B, and RAB11B AS genes in cluster. In cells treated with Dex, ANGPTL4 mRNA was only induced by Dex. **P < 0.01. (B) Western blot analysis of ANGPTL4 expression in HepG2 cells under the presence or absence of Dex.
(TIF)
(A) Analysis of the expression of glucocorticoid receptor (GRα) and CTCF mRNAs in LTDT cells. In the presence of Dex, there were no significant differences in the levels GRα and CTCF transcripts in control and LTDT cells. *P < 0.05. (B) Western blot analysis of GR expression in LTDT cells. Uncropped image of western blot analysis is shown in S6 Fig.
(TIF)
(A) qRT-PCR analysis of GRα (left) and FKBP5 (right) in siRNA-transfected HepG2 cells treated with Dex. (B) The transcribed sequence of RAB11B and RAB11B AS in the human ANGPTL4 locus. The arrow indicates the transcriptional start site. (C) Close localization of AC3/AG2 with the 3´-region of RAB11B AS. (D) qRT-PCR analysis of four genes within human ANGPTL4 locus in siRNA-transfected HepG2 cells treated with Dex. The relative expression level is indicated as a value normalized to the level of 36B4 mRNA. Asterisks indicate statistically significance between control and CTCF-knockdown cells at each time point. *P < 0.05, **P < 0.01, ***P < 0.005.
(TIF)
(A) Uncropped images of Fig 3E. (B) Uncropped image of S4B Fig.
(TIF)
(PDF)
Data Availability Statement
ChIP-seq datasets of GR are available from the Gene Expression Omnibus (GEO) database (accession number GSE85343).
ChIP-seq datasets of GR are deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE85343.