Abstract
CD4+ effector T cells (Teff cells) and regulatory T cells (Treg cells) undergo metabolic reprogramming to support proliferation and immunological function. Although signaling via the lipid kinase PI(3)K (phosphatidylinositol-3-OH kinase), the serine-threonine kinase Akt and the metabolic checkpoint kinase complex mTORC1 induces both expression of the glucose transporter Glut1 and aerobic glycolysis for Teff cell proliferation and inflammatory function, the mechanisms that regulate Treg cell metabolism and function remain unclear. We found that Toll-like receptor (TLR) signals that promote Treg cell proliferation increased PI(3)K-Akt-mTORC1 signaling, glycolysis and expression of Glut1. However, TLR-induced mTORC1 signaling also impaired Treg cell suppressive capacity. Conversely, the transcription factor Foxp3 opposed PI(3)K-Akt-mTORC1 signaling to diminish glycolysis and anabolic metabolism while increasing oxidative and catabolic metabolism. Notably, Glut1 expression was sufficient to increase the number of Treg cells, but it reduced their suppressive capacity and Foxp3 expression. Thus, inflammatory signals and Foxp3 balance mTORC1 signaling and glucose metabolism to control the proliferation and suppressive function of Treg cells.
The activation of CD4+ T cells is accompanied by increased growth, proliferation and specification into functional effector T cell (Teff cell) or regulatory T cell (Treg cell) subsets that promote or suppress immunity and subsequent inflammatory resolution. To support this transition from quiescent naive T cells to Teff cells and Treg cells, activated CD4+ T cells reprogram their cellular metabolism1. Resting T cells utilize a catabolic oxidative metabolism of glucose, lipids and amino acids. After being activated via co-stimulation, however, T cells upregulate the expression of glucose and amino acid transporters and increase their metabolic rate. Although both mitochondrial oxidative phosphorylation and glycolytic metabolism increase, glycolytic flux is elevated to a greater degree. In Teff cells, this leads to a predominantly glycolytic phenotype that is reminiscent of the proliferative metabolism of cancer cells, aerobic glycolysis, that is thought to provide biosynthetic intermediates for anabolic cell growth2. CD4+ Teff cells are dependent on the glucose transporter Glut1 and aerobic glycolysis for proliferation and inflammatory functions, as inhibition of glycolysis or deletion of Glut1 impairs Teff cell function in vivo3,4. In contrast, induced Treg cells have been shown to utilize mainly a distinct metabolic program based on mitochondrial oxidation of lipid and pyruvate5–8, and Treg cells can act independently of Glut1 in vivo4. This oxidative metabolism provides inefficient support for biosynthesis for cell growth. However, Treg cells can be highly proliferative and glycolytic in vivo9,10. Although Treg cell are important in autoimmunity9 and inflammatory resolution and healing11–13, the mechanisms that regulate Treg cell metabolism and how these metabolic pathways affect Treg cell proliferation and function remain poorly understood.
Signals through the transcription factors Myc and HIF1α, or the PI(3)K-Akt-mTORC1 pathway1 and transcription factors of the CD4+ T cell subset14,15, facilitate the induction of Glut1 expression and aerobic glycolysis in Teff cells. The PI(3)K-Akt-mTOR pathway has also been suggested to be a regulator of Treg cell metabolism. However, Treg cells do not require mTOR kinase16. Furthermore, although PI(3)K-Akt-mTOR signaling can be activated in Treg cells17,18, selective loss of mTORC1 (ref. 18) or constitutive activation of mTORC1 as a result of deficiency in the lipid phosphatase PTEN or the tumor suppressor TSC2 leads to increased numbers of Treg cells, but those Treg cells have impaired capacity for immunosuppression19–21. Although mTORC1 regulates protein translation and other aspects of cell physiology beyond anabolic metabolism, Treg cell metabolism might be heterogeneous or oscillatory17, and these changes could affect Treg cell function and proliferation.
Another signaling mechanism that modulates Treg cell proliferation and function is signaling through TLRs. Treg cells express several TLRs, including TLR1, TLR2, TLR4, TLR5 and TLR8 (refs. 22–25). In each case, ligation of the TLR leads to increased Treg cell proliferation but reduced suppressive function23–26. Pattern recognition via TLR signaling appears to be critical for Treg cell homeostasis and function, as microbiota can promote the loss of Foxp3 expression on intestinal Treg cells27 and Treg cells deficient in the TLR adaptor-transducer MyD88 are reduced in number in the gut, which leads to the onset of inflammatory bowel disease (IBD)28. Similarly, microbiota that provide TLR ligands have been shown to be important for driving Treg cell proliferation in the skin29. TLR ligation has also been shown to activate PI(3)K-Akt-mTORC1 signaling through modulation of PI(3)Kγ in macrophages30 and to induce glycolysis in B cells31 and dendritic cells32. However, the role of TLR signals as modulators of Treg cell metabolism and how metabolic pathways balance the anabolic growth and proliferation of Treg cells with suppression remain unknown.
We investigated mechanisms that regulate Treg cell metabolism and how changes in glucose metabolism affect Treg cell proliferation and function. Although most Treg cell used a catabolic metabolism with low Glut1 expression, proliferating Treg cell showed elevated Glut1 levels and mTOR activity. Ligation of TLR1 and TLR2 on activated Treg cells further increased glycolysis and proliferation, but the cells’ suppressive capacity was reduced in an mTORC1-dependent manner. Conversely, Foxp3 opposed PI(3)K-Akt-mTOR signaling, reducing glycolytic and anabolic metabolism while increasing the activity of oxidative and catabolic pathways. These metabolic changes directly modified Treg cell function, as transgenic expression of Glut1 in T cells33 reduced Treg cell suppressive capacity and led to the downregulation of Foxp3. Thus, TLR signals and Foxp3 counter-regulate Treg cell metabolism to balance proliferation and suppressive function.
RESULTS
Proliferative Treg cells activate mTORC1 and glucose uptake
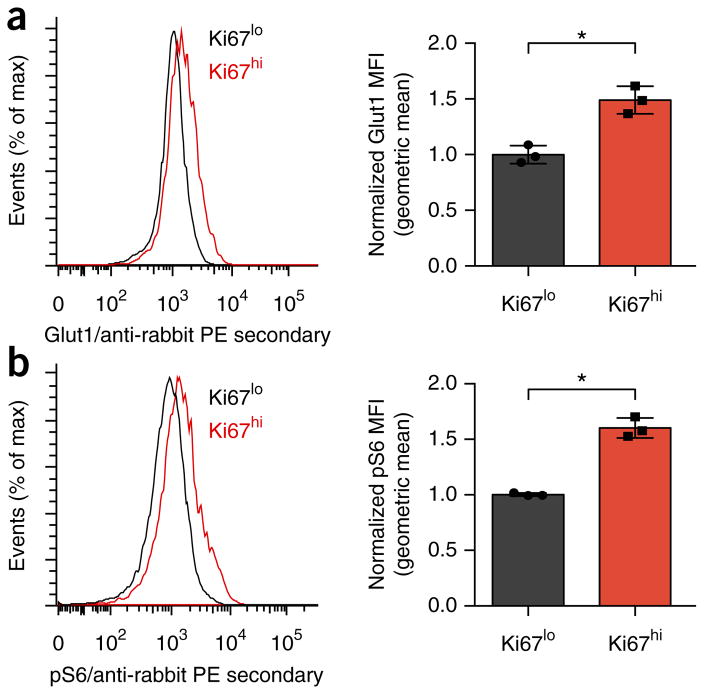
To assess the regulation and role of Treg cell metabolism in proliferation and function, we first investigated the metabolic programs of proliferative and non-proliferative Treg cell34. Foxp3+ thymus-derived Treg cells (tTreg cells) were more proliferative than Foxp3− CD4+ T cells were under homeostatic conditions, as determined by staining for the proliferation marker Ki67 (Supplementary Fig. 1a). The tTreg cells with high Ki67 expression (Ki67hi), however, were phenotypically distinct from those with low Ki67 expression (Ki67lo). Consistent with glycolysis being an anabolic metabolic program that supports proliferation, Ki67hi tTreg cells had increased Glut1 expression (Fig. 1a). mTORC1 activity was also elevated in Ki67hi tTreg cells, as measured by phosphorylated ribosomal protein S6 (Fig. 1b). Thus, tTreg cells were not metabolically uniform, and those cells that had undergone proliferation showed increased expression of markers of glycolysis.
Figure 1.
Treg cell proliferation is associated with increased glycolysis and anabolic signaling. (a,b) Flow cytometry analyzing the expression of Glut1 (a) and phosphorylated S6 (p-S6) (b) in Ki67high and Ki67low populations of CD4+Foxp3+ tTreg cells from the spleens of wild-type mice (gating strategy, Supplementary Fig. 1a). MFI, mean fluorescent intensity. *P < 0.05 (two-tailed Student’s t test). Data in a and b are each representative of three independent experiments (left) or are pooled and normalized to Ki67low from three independent experiments (right; mean + s.d.).
TLR1, TLR2 promote iTreg glycolysis but impair suppression
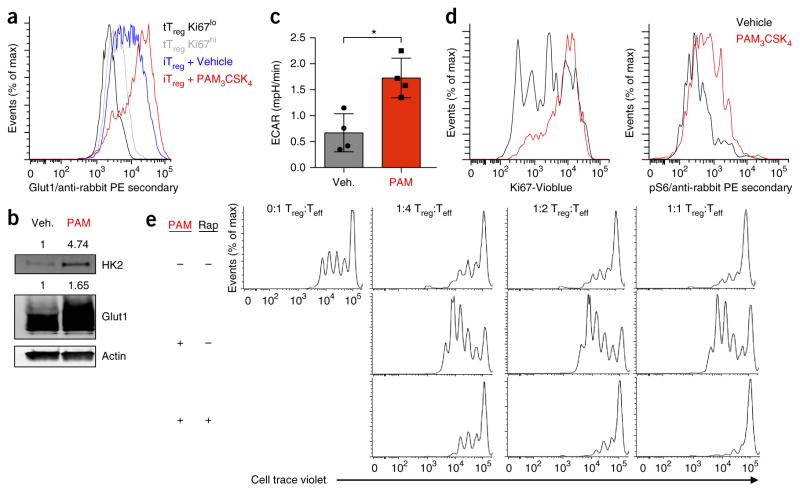
We next assessed the effect of Treg cell activation on glucose metabolism. Although proliferative tTreg cells had higher surface expression of Glut1 protein than that of non-proliferative tTreg cells, Glut1 expression was even higher in activated induced Treg cells (iTreg cells) (Fig. 2a). Treg cells express TLR1 and TLR2 at high levels22 and can respond to TLR ligation with increased proliferation24. Treatment of activated iTreg cells with the TLR1 and TLR2 agonist Pam3CSK4 further increased the expression of Glut1 and hexokinase 2 (HK2), an enzyme that phosphorylates intracellular glucose (Fig. 2a,b). This was accompanied by a sharp increase in Treg cell size (Supplementary Fig. 1b), glycolytic lactate production (Fig. 2c and Supplementary Fig. 1c), activation of the mTORC1 pathway, and proliferation, as determined by Ki67 expression (Fig. 2d). To determine how TLR1- and TLR2-induced changes in Treg cell signaling and metabolism affect Treg cell function, we treated iTreg cells overnight with vehicle or Pam3CSK4 and then re-purified the cells. TLR-ligand-treated iTreg cells showed decreased capacity to suppress effector CD8+ T cells and instead increased Teff cell proliferation (Fig. 2e). Activation of mTORC1 signaling by Pam3CSK4 was critical for the subsequent decrease in Treg cell suppressive capacity, as treatment with the mTORC1 inhibitor rapamycin together with ligation of TLR1 and TLR2 before re-purification restored the ability of iTreg cells to suppress the proliferation of Teff cells (Fig. 2e).
Figure 2.
Signaling via TLR1 and TLR2 drives Treg cell glycolysis and proliferation, yet reduces suppressive capacity. (a) Glut1 expression (as in Fig. 1) of Ki67high and Ki67low tTreg cells derived from the spleens of wild-type mice an of iTreg cells derived from CD4+CD25− T cells isolated from the spleens of wild-type mice and polarized for 5 d under Treg cell–skewing conditions and treated with H2O vehicle (Veh) or Pam3CSK4 (PAM; 5 μg/ml) for the final 24 h. (b) Immunoblot analysis of HK2, Glut1 and actin (loading control) in iTreg cells as in a. (c,d) ECAR of vehicle or Pam3CSK4 treated iTreg (c), and Ki67 expression and phosphorylated S6, determined by flow cytometry (d), in iTreg cells as in a. (e) Inhibition of the proliferation of Teff cells by Treg cells treated for 24 h with DMSO (vehicle (−)) or 20 nM rapamycin (Rap) along with Pam3CSK4 (5 μg/ml) (left margin), then repurified by magnetic selection and functionally assessed an in vitro suppression assay with various ratios (above plots) of Treg cells to Teff cells. *P < 0.05 (Two-tailed Student’s t-test). Data are representative of three independent experiments (a,b,d,e) or two independent experiments with four technical replicates per group (c; mean + s.d.).
Foxp3 promotes oxidative metabolic gene expression
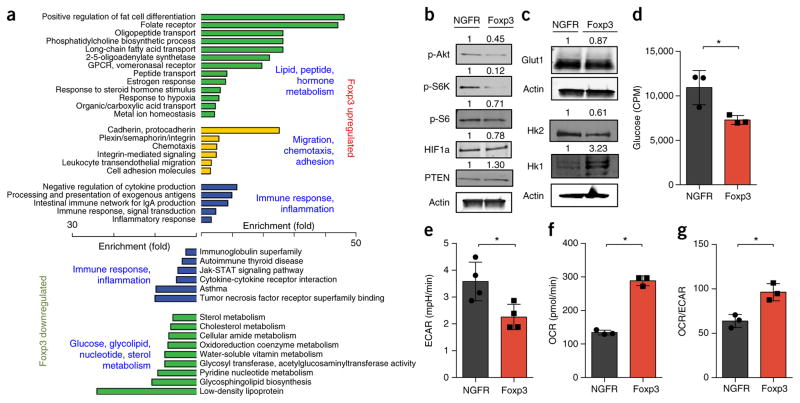
Genetic deletion of Foxp3 leads to a loss of Treg cell suppressive function and a broad inflammatory autoimmune disorder35. Foxp3 expression has also been reported to diminish Glut1 expression and Akt activity36. We therefore examined published data sets to determine whether deletion of Foxp3 also altered T cell metabolism and markers of mTORC1 signaling. Following deletion of Foxp3, tTreg cells were reported to induce widespread changes in gene expression that included not only genes encoding immunological products but a large number of genes encoding products that regulate or participate in varied metabolic pathways (Supplementary Fig. 1d). Alterations in the expression of genes that regulate primary metabolic process included protein products involved in PI(3)K-Akt-mTORC1 signaling as well as glucose metabolism, such as PTEN, phospho-fructokinase, enolase 2, transketolase, triosephosphate isomerase and others (Supplementary Table 1). Consistent with the inverse relationship of Foxp3 and PI(3)K signaling, it has also been reported that tTreg cell activation in vivo correlates with a diminished Akt-related gene-expression signature37.
To directly test how Foxp3 affects the expression of genes encoding products involved in metabolism, we stimulated CD4+ T cells in vitro under neutral conditions and transduced them to express Foxp3. Pathway analysis revealed that the group of genes with significantly altered expression in Foxp3-expressing cells showed enrichment for those encoding products involved in metabolic pathways (Fig. 3a). In particular, genes encoding products involved in lipid and peptide hormone metabolism pathways were upregulated, whereas those encoding products involved in glucose and nucleotide-metabolism pathways were downregulated. A comparison of genes altered by increased Foxp3 and those reported to change following genetic deletion of Foxp3 (ref. 35) revealed an overlap of numerous genes encoding products involved in metabolism (Supplementary Table 2). Both metabolic and immune regulatory pathways were broadly altered (Supplementary Table 3). Although the regulatory mechanisms were unclear, analysis of Foxp3-bound loci by chromatin immunoprecipitation and sequencing showed that Foxp3 associated with genes encoding the pyruvate dehydrogenase kinase PDK3 and the catalytic γ-subunit of PI(3)K (PI(3)Kcγ) (Supplementary Fig. 1e), which correlated with decreased expression (Supplementary Figs. 1f and 2a). Together these data indicated that Foxp3 regulated metabolic gene expression both directly and indirectly through modulation of the PI(3)K-Akt-mTORC1 pathway.
Figure 3.
Foxp3 reduces aerobic glycolysis and promotes mitochondrial oxidative metabolism. (a) Gene-expression microarray analysis and pathway analysis (with the DAVID bioinformatics database) of RNA from primary mouse CD4+CD25− T cells stimulated and transduced with control or Foxp3-expressing retrovirus, presented as enrichment for selected pathways (left and right margins) among genes upregulated (top) or downregulated (bottom) by Foxp3. (b–g) Immunoblot analysis (b,c), glucose uptake (d), glycolysis (as ECAR after glucose injection) (e) basal OCR (f) and basal OCR/ECAR (g) of primary mouse CD4+CD25− T cells stimulated and transduced with control (NGFR) or Foxp3-expressing retrovirus. Numbers above lanes (b,c) indicate band intensity relative to that of actin. CPM, counts per minute. *P < 0.05 (Two-tailed Student’s t-test). Data are representative of an experiment with biological triplicates for each group (a), three independent experiments (b,c) or two independent experiments with technical triplicates (d–g; mean + s.d.).
Foxp3 inhibits PI(3)K-Akt-mTORC1 signaling and glycolysis
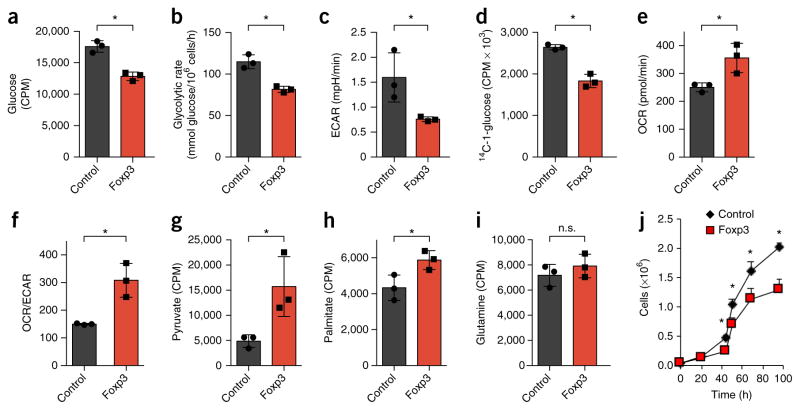
We next assessed the effect of Foxp3 expression on metabolic proteins and pathways. Consistent with reduced PI(3)Kcγ expression and PI(3)K activity, Foxp3 expression led to diminished phosphorylation of Akt, S6 kinase and S6 relative to that of T cells transduced with a control vector (Fig. 3b). In addition, Foxp3 expression was sufficient to reduce the expression of Glut1 and HK2 and to increase the level of HK1 protein (Fig. 3c), similar to the expression pattern of Treg cells6. Foxp3 expression in T cells led to lower glucose uptake and glycolysis, but the oxygen consumption rate (OCR) and the ratio of oxygen consumption to lactate production (OCR/extracellular media acidification rate (ECAR)) increased (Fig. 3d–g). We next transduced the mouse pro-B cell line FL5.12 to express a conditionally active Foxp3; activated Foxp3 led to reduced phosphorylation of S6 kinase, Akt and mTOR, and we observed lower amounts of PI(3)Kcγ and HIF1α protein in this setting (Supplementary Fig. 2a). Conversely, PTEN protein was modestly increased by activation of Foxp3 (Supplementary Fig. 2a). Expression of the mitochondrial proteins cytochrome C and CPT1a increased, whereas the expression of Glut1, HK2 and the majority of glycolytic enzymes decreased (Supplementary Fig. 2b,c and Supplementary Table 4). Activation of Foxp3 in FL5.12 cells was also sufficient to promote an oxidative shift, with reprogramming of the metabolome to increase catabolic metabolites and decrease many anabolic metabolites (Supplementary Fig. 2d and Supplementary Table 5). Altered metabolites included increased succinyl-carnitine, which is indicative of mitochondrial metabolism, and decreased glucoseamine-6-phosphate, glyceraldehyde-3-phosphate and N-carbamoyl-aspartate, which are indicative of glycosylation, glycolysis and pyrimidine synthesis, respectively (Supplementary Fig. 2e). Foxp3 activation led to decreased glucose uptake, glycolysis, lactate production and pentose-phosphate-pathway flux (Fig. 4a–d). Conversely, oxygen consumption, the OCR/ECAR ratio, pyruvate oxidation and palmitate oxidation all increased (Fig. 4e–h). Not all pathways were altered, however, as glutamine oxidation remained unchanged (Fig. 4i). Consistent with diminished anabolic metabolism, Foxp3 slowed the rate at which cells accumulated in vitro (Fig. 4j). Together these data indicated that TLR signals promoted Treg cell proliferation, PI(3)K-Akt-mTORC1 activation and glycolysis, whereas Foxp3 promoted oxidative metabolism and slowed anabolic metabolism and growth.
Figure 4.
Foxp3 expression is sufficient to suppress glycolysis and promote oxidative metabolism. (a–i) Glucose uptake (a), glycolytic rate (assessed by a radiolabeled-glucose-tracer assay) (b), ECAR (c), flux of glucose into the pentose phosphate pathway (PPP) (d), OCR (e), OCR/ECAR (f), pyruvate oxidation (g), fatty-acid oxidation of palmitate (h) and glutamine oxidation (i) of vector-control-transduced FL5.12 pro-B cells or FL5.12 pro-B cells transduced with tamoxifen-inducible expression of Foxp3, treated with 4-hydroxytamoxifen. (j) Quantification of live cells as in a–i to assess cell accumulation over time. *P < 0.05 (two-tailed Student’s t test). Data are representative of three (a,b,j) or two (c–i) independent experiments with three independent clones per group (mean + s.d. in a–i or (mean ± s.d. in j).
Akt promotes Treg cell growth but inhibits suppressive capacity
To directly test how the metabolic transitions noted above affect Treg cell phenotype and function, we modified the Akt-mTORC1 signaling and metabolism of Treg cells through the use of mice with transgenic T cell–specific expression of constitutively active Akt (Akt-tg mice) or Glut1 (Glut1-tg mice)7. tTreg cells isolated from Akt-tg mice had increased glucose uptake and diminished reactive oxygen species (ROS) than that of tTreg cells from wild-type mice (Supplementary Fig. 3a,b), and were more abundant in Akt-tg mice than in wild-type mice (Supplementary Fig. 3c,d). Akt-tg Treg cell were also enlarged and had altered cell surface markers that were consistent with activation (Supplementary Fig. 3e–i). Similar to TLR1- and TLR2-stimulated Treg cells and consistent with poor suppression by Treg cells deficient in the tumor suppressors PTEN or TSC2 (refs. 19–21), Akt-tg Treg cells were less able to suppress Teff cell proliferation than were control wild-type Treg cells (Supplementary Fig. 3j).
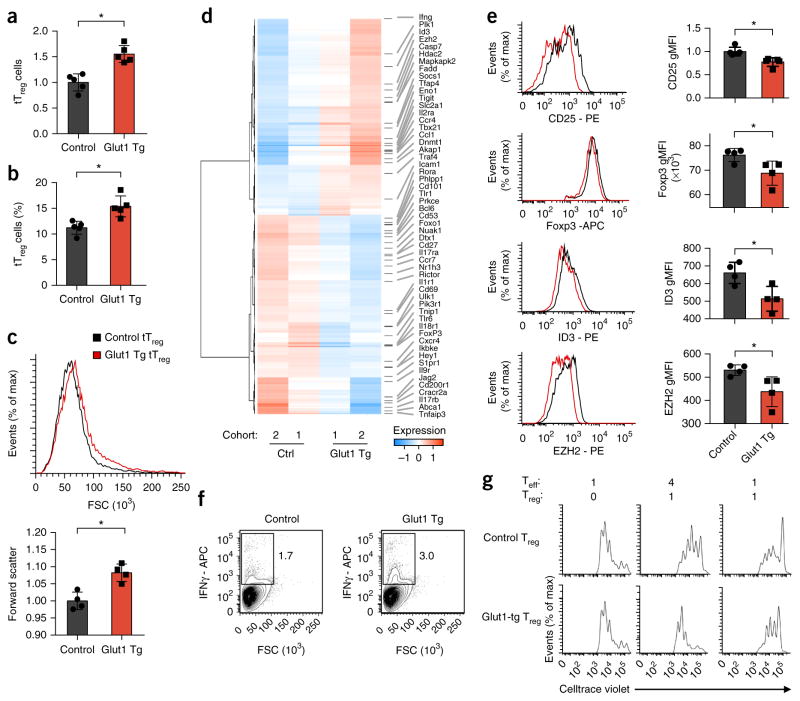
Glut1 enhances Treg cell growth, but reduces function
Foxp3 and TLR signals might, in principle, modify Treg cell metabolism and function through control of Glut1 expression and glycolysis. To directly assess the role of Glut1 in the regulation in Treg cell function, we studied tTreg cells from Glut1-tg mice ex vivo33. Glut1-tg CD4+ T cells showed increased Glut1 expression and glucose uptake, whereas ROS were diminished (Supplementary Fig. 4a–c), relative to that of wild-type cells. Glut1-tg Treg cells also had higher ECAR and OCR than that of wild-type control Treg cells (Supplementary Fig. 4d). Glut1-tg tTreg cells accumulated in terms of both numbers and frequency (Fig. 5a,b) and were modestly enlarged (Fig. 5c). However, Glut1-tg mice develop autoimmunity with age7, which suggests that their Treg cell function is impaired. Consistent with altered function, tTreg cells from Glut1-tg mice had a greater proportion of CD25lo Foxp3+ cells than did those from wild-type mice (Supplementary Fig. 4e). We also observed a modestly increased frequency of Helios− Foxp3+ CD4+ T cells in Glut1-tg mice (Supplementary Fig. 4f), which might indicate a greater frequency of peripherally induced Treg cells or lower Treg cell suppressive capacity.
Figure 5.
Glut1 expression increases the number and size of Treg cells but reduces their suppressive capacity. (a–c) Number (a), frequency (b) and size (by forward scatter (FSC)) (c) of Foxp3+ tTreg cells from the spleen of control and Glut1-tg mice, assessed by flow cytometry. (d) RNA-sequencing analysis of Treg cells from age-matched control and Glut1-tg mice (two independent cohorts), presented as normalized data. (e,f) Flow cytometry analyzing the expression of CD25, Foxp3, ID3 and EZH2 (e) or IFN-γ (f) by CD4+CD25− T cells isolated from the spleen of control and Glut1-tg mice and polarized under Treg cell–skewing conditions; results presented as the geometric mean fluorescent intensity (MFI) in e, right. Numbers adjacent to outlined areas (f) indicate percent IFN-γ+ cells. (g) Inhibition of Teff cell proliferation by control and Glut1-tg Treg cells as in e,f (presented as in Fig. 2e). *P < 0.05 (two-tailed Student’s t test). Data are representative of five independent experiments (a,b; mean + s.d.), four independent experiments (c; mean + s.d.), an experiment with two independent cohorts (two control mice with three Glut1-tg mice, and three control mice with one Glut1-tg mouse) (d), an experiment with four biological replicates per genotype (e,f; mean + s.d. in e) or three independent experiments (g).
To better assess the effect of Glut1 on Treg cells, we activated and induced wild-type control and Glut1-tg T cells to generate Glut1-tg iTreg cells. Similar to tTreg cells, Glut1-tg iTreg cells were generated at an increased frequency, were enlarged and had reduced expression of the cytokine receptor CD25 (IL-2Rα), relative to that of wild-type control iTreg cells (Supplementary Fig. 5a–c). Unlike tTreg cells, Glut1-tg iTreg cells also showed reduced expression of the co-stimulatory receptor ICOS, relative to that of control iTreg cells (Supplementary Fig. 5d). In two cohorts of control and Glut1-tg iTreg cells, we found, by RNA sequencing, that Glut1 expression altered the expression of genes encoding products involved in both metabolism and immunological function (Fig. 5d). These changes included upregulation of Ifng and Tbx21 (which encodes the transcription factor T-bet), and downregulation of Foxp3, Il2ra (CD25), Foxo1 and Bcl6. Gene-ontology analysis of significantly altered genes revealed enrichment for the expression of genes encoding products involved in T cell activation, cytokine signaling and regulation of epigenetic mechanism pathways (Supplementary Table 6). Similarly, quantitative-PCR array analysis of wild-type control and Glut1-tg iTreg cells also showed numerous differences in gene expression, including downregulation of Foxp3, Icos, Pdcd1 (PD-1), Gata3 and Tnfrs8 (CD30) (Supplementary Fig. 5e and Supplementary Table 6). We assessed expression of CD25, Foxp3, the transcription factor ID3 and the histone methyltransferase EZH2 by flow cytometry and found that expression of all of them was reduced in Glut1-tg iTreg cells (Fig. 5e and Supplementary Fig. 4e). A portion of Glut1-tg Treg cells also showed altered cytokine expression, with decreased Il-10 (Supplementary Fig. 5e) and an increase in the population of IFN-γ-producing cells and a decrease in the population of IL-2-producing cells (Fig. 5f and Supplementary Fig. 5f). Consistent with reduced expression of functional Treg cell markers, induced Glut1-tg Treg cells had reduced in vitro suppressive capacity relative to that of control iTreg cells (Fig. 5g and Supplementary Fig. 6a).
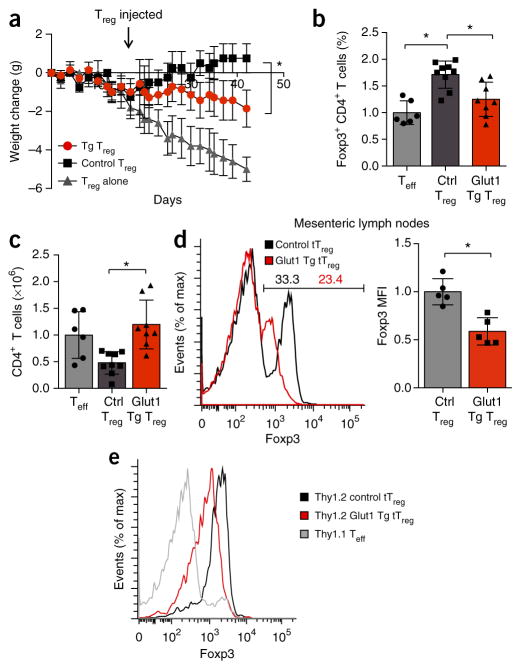
Lower stability of Glut1-expressing Treg cells in vivo in IBD
The phenotypic changes noted above suggested that increased glucose metabolism altered or reduced the suppressive function or stability of Treg cells in vivo. To test whether the Glut1-tg Treg cell were functionally impaired in vivo, we depleted naive T cell populations of Treg cells and adoptively transferred the resultant cells into lymphopenic mice to induce IBD. After disease onset, we gave some of the host mice injection of CD4+CD25+CD45Rblo tTreg cells sorted from wild-type control or Glut1-tg mice (Supplementary Fig. 6b). Despite potential alterations from transgenic Glut1 expression, we found no substantial difference between the CD4+CD25+CD45Rblo tTreg cell populations in their frequency of Foxp3-expressing cells (Supplementary Fig. 6c). As expected, control tTreg cells reversed disease progression, and the mice began to gain weight (Fig. 6a). Glut1-tg tTreg cells, however, slowed disease progression only partially (Fig. 6a). At the termination of the experiment, we found that Glut1-tg tTreg cells had restrained Teff cell accumulation less effectively than control tTreg cells had (Fig. 6b). Despite the transfer of similar initial numbers of tTreg cells, CD4+ Foxp3+ T cells from Glut1-tg donors were also significantly less frequent than were control Treg cells (Fig. 6c). Foxp3 expression was significantly reduced (P = 0.003) in transferred Glut1-tg Treg cells in both mesenteric lymph nodes (Fig. 6d) and spleen (Supplementary Fig. 6d). Reduced Foxp3 expression was confirmed by transfer of congenically marked control and Glut1-tg Treg cells; this revealed that increased Treg cell glycolysis in IBD led to Treg cell downregulation of Foxp3 expression (Fig. 6e and Supplementary Fig. 6e). Together these data showed that increased glucose uptake altered Treg cell phenotype and Foxp3 expression in vivo. Foxp3 and TLR signals therefore act to balance Treg cell metabolism and regulate the proliferation, function and potential stability of Treg cells (Supplementary Fig. 6f).
Figure 6.
Increased glucose uptake impairs the suppressive ability of Treg cells. (a) Body weight of T cell– and B cell–deficient Rag1−/− host mice given injection of sorted naive Teff cells (CD4+CD25−CD45RBhi) for the induction of colitis, then (after weight loss indicated active disease), given injection of no additional cells (Teff alone) or control or Glut1-Tg CD4+CD25+CD45RBlo Treg cells (downward arrow), assessed by weighing three times per week and presented as change from initial weight. (b–e) Frequency of CD4+Foxp3+ T cells (b), number of CD4+ T cells (c) in each Treg cell recipient group, and expression of Foxp3 and the MFI of Foxp3 expression of CD4+Foxp3+ T cells in the mesenteric lymph nodes (d,e) of mice as in a (with naive effector cells expressing Thy1.1 and Treg cells expressing Thy1.2 in e, to allow distinction of cells by these congenic markers). *P < 0.05 (b–d; two-tailed Student’s t test, a; two-way ANOVA followed by Tukey’s post hoc test). Data are representative of at least three independent experiments with over four mice per group in each (a; mean ± s.e.m.), two independent experiments with n = 6 mice (Teff cells alone), n = 9 mice (control Treg cells) or n = 8 mice (Glut1-Tg Treg cells) (b,c; mean + s.d.), two experiments with at least four mice per group (c; mean + s.d.), or one experiment with n = 4 mice (Teff cells alone), n = 4 mice (control Treg cells) or n = 5 mice (Glut1-Tg Treg cells) (e).
DISCUSSION
Treg cells have been shown to rely mainly on the mitochondrial oxidative metabolic pathways of lipid and pyruvate5–8. However, this mode of metabolism supports anabolic growth pathways only inefficiently, and Treg cell can be highly proliferative in vivo9. Treg cell have also been found to be capable of inducing glycolysis17,18, but the mechanisms by which this occurs remain unclear. We found that Treg cell metabolism was dynamically regulated by TLR signals and Foxp3 to balance the anabolic metabolism and proliferation of Treg cells with suppressive capacity. Treg cells responded to activation and TLR ligation by increasing Glut1 expression and glycolysis. Conversely, Foxp3 decreased Glut1 expression and glycolytic and anabolic metabolism to instead favor mitochondrial oxidative pathways. Similar inhibition of glycolysis has also been described for Foxo1 (ref. 38), which suggests that these two Treg cell transcription factors might induce a barrier to aerobic glycolysis that favors suppression. In contrast, inflammatory signals via TLR1 and TLR2 ligation stimulated glucose uptake and downregulated Foxp3 expression. This model suggests that metabolic changes in acute infection or inflammation provide signals that promote Treg cell glycolysis and proliferation while limiting suppressive capacity to allow Teff cells to eliminate the invading pathogen. As inflammatory signals decrease, Foxp3 levels might increase and tilt the metabolic balance away from glycolysis to favor mitochondrial oxidative pathways instead. Although this would decrease proliferation, this shift might increase suppressive capacity and promote wound healing and inflammatory resolution.
The metabolic heterogeneity of Treg cell that we observed might reflect the breadth of Treg cell phenotypes observed in vivo. Treg cells derived from the thymus or periphery, resting or activated by antigen or distinct stimuli, or circulating or tissue resident have each been shown to have distinct immunological and gene-expression phenotypes9,39. Increased glucose metabolism resulting from Glut1 expression leads to an accumulation of Treg cells that is reminiscent of effector Treg cells40, but with several key differences. In particular, Treg cells with elevated Glut1 downregulated Foxp3 expression and suppressive capacity and seemed to have potentially reduced stability. Thus, although glucose metabolism contributes to Treg cell phenotypes and heterogeneity, Treg cell function and stability also depend on other factors and stimuli.
The direct regulation of metabolism by the central Treg cell transcription factor Foxp3 ensures that metabolic pathways are matched to functional demands. Similar regulation of T cell metabolism by key subset transcription factors has been described for the transcription factor RORγt in the TH17 subset of helper T cells14 and the transcription factor Bcl6 in follicular helper T cells15. Foxp3 can suppress Treg cell glycolysis through several mechanisms, including both direct metabolic targets and inhibition of PI(3)K-Akt-mTORC1 signaling. Indeed, Foxp3 has been shown to lead to reduced activation of Akt36, and Foxp3 activation leads to a gene-expression signature of diminished Akt function37. In addition to potential regulation of the Akt phosphatase PHLPP36, Foxp3 might repress this pathway by suppressing PI(3)K signaling and PI(3)Kcγ expression. We also found that Foxp3 associated with the locus encoding Pdk3, which led to reduced expression of Pdk3; this suggested that Foxp3 might repress expression of this kinase to allow increased mitochondrial oxidation of pyruvate in direct suppression of glycolysis. Consistent with a potential role for Pdk3 in regulating Treg function, inhibition of Pdk1 has been shown to lead to increased generation and suppressive activity of Treg cells in vivo, which improves protection from asthma and experimental autoimmune encephalomyelitis6,41.
Treg cell are stimulated by antigens, cytokines and inflammatory signals. We found that Treg cells increased their PI(3)K-Akt-mTORC1 signaling when proliferating under homeostatic conditions and after activation. Ligation of TLR1 and TLR2 on activated Treg cells led to the greatest upregulation of Glut1 expression and glycolysis. Those findings are consistent with the proposal of a role for PTEN in suppressing PI(3)K-Akt-mTORC1 signaling in Treg cells following stimulation with antigen and cytokines19,20. The mechanism by which ligation of TLR1 and TLR2 led to activation of the PI(3)K-Akt-mTORC1 pathway in Treg cells is not clear but might involve activation through PI(3)Kcγ30, which we identified as a potential target of Foxp3 repression. In addition to activating PI(3)K-Akt-mTORC1, TLR signaling also substantially activates the transcription factor NF-κB42, which might also modulate metabolism and contribute to the regulation of Treg cell proliferation and function. Consistent with a role for alternative pathways, TLR activation in dendritic cells leads to Akt activation and glycolysis, in part through activation of the kinase TBK1–kinase IKKε pathway in a mechanism independent of PI(3)K32,43.
Increased glucose uptake on Treg cells involved in an IBD response resulted in a sharp reduction in Foxp3 expression. Although these cells did not widely lose Foxp3 expression and convert to Teff cells, as formerly Treg cells, an increased portion of Glut1-tg Treg cells were found to secrete IFN-γ, which suggested that Treg cell stability was decreased. Similarly, Glut1-tg Treg cells had downregulated expression of CD25, which is also a marker of reduced Treg cell stability19. Prior to IBD, however, resting Glut1-tg Treg cells showed only modestly lower Foxp3 expression, which indicated that activation and inflammatory signals were needed to more substantially downregulate Foxp3. Our finding that Glut1-tg Treg cells had normal or modestly enhanced mitochondrial function but much greater glycolysis suggested that increased glycolysis itself might promote lower Foxp3 expression. The elevated glycolysis that occurs in PTEN- or TSC1-deficient Treg cells19–21 might therefore directly lead to the downregulation of Foxp3 mRNA and protein.
Together our data have described a dynamic regulation of Treg cell metabolism with a glycolytic switch influencing anabolic metabolism for growth and proliferation or oxidative metabolism for maximal suppressive capacity. Metabolic separation of Treg cell proliferation and Treg cell suppression might allow kinetic resolution of T cell function. Treg cells could accumulate in damaged or inflamed tissues without substantially impairing the protective immune response and are involved in inflammatory resolution and healing11–13. Although multiple signals also strongly influence Treg cell proliferation and function, metabolic regulation is a key contributor. The role of TLR signals in increasing Treg cell glycolysis and proliferation while reducing suppression is further demonstrated in the interactions of Treg cells with microbiota23–28,39. Microbe-derived metabolites can drive Treg cells to maintain homeostasis and tolerance, but physical contact and TLR ligation that indicate invading pathogenic bacteria instead allow proliferation and inflammatory function. Similarly, Treg cells often accumulate in but fail to suppress inflammation in chronic autoimmune diseases, such as systemic lupus erythematosus and rheumatoid arthritis44,45. Conversely, these pathways might promote the suppressive function of Treg cells in tumors, in which glycolysis might be limited by reduced glucose or elevated lactate concentrations. Understanding the metabolic signaling and roles in Treg cells might define new regulatory mechanisms and potential targets to modulate immunity.
ONLINE METHODS
Mice
6–8-week-old C57BL/6J mice from Jackson Laboratory were used for all experiments unless otherwise indicated, and in these cases animals were littermates and control and test animals were co-housed. Glut1 and MyrAkt transgenic mice were previously described7,33,46. All procedures were performed under protocols approved by the Duke University Medical Center Institutional Animal Care and Use Committee.
Cell culture
Primary murine CD4+ and CD8+ T cells were isolated by magnetic bead negative (Miltenyi #130-104-454 or #130-104-075) and cultured in RPMI 1640 media supplemented with 10% FBS, 2 mM glutamine, 10 mM HEPES and 55 μM β-mercaptoethanol. Foxp3ER-NGFR and vector control-NGFR expressing FL5.12 cells were cultured in RPMI 1640 media lacking phenol red supplemented with 10% FBS, 2 mM glutamine, 10 mM HEPES and 55 μM β-mercaptoethanol.
Drug treatments
Foxp3ER and vector control-NGFR expressing cells were treated with 100 nM 4-hydroxytamoxifen (Sigma H6278) dissolved in 100% ethanol. Pam3CSK4 (Invivogen #tlrl-pms) was dissolved in DI H2O and was used at a final concentration of 5 μg/ml. Rapamycin was dissolved in DMSO and was used at a final concentration of 20 nM.
iTreg differentiation
CD4+CD25− T cells were cultured on irradiated splenic feeder cells (30 Gy) with 2.5 μg/ml of anti-CD3 antibody at a ratio of 5:1 in RPMI supplemented with 10% FBS, sodium pyruvate, penicillin/streptomycin, HEPES and beta-mercaptoethanol. 3 ng/ml TGFβ was added to induce Foxp3+ iTreg. On day 3 post-stimulation, cells were split 1:2 and re-plated with IL-2 alone for an additional 2 d. In some cases, cells were treated with 5 μg/ml Pam3CSK4 for 24 h beginning on day 4 post-stimulation.
Treg suppression assay
Treg cells were differentiated as described above and cultured at ratios of 1:1, 1:2 and 1:4 with CellTrace Violet (CTV) labeled CD8+ T cells along with 1 μg/mL of anti-CD3 antibody and irradiated feeder splenocytes (30Gy). Treg suppression of effector T cell proliferation was determined 72 h post stimulation by CTV dilution of target population using flow cytometry.
Foxp3-ER cell lines
FL5.12 cells were infected with control or Foxp3-ER expressing lentivirus and selected with NGFR. The Foxp3-ER-NGFR construct was generously provided by M. Levings (University of British Columbia) and has been described previously47. Three clones of each were selected based on equivalent NGFR expression and were Mycoplasma negative. To activate Foxp3-ER, 100 nM 4-hydroxytamoxifen (4-OHT) was added 36 h before experimental procedures.
Retroviral packaging
Retroviral constructs were packaged in Plat-E cells. Plat-E cells were maintained in DMEM supplemented with 10% FBS, with 1 μg/mL puromycin and 10 μg/mL blasticidin. To produce virus, Plat-E cells at 70% confluency in a six-well dish were transfected with construct plasmid along with Lipofectamine 2000 Transfection Reagent (Life Technologies). Media supernatant was removed 24 h post transfection, and fresh virus was collected at 72 and 96 h post transfection.
Retroviral Foxp3 expression
Primary CD4+CD25− T cells were isolated from mice and stimulated with 3 ng/ml PMA and 1 μM ionomycin overnight. Cells were then treated with MSCV-Foxp3-NGFR or MSCV-NGFR control retroviral supernatant containing 8 μg/ml polybrene and centrifuged for 90 min at 2,500 rpm. Cells were rested for 5 h and then plated with fresh media and 10 U/ml interleukin-2 for 72 h. Foxp3-NGFR or NGFR control cells were positively selected with PE magnetic beads following NGFR-PE labeling (≥90% purity, Miltenyi Biotec).
Microarray gene expression analysis
Total RNA from activated transduced cells was isolated using TRIzol reagent (Invitrogen), and dissolved in RNase-free water. Total RNA treated with RNase-free DNase and cleaned up using Qiagen RNeasy columns. Biotinylated antisense cRNA was prepared by two rounds of amplification using the BioArray RNA Amplification and Labeling system according to the protocol for 10–1,000 ng of input RNA. cDNA libraries were amplified and hybridized to Affymetrix GeneChip Mouse Gene 1.0 ST array at the Nucleic Acid and Protein core facility of the Children’s Hospital of Philadelphia Research Institute. Array data were normalized and differential expression was determined using Utils, Stats, Samr, RankProd, topGO and Bioconductor. Significance was assessed using significance analysis of microarrays (SAM). Data are deposited in ArrayExpress (accession E-MTAB-4561). Genes differing in intensity by at least 1.5-fold (up or down) with P < 0.001 were considered Foxp3-regulated. Functional annotation of the top 200 FOXP3-regulated genes was achieved using DAVID software48, and network analyses of the same gene sets were generated using Ingenuity software. Williams and Rudensky35 gene expression array data from wild-type and Foxp3-deficient Treg were analyzed using Geo Data set Browser Data Analysis Tools to select genes with 1.5 fold expression change (P < 0.1) for gene ontology with PANTHER49. Genes overlapping between Foxp3 expression or deficient Treg were analyzed for gene ontology and pathway enrichment with DAVID.
ChIP-seq
Foxp3-chromatin complexes were immunoprecipitated from Foxp3- or empty-vector-transduced CD4+ T cells as described previously50. Genomic DNA libraries were constructed from these samples using NEBNext Ultra DNA Library Prep kits (NEB) and sequenced using the HiSeq platform (Illumina). Sequence reads were mapped to mm9 using Bowtie, and HOMER software was used to identify read densities (peaks) enriched significantly (>5-fold) in ChIP samples over input controls. The top 30% of peaks were highly enriched for the Foxp3 consensus binding sequence (>85%), and were used as the set of high confidence, genome-wide Foxp3 binding sites.
RNA-seq of iTreg
Glut1-tg (n = 4) and wild-type (control; n = 5) mice were used as naive T cell donors to generate iTreg in two age-matched groups. Group 1 included control 1, 3 and Glut1-tg 2, 3, and 5 and Group 2 included control 2, 4, 5 and Glut1-tg 1. RNA was isolated from Glut1-transgenic and control Treg cells using the RNeasy Plus Mini kit (Qiagen) following the manufacturers’ instructions. Libraries were prepared with Illumina TruSeq Stranded mRNA sample kits using indexed adaptors (Illumina). Pooled libraries were subjected to 75-bp paired-end sequencing according to the manufacturer’s protocol (Illumina HiSeq3000). Targeted sequencing depth was 30 million paired-end reads per sample. Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files. The RNA reads obtained from sequencing were mapped with the TopHat 2 aligner using the default parameters to the UCSC mm10 mouse reference genome. Reads were examined for quality control and quantified using FeatureCounts. Subsequently, raw counts were normalized taking into account library sizes, filtering out genes with near zero expression, and the variance-stabilizing transformation was applied to the remaining genes (n = 13700) using the DESeq2 software package. Data are deposited in Geo Database (accession GSE84919). Statistical analyses were performed in the R Statistical Computing Environment. Differential gene expression was assessed between Glut1-tg and wild-type iTreg samples, while at the same time adjusting for animal age groups. Benjamini-Hochberg false discovery rate (FDR) correction was subsequently used to account for multiple testing, and genes were considered to be differentially expressed (DE) if the corrected P < 0.05. To visualize gene expression differences between experimental groups, a heatmap was generated using the top 1000 most variably expressed genes. The mean expression values Glut1-tg and wildtype iTreg blocked by age groups were used as heatmap input. Functional analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources48 and PANTHER49. For this purpose, all differentially expressed genes (corrected p-values < 0.05) between Glut1-tg and wild-type iTreg cell groups were split into two groups according to whether genes were downregulated or upregulated in the Glut1-tg experimental groups relative to the wild-type groups.
Flow cytometry analyses
Flow cytometry was performed on a Miltenyi Macsquant Analyzer. Antibodies used included anti-CD4 eFluor450 (eBioscience 48-0041-82), anti-CD25 PE (BD Biosciences 553866), anti-CD62L PE (eBioscience 12-0621-82), anti-CD69 PE (BD Biosciences 553237), anti-EZH2 PE (BD 562478), anti-FoxP3-APC (eBioscience 17-5773-80), anti-FoxP3-PE (eBioscience 12-5773-80), anti-Helios PE (eBioscience 12-9883-41), anti-ICOS PE (eBioscience 12-9942-81), anti-ID3 PE (BD Biosciences 564564), anti-mouse IFN gamma APC (eBioscience 17-7311-81), anti-mouse IL2 PE (BD Bioscience 561061), anti-KI67 eFluor450 (eBioscience 48-5698-80). Flow cytometry antibodies were used at a 1:100 dilution except for anti-EZH2 PE, anti-FoxP3-APC, anti-FoxP3-PE, anti-Helios PE, anti-ICOS PE, anti-ID3 PE, anti-mouse IFN gamma APC, anti-mouse IL2 PE, anti-KI67 eFluor450, which were all used at a 1:50 dilution. Cell trace violet (Invitrogen #C34557) was used following manufacturer’s specifications. Flow cytometry data analysis was performed using FlowJo.
PCR arrays
RNA was extracted as above and 1 μg total RNA was subjected to single-strand cDNA synthesis using the RT2 first strand kit (Qiagen). The cDNA was used in the Glucose Metabolism and T cell Anergy and Immune Tolerance SuperArray RT2 Profiler PCR arrays according to the manufacturer’s instructions and assayed on a ViiA 7 (Applied Biosystems). Data were analyzed using the RT2 Profiler program supplied by Qiagen and normalized to the geometric mean of the housekeeping genes beta-actin, beta-2 microglobulin, beta-glucuronidase and heat shock protein 90 kDa alpha.
Immunoblotting
Immunoblotting was performed as described previously6. Primary antibodies were followed by mouse- or rabbit-conjugated horseradish peroxidase (HRP). HRP-conjugated antibodies (anti-mouse or anti-rabbit IgG HRP conjugate, Promega) were detected by enhanced chemiluminescence detection (Thermofisher). This included the following antibodies: Akt (9272, Cell Signaling), Cpt1a (15184-1-AP, Proteintech group), Hif-1α (10006421, Cayman Chemical Company), phosphor-Akt (4060, Cell Signaling), PIK3cγ (ab140310, abcam), phosphor-mTOR (5536, Cell Signaling), PTEN (9552, Cell Signaling), phosphor-p70 S6K (9204, Cell Signaling). Alternatively, primary antibodies were followed by fluorescently labeled anti-mouse or rabbit antibodies (LiCor) and imaged using the Odyssey infrared imaging system (LiCor). This included the following antibodies: Glut1 (ab652, Abcam), hexokinase 2 (2867, Cell Signaling), hexokinase 1 (ab104835, Abcam), cytochrome C (556432, BD Biosciences), β-actin (A5441, Sigma) and phosphor-S6 (4858, Cell Signaling). All primary antibodies were used at a 1:1,000 dilution, with secondary HRP antibodies used at 1:8000. Secondary LiCor antibodies were used at 1:10000 (anti-mouse) or 1:8000 (anti-rabbit).
Metabolic assays
Glycolysis and glucose uptake assays using 3H-glucose or 3H-2-deoxyglucose have been described previously6. Glutamine oxidation, pentose phosphate pathway (PPP) flux and glucose oxidation were determined by the rate of 14CO2 release from U-14C-glutamine, 1-14C-glucose and 6-14C-glucose as described6. All values were normalized to cell number. OCR and ECAR were measured with an XF24 extracellular flux analyzer (Seahorse Bioscience) as described31. Suspension cells were attached to culture plates using Cell-Tak (BD Bioscience). OCR and ECAR were measured in unbuffered DMEM (Sigma-Aldrich) supplemented with 10 mM D-glucose (Sigma-Aldrich) and 10 mM L-glutamine, as indicated. OCR and ECAR values were normalized to cell number. For certain experiments, ECAR was measured over time following injection of 10mM D-glucose, oligomycin and 2-deoxy glucose (2DG). Glycolytic capacity is defined as the difference between the ECAR following the injection of oligomycin and ECAR following glucose injection. Nontargeted metabolomic analyses were performed as described using LC Q Exactive Mass Spectrometer (LC-QE-MS) (Thermo Scientific)6.
T cell transfer model of colitis
Splenic CD4 T cells were isolated as described above, and naive effector (CD4+CD25−CD45RBhi) T cells were sorted (FACSVantage; BD Bioscience). Naive effector T cells were injected intraperitoneally into 6–8-week-old C57BL/6 RAG1−/− recipients (106 cells per mouse). Recipient animals were purchased from Jackson Labs and were assigned to groups randomly and co-housed in blinded fashion. Thy1.1 naive effector T cells were used in select experiments. Because the mice were H. pylori negative, and colitis does not occur spontaneously in this setting, disease was initiated 2 weeks following T cell injection with 200 p.p.m. piroxicam (Sigma-Aldrich) in powdered rodent chow for 3 d to enhance mucosal exposure to enteric bacteria and induce colitis6. 21 d following piroxicam treatment, Glut1 transgenic or littermate control Treg cells (sorted CD4+CD25+CD45RBlo, 2 × 105 cells per mouse) were injected as indicated. Mouse weights were monitored tri-weekly. At the end of the experiment, total CD4+ and CD4+Foxp3+ Treg in the spleen and mesenteric lymph nodes were analyzed by flow cytometry.
Statistical analysis
Sample sizes were determined by prior experience and estimated power calculations. Statistical analyses were performed with Prism software (GraphPad). Data were analyzed using a two-tailed Student’s t-test, and P < 0.05 was considered significant. Longitudinal data was analyzed by two-way ANOVA followed by Tukey’s post hoc test.
Supplementary Material
Acknowledgments
We thank members of the Rathmell and Wells laboratories for discussions, and the Immunological Genome Project. Supported by the Crohn’s and Colitis Foundation of America (Senior Research Grant to J.C.R.), the Alliance for Lupus Research (J.C.R.), the US National Institutes of Health (R01HL 108006 and R01105550DK to J.C.R.; P01HL018646 to L.A.T. and J.C.R.; F31CA183529 to R.J.K.; R00CA168997 to J.W.L.; and R01AI070807 and P01AI073489 to A.D.W.), and the German Research Foundation (Deutsche Forschungsgemeinschaft; P.J.S.).
Footnotes
Accession codes. GEO: RNAseq data, GSE84919; ArrayExpress: microarray data, E-MTAB-4561.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
V.A.G., R.J.K., A.D.W. and J.C.R. designed the study, interpreted data and wrote the manuscript. V.A.G. and R.J.K. performed most of the experiments. M.O.J., S.C. and P.J.S. performed experiments to analyze Foxp3 regulation of metabolism. A.G.N. assisted V.A.G. and R.J.K. and maintained animals that were essential for the study. M.O.W. performed mass spectrometry. A.A.d.C. analyzed RNAseq data. J.W.L. assisted in metabolomics analysis. N.J.M. and L.A.T. assisted with data analysis and interpretation. A.D.W. performed microarray analysis.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212:1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang CH, et al. Post-transcriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macintyre AN, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beier UH, et al. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J. 2015;29:2315–2326. doi: 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerriets VA, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smigiel KS, Srivastava S, Stolley JM, Campbell DJ. Regulatory T-cell homeostasis: steady-state maintenance and modulation during inflammation. Immunol Rev. 2014;259:40–59. doi: 10.1111/imr.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng H, Chi H. Metabolic control of regulatory T cell development and function. Trends Immunol. 2015;36:3–12. doi: 10.1016/j.it.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laidlaw BJ, et al. Production of IL-10 by CD4+ regulatory T cells during the resolution of infection promotes the maturation of memory CD8(+) T cells. Nat Immunol. 2015;16:871–879. doi: 10.1038/ni.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nosbaum A, et al. Cutting edge: regulatory T cells facilitate cutaneous wound healing. J Immunol. 2016;196:2010–2014. doi: 10.4049/jimmunol.1502139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oestreich KJ, et al. Bcl-6 directly represses the gene program of the glycolysis pathway. Nat Immunol. 2014;15:957–964. doi: 10.1038/ni.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Procaccini C, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng H, et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh A, et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol. 2015;16:188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrestha S, et al. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol. 2015;16:178–187. doi: 10.1038/ni.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park Y, et al. TSC1 regulates the balance between effector and regulatory T cells. J Clin Invest. 2013;123:5165–5178. doi: 10.1172/JCI69751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 23.Peng G, et al. Toll-like receptor 8-mediated reversal of CD4+ regulatory T cell function. Science. 2005;309:1380–1384. doi: 10.1126/science.1113401. [DOI] [PubMed] [Google Scholar]
- 24.Sutmuller RP, et al. Toll-like receptor 2 controls expansion and function of regulatory T cells. J Clin Invest. 2006;116:485–494. doi: 10.1172/JCI25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voo KS, et al. Targeting of TLRs inhibits CD4+ regulatory T cell function and activates lymphocytes in human peripheral blood mononuclear cells. J Immunol. 2014;193:627–634. doi: 10.4049/jimmunol.1203334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyirenda MH, et al. TLR2 stimulation drives human naive and effector regulatory T cells into a Th17-like phenotype with reduced suppressive function. J Immunol. 2011;187:2278–2290. doi: 10.4049/jimmunol.1003715. [DOI] [PubMed] [Google Scholar]
- 27.Sujino T, et al. Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science. 2016;352:1581–1586. doi: 10.1126/science.aaf3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, et al. MyD88 adaptor-dependent microbial sensing by regulatory T cells promotes mucosal tolerance and enforces commensalism. Immunity. 2015;43:289–303. doi: 10.1016/j.immuni.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scharschmidt TC, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo L, et al. Rab8a interacts directly with PI(3)Kγ to modulate TLR4-driven PI(3)K and mTOR signalling. Nat Commun. 2014;5:4407. doi: 10.1038/ncomms5407. [DOI] [PubMed] [Google Scholar]
- 31.Caro-Maldonado A, et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol. 2014;192:3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everts B, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKε supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs SR, et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisson S, et al. Continuous activation of autoreactive CD4+ CD25+ regulatory T cells in the steady state. J Exp Med. 2003;198:737–746. doi: 10.1084/jem.20030686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 36.Basu S, Hubbard B, Shevach EM. Foxp3-mediated inhibition of Akt inhibits Glut1 (glucose transporter 1) expression in human T regulatory cells. J Leukoc Biol. 2015;97:279–283. doi: 10.1189/jlb.2AB0514-273RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arvey A, et al. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2014;15:580–587. doi: 10.1038/ni.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilhelm K, et al. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature. 2016;529:216–220. doi: 10.1038/nature16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panduro M, Benoist C, Mathis D. Tissue Tregs. Annu Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cretney E, Kallies A, Nutt SL. Differentiation and function of Foxp3+ effector regulatory T cells. Trends Immunol. 2013;34:74–80. doi: 10.1016/j.it.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Ostroukhova M, et al. The role of low-level lactate production in airway inflammation in asthma. Am J Physiol Lung Cell Mol Physiol. 2012;302:L300–L307. doi: 10.1152/ajplung.00221.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leifer CA, Medvedev AE. Molecular mechanisms of regulation of Toll-like receptor signaling. J Leukoc Biol. doi: 10.1189/jlb.2MR0316-117RR. http://dx.doi.org/10.1189/jlb.2MR0316-117RR (published online 24 June 2016) [DOI] [PMC free article] [PubMed]
- 43.Xie X, et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc Natl Acad Sci USA. 2011;108:6474–6479. doi: 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohl K, Tenbrock K. Regulatory T cells in systemic lupus erythematosus. Eur J Immunol. 2015;45:344–355. doi: 10.1002/eji.201344280. [DOI] [PubMed] [Google Scholar]
- 45.Byng-Maddick R, Ehrenstein MR. The impact of biological therapy on regulatory T cells in rheumatoid arthritis. Rheumatology (Oxford) 2015;54:768–775. doi: 10.1093/rheumatology/keu487. [DOI] [PubMed] [Google Scholar]
- 46.Rathmell JC, Elstrom RL, Cinalli RM, Thompson CB. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. Eur J Immunol. 2003;33:2223–2232. doi: 10.1002/eji.200324048. [DOI] [PubMed] [Google Scholar]
- 47.Allan SE, Song-Zhao GX, Abraham T, McMurchy AN, Levings MK. Inducible reprogramming of human T cells into Treg cells by a conditionally active form of FOXP3. Eur J Immunol. 2008;38:3282–3289. doi: 10.1002/eji.200838373. [DOI] [PubMed] [Google Scholar]
- 48.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Rowell EA, Thomas RM, Hancock WW, Wells AD. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J Biol Chem. 2006;281:36828–36834. doi: 10.1074/jbc.M608848200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.