Abstract
Although the presence of cryptic host specificity has been documented in Blastocystis, differences in infection rates and high genetic polymorphism within and between populations of some subtypes (ST) have impeded the clarification of the generalist or specialist specificity of this parasite. We assessed the genetic variability and host specificity of Blastocystis spp. in wild howler monkeys from two rainforest areas in the southeastern region of Mexico. Fecal samples of 225 Alouatta palliata (59) and A. pigra (166) monkeys, belonging to 16 sylvatic sites, were analyzed for infection with Blastocystis ST using a region of the small subunit rDNA (SSUrDNA) gene as a marker. Phylogenetic and genetic diversity analyses were performed according to the geographic areas where the monkeys were found. Blastocystis ST2 was the most abundant (91.9%), followed by ST1 and ST8 with 4.6% and 3.5%, respectively; no association between Blastocystis ST and Alouatta species was observed. SSUrDNA sequences in GenBank from human and non-human primates (NHP) were used as ST references and included in population analyses. The haplotype network trees exhibited different distributions: ST1 showed a generalist profile since several haplotypes from different animals were homogeneously distributed with few mutational changes. For ST2, a major dispersion center grouped the Mexican samples, and high mutational differences were observed between NHP. Furthermore, nucleotide and haplotype diversity values, as well as migration and genetic differentiation indexes, showed contrasting values for ST1 and ST2. These data suggest that ST1 populations are only minimally differentiated, while ST2 populations in humans are highly differentiated from those of NHP. The host generalist and specialist specificities exhibited by ST1 and ST2 Blastocystis populations indicate distinct adaptation processes. Because ST1 exhibits a generalist profile, this haplotype can be considered a metapopulation; in contrast, ST2 exists as a set of local populations with preferences for either humans or NHP.
Introduction
Blastocystis spp. is a common parasite that colonizes the intestines of many animals, including mammals, reptiles and birds. Although this microorganism is the most common eukaryotic parasite identified in human feces, its pathogenic role remains controversial. While some studies have reported that this parasite is associated with the development of cutaneous and intestinal disorders, other studies report no clinical manifestations [1–5]. In addition, Blastocystis exhibits high genetic polymorphism, presenting at least 17 subtypes (ST). The ST1-ST4 are frequently observed in humans but can also occur in birds, pigs, cows, dogs, rats and non-human primates (NHP). ST5 is common in pigs, and ST6 and ST7 are common in birds, while ST8 is documented in NHP and ST9 is only detected in humans. The ST10-ST17 have never been identified in people [2, 6, 7]; although, a recent study showed that ST12 may occur in humans [8]. The potential zoonotic transmission of Blastocystis has been under debate, as studies have reported dissimilar results [9–15]. In Kathmandu, Nepal, the potential transmission of Blastocystis between local rhesus monkeys (Macaca mulatta) and children was assessed; the authors detected three subgroups of ST2 shared between the children and the monkeys, suggesting that the local rhesus monkeys might serve as a potential source for human ST2 infections [12]. Studies primarily concentrating on the genetic variability of Blastocystis recovered from humans and several primate genera, including Old and New World monkeys and prosimians, have confirmed the cryptic host specificity of ST1 and ST3. These parasites exhibit only minimal overlap between the sources, indicating that some ST3 isolates have adapted to NHP, while others have adapted to humans. Furthermore, reflecting considerable overlap in ST2, haplotypes exists among humans and NHP, and transmission among these species may occur in zoological parks and animal sanctuaries [11, 14, 16]. In contrast, a study performed in the Rubondo Island National Park, Tanzania, where different autochthonous and introduced NHP live together, ST1-ST3 and ST5 were detected. Interestingly, the chimpanzees (Pan troglodytes) were parasitized only by ST1, which formed a unique phylogenetic clade. This finding suggested that Rubondo chimpanzees were colonized by a single, host-specific Blastocystis strain that circulates among the members of the group and that transmission of this genotype does not occur between primate populations and thus does not constitute a reservoir for human infections [13]. Recently, Helenbrook et al. [15] studied fecal samples from humans and howler monkeys (Alouatta palliata aequatorialis) from Ecuador living in allopatric areas at close proximity. Blastocystis ST1-ST3 were detected in the human samples, while all monkeys had ST8, thus questioning the zoonotic potential of ST8. Furthermore, howlers are particularly interesting because these animals appear to be more sensitive to infectious diseases, such as yellow fever and gastrointestinal parasites [17–19]. Different howler species live in forest fragments, disturbed habitats, and in close proximity to human populations [17, 19].
For many pathogens, the presence of multiple host species has important epidemiological and evolutionary implications, i.e., alternative host species might be reservoirs of a disease extinct in one host. In addition, parasites that interact with multiple host species may be less locally adapted, and consequently, these organisms are expected to be less specific. In addition, the assessment of host ranges can be hindered by the presence of cryptic species: even if a parasite is able to infect different host species, differences in infection rates among the alternative hosts might be interpreted as a consequence of a local adaptation process, leading to the preference for certain host species over others [20]. Hence, the identification of a generalist or specialist profile of a parasite for its host is relevant for understanding its prevalence, transmission and other biological features. Therefore, the aim of the present study was to assess the genetic variability and host specificity of Blastocystis spp. populations in howler monkey species A. palliata and A. pigra from two sylvatic areas of Mexico.
Materials and Methods
Study population
The genus Alouatta comprises 6 species widely distributed between Mexico and Argentina in allopatric or sympatric patterns. A. palliata and A. pigra howler monkeys are important inhabitants of some tropical rainforest areas in the southeastern region from Mexico, and both species are critically endangered and primarily distributed in states of Chiapas, Campeche, Quintana Roo, Tabasco, Veracruz and Yucatan. These arboreal monkeys live in troops, and their population densities vary considerably, ranging from <10 to approximately 100 individuals per km2 [17]. Although these monkeys are largely folivorous, their indiscriminate diet is likely one of the main reasons that these animals are adaptable to changing ecological landscapes, whether natural or anthropogenic [19].
Sample strategy and study area
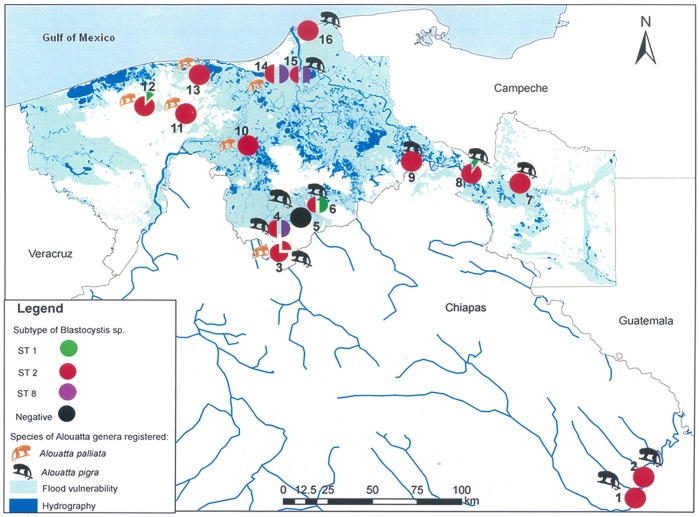
The present study was conducted in Tabasco and Chiapas states in the southeastern region of Mexico. Tabasco is a large flat coastal alluvial plain characterized by poor drainage and large areas of permanently or seasonally inundated terrains. The Grijalva and lower Usumacinta Rivers water the eastern and central parts of the plain. The weather is warm and moist, with a high constant temperature of ~26°C and an annual range of rainfall of 2000 to 4000 mm H2O. Chiapas, the neighboring state, has areas with different climates, abundant rainfall and diverse topology. Furthermore, the study region is a mosaic formed by native vegetation patches and extensive urban areas, grasslands, crops, shrubs, flood areas, and riparian forests. The native vegetation comprises patches of evergreen forest and riparian forest, mangle and marsh areas [21]. Field collection occurred in 2014 and 2015. The sampling sites were selected among vegetation areas used by Alouatta species for feeding, and there is information available on the general landscape, hydrography and flood vulnerability recorded from Tabasco's Ecological Planning Program [22]. An arc-tool called “neighbor joining” in the Arcview 10.0 program was used to discriminate overlapping areas. Initially, we established a random minimum separation of 5 km between every population to avoid resampling the same population. This distance was established after considering the mobility reported for a group of A. palliata, which traveled less than 50 m on 12 of the 34 days during that these animals were followed. The A. pigra group traveled the same distance but on 15 of the 34 days [23]. After interviews with local people, subdivisions and fieldwork, the presence of these monkeys was corroborated when howler troops were visually located using binoculars or based on their vocalizations. Sampling sites were selected considering 16 buffer areas, ranging from 7,852 m as minimum to 13,230 m as maximum, with distances of 10 to 357 km between each buffer. These sites included 14 sites in Tabasco and two sites in Chiapas (Table 1). The sites where A. palliata and A. pigra populations showed allopatric distribution were separated by the Grijalva River, living on the west and east sides of the river, respectively, except for one site in Tabasco (site 3, corresponding to the Tapijulapa, Cascadas de Villa Luz and Kolenchen localities) where both monkey species live in a sympatric area. The municipality Emiliano Zapata in Tabasco (site 8a) was of particular interest because, at this site, there is a group of small islands in the middle of the Chaschoc-Seja Lagoon (with some howlers living on two of the islands), with only boat access from the Bertollini Ranch (site 8b). Although some areas showed different grades of anthropogenic disturbance, among the 16 sites, 9 sites are considered as conserved zones, including forest and wetlands, while the other sites are meadows without human interactions, except for site 10 (Parque La Venta/ Tabasco), in which there is a proximity to humans and their homes.
Table 1. Infection rates of Blastocystis subtype (ST) for howler monkey populations.
| Site ID | Localities/State | Species | N1 | % Blastocystis infection2 | % Blastocystis ST3 | Accession number of GenBank | ||
|---|---|---|---|---|---|---|---|---|
| ST1 | ST2 | ST8 | ||||||
| 1 | Playon de la Gloria/CHIAPAS | A. pigra | 9 | 11.1 (1/9) | - | 100 (1/1) | - | KT591833 |
| 2 | Reforma Agraria/CHIAPAS | A. pigra | 40 | 60 (24/40) | - | 100 (24/24) | - | KT591789-94, KT591804-05, KT591814-17, KT591799-803, KT591806-09, KT591811-13 |
| 3 | Tapijulapa, Cascadas de Villa Luz, Kolenchen/ TABASCO | A. pigra | 5 | 20 (1/5) | - | 100 (1/1) | - | KT591831, KT591839, KT591769 |
| A. palliata | 8 | 37.5 (3/8) | 100 (3/3) | |||||
| 4 | Poana, Xicotencatl/ TABASCO | A. pigra | 22 | 18.2 (4/22) | - | 50 (2/4) | 50 (2/4) | KT591786-87, KT591852-53 |
| 5 | Pochitocal/ TABASCO | A. pigra | 5 | 0 | - | - | - | - |
| 6 | Cascadas de Agua Blanca/ TABASCO | A. pigra | 3 | 66.6 (2/3) | 50 (1/2) | 50 (1/2) | - | KT591851 |
| 7 | Rancheria Josefa Ortiz de Dominguez/ TABASCO | A. pigra | 5 | 80 (4/5) | - | 100 (4/4) | - | KT591795-98 |
| 8a | Islands, Xeha Lagoon, Emiliano Zapata/ TABASCO | A. pigra | 34 | 47(16/34) | 6.3 (1/16) | 93.7 (15/16) | - | KT591850, KT591770-75, KT591777-85 |
| 8b | Bertollini Ranch, Emiliano Zapata/ TABASCO | A. pigra | 16 | 37.5 (6/16) | 16.6 (1/6) | 83.3 (5/6) | - | KT591848, KT591820-23,KT591838 |
| 9 | Los Pajaros/ TABASCO | A. pigra | 4 | 25 (1/4) | - | 100 (1/1) | - | KT591788 |
| 10 | Parque La Venta/ TABASCO | A. palliata | 8 | 100 (8/8) | - | 100 (8/8) | - | KT591840-47 |
| 11 | Cocoa plantation and Ranch Cali/ TABASCO | A. palliata | 3 | 33.3 (1/3) | - | 100 (1/1) | - | KT591835 |
| 12 | Carlos Greene/ TABASCO | A. palliata | 30 | 26.6 (8/30) | 12. 5 (1/8) | 87.5 (7/8) | - | KT591849, KT591824-30 |
| 13 | Palestina/ TABASCO | A. palliata | 4 | 25 (1/4) | - | 100 (1/1) | - | KT591768 |
| 14 | Tabasquillo/ TABASCO | A. palliata | 7 | 28.6 (2/7) | - | 50 (1/2) | 50 (1/2) | KT591837, KT591854 |
| 15 | San Juanito and Tres Brazos/ TABASCO | A. pigra | 11 | 18.2 (2/11) | - | 100 (2/2) | - | KT591818-19 |
| 16 | Nueva Alianza and La Victoria /TABASCO | A. pigra | 11 | 27.3 (3/11) | - | 100 (3/3) | - | KT591834, KT591836, KT591832 |
| A. pigra | 166 | 38.5 (64/166) | 4.6 (3/64) | 92.1 (59/64) | 3.1 (2/64) | |||
| A. palliata | 59 | 38.9 (23/59) | 4.3 (1/23) | 91.3 (21/23) | 4.3 (1/23) | |||
| Total | 225 | 38.7 (87/225) | 4.6 (4/87)4 | 91.9 (80/87)5 | 3.5 (3/87)6 | |||
1N, number of analyzed howlers.
2% Blastocystis infection, obtained as all positive samples for Blastocystisx100/N.
3% Blastocystis ST, obtained as positive samples for each STx100/all positive samples for Blastocystis in a specific site.
4ST1 vs ST2+ST8 for A. palliata or A. pigra; p = 0.9471, 95% Confidence interval (95%IC) = 0.09–28.5.
5ST2 vs ST1+ST8 for A. palliata or A. pigra; p = 0.4981, 95%IC = 0.29–9.17.
6ST8 vs ST1+ST2 for A. palliata or A. pigra; p = 0.7840, 95%IC = 0.71–20.85.
Species identification and fecal sample collection
Species identification was performed using morphological characteristics: A. pigra is larger, and their hair color is completely black. Additionally, testes are evident in male A. pigra infants, unlike A. palliata males, in which the testicles do not descend until sexual maturity. The fur of A. palliata is not uniform in color and is typically dense, with golden flecks in the underarm region and areas with no pigment on the hands, feet and tail [24]. Fecal samples were collected using a non-invasive technique: waiting until the monkeys defecated. Only one sample per animal was collected. To this end, at least two persons monitored the howler monkey troops: one person recovered the fecal sample and the other person followed the howlers to obtain their approximate age, sex and some identification features of each individual, thereby guaranteeing no resampling. Additionally, we verified the species identification using mitochondrial and nuclear markers [25] and determined whether each sample and its corresponding monkey matched. All samples were fresh and carefully collected using gloves to avoid contamination from the soil, arthropods and vegetal detritus; only the top of each sample was saved and stored in 70% ethanol for further molecular analysis.
DNA extraction and PCR assays
DNA was extracted from approximately 100 mg of each fecal sample using the ZR Fecal DNA MiniPrep Kit (Zymo Research, Irvine, USA). The primers used for end-point PCR assays amplified an ~500 bp region of the small subunit rDNA (SSUrDNA) gene [26], [27]. PCR amplifications were performed in a final volume of 25 μL, containing 6.25 pmol of each primer, 1X PCR buffer (8 mM Tris-HCl, pH 8, and 20 mM KCl), 2.4 mM MgCl2, 0.5 mM dNTPs, 0.01 mg BSA, and 1 U Taq DNA Polymerase (Promega). Up to 50 ng of DNA (~2 μL) was used as a template to amplify the genomic sequences. The following amplification conditions were used: 94°C for 5 min, followed by 36 cycles at 94°C-30 s, 54°C-30 s and 72°C-30 s, with a final extension step at 72°C for 10 min. The amplicons were assessed using electrophoresis on 1.2% agarose gels, after which the bands were purified using the AxyPrep PCR Clean-up Kit (Axigen Biosciences, CA, USA) and sequenced on both strands by a commercial supplier. Chromatograms were evaluated with Mesquite software using the Chromaseq package [28–29], with phred and phrap algorithms for base calling, assigning quality values to each one and assembling contigs [30–32].
Phylogenetic reconstruction and genetic variation analysis
All sequences were subjected to BLAST search in the GenBank database, and the sequences obtained in this work were accessed with the numbers KT591768-KT591854. Multiple alignments were performed using the CLUSTAL W [33] and Muscle [34] programs, with manual adjustment in MEGA 5.05 software [35]. The best-fit model of nucleotide substitution was determined using the Akaike Information Criterion in Modeltest software, version 3.7 [36] and the Hasegawa Kishino Yano model with gamma distribution and invariable sites. Phylogenetic reconstruction using Bayesian inference was performed using Mr. Bayes 3.1.2 [37]. The analysis was performed over 10 million generations, with sampling trees every 100 generations. Trees with scores lower than those at the stationary phase (burn-in) were discarded. The trees that reached the stationary phase were collected and used to build consensus trees. Other sequences were collected from GenBank and used as subtype references and for population genetics analysis (S1 Table).
A Median Joining Network analysis [38] was performed using NETWORK 4.611(fluxus-engineering.com) with default settings and assumptions. Genetic diversity analyses within and between populations were performed using DnaSPv4 [39], and indices, such as nucleotide diversity (π), haplotype polymorphism (θ), gene flow (Nm),genetic differentiation index (FST) and Tajima’s D test were obtained. These indices have been previously applied for population genetics studies in Blastocystis [40, 41]. They denote the average proportion of nucleotide differences between all possible pairs of sequences in the sample (π); the proportion of nucleotide sites expected to be polymorphic in any suitable sample from this region of the genome (θ); the movement of organisms among subpopulations (Nm); and the differentiation between or among populations (FST). To support the data interpretation, populations strongly differentiated have an Nm < 1, whereas those with an Nm > 4 behave as a single panmictic unit. For FST, the following commonly used values for genetic differentiation were considered: 0 to 0.049, small; 0.05 to 0.149, moderate; 0.150 to 0.25, great; and values above 0.26 indicate enormous genetic differentiation. Negative values for Tajima’s D test suggest a recent expansion process or an effect of purifying selection [42].
Statistical analysis
Descriptive statistics are expressed as the mean and standard deviation (SD). Analysis using Student's t test for unrelated samples and Mantel–Haenszel test were applied, and 95% confidence intervals (95%CI) were also obtained. Data analysis was performed using SPSS software, version 15.0 (SPSS Institute, Chicago, IL).
Ethics statement
All fecal samples were collected using a non-invasive technique after the monkeys defecated. Sampling and procedures were performed in accordance with the provisions of the Regulations of the Environment and Natural Resources Ministry and the Under-Secretary of Management for Environmental Protection NOM-059-SEMARNAT-2010; reference SGPA/DGVS/04725/13. Academic authorities of the Universidad Juarez Autonoma de Tabasco authorized the present study (reference number UJAT-2013-IB-43).
Results
Frequency of Blastocystis spp. in A. palliata and A. pigra
In 14 sites from Tabasco and two sites from Chiapas, fecal samples from 225 howler monkeys, A. palliata (59) and A. pigra (166) were collected and analyzed. Table 1 summarizes the positive Blastocystis ST results for the animals from each site; only at site 5 (Pochitocal, Tabasco) were the monkeys not parasitized. The frequency of Blastocystis spp. was 38.7% (n = 87), 39% for A. palliata and 38% for A. pigra; ST2 was the most abundant (91.9%), followed by ST1 and ST8 with 4.6% and 3.5%, respectively. No association between Blastocystis ST and Alouatta species was observed. In addition, we identified infected howlers with similar frequencies and ST distributions in flooded and non-flooded areas (Fig 1).
Fig 1. Field sampling sites, hydrography and frequency of Blastocystis ST in Alouatta palliata and A. pigra from Tabasco and Chiapas.
The pie charts indicate those localities with positive samples and each colour represents a different ST; proportions of colours are according to ST frequencies.
Phylogenetic reconstruction and genetic variation analysis
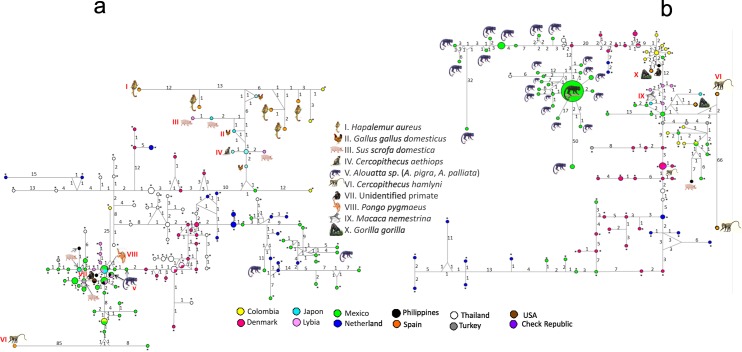
The contigs were assembled using both forward and reverse chromatograms from each sample, and analysis using Mesquite/Chromaseq software showed well-defined peaks without significant background noise. A Bayesian phylogenetic tree was built for SSUrDNA using all worldwide available sequences recorded in GenBank (S1 Fig). The sequences were grouped into the ST1, 2, and 8 clusters. The haplotype networks for ST1 and ST2 showed contrasting distributions: for ST1 (Fig 2A), several haplotypes from different mammals (humans, NHP, and pigs) and birds are homogeneously distributed in different countries, and in general, few mutational differences are present among them. Conversely, for ST2 (Fig 2B), a principal dispersion center grouped most of the Mexican howlers together, with the remaining sequences also closely distributed. Interestingly, some haplotypes showed high mutational differences, particularly in the NHP vs. human haplotypes. For example, for the Mexican sequences, three howler haplotypes diverged from the principal dispersion center by over 40 mutational changes, with the most divergent being a sample from one of the islands (site 8a). A similar phenomenon was observed for the ST2 sequences previously reported for Cercopithecus hamlyni from Spain [26], with one haplotype close to the human haplotypes, and another haplotype markedly distant, with up to 76 mutational changes.
Fig 2. Haplotype networks for Blastocystis.
Haplotype network trees using SSUrDNA sequences from different countries and hosts for ST1 (a) and ST2 (b). Numbers in branches refer to mutational changes; sizes of circles and colors are proportional to haplotype frequencies. For those animal haplotypes, an image and Roman reference numbers were included, while for human haplotypes, asterisks were added.
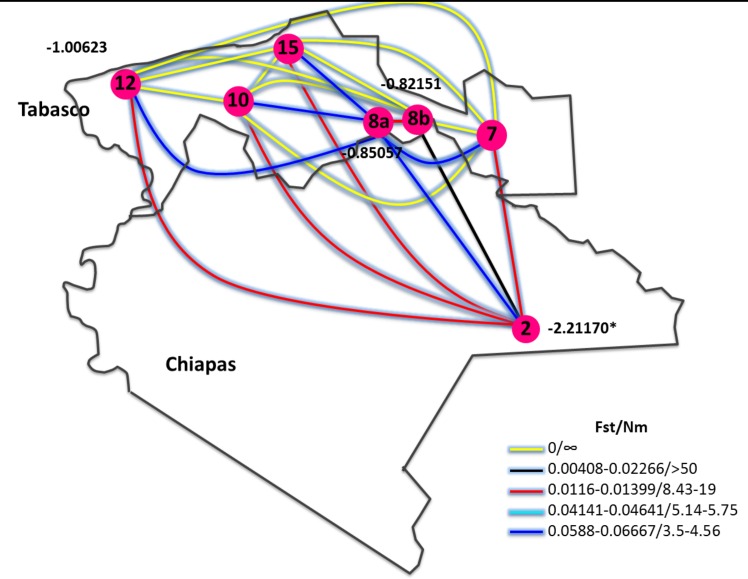
Fig 3 shows the ranges of FST and Nm among the different sampling sites in Tabasco and Chiapas. Despite the great geographic distances between some of the populations, minimal differentiation and high gene flow were observed. As expected, the population with the lowest differentiation and gene flow was in the islands (site 8a), in contrast with site 8b, which showed FST and Nm values similar to those of the other sampling sites. The populations at sites 2, 8a, 8b and 12 yielded negative Tajima’s D values, but only for site 2, and this result was statistically significant (p<0.01).
Fig 3. Schematic representation of interactions among population indexes.
The gene flow (Nm), genetic differentiation index (FST), and Tajima’s D values of Blastocystis ST by SSUrDNA analysis, according to different sampling sites; only those sites in which there were enough infected howlers to obtain the indexes are shown. The number together the sampling size circle, mean the Tajima’s D value. * p<0.01
Table 2 shows the comparisons between the indexes obtained for ST1 and ST2 of humans and NHP. The ST8 haplotype was not analyzed because of an insufficient number of sequences. These indexes support the topologies of the contrasting haplotype networks (Fig 2A and 2B). In the intra-ST comparison between humans and NHP, human Blastocystis ST1 populations had lower π values than those of NHP, whereas for ST2, human and NHP parasites had similar variability; despite the disparity between the number of available sequences in the GenBank of humans and NHP for ST1, the genetic diversity values were statistically significant. The Nm and FST values for ST1 and ST2 were inverted, suggesting that Blastocystis ST2 populations from humans and NHP are strongly differentiated with occasional gene flow, while scarcely any differentiation between ST1 parasite populations from humans and NHP was detected.
Table 2. Values of nucleotide diversity (π), haplotype polymorphism (θ), gene flow (Nm) and genetic differentiation index (FST) for sequences of humans and HNP populations of Blastocystis ST1 and ST2.
| N | π±SD | p* | θ±SD | p* | FST | Nm | |
|---|---|---|---|---|---|---|---|
| ST1 | |||||||
| Humans | 147 | 0.0241±0.0030 | 0.0001 | 0.0298±0.0080 | 0.0001 | 0.071 | 3.25 |
| NHP | 14 | 0.061±0.0169 | 0.0694±0.00026 | ||||
| ST2 | |||||||
| Humans | 94 | 0.0211±0.0022 | 0.247 | 0.0238±0.0070 | 0.0001 | 0.644 | 0.14 |
| NHP | 83 | 0.0204±0.0059 | 0.0774±0.0204 |
*Student’s t-test, for independent samples.
Discussion
The few reports on the molecular identification of Blastocystis ST in Alouatta monkeys [14, 15, 43] have shown dissimilar results for this NHP genus. The results obtained in the present study not only strengthen the existence of cryptic host specificity for ST1 and ST2 but also show that these ST have different population structures, with certain haplotypes of ST2 showing a preference for either humans or NHP. Generalist parasites infecting several host species can actually be cryptic parasite species, each characterized by a different degree of host specificity, particularly influenced by local adaptations that have led to a preference for certain host species over others and have zoonotic implications. Therefore, the generalist profile of some parasites may be a key process for the maintenance of their genetic diversity and population diversification [16, 20, 44–46].
In the present study, we observed that ST1 exhibits a generalist profile because there was minimal differentiation between parasite populations from humans and NHP, as well as substantial gene flow between them, suggesting that ST1 landscapes resemble a metapopulation (ensemble of interacting populations with a finite lifetime) [47–50]. In contrast, some Blastocystis ST2 haplotypes have diverged similar to a set of local populations with preferences towards certain host species (humans or NHP). Both population structures have relevance for understanding the prevalence and transmission of Blastocystis ST. According to these findings, the putative metapopulation structure of ST1 implies a link with the processes of population turnover, extinction and establishment of new populations [48], i.e., Blastocystis ST1 may have the capacity to recolonize vacant niches, such as new hosts, even of different species, supporting the zoonotic transmission of this parasite [6–9, 11–14]. In addition, Sanchez-Aguillon et al. [27] documented reinfection by Blastocystis ST1 in an asymptomatic patient who received anti-parasitic treatment and follow-up three months earlier.
However, the existence of ST2 as a set of resident populations locally adapted by each parasite population would support the preference for infecting either humans or NHP. Such a process of local adaptation is supported by Helenbrook et al. [51] who observed that Blastocystis was the unicellular parasite most frequent (60%) in samples of A. palliata aequatorialis howlers from Ecuador. Subsequently, in another study performed in similar populations of these howler monkeys, 68% of these animals were infected with Blastocystis ST8, while humans living in close proximity were infected with ST1, ST2 and ST3 [15]. Interestingly, although these subtypes (ST1-ST3) are distributed worldwide, they are common in America [8, 40, 41]. In the present study, 39% and 38% of A. palliata and A. pigra howlers, respectively, were primarily infected with Blastocystis spp., and of these, 91.3% of A. palliata and 92.1% of A. pigra howler monkeys were harboring Blastocystis ST2. Other relevant findings regarding the host specificity of ST2 were derived from the phylogenetic tree of parasites from Rubondo Island, Tanzania [13], in which the cluster for ST2 showed two clades for Blastocystis, those from humans and those from NHP.
In addition, as cited above, local adaptations are relevant for the maintenance of genetic diversity and population diversification. The high degree of intra-ST genetic polymorphism has been reliably documented, particularly for ST1 and ST3 [12, 14, 16, 41, 43–44], suggesting that populations of recent origin have undergone a radiation process [52]. However, the great divergence of some haplotypes for ST1 and ST2 observed in the present study deserves special attention. The presence of divergent haplotypes with more than 40 mutational differences between sequences from NHP of Spain and Mexico, particularly in a monkey from an island (site 8a), suggests the island-continent model described by Wright [53], in which many finite subpopulations (equivalent to the continent) are present, including the source of migrants to the island. When the amount of gene flow and the population size on the islands are both large, the allele frequency on the islands will soon become similar to that of the continent. However, if the population size on the island is small or if the rate of gene flow is low (as in the present study), then genetic drift could lead to random changes in allele frequency. As a result, the allele frequency on the islands may differ significantly from that on the continent and that of the migrants [54]. Alternatively, the population size could correspond to an epidemic population structure such as that depicted by Maynard-Smith et al. [55], in which there is frequent recombination within all members of the population, such that the structure is a net rather than a tree.
It has been observed that the use of more than one genetic marker has facilitated the clarification of the differences between organism populations. However, some studies of genetic variability focused only on the SSUrDNA gene analysis have shown that this marker is sensitive enough to resolve phylogenetic relationships, population differentiation events and cryptic infections in both Blastocystis and other parasites [8, 9, 14, 36, 56–58].
Since mixed ST infections are common [43, 59–60], we used Mesquite/Chromaseq software to identify co-infections in the howler monkeys as this technique has been used to reduce bias and misinterpretations during sequence analysis [28–32]; in this way, this analysis could distinguish a double profile in the chromatogram sequences, suggesting mixed infections based on different ST. However, future use of ST-specific primers, cloning, and SSCP analysis should be performed to confirm the presence or absence of ST co-infections.
Although a definitive mode of transmission for Blastocystis has not been identified, it has been suggested that transmission occurs orally through water, food or direct contact with the parasite; here, Blastocystis transmission apparently occurs through water [61], but wild howler monkeys have rarely been observed drinking river water. Instead, these animals use arboreal water cisterns [62], suggesting that transmission may occur when monkeys are in contact with contaminated leaves or other non-arboreal elements (i.e., lianas, vines and epiphytes) [63–64], wet ground or sewage during their movement between trees. Nevertheless, transmission may occur when monkeys drink water directly from the ground; Serio-Silva and Rico-Gray observed this behavior for A. palliata and A. pigra feeding and traveling on the ground, in small patches in locations with scarce trees, and during the dry and wet seasons in Balancan, Tabasco [65].
The results of the present study demonstrate that howler monkey populations of A. palliata and A. pigra with locally adapted Blastocystis ST1 and ST2 populations exhibit distinct generalist and specialist types of host specificity. In addition, these data suggest that the Blastocystis ST2 populations in humans are highly differentiated from those of NHP, while the ST1 populations are only minimally differentiated. The host generalist and specialist specificities exhibited by ST1 and ST2 Blastocystis populations are thus distinct processes. Because ST1 exhibits a generalist profile, this haplotype can be considered a metapopulation; in contrast, Blastocystis ST2 exists as a set of local populations with preferences for either humans or NHP.
Furthermore, it is important to highlight the use of population genetics studies in the epidemiology of this eukaryote because these studies may clarity some aspects of host-parasite relations; for example, how genetic variability within a subtype is reflected in phenotypic and functional variability and its potential role on the symptoms of the parasite carriers [66].
Supporting Information
Bayesian phylogenetic tree using a fragment of SSUrDNA sequences; the values of the nodes indicate posterior probabilities values using 10 million generations. ST and GenBank accession numbers are shown, as well as identification of each sample.
(TIF)
(XLSX)
Acknowledgments
The authors thank Carlos Baños-Ojeda, Guadalupe Hernandez and staff of Parque Museo de la Venta en Villahermosa, Tabasco. We also thank Estacion Biologica “La Florida”, Tacotalpa, Tabasco. To SERNAPAM and Grupo Etnico Chinanteco, Centro Ecoturístico “Las Guacamayas”, Chiapas, for their support and facilities.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was sponsored by Consejo Nacional de Ciencia y Tecnologia (CONACYT) grant numbers 182089 and 168619.
References
- 1.Poirier P, Wawrzyniak I, Vivarès CP, Delbac F, El Alaoui H. New insights into Blastocystis spp.: a potential link with irritable bowel syndrome. PLoS Pathog. 2012;8: e1002545 10.1371/journal.ppat.1002545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scanlan PD, Stensvold CR. Blastocystis: getting to grips with our guileful guest. Trends Parasitol. 2013;29: 523–529. 10.1016/j.pt.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 3.Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S, Islam M, et al. Irritable bowel syndrome: is it associated with genotypes of Blastocystis hominis. Parasitol Res. 2010;106: 1033–1038. 10.1007/s00436-010-1761-x [DOI] [PubMed] [Google Scholar]
- 4.Tan KS. Clin Microbiol Rev. 2008;21: 639–665. 10.1128/CMR.00022-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scanlan PD, Stensvold CR, Rajilić-Stojanović M, Heilig HG, De Vos WM, O'Toole PW, et al. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol Ecol. 2014;90: 326–330. 10.1111/1574-6941.12396 [DOI] [PubMed] [Google Scholar]
- 6.Yoshikawa H, Abe N, Wu Z. PCR-based identification of zoonotic isolates of Blastocystis from mammals and birds. Microbiology 2004;150: 1147–1151. 10.1099/mic.0.26899-0 [DOI] [PubMed] [Google Scholar]
- 7.Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, et al. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol. 2009;39: 473–479. 10.1016/j.ijpara.2008.07.006 [DOI] [PubMed] [Google Scholar]
- 8.Ramírez JD, Sánchez A, Hernández C, Flórez C, Bernal MC, Giraldo JC, et al. Geographic distribution of human Blastocystis subtypes in South America. Infect Genet Evol. 2016;41: 32–35. 10.1016/j.meegid.2016.03.017 [DOI] [PubMed] [Google Scholar]
- 9.Abe N. Molecular and phylogenetic analysis of Blastocystis isolates from various host. Vet Parasitol. 2004;120: 235–242. 10.1016/j.vetpar.2004.01.003 [DOI] [PubMed] [Google Scholar]
- 10.Parkar U, Traub RJ, Kumar S, Mungthin M, Vitali S, Leelayoova S, et al. Direct characterization of Blastocystis from faeces by PCR and evidence of zoonotic potential. Parasitology. 2007;134: 359–367. 10.1017/S0031182006001582 [DOI] [PubMed] [Google Scholar]
- 11.Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, et al. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol. 2010;169: 8–17. 10.1016/j.vetpar.2009.12.032 [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa H, Wu Z, Pandey K, Pandey BD, Sherchand JB, Yanagi T, et al. Molecular characterization of Blastocystis isolates from children and rhesus monkeys in Kathmandu, Nepal. Vet Parasitol. 2009;160: 295–300. 10.1016/j.vetpar.2008.11.029 [DOI] [PubMed] [Google Scholar]
- 13.Petrašova J, Uzlikova M, Kostka M, Petrzelkova KJ, Huffman MA, Modrý D. Diversity and host specificity of Blastocystis in syntopic primates on Rubondo Island, Tanzania. Int J Parasitol. 2011;41: 1113–1120. 10.1016/j.ijpara.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 14.Alfellani MA, Jacob AS, Ortiz Perea N, Krecek RC, Taner-Mulla D, Verweij JJ, et al. Diversity and distribution of Blastocystis sp. subtypes in non-human primates. Parasitology. 2013;140: 966–971. 10.1017/S0031182013000255 [DOI] [PubMed] [Google Scholar]
- 15.Helenbrook WD, Shields WM, Whipps CM. Characterization of Blastocystis species infection in humans and mantled howler monkeys, Alouatta palliata aequatorialis, living in close proximity to one another. Parasitol Res. 2015;114: 2517–2525. 10.1007/s00436-015-4451-x [DOI] [PubMed] [Google Scholar]
- 16.Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12: 263–273. 10.1016/j.meegid.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 17.Crockett CM. Conservation biology of the genus Alouatta. Int J Primatol. 1998;19: 549–578. [Google Scholar]
- 18.Sallis ES, de Barros VL, Garmatz SL, Fighera RA, Graça DL. A case of yellow fever in a brown howler (Alouatta fusca) in Southern Brazil. J Vet Diagn Invest. 2003;15: 574–576. [DOI] [PubMed] [Google Scholar]
- 19.Helenbrook WD, Wade SE, Shields WM, Stehman SV, Whipps CM. Gastrointestinal parasites of Ecuadorian mantled howler monkeys (Alouatta palliata aequatorialis) based on fecal analysis. J Parasitol. 2015;101: 341–350. 10.1645/13-356.1 [DOI] [PubMed] [Google Scholar]
- 20.Westram AM, Baumgartner C, Keller I, Jokela J. Are cryptic host apecies also cryptic to parasites? Host specificity and geographical distribution of acanthocephalan parasites infecting freshwater Gammarus. Infect Genet Evol. 2011;11: 1083–1090. 10.1016/j.meegid.2011.03.024 [DOI] [PubMed] [Google Scholar]
- 21.INEGI. Red hidrografica 30 Grijalva Usumacinta ESCALA 1:50 000 Edicion: 2.0. Instituto Nacional de Estadistica y Geografía; 2010. Aguascalientes, Aguascalientes, Mexico. [Google Scholar]
- 22.Gama L. Programa de Ordenamiento Ecologico del Estado de Tabasco (POEET). SERNAPAM-TABASCO. Division Academica de Ciencias Biologicas. Universidad Juarez Autónoma de Tabasco; Villahermosa, Tabasco, Mexico: 2013. Available: http://sernapam.tabasco.gob.mx/sites/all/files/sites/sernapam.tabasco.gob.mx/files/POET2013.pdf [Google Scholar]
- 23.Amato KR, Estrada A. Seed dispersal patterns in two closely related howler monkey species (Alouatta palliata and A. pigra): a preliminar report of diferences in fruit consumption, traveling behavior, and associated dung beetle assemblages. Neotropical Primates. 2010;17: 59–66. [Google Scholar]
- 24.Oropeza-Hernandez P, Rendon Hernandez E. Programa de Accion para la Conservacion de las Especies: Primates, Mono Araña (Ateles geoffroyi) y Monos Aulladores (Alouatta palliata, Alouatta pigra). 1st ed Mexico: Secretaria de Medio Ambiente y Recursos Naturales/Comision Nacional de Areas Naturales Protegidas; 2011. [Google Scholar]
- 25.Cortés-Ortiz L, Mondragón E, Cabotage J. Isolation and characterization of microsatellite loci for the study of Mexican howler monkeys, their natural hybrids, and other Neotropical primates. Conserv Genet Resour. 2009;2: 21–26. [Google Scholar]
- 26.Santín M, Gómez-Muñoz MT, Solano-Aguilar G, Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol Res. 2011;109: 205–212. 10.1007/s00436-010-2244-9 [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Aguillon F, Lopez-Escamilla E, Velez-Perez F, Martinez-Flores WA, Rodriguez-Zulueta P, Martinez-Ocaña J, et al. Parasitic infections in a Mexican HIV/AIDS cohort. J Infect Dev Ctries. 2013;7: 763–766. 10.3855/jidc.3512 [DOI] [PubMed] [Google Scholar]
- 28.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 3.04 http://mesquiteproject.org
- 29.Maddison WP, Maddison DR. Chromaseq: a Mesquite package for analyzing sequence chromatograms. [Internet]. 2016. Available: http://mesquiteproject.org/packages/chromaseq
- 30.Ewing B, Hillier L, Wendl MC, Green P. Base-Calling of Automated Sequencer Traces Using Phred. I. Accuracy Assessment. Genome Res. 1998;8: 175–185. [DOI] [PubMed] [Google Scholar]
- 31.Ewing B, Green P. Base-Calling of Automated Sequencer Traces Using Phred. II. Error Probabilities. Genome Res. 1998;8: 186–194. [PubMed] [Google Scholar]
- 32.Gordon D, Abajian C, Green P. Consed: A Graphical Tool for Sequence Finishing. Genome Res. 1998;8: 195–202. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity and progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, et al. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14: 817–818. [DOI] [PubMed] [Google Scholar]
- 37.Ronquis F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 38.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16: 37–48. [DOI] [PubMed] [Google Scholar]
- 39.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- 40.Vargas-Sanchez GB, Romero-Valdovinos M, Ramirez-Guerrero C, Vargas-Hernandez I, Ramirez-Miranda ME, Martinez-Ocaña J, et al. Blastocystis isolates from patients with irritable bowel syndrome and from asymptomatic carriers exhibit similar parasitological loads, but significantly different generation times and genetic variability across multiple subtypes. PLoS One. 2015;10: e0124006 10.1371/journal.pone.0124006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villalobos G, Orozco-Mosqueda GE, Lopez-Perez M, Lopez-Escamilla E, Córdoba-Aguilar A, Rangel-Gamboa L, et al. Suitability of internal transcribed spacers (ITS) as markers for the population genetic structure of Blastocystis spp. Parasit Vectors. 2014;7: 461 10.1186/s13071-014-0461-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartl DL, Clark AG. Principles of Population Genetics. 3th ed Sunderland, Massachusetts: Sinauer Associates Inc. Publishers; 1997. [Google Scholar]
- 43.Ramírez JD, Sánchez LV, Bautista DC, Corredor AF, Flórez AC, Stensvold CR. Blastocystis subtypes detected in humans and animals from Colombia. Infect Genet Evol. 2014;22: 223–228. 10.1016/j.meegid.2013.07.020 [DOI] [PubMed] [Google Scholar]
- 44.Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12: 263–273. 10.1016/j.meegid.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 45.Kankare M, Van Nouhuys S, hanski I. Genetic divergence among host-specific cryptic species in Cotesia melitaearum aggregate (Hymenoptera: Braconidae), parasitoids of checkerspot butterflies. Ann Entomol Soc Am. 2005;98: 382–394. [Google Scholar]
- 46.Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc Natl Acad Sci USA. 2006;103: 3657–3662. 10.1073/pnas.0511318103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Gac M, Hood ME, Fournier E, Giraud T. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution. 2007;61: 15–26. 10.1111/j.1558-5646.2007.00002.x [DOI] [PubMed] [Google Scholar]
- 48.Roulin AC, Mariadassou M, Hall MD, Walser JC, Haag C, Ebert D. High genetic variation in resting-stage production in a metapopulation: is there evidence for local adaptation?. Evolution. 2015;69: 2747–2756. 10.1111/evo.12770 [DOI] [PubMed] [Google Scholar]
- 49.Levins R. Some demographic and genetic consequences of environmental heterogeneity for biological control. Bull Ent Soc Amer. 1969;15: 237–240. [Google Scholar]
- 50.Hanski I, Gilpin M. Metapopulation dynamics: brief history and conceptual domain. Biol J Linn Soc. 1991;42: 3–16. [Google Scholar]
- 51.Helenbrook WD, Wade SE, Shields WM, Stehman SV, Whipps CM. Gastrointestinal parasites of Ecuadorian mantled howler monkeys (Alouatta palliata aequatorialis) based on fecal analysis. J Parasitol. 2015;101: 341–50.48. 10.1645/13-356.1 [DOI] [PubMed] [Google Scholar]
- 52.Tomasini N, Lauthier JJ, Ayala FJ, Tibayrenc M, Diosque P. How often do they have sex? A comparative analysis of the population structure of seven eukaryotic microbial pathogens. PLoS One. 2004;9: e103131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright S. Breeding structure of populations in relation to speciation. Am Nat. 1940;74: 232–248. [Google Scholar]
- 54.Hedrick PH. Genetics of Populations. 4th ed Sudbury, Massachusetts: Jones and Bartlett Publishers; 2011. [Google Scholar]
- 55.Maynard Smith J, Smith NH, O’Rourke M, Spratt G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chambouvet A, Gower DJ, Jirku M, Yabsley MJ, Davis AK, Leonard G, et al. Cryptic infection of a broad taxonomic and geographic diversity of tadpoles by Perkinsea protists. Proc Natl Acad Sci USA. 2015;112: E4743–E4751. 10.1073/pnas.1500163112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aurahs R, Grimm GW, Hemleben V, Hemleben C, Kucera M. Geographical distribution of cryptic genetic types in the planktonic foraminifer Globigerinoides ruber. Mol Ecol. 2009;18: 1692–1706. 10.1111/j.1365-294X.2009.04136.x [DOI] [PubMed] [Google Scholar]
- 58.Pillet L,Fontaine D, Pawlowski J. Intra-genomic ribosomal RNA polymorphism and morphological variation in Elphidium macellum suggests inter-specific hybridization in Foraminifera. PLoS One 2012; 7: e32373 10.1371/journal.pone.0032373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scanlan PD, Stensvold CR, Cotter PD. Development and application of a Blastocystis subtype-specific PCR assay reveals that mixed-subtype infections are common in a healthy human population. Appl Environ Microbiol. 2015;81: 4071–4076. 10.1128/AEM.00520-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jimenez-Gonzalez DE, Jimenez-Gonzalez DE, Martinez-Flores WA, Reyes-Gordillo J, Ramirez-Miranda ME, Arroyo-Escalante S, et al. Blastocystis infection is associated with irritable bowel syndrome in a Mexican patient population. Parasitol Res. 2012;110: 1269–1275. 10.1007/s00436-011-2626-7 [DOI] [PubMed] [Google Scholar]
- 61.Moe KT, Singh M, Howe J, Ho LC, Tan SW, Ng GC, et al. Observations on the ultrastructure and viability of the cystic stage of Blastocystis hominis from human feces. Parasitol Res. 1996;82: 439–444. [DOI] [PubMed] [Google Scholar]
- 62.Glander KE. Drinking from arboreal water sources by mantled howling monkeys (Alouatta palliata Gray). Folia Primatol. 1978;29: 206–217. [DOI] [PubMed] [Google Scholar]
- 63.Vitone ND, Altizer S, Nunn CL. Body size, diet and sociality influence the species richness of parasitic worms in anthropoid primates. Evol Ecol Res. 2004;6: 183–199. [Google Scholar]
- 64.Gonzalez-Hernandez M, Rangel-Negrin A, Schoof VA, Chapman CA, Canales-Espinosa D, Dias PAD. Transmission patterns of pinworms in two sympatric congeneric primate species. Int J Primatol. 2014;35: 445–462. [Google Scholar]
- 65.Serio-Silva JC, Rico-Gray V. Howler monkeys (Alouatta palliata mexicana) as seed dispersers of strangler figs in disturbed and preserved habitat in southern Veracruz, Mexico In: Marsh LK, editor. Primates in Fragments: Ecology and Conservation. New York: Kluwer Academic/Plenum Press; 2003. pp. 267–281. [Google Scholar]
- 66.Stensvold CR, Clark CG. Current status of Blastocystis: A personal view. Parasitol Int. 2016; S1383-5769(16)30154-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian phylogenetic tree using a fragment of SSUrDNA sequences; the values of the nodes indicate posterior probabilities values using 10 million generations. ST and GenBank accession numbers are shown, as well as identification of each sample.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.