Abstract
Site-specific incorporation of non-natural amino acids into proteins, via genetic code expansion with pyrrolysyl tRNA synthetase (PylRS) and tRNAPylCUA pairs (and their evolved derivatives) from Methanosarcina sp., forms the basis of powerful approaches to probe and control protein function in cells and invertebrate organisms. Here we demonstrate that adeno-associated viral delivery of these pairs enables efficient genetic code expansion in primary neuronal culture, organotypic brain slices and the brains of live mice.
The site-specific incorporation of non-natural amino acids into proteins, via genetic code expansion, provides new approaches for imaging, controlling and understanding protein function in cells1–3. This approach uses orthogonal aminoacyl tRNA synthetase and tRNACUA pairs to direct the incorporation of a non-natural amino acid into a target protein in response to an amber stop codon introduced at a desired site in the corresponding gene1,2. The PylRS (encoded by PylS) and tRNAPylCUA (encoded by PylT) pair from Methanosarcina species (most commonly M. barkeri or M. mazei, here abbreviated Mb and Mm, respectively) is a particularly useful pair for genetic code expansion2. This pair and its evolved variants have been used to direct the incorporation of numerous non-natural amino acids with diverse structures and functions into proteins produced in Escherichia coli4, Salmonella typhimurium5, Saccharomyces cerevisiae6, mammalian cells7,8 and certain invertebrates and plants, including Caenorhabditis elegans9, Drosophila melanogaster10 and Arabidopsis thaliana11. Genetic code expansion in these model organisms is facilitating new approaches to manipulate and understand the molecular basis of biology1. However there are no reports of developing this pair for genetic code expansion in a live vertebrate.
The mouse (Mus musculus) is a powerful mammalian, vertebrate model organism because it captures many key features of human biology, including an adaptive immune system and a complex brain. Thus, mice are commonly used to study human disease and infection12 and to illuminate the principles of mammalian cognition, learning and memory13. There are limited reports of nonsense suppression in mice. Previous work has demonstrated direct injection of plasmids encoding human tRNASerUUA into cardiac and skeletal muscle and reported ochre suppression, leading to the incorporation of serine, with an efficiency of ~1% (ref. 14). Electroporation of mouse embryos with a plasmid encoding a mutant of the E. coli LeuRS/tRNACUA pair had demonstrated amber suppression in a small number of embryonic brain cells15; however, that work used components that are orthogonal in a limited number of hosts, cannot be evolved in E. coli and have been used to incorporate a limited number of non-natural amino acids with low and uncharacterized efficiency. Thus, previous work has not addressed the challenges of efficient genetic code expansion in live mice. Here we develop adeno-associated viral (AAV) vectors for non-natural amino acid incorporation and demonstrate that the use of those vectors enables the efficient, site-specific incorporation of several non-natural amino acids in dissociated neurons, brain slices and finally live mice.
We focused on using AAV vectors for non-natural amino acid incorporation, as these are among the most widely used platforms for introducing genes into the mouse16,17. Because AAV DNA can reach a high copy number in infected cells, we anticipated that AAVs could deliver many copies of PylT, as required for efficient non-natural amino acid incorporation18. We first created an AAV in which the neuron-specific human synapsin I promoter drives expression of N-terminally tagged MmPylS, and the human U6 promoter drives expression of PylT (Supplementary Results, Supplementary Fig. 1). The MmPylS gene is preceded by the gene for a red fluorescent protein, mCherry, and a P2A sequence that directs co-translational cleavage of the mCherry protein from PylRS19. As a result of this arrangement, all cells that express PylS and contain PylT are marked red. We created a second AAV containing the gene of interest, bearing an amber codon, driven from the synapsin promoter (in this case sfGFP with an amber codon at codon position 150, encoding sfGFP(150TAG)) and containing a second copy of PylT.
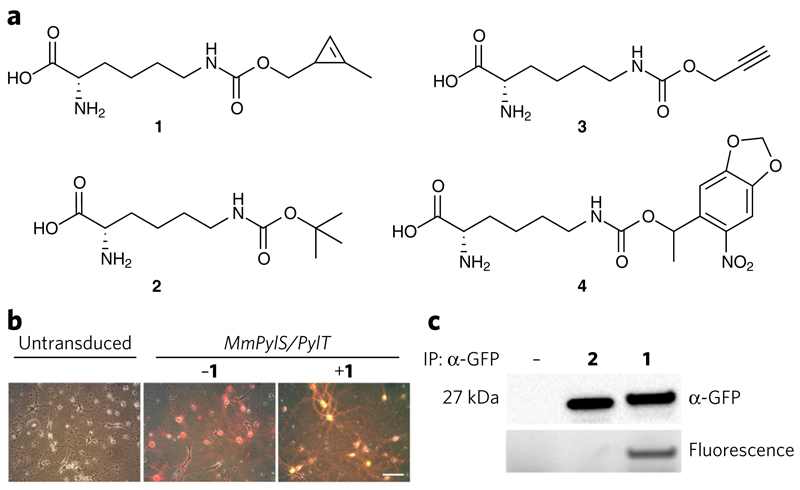
To test the AAV-mediated approach for non-natural amino acid incorporation we infected primary dissociated neurons from rat neocortex with AAV vectors containing MmPylS, PylT and sfGFP(150TAG). After 48 h, we transferred the cultures to media with or without 1 mM Nε-(((2-methylcycloprop-2-en-1-yl) methoxy)carbonyl)-l-lysine (1) (Fig. 1a). Non-natural amino acid-dependent sfGFP production was visible after 6 d (Fig. 1b and Supplementary Fig. 2). We detected full-length sfGFP from transduced neurons cultured in the presence of 1 or Nε-(tert-butyloxycarbonyl)-l-lysine (2) but not from neurons cultured in the absence of non-natural amino acid (Fig. 1c and Supplementary Fig. 3). sfGFP incorporating 1, but not 2, was specifically labeled via an inverse electron demand Diels-Alder reaction with a tetrazine-Alexa Fluor 647 conjugate (5)20,21. The amount of full-length protein produced from sfGFP(150TAG) in the presence of non-natural amino acids, as a percentage of the amount of full length protein from an sfGFP gene, was 23.4% ± 4.8% for 1, and 33.3% ± 17.3% (mean ± s.e.m.) for 2 (Supplementary Fig. 4). This compares favorably to most non-natural amino acid incorporation efficiencies reported18 and underscores the utility of the system reported here. These experiments demonstrated that our AAVs enabled efficient incorporation of non-natural amino acids with new functionality in mammalian neurons.
Figure 1. AAV-mediated incorporation of non-natural amino acids into proteins in dissociated neurons and bioorthogonal labeling.
(a) Non-natural amino acids used in this study. (b) Fluorescence microscopy images of untransduced cells and cells transduced as indicated to determine non-natural amino acid incorporation in dissociated rat cortical neurons. Scale bar, 200 μm. (c) Western blot analysis of non-natural amino acid-dependent production of sfGFP from sfGFP(150TAG). sfGFP incorporating 1, but not 2, was selectively labeled with 5 in an inverse electron demand Diels-Alder reaction. IP, immunoprecipitation.
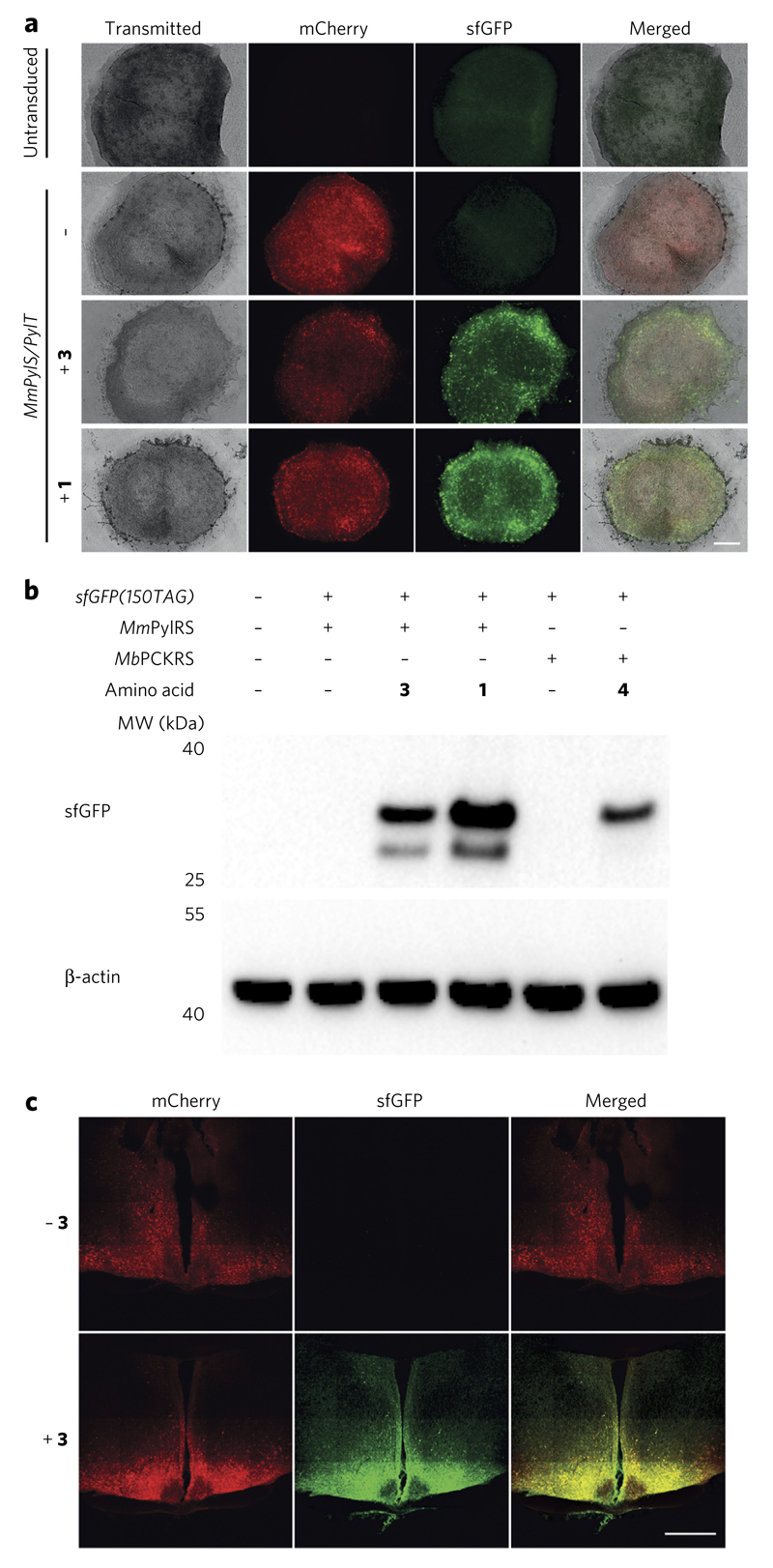
To demonstrate that the AAV mediated approach can be used for genetic code expansion in tissue, we used organotypic slices of the mouse suprachiasmatic nucleus (SCN), the region of the brain that contains the biological clock responsible for coordinating circadian rhythms in mammals22. These slices are a well-established system for studying the circadian clock and persist in culture for over two months23. We infected SCN slices with the AAVs containing MmPylS, PylT and sfGFP(150TAG) and added 1 or Nε-((prop-2-yn-1-yloxy)carbonyl)-l-lysine (3) after 7 d. Within an additional 7 d, we observed non-natural amino acid-dependent production of sfGFP (Fig. 2a), which increased for 26 d after AAV transduction (Supplementary Fig. 5a). We used an AAV in which a gene encoding PCKRS, a variant of MbPylRS evolved to incorporate Nε-((1-(6-nitrobenzo[d][1,3]dioxol-5-yl)ethoxy)carbonyl)-l-lysine (4)8, replaced MmPylS to demonstrate the incorporation of photocaged lysine in response to the amber codon in mouse SCN slices (Supplementary Fig. 5b). We detected full-length sfGFP, by western blot, from transduced organotypic brain slices cultured in the presence of amino acids 1, 3 or 4. We observed a lower-molecular-weight immunoreactive species, which may arise through proteolysis of sfGFP when it is expressed in the organotypic slices for 26 d. We did not detect sfGFP from slices cultured in the absence of non-natural amino acid (Fig. 2b). sfGFP incorporating 1 was selectively labeled with 5 (Supplementary Fig. 5c).
Figure 2. Incorporation of non-natural amino acids into proteins in SCN slices and live mice.
(a) Non-natural amino acid-dependent production of sfGFP in SCN slices. Scale bar, 200 μm. (b) Western blot analysis of lysates from brain slices in a, confirm non-natural amino acid-dependent read-through of the amber codons in sfGFP(150TAG). (c) Fluorescence images showing non-natural amino acid-dependent expression of sfGFP in coronal sections of the hypothalamus and SCN. Scale bar, 500 μm.
To investigate the effects of amber suppression and non-natural amino acid incorporation on neuronal function, we transduced organotypic slices of the mouse SCN derived from Period2∷LUC mice with the AAVs containing MmPylS,PylT and sfGFP(150TAG) and added 1. Period2∷LUC mice express a knock-in fusion protein showing circadian oscillations by bioluminescence levels that can be followed in organotypic slices (Supplementary Fig. 6)24. It is well established that these circadian oscillations are exquisitely sensitive to cellular perturbations. Indeed, perturbations to the electrical firing of neurons, or to synaptic signaling, compromise circadian oscillations in the SCN17,25. We found that transduced Period2∷LUC organotypic SCN slices supported non-natural amino acid incorporation, as expected (Supplementary Fig. 6a). Moreover, we observed robust circadian oscillations in Period2∷LUC organotypic SCN slices that incorporated the non-natural amino acid for more than a month (Supplementary Fig. 6b). This observation demonstrated that the physiological functions of neurons, crucial for the circadian circuit, were not perturbed by non-natural amino acid incorporation using our system.
We conclude that efficient genetic code expansion with diverse non-natural amino acids may be implemented in mouse brain slices using the AAV strategy we report, and that amber suppression and non-natural amino acid incorporation does not affect key neuronal functions. These experiments demonstrate genetic code expansion using our AAV system in a mammalian tissue.
Next we investigated whether the strategy we developed for non-natural amino acid incorporation in dissociated neurons and SCN slices can be extended to live mice. We performed stereotactic injections of the MmPylS,PylT AAV and the sfGFP(150) AAV into the region of the SCN in adult mice. One week later we intracranially perfused 3, or vehicle that did not contain 3 for control mice, into the third ventricle. After two weeks we examined the coronal sections from the brains of the mice and observed clear non-natural amino acid-dependent sfGFP production in cells (Fig. 2c). The incorporation of the non-natural amino acid in the SCN in vivo was consistent with our data on non-natural amino acid incorporation in organotypic SCN slices in vitro. Moreover, we observed extensive incorporation in neurons in the hypothalamus surrounding the SCN, suggesting that AAV-mediated non-natural amino acid incorporation may be even more efficient in other regions of the brain (Fig. 2c and Supplementary Fig. 7). AAV-mediated genetic code expansion did not have substantive effects on the weight of the mice (Supplementary Fig. 8), consistent with the approach being not toxic. Pharmacokinetic analysis suggested that 3 was bioavailable (Supplementary Fig. 9), and we obtained similar non-natural amino acid incorporation results when we delivered 3 to the mice through their drinking water (Supplementary Fig. 10). These experiments demonstrated that 3 can enter the blood and reach the brain, and suggest that this amino acid should be bioavailable for incorporation into proteins in diverse tissues.
We labeled sections of the brain from mice incorporating 3 into sfGFP with an azide-sulfo-Cy5 fluorophore conjugate (6) in a Cu(I)-catalyzed 3 + 2 cycloaddition20 (Supplementary Fig. 11). We observed that cells in the brain producing sfGFP containing 3 were selectively labeled with 6, whereas neighboring cells that did not produce sfGFP containing 3 were not labeled.
We performed unilateral injection of the MmPylS,PylT virus and the sfGFP(150TAG) virus, and intracranial perfusion of 3, both into the lateral ventricle. This enabled non-natural amino acid incorporation in the hippocampus (Supplementary Fig. 12). These experiments demonstrated the generality of the approach for targeting different regions of the brain. Quantifying the levels of sfGFP incorporating 3 in neurons in the mouse brain (Supplementary Fig. 13) demonstrated that protein containing the non-natural amino acid was present at concentrations of approximately 50 μM (1.4 mg/mL).
In conclusion, we created AAV vectors for genetic code expansion and demonstrated that they can be used for non-natural amino acid incorporation into neurons in primary culture, mouse organotypic tissue slices and the brain of live mice. Although we focused on the incorporation of non-natural amino acids in neurons, AAVs have been used extensively to target diverse tissues16, and we anticipate that extensions of our approach will enable other cells and other tissues to be targeted for genetic code expansion in mice. Extensions of the approach we report will also enable cell-specific proteomics in mouse tissue and live mice via stochastic orthogonal recoding of translation21. We anticipate that our approach will enable the extension of exciting and emerging genetic code expansion-based strategies to probe and control protein function in experiments from cells to live mice1,2.
Online Methods
Materials
AAV serotype 1 viral vectors were obtained commercially from the Penn Vector Core. All animal work was licensed under the UK Animals (Scientific Procedures) Act of 1986 with local ethical approval (MRC AWERB). At least two mice were used for every condition tested, mice were not excluded, no randomization or blinding was used.
Statistical methods
Error bars represent the s.d. or s.e.m. of at least three biological replicates, as indicated. Regression analysis was calculated by the least-squares method.
Chemical synthesis
All solvents and chemical reagents were purchased from Sigma-Aldrich or Fisher Scientific, and used without further purification. Nε-(tert-butyloxycarbonyl)-l-lysine (amino acid 2) was purchased from Chem-Impex International Inc., and used without further purification. The synthesis and purification of the tetrazine-Alexa Fluor 647 conjugate, compound 5, has been previously reported21. Briefly, the 3,6-di-2-pyridyl-1,2,4,5-tetrazine amine (2-amino-N-(6-(6-(pyridin-2-yl)-1,2,4,5-tetrazin-3-yl) pyridin-3-yl)acetamide) was coupled with commercially available Alexa Fluor 647 succinimidyl ester (ThermoFisher Scientific, A37573) using the manufacturer’s protocols. The resultant tetrazine-dye conjugate 5 was purified by semipreparative HPLC. The structure of the Alexa Fluor 647 succinimidyl ester is not available, and so we are not able to provide a molecular structure for 5.
Nε-(((2-methylcycloprop-2-en-1-yl)methoxy)carbonyl)-l-lysine, (amino acid 1)21, and photocaged lysine, Nε-((1-(6-nitrobenzo[d][1,3]dioxol-5-yl) ethoxy)carbonyl)-l-lysine, (amino acid 4)8 were synthesized as previously described. Analytical thin-layer chromatography (TLC) was performed on silica 60F-254 plates, which were visualized by UV light and by Vanillin staining. Electrospray ionization mass spectrometry (ESI-MS) was acquired using an Agilent 1200 LC-MS system equipped with a 6130 quadrupole spectrometer. The solvent used for the liquid chromatography (LC)-MS was 0.02% formic acid in water as buffer A and 0.02% formic acid in acetonitrile as buffer B. A Poroshell 120 (Agilent) C18 column (3.0 mm × 50 mm, 2.7 μm) was used for liquid chromatography and mass spectra were acquired in both positive and negative modes. NMR spectra were acquired on a 400 MHz Bruker instrument.
Nε-((prop-2-yn-1-yloxy)carbonyl)-L-lysine HCl salt (3). The Nε-((prop-2-yn-1-yloxy)carbonyl)-l-lysine was synthesized from Boc-Lys-OH and characterized as previously reported26. To a solution of the tert-Boc protected propargyl carbamate (45.8 g, 140 mmols) in dry DCM (200 mL) was added 4 N HClin 1,4-dioxane (200 mL) drop-wise. The reaction was allowed to stir at room temperature for 8 h. The product was filtered and washed with 800 mL of DCM and dried in vacuo to obtain the amino acid 2 as white solid in 96% yield (35.6 g, 135 mmols).
Residual 1,4-dioxane was removed by ion exchange chromatography. Fifty grams of Amberlite IRA-400(Cl) were charged to a glass column and washed with five bed volumes of water, followed by five bed volumes of 0.2 N NaOH and then five bed volumes of water, allowing the buffers to elute under gravity. An aqueous solution of 5 g of amino acid 3 was adjusted to pH 10 with NaOH and loaded three times onto the washed resin. The charged resin was then washed with five bed volumes each of the following: water, 25% methanol, 50% methanol, 100% methanol, water, and then the product was eluted with 1 N HCl (200 mL). The eluate was concentrated under reduced pressure and then dried in vacuo, yielding the dioxane-free amino acid 3 as a white solid in 67% yield (3.35 g, 12.7 mmols). 1H NMR (D2O): δ = 1.35−1.55 (m, 4H), 1.84−2.00 (m, 2H), 2.84 (s, 1H), 3.11 (t, 2H, J = 6.6 Hz), 3.99 (t, 1H, J = 6.4 Hz), 4.62 (s, 2H). 13C NMR (D2O): δ = 21.4, 28.3, 29.4, 40.0, 52.7, 53.0, 75.6, 78.6, 157.7, 172.4. ESI/APCI-MS: m/z calculated for C10H16N2O4 [M+H]+: 229.1; found: 229.2.
Plasmid construction
First, the human U6-PylT gene was cloned by standard molecular biology methods in a divergent orientation at the XbaI site upstream of the human Synapsin promoter in pAAV-hSyn-hChR2(H134R)-EYFP (Addgene #26973). Into this backbone, either the MmPylRS, MbPCKRS, sfGFP or sfGFP(150TAG) coding sequences were cloned between the BamHI and EcoRI sites.
HEK293 cell culture
HEK293 cells from ATCC were cultured in DMEM supplemented with 10% FBS and incubated at 37 °C and 5% CO2 atmosphere. All transfections were performed with PEI. Lysates were prepared in RIPA (Sigma) buffer supplemented with protease inhibitor tablets (Roche).
Primary culture of dissociated rat cortical neurons
Dissociated rat cortical neurons (RCNs) were prepared using a modified version of the standard protocol27. Briefly, embryonic day 18 (E18) timed pregnant rat was euthanized, and brains of fetuses were dissected and cleared of meninges. Isolated cortices were digested in 0.5% trypsin for 15 min at 37 °C. The cortices were resuspended in Neurobasal media with B27 Supplement (Invitrogen) and 2.5% foetal bovine serum (Labtech International) and subsequently washed three times in 5 mL of HBSS (Ca/Mg-free) with 10 mM Hepes (both from Invitrogen). The washed cortices were resuspended in 1 mL of HBSS (Ca/Mg-free) and triturated with a 1 mL Gilson pipette until homogeneous solution was achieved. The dissociated cells were then seeded onto poly(l-lysine)-coated coverslips at density of 20,000–50,000 cells/cm2. The cells were cultured at 37 °C under 5% CO2 atmosphere in Neurobasal medium supplemented with B27 and GlutaMAX (ThermoFisher), exchanging half of the medium every 5 d.
Genetic code expansion in dissociated RCNs
After ~72 h, dissociated cultures were transduced by addition of 1 μL (~1012–1013 transducing units) of a 1:1 mixture of pSynI-sfGFP150TAG and pSynI-mCherry∷P2A∷MmPylRS AAV particles in 200 μL of conditioned medium. An additional 48 h after transduction, medium was changed completely to 800 μL of 1:1 complete:conditioned media, with or without 0.5–1 mM amino acid 1 or 1 mM amino acid 2. Fluorescence images were recorded on a Leica DM IL LED inverted fluorescent microscope with QCapture Suite PLUS software. Lysates from cultured neurons were prepared in ice-cold RIPA buffer (10 mM Tris-Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.1% SDS; 1% Triton X-100; 1% deoxycholate) for 30 min on ice with intermittent pipetting. The lysate was centrifuged at 20,000g for 10 min at 4 °C, isolating the supernatant for subsequent analysis.
Determination of amber suppression efficiency in RCNs
RCNs were transduced with a 1:1 mixture of the two AAV vectors containing MmPylS,PylT and sfGFP(150TAG) in conditioned medium. Protein extracts were prepared from the cultures 8 d after transduction and 6 d after addition of 1 mM amino acid 1 or 2. The extracts were analyzed by SDS-PAGE and immunoblot against sfGFP (Roche 11814460001 primary 1,000:1, Cell Signal Technology 7076 secondary 2,000:1) and the N-terminal FLAG epitope tag (Sigma A8592 1,000:1) of MmPylRS. Levels of sfGFP produced from sfGFP(150TAG) in RCNs relative to the level of MmPylRS was normalized to the levels in HEK293 of wild-type sfGFP produced from an sfGFP gene relative to the level of MmPylRS. The wild-type calibration curve was prepared by serial dilutions (1-, 2-, 5-, 10-, 20-, 50-, 100-, 200- and 500-fold) of an arbitrary concentration of protein extracts from HEK293 cells transiently transfected with a 1:1 mixture of plasmids encoding the wild-type sfGFP and MmPylS transgenes under the human SynI promoter.
Genetic code expansion in SCN slices
SCN organotypic slices were prepared as described from 7–9 d old C57/BL6 or Period2∷LUC pups23. Slices were transduced with 1 μL (~1012–1013 transducing units) of a 1:1 mixture of pSynI-sfGFP150TAG and pSynI-mCherry∷P2A∷MmPylRS or pSynI-mCherry∷P2A∷MbPCKRS AAV particles. After one or two weeks, slices were transferred to either fresh organotypic culture media (OCM) or fresh OCM supplemented with 1 mM amino acid 1, 1 mM amino acid 2, 10 mM amino acid 3 or 2 mM amino acid 4. After weekly medium changes, the slices were imaged for 4 weeks on a Leica DM IL LED inverted fluorescent microscope with QCapture Suite PLUS software. After the 4-week imaging period, the slices were lysed by sonication in 2% SDS. Protein concentrations were determined by BCA assay (Pierce). Equal amounts of protein were analyzed by PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% (w/v) milk in PBS-T and probed with primary antibodies to GFP (Roche, 11814460001; 1:1,000), FLAG (Sigma, A8592; 1:1,000), or β-actin (Cell Signaling, 12620; 1:1,000) overnight at 4 °C. Membranes were rinsed with PBS-T and incubated with secondary antibody where appropriate (Cell Signaling, 7076; 1:2,000). After final rinses with PBS-T, Luminata Forte HRP substrate (Millipore) was applied and the blots were imaged on a ChemiDoc XRS+ system (Bio-Rad).
Bioluminescence recordings of Period2∷Luciferase SCN slices
Seven days after dissection, two Period2∷LUC SCN slices from the same litter were changed to air medium with 1 mM luciferin (AM-Luci) and placed into photo-multiplier tubes to record circadian oscillation as a bioluminescence signal24. After ~11 d, one slice was change to fresh AM-Luci with 1 mM UAA 1 and transduced with pSynI-sfGFP150TAG and pSynI-mCherry∷P2A∷MmPylRS as described earlier. The control slice was likewise changed to fresh AM-Luci but treated with a vehicle. After ~15 d, the slices were changed to fresh AM-Luci with or without 1 mM 1 UAA and then finally imaged 25 d after transduction using the fluorescence microscope setup as described earlier. Oscillation amplitude was normalized to the last peak before transduction.
Genetic code expansion in live mice
Adult male C57/BL6 mice aged 8–9 weeks were individually housed in cages with ad libitum access to food and water. The mice received bilateral stereotactic injections of a 1:1 mixture of pSynI-mCherry∷P2A∷MmPylRS and pSynI-sfGFP150TAG AAV particles (0.3 μL of ~5 × 1012 genome copies per mL; ~2 × 109 viral particles) into the region of the SCN (0.3 mm medio-lateral to Bregma, 5.5 mm deep to dural surface) under halothane anesthesia. Following surgery, the mice were allowed to recover for 1 week before subcutaneous implantation (under halothane anesthesia) of an Alzet mini-pump (model 1002) attached to a cannula (brain infusion kit II) for targeted delivery of 3 (aqueous solution of 200 mM 3, partially neutralized with 125 mM Na2CO3 to pH 4 and a final osmolality of 0.570 osmol/kg) or vehicle (0.26 μL/h) into the third ventricle close to the SCN. Mice received 3 administered orally in the drinking water (black currant-flavored water containing 30 mg/mL 3), through intracranial perfusion, or not at all (vehicle in both drinking water and mini-pump). After 14 d of amino acid treatment, mice were killed, brains were fixed in 4% paraformaldehyde (~4 h), cryoprotected in 20% sucrose overnight before cutting sections on a freezing microtome (60 μm) and immunostained with rabbit anti-GFP (Abcam, AB6556; 1:1,000), and mounted with DAPI (Vectorlabs). Confocal images were acquired on a Zeiss LSM 780 confocal microscope and processed in ImageJ (NIH).
sfGFP immunoprecipitation and fluorescence labeling
sfGFP was immunoprecipitated from clarified lysates using GFP-Trap-M beads and associated buffers in kit format (Chromotek) according to manufacturer’s instructions. For RCN lysates, sfGFP was eluted using glycine-elution buffer (200 mM glycine pH 2.5), followed immediately by neutralization with 1 M Tris base (pH 10.4), and overnight labeling with 5 (100 nM final concentration) at room temperature28. For SCN slices, the sfGFP was labeled after binding but before elution from the beads. For labeling of sfGFP containing amino acid 1 with the fluorescent probe 5, the beads were incubated with 40 μM of the dye conjugate overnight at room temperature. After labeling, the beads were washed three times with ice-cold wash buffer, and sfGFP was eluted by boiling in 2× SDS loading buffer. All labeled samples were separated on 4–12% Bis-Tris SDS-Page gel, imaged on Typhoon gel imager (excitation 633 nm/emission 670 nm), and transferred onto nitrocellulose membranes for Western Blot analysis against GFP (Roche 11814460001 primary, Cell Signal Technology 7076 secondary). After final rinses with PBS-T, Luminata Forte HRP substrate (Millipore) was applied and the blots were imaged on a ChemiDoc XRS+ system (Bio-Rad).
In vivo pharmacokinetics
A cohort of 5 male C57BL/6 mice aged 6–8 weeks were used in each single dose pharmacokinetic experiment, and replicated 3 times. For iv administration, 100 mg/kg (380 μmol/kg) were injected as a bolus in the tail vein. The dose was delivered in 150 μL in a pH neutral and osmotically balanced solution (a filter-sterilized stock solution of 100 mM (26.4 mg/mL) 3 in 75 mM Na2CO3 was diluted according to the weight of each mouse in sterile saline). For oral administration (po) animals were gavaged with 250 mg/kg (950 μmol/kg) delivered in 300 μL of aqueous solution at pH 4 (a filter-sterilized stock solution of 200 mM (52.8 mg/mL) 3 in 125 mM Na2CO3 was diluted according to the weight of each mouse in sterile water). A batch design was used where a 60 μL pre-dose blood sample was collected in Li-heparin-coated tubes from the saphenous vein from all animals. Then a further two 60 μL blood samples were taken at a defined time after dosage from each animal. Plasma was separated by centrifugation (15 min, 2,000g). Plasma proteins were precipitated by adding an equal volume of 5% (w/v) 5-sulfosalicylic acid (Sigma) to each plasma sample, which included 500 μM norleucine (NorL) (Sigma), as internal standard. Samples were incubated at 4 °C for 30 min, and then centrifuged at 10,000g for 10 min at 4 °C. The supernatant was collected, filtered through 0.22 μm filters (Nanosep, Pall) and analyzed on a Biochrom 30+ automated amino acid analyzer, using the manufacturer’s accelerated physiological fluids method. The retention time for 3 was found to be 53.233 min, falling between that for isoleucine (52.97 min) and leucine (53.77 min). To determine the concentration of 3, the internal standardized areas (to the norleucine peak) for isoleucine and leucine in the pre-dose plasma samples were subtracted to the combined areas for the isoleucine and leucine peaks in the post-dosage samples. The residual internal standardized area was then used to determine the concentration of 3 from a standard curve; below the quantification limit concentrations were assigned with zero value29. The pharmacokinetic parameters were estimated using the PK package in R 3.1.0 (ref. 30). and the graphs were plotted with Graphpad Prism 6.
Cu(I)-catalyzed azide-alkyne labeling of brain sections
Sulfo-Cy5-azide (Jena Bioscience) 6 was conjugated to 3 using a modified version of a previously reported protocol31. Briefly, 4% PFA fixed brain sections (~60 μm) were permeabilized in 0.3% Triton-X 100, 1% BSA in PBS overnight. The detergent was then removed by washing the sections in PBS (five times, 5 min). Sections were then incubated in 1 mM CuSO4, 100 μM TBTA, 5 μM Cy5-azide and 5 mM sodium ascorbate in PBS supplemented with 1% BSA at 37 °C for 1 h. The staining solution was removed and brain sections were rinsed in 1 mM EDTA in PBS. Then the sections were washed in 0.3% Triton-X in PBS (ten times, 30 min), counterstained with DAPI and then mounted in Fluoromount G (SouthernBiotech). Confocal images were acquired on a Zeiss LSM 780 confocal microscope and processed in ImageJ (NIH).
Direct determination of sfGFP concentration in hippocampal brain sections
A stock solution of recombinant sfGFP at a concentration of 17 μM (0.5 mg/ml) was serially diluted in PBS by factors of two from 17 μM to 266 nM and imaged at 25 μm above the coverglass on a Leica SP8 confocal microscope operated in accordance with manufacturers’ recommendations. Fixed brain sections were mounted in PBS on a standard glass slide, covered with a coverglass, and imaged at the same instrument settings as the GFP solution measurements (40×, 1.1 NA water immersion objective, 1 AU pinhole at 530 nm, excitation 0.11% power at 488 nm, emission detection 494 nm to 553 nm, pixel spacing 90 nm in x by 90 nm in y by 390 nm in z). Pixel intensity values were taken as the average of the center 80% of the image. Linear regression of the pixel intensity values (PIV) gave a fit of PIV = 0.047 + 5.22 × [sfGFP].
Supplementary Material
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
Acknowledgments
This work was supported by the Medical Research Council (MRC), UK (MC_U105181009 and MC_UP_A024_1008, to J.W.C., MC_U105170643 to M.H.H.). V.B. is supported by an MRC case studentship (Nikon). We thank the MRC biomedical facility staff at ARES for their help.
Footnotes
Author contributions
J.W.C. defined the direction of research. R.J.E. designed the AAV vectors. V.B. and R.J.E. performed the experiments in rat cortical neurons. R.Z., E.S.M. and R.J.E. defined the amino acid delivery conditions in live mice. R.J.E. and T.P.K. performed the SCN slice experiments under the direction of M.H.H. R.Z. performed the labeling of brain sections containing 3 with 6 and performed microscopy. E.S.M. performed the experiments in live mice. T.S.E. provided amino acid 1. R.Z. provided amino acid 3, designed pharmacokinetic experiments, determined the plasma concentrations of 3 and analyzed pharmacokinetic data. N.P.B. performed quantitative microscopy and assisted imaging experiments. J.W.C. and R.J.E. wrote the paper with input from all authors.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Davis L, Chin JW. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 2.Chin JW. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 3.Xue L, Karpenko IA, Hiblot J, Johnsson K. Nat Chem Biol. 2015;11:917–923. doi: 10.1038/nchembio.1959. [DOI] [PubMed] [Google Scholar]
- 4.Neumann H, Peak-Chew SY, Chin JW. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 5.Lin S, et al. J Am Chem Soc. 2011;133:20581–20587. doi: 10.1021/ja209008w. [DOI] [PubMed] [Google Scholar]
- 6.Hancock SM, Uprety R, Deiters A, Chin JW. J Am Chem Soc. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukai T, et al. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 8.Gautier A, et al. J Am Chem Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 9.Greiss S, Chin JW. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Nat Chem Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- 11.Li F, et al. Angew Chem Int Ed Engl. 2013;52:9700–9704. doi: 10.1002/anie.201303477. [DOI] [PubMed] [Google Scholar]
- 12.Rosenthal N, Brown S. Nat Cell Biol. 2007;9:993–999. doi: 10.1038/ncb437. [DOI] [PubMed] [Google Scholar]
- 13.Fossella JA, Casey BJ. Cogn Affect Behav Neurosci. 2006;6:1–8. doi: 10.3758/cabn.6.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Buvoli M, Buvoli A, Leinwand LA. Mol Cell Biol. 2000;20:3116–3124. doi: 10.1128/mcb.20.9.3116-3124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang J-Y, et al. Neuron. 2013;80:358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 17.Brancaccio M, Maywood ES, Chesham JE, Loudon ASI, Hastings MHA. Neuron. 2013;78:714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmied WH, Elsässer SJ, Uttamapinant C, Chin JW. J Am Chem Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, et al. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang K, Chin JW. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 21.Elliott TS, et al. Nat Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 23.Hastings MH, Reddy AB, McMahon DG, Maywood ES. Methods Enzymol. 2005;393:579–592. doi: 10.1016/S0076-6879(05)93030-9. [DOI] [PubMed] [Google Scholar]
- 24.Yoo SH, et al. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colwell CS. Nat Rev Neurosci. 2011;12:553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. J Am Chem Soc. 2009;131:14194–14195. doi: 10.1021/ja906603s. [DOI] [PubMed] [Google Scholar]
- 27.Kaech S, Banker G. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 28.Lang K, et al. J Am Chem Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beal SL. J Pharmacokinet Pharmacodyn. 2001;28:481–504. doi: 10.1023/a:1012299115260. [DOI] [PubMed] [Google Scholar]
- 30.Jaki T, Wolfsegger MJ. Pharm Stat. 2011;10:284–288. doi: 10.1002/pst.321. [DOI] [PubMed] [Google Scholar]
- 31.Uttamapinant C, Sanchez MI, Liu DS, Yao JZ, Ting AY. Nat Protoc. 2013;8:1620–1634. doi: 10.1038/nprot.2013.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.