Abstract
Schizophrenia (SZ) and HIV infection are serious disorders with a complex phenotypic relationship. Observational studies have described their comorbidity; their genetic correlation is not well studied. We performed extensive analysis in search of common genetic factors for SZ and HIV, and their relationship with risky sexual behavior (RSB). Summary statistics from genome-wide association studies of HIV infection and schizophrenia were obtained and 2379 European Americans were genotyped and assessed for RSB score. Genetic relationships between traits were analyzed in three ways: linkage disequilibrium (LD) score regression to estimate genetic correlation; GPA (Genetic analysis incorporating Pleiotropy and Annotation) to test pleiotropy and identify pleiotropic loci; polygenic risk scores (PRS) of SZ and HIV to predict RSB using linear regression. We found significant pleiotropy (p = 5.31E − 28) and a positive genetic correlation (cor = 0.17, p = 0.002) for SZ and HIV infection. Pleiotropic SNPs with opposite effect directions (antagonistic) and SNPs with the same effect direction (synergistic) were enriched for distinctly different biological functions. SZ PRS computed with antagonistically pleiotropic SNPs consistently predicted RSB score with nominal significance, but SZ PRS based on either synergistically pleiotropic SNPs or all SNPs did not predict RSB. The epidemiologic correlation between schizophrenia and HIV can partly be explained by overlapping genetic risk factors, which are related to risky sexual behavior.
Introduction
Schizophrenia (SZ) is a debilitating psychiatric disorder that—among its many other symptoms—impairs social and interpersonal skills (Seeman 1997). In 2002, the cost associated with SZ was estimated to be $62.7 billion in the United States (Wu et al. 2005). Infection by the human immunodeficiency virus (HIV) also poses a serious threat to public health and social and economic development. In 2013, there were about 2.1 million new HIV cases, 35 million people living with HIV, and 1.5 million acquired immunodeficiency syndrome (AIDS)-related deaths worldwide (Hall et al. 2015). In the United States, it was estimated that 1.2 million people above age 13 were infected by HIV at the end of 2012 (Hall et al. 2015). Risky sexual behavior (RSB) is a major determinant of HIV transmission (Marks et al. 2005).
The relationship between SZ and human immunodeficiency virus (HIV) infection is complicated epidemiologically and genetically and has long been studied. For instance, schizophrenia patients were found to be 1.5 times as likely to be HIV infected as non-schizophrenics among Medicaid recipients (Blank et al. 2002). The relationship between SZ and HIV infection is further complicated by substance dependence (Walkup et al. 2008). Helleberg et al. (Helleberg et al. 2015) found increased HIV diagnoses in SZ cases with a history of substance dependence and that compared with non-HIV-infected people, individuals infected with HIV had an incidence rate ratio (IRR) of 2.26 for SZ with a history of substance dependence and 4.09 IRR without a history of substance dependence (Helleberg et al. 2015). Apart from the possibility that one of these traits sometimes leads to another (by virtue of behavior), the correlated incidence rates also signal the possibility of common genetic or environmental factors for HIV and SZ. Besides epidemiological studies, considerable effort has been made from genetic and molecular studies towards understanding the role played by the immune system in SZ (Muller and Schwarz 2010). For example, a mega-analysis on SZ carried out by the Psychiatric Genomics Consortium (PGC) detected 108 loci associated with SZ; among these, genes with important roles in immunity are enriched (Schizophrenia Working Group of the Psychiatric Genomics 2014). The genetic relationship of immune system regulation and SZ further complicates understanding the comorbidity of SZ and HIV infection.
People with SZ are less likely to be sexually active (Carey et al. 1999, 2001) and have lower fertility than the general population, especially males (Bundy et al. 2011). However, SZ patients also have impaired judgment, and those who are sexually active have a higher rate of risky sexual behaviors than sexually active comparison subjects (Coverdale and Turbott 2000; Ramrakha et al. 2000). A study of 95 SZ patients found that 44 % were sexually active in the preceding 6 months, and of these, 62 % had multiple sexual partners (Cournos et al. 1994). Finding a partner is prerequisite to engaging in risky sexual behavior, and to the extent that behaviors seen in SZ decrease this possibility, SZ may decrease the risk of HIV exposure and infection. On the other hand, because SZ is also associated with decreased judgment, a situation that might result in sexual activity could result in a greater incidence of risky behavior. Finally, HIV is also associated strongly with drug use, another behavior that is elevated in SZ.
While there are many reports on the epidemiological relationships between HIV, SZ, and RSB, their genetic relationships are not well studied. There has been a recent accumulation of genomics datasets and the development of increasingly powerful statistical methods designed to perform integrative genomics analysis. In this study, we obtained publically accessible GWAS summary statistics relevant to SZ and HIV infection and estimated their genetic correlation and level of pleiotropy using two independent methods, linkage disequilibrium (LD) score regression (Bulik-Sullivan et al. 2015) and Genetic analysis incorporating Pleiotropy and Annotation (GPA) (Chung et al. 2014). We also used data regarding RSBs collected by us in a sample recruited for study of the genetics of drug and alcohol dependence (Polimanti et al. 2016). Using these data, we explored the shared genetic relationships between SZ and HIV infection from two perspectives: genetic correlation, which characterizes the global correlation between the genetic effect sizes of two phenotypes, and pleiotropy, which refers to the phenomenon that a single locus may influence multiple phenotypes, regardless of the effect directions. We also studied the genetic relationship of SZ and HIV with RSB. The results of our study could help to increase our understanding of the biology of these traits.
Materials and methods
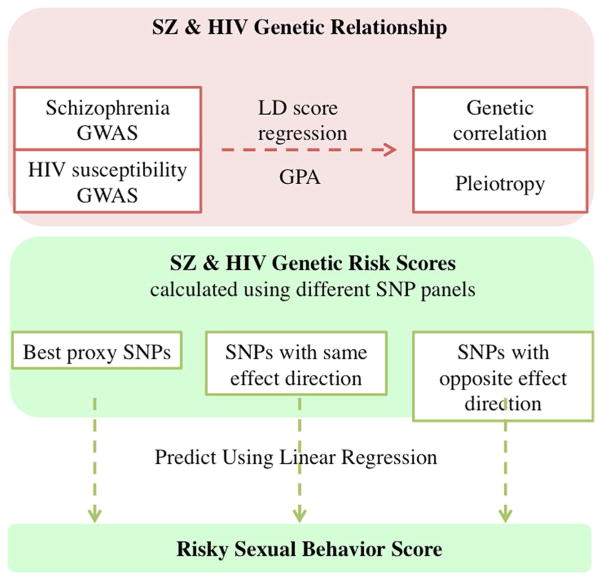
Our overall analysis procedure is summarized in Fig. 1. We first explored the shared genetic etiology of SZ and HIV infection through LD score regression and GPA using summary statistics from publically available GWAS data. We then tested whether HIV or SZ risk could independently predict RSB using our own cohort comprising 2379 European Americans (EAs) for whom genotype and RSB data were available. 2321 of these individuals passed genotype quality control and 1057 individuals had non-missing RSB scores. We also removed 55 individuals with SZ or HIV infection, leaving 1002 individuals for inclusion in polygenic risk score analysis. Detailed analysis procedures are described below.
Fig. 1.
Flowchart of study pipeline. Datasets and analysis procedures for this study are shown in the flowchart. Red (upper section) indicates analysis using public datasets, and green (lower section) indicates procedures used with our own dataset (color figure online)
Study recruitment and genotyping
A total of 2379 EA subjects (1400 males and 979 females) were included in this study. Recruitment was conducted in five centers in the Eastern United States, as described elsewhere (Gelernter et al. 2014a; Xie et al. 2013). The institutional review board at each center approved the study and written informed consent was obtained from all participants. DNA was extracted from blood or cell lines (or saliva for some subjects whose blood could not be obtained), and genotyped on the Illumina HumanOmni-Quad v1.0 micro-array. Detailed information about the genotyping and quality control pipeline is available in our published GWAS (e.g. Gelernter et al. 2014a, b). Multiple sample quality control procedures were performed, such that ten individuals were removed due to discordant sex information between self-reported and genotypic information; 46 subjects were removed due to excessive heterozygosity; two subjects were removed for genotype missing rates >0.02. Subjects that passed these quality control procedures (2321 individuals) were later used to calculate polygenic risk scores for SZ and HIV infection.
Assessment of risky sexual behaviors (RSB)
Subjects were evaluated using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al. 2005). Details about RSBs were derived from the SSADDA section on antisocial personality disorder: item I35B (Have you ever had sex with 10 different people within a single year?) and item I37 [Have you more than once had unprotected sex (without a condom) with someone you believed could give you a disease, or when you had a disease that could be spread that way?]. We modeled the RSB score as a quantitative measure of unprotected sex and multiple sexual partners that ranged from 0 to 2 based on the number of affirmative responses. Non-missing RSB scores were obtained for 1079 of the 2379 EA subjects, of which 1057 subjects passed genotype quality control, and 1002 subjects were free from SZ and HIV infection. For the 1057 subjects that passed phenotype and genotype quality control, demographics and SZ and HIV affection are shown in Table 1. To assess the potential for ascertainment bias, we tested correlation of RSB score availability with SZ and HIV infection. In 2379 EA subjects, there are 4 SZ and 113 HIV patients, of which 1 and 57 have missing RSB scores, respectively. Fisher’s exact test demonstrated that neither SZ (p = 0.335) nor HIV (p = 0.384) rate is biased by RSB ascertainment.
Table 1.
Demographics of 1057 subjects that passed genotype and phenotype quality control
| Risky sexual behavior (RSB) | 0 | 1 | 2 |
|---|---|---|---|
| Male | 346 (69.2 %) | 273 (66.9 %) | 86 (57.7) |
| Female | 154 (30.8 %) | 135 (33.1 %) | 63 (42.3 %) |
| Age | 39.2 (10.1) | 38.3 (9.8) | 38.5 (8.9) |
| Weight (lbs) | 180.5 (40.8) | 182.4 (40.4) | 184.9 (42.9) |
| Schizophrenia (SZ) | 2 (0.4 %) | 1 (0.2 %) | 0 |
| HIV | 17 (3.4 %) | 21 (5.1 %) | 15 (10.1 %) |
| SZ and HIV | 0 | 1 (0.2 %) | 0 |
| Drug injection (DI) | 241 (48.2 %) | 234 (57.4 %) | 96 (64.4 %) |
| Needle sharing (Nsh) | 129 (25.8 %) | 145 (35.5 %) | 75 (50.3 %) |
| Total sample size | 500 | 408 | 149 |
Numbers in parentheses are sample standard deviations for continuous measurements age and weight, and percentages for binary measurements sex, schizophrenia, HIV, drug injection, and needle sharing
Assessment of injection drug use (DI) and needle sharing (NSh)
DI and NSh were evaluated using self-report information from the SSADDA. DI is a binary measure, with “1” representing any self-reported DI experience assessed by six DI-related items from the SSADDA sections on cocaine use, opioid use, and “other drug” use. NSh is a binary measurement, with “1” representing any self-reported NSh assessed by the SSADDA item “Have you ever shared a needle” from the same three sections. Among the 1002 subjects used in the risk score prediction analysis, 527 subjects reported DI and of these, 317 also reported NSh. DI and Nsh are used in our polygenic risk score analysis as covariates.
GWAS data resources from studies by other investigators
HIV susceptibility GWAS summary statistics were obtained from McLaren et al. (2013), where genetic risk of HIV-1 infection was assessed in 6300 infected cases and 7200 controls.
Results files from the second Psychiatric Genomics Consortium (PGC) SZ mega-analysis (Schizophrenia Working Group of the Psychiatric Genomics 2014) were downloaded. The multi-stage SZ mega-analysis included up to 36989 cases and 113075 controls. Summary statistics of bipolar disorder GWAS (Psychiatric 2011) on 7481 cases and 9250 controls were also acquired from the PGC website.
GWAS summary statistics for height were obtained from a study with 253,288 participants (Wood et al. 2014) from the GIANT consortium.
GWAS summary statistics for body mass index (BMI) were obtained from a study with 234,069 participants (Locke et al. 2015) from the GIANT consortium.
Statistical analysis
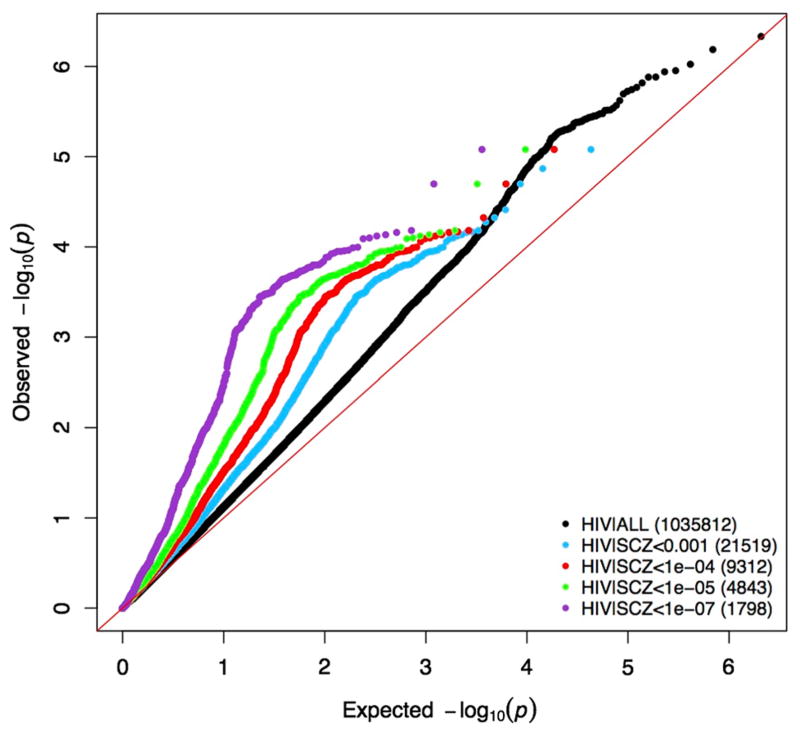
Conditional Q–Q plots were used to show the enrichment of GWAS signals between SZ and HIV. First, a standard Q–Q plot (observed log-transformed p value against expected log-transformed p value) based on HIV GWAS was generated using all 1,035,812 SNPs in the HIV GWAS. Then, conditional level of significance in SZ GWAS, SNP sets was selected with a series of cutoffs (p < 0.001, 1e-4, 1e-5, 1e-7). For each of the SNP sets, we plotted observed log-transformed HIV GWAS p values against expected log-transformed p values. The level of deviation of conditional Q–Q plot lines from the standard Q–Q plot line indicates strength of signal enrichment between the two traits, with respect to corresponding cutoffs.
LD score regression (Bulik-Sullivan et al. 2015) was used to estimate the overall genetic correlation of SZ and HIV by regressing the product of z-scores from the two studies onto LD scores. GPA (Chung et al. 2014) was used to test the significance of pleiotropy of HIV and SZ and to estimate the posterior probability of each SNP being associated with both traits. GPA adopts a mixture model, and estimates the underlying true association status via the expectation–maximization (EM) algorithm. Both of these methods require only GWAS summary statistics. DAVID (Huang et al. 2009a, b) was used to perform Gene Ontology (GO) term enrichment for the top SZ-HIV pleiotropic genes. Only biological process GO terms were used in our analysis.
We calculated SZ and HIV polygenic risk scores (PRS) using publically available summary statistics. PRS were computed using Polygenic Risk Score software (PRSice) (Euesden et al. 2015). Our analysis was limited to the 539,034 SNPs with available summary statistics for both SZ and HIV. To calculate SZ risk score, we first selected index SNPs with a series of p value thresholds, ranging from 0 to 0.5 in increments of 0.001, and then performed linkage disequilibrium-based clumping to select independent GWAS signals. For each index SNP, a best proxy SNP was selected that was within a distance of 250 kb, had r2 >0.1, and had the smallest p value for the interval. The SZ infection risk score of the 2379 European Americans from our GWAS was then calculated as the sum of the number of risk alleles weighted by an SZ log-transformed odds ratio. The HIV risk score of our cohort was calculated similarly.
Examination of same-direction vs. opposite-direction SNP effects
To examine the effect of HIV-SZ pleiotropic SNPs on RSB, we split the SNP panel for PRS calculation into two groups, one with the same effect direction for SZ and HIV and the other with the opposite effect direction. For all p value cutoffs, SZ and HIV risk scores were calculated separately for the same-direction group and the opposite-direction group. After quality control, we had 1002 European Americans with a non-missing RSB score, who had neither SZ nor HIV infection.
To study the relationship between SZ risk score and RSB score, the mean SZ risk score was calculated for three RSB groups (RSB scores = 0, 1, 2) and compared. We also used linear regression with age, sex, DI and NSh as covariates, as well as the first two ancestry-informative covariates calculated from our genotype data, to assess whether SZ risk score predicts RSB score in our cohort. The strength of association between HIV infection risk score and RSB score was assessed similarly.
Results
Genetic correlation and pleiotropy of SZ and HIV risk
The conditional Q–Q plot shows an enriched HIV GWAS signal conditional on SZ GWAS (Fig. 2). Using LD score regression, the estimated genetic correlation of SZ and HIV was 0.202 (se = 0.062) genome wide and 0.172 (se = 0.057) genome wide after exclusion of the major histocompatibility complex (MHC) region, with p values 0.001 and 0.002, respectively. Using this same approach to ascertain whether this signal had specificity, the genetic correlation between HIV and bipolar disorder was estimated to be 0.009 (se = 0.081), which was non-significant (p = 0.91). To rule out the alternative explanation that the large sample size of the schizophrenia GWA study contributed decisively to the observed significant genetic correlation, we used height and BMI as negative controls, as GWAS summary statistics from even larger studies are available for those two traits. Neither of them were found to have significant genetic correlation, with genetic correlation between HIV and height estimated to be 0.0183 (se = 0.041, p = 0.656), and genetic correlation between HIV and BMI estimated to be −0.0165 (se = 0.0439, p = 0.7064).
Fig. 2.
Conditional Q–Q plots of HIV GWAS p value conditional on SZ GWAS p values. Black dots include all overlapping SNPs, and the other four colors are SNP subsets conditional on p value of SZ GWAS below cutoffs (1e-3, 1e-4, 1e-5, 1e-7). X-axis is the expected log-transformed p value; y-axis is the observed log-transformed p value (color figure online)
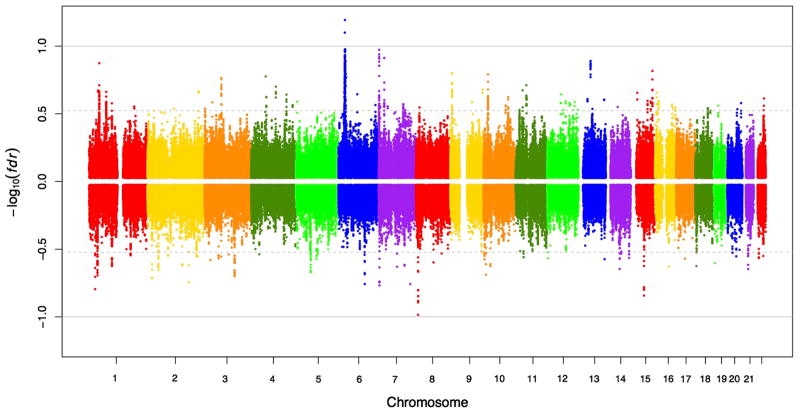
Consistent with the findings from LD score regression, we observed significant pleiotropy of SZ and HIV using GPA, with a p value close to 0 with all the overlapping SNPs included and p = 8.96E − 257 with only Hapmap3 SNPs. When the MHC region is excluded, the significance of pleiotropy is p = 6.31E − 62 with all overlapping SNPs, and p = 5.31E − 28 with only Hapmap3 SNPs. The posterior probabilities of SNPs being associated with both HIV and SZ were plotted across the genome and split into two panels, with the upper panel showing SNPs with same effect direction and the lower panel showing SNPs with the opposite effect direction (Fig. 3). There are 191 SNPs (Online Resource 1) acting in the same effect direction and 24 SNPs (Online Resource 2) acting in the opposite effect direction that have posterior probabilities above 0.8. Top genes hosting SNPs acting in the same effect direction include ZKSCAN3, SNX8, and TRIM13. The top gene hosting SNPs (rs678892, rs12902710, etc.) acting in the opposite effect direction is PIGB, which has mannosyltransferase activity and is universally expressed in many tissues.
Fig. 3.
Manhattan plot of posterior with effect directions. Absolute value of y-axis is the log-transformed false discovery rate (fdr) of SNPs to be associated with both SZ and HIV. Points above x-axis are SNPs with the same effect direction, while those below x-axis are SNPs with the opposite effect direction. 536 (73 %) of 730 SNPs with fdr <0.3 have the same direction
Different biological functions enriched for pleiotropic SNP sets with different effect directions
To study the biological function of SNPs pleiotropic for SZ and HIV, we selected SNPs with pleiotropy posterior probability greater than 0.5 and split the SNP sets into two groups, one with the same effect direction and the other with the opposite effect direction for HIV and SZ. Next, GO enrichment analysis was performed for each group using DAVID (Huang et al. 2009a, b). The 5942 SNPs with the same effect direction were mapped to 3853 genes with a 10-kb extension at each end and were primarily enriched for chromatin assembly and nucleosome organization, as well as protein–DNA complex assembly (Online Resource 3: Table S1). There were 4303 SNPs with opposite effect direction, which mapped to 3250 genes with a 10-kb extension at each end. This set of SNPs was enriched for the regulation of synaptic transmission and neurotransmitter transport, neuron differentiation, as well as the regulation of transmission of nerve impulse (Online Resource 3: Table S2).
SZ risk score predicts risky sexual behavior
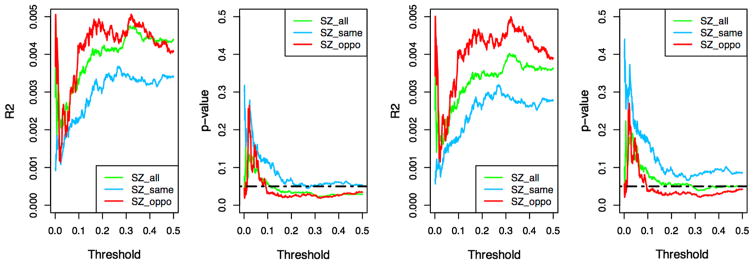
To study the genetic relationship between SZ-RSB and HIV-RSB, we calculated SZ and HIV polygenic risk scores and used them to predict RSB score in our substance dependence GWAS sample. To calculate the risk scores, p value thresholds ranging from 0 to 0.05 at increments of 0.001 were used to select index SNPs, as detailed in Methods. To control for confounding factors, age, sex, and the two top ancestry-informative covariates were included in linear regression analyses. To account for potential influence of drug use and to evaluate the consequences of adjusting for those influences, we performed tests with and without adjusting for DI and Nsh (Fig. 4). Throughout the range of p value thresholds we used, SZ risk scores computed with SZ-HIV antagonistically pleiotropic SNPs consistently outperformed SZ risk scores calculated with all SNPs regardless of HIV-SZ pleiotropy direction, and SZ-HIV synergistically pleiotropic SNPs. Using SZ scores calculated with SZ-HIV antagonistically pleiotropic SNPs and with DI and Nsh adjusted, the highest R2 observed is 0.005 (p = 0.019) achieved at two very different p value thresholds, 0.32 and 0.002. HIV risk scores did not predict RSB with a consistent pattern (Online Resource 3: Figures S1, S2), probably due to limited sample size. In all cases, NSh was a significant predictor of RSB score (p = 0.0002).
Fig. 4.
Effect sizes and p values in SZ polygenic risk score analyses. SZ risk scores were calculated with p value thresholds from 0 to 0.5 at increments of 0.001 (x-axis). Green, blue, and red colors represent SZ risk scores calculated using all SNPs, SNPs with the same effect directions for SZ and HIV, and SNPs with the opposite effect directions, respectively. SZ risk scores were used to predict RSB score via linear regression with sex, age and ancestry-informative covariates. From left to right: R2 with DI and Nsh adjusted, p values with DI and Nsh adjusted, R2 without DI and Nsh adjusted, p values without DI and Nsh adjusted. SZ risk scores from opposite SNPs consistently outperform SNPs with the same effect direction (color figure online)
Discussion
Our study shows that SZ and HIV have a positive genetic correlation, which is highly significant with or without inclusion of the MHC region. Consistently, our pleiotropy analysis found that a majority of the genetic factors they share have the same effect direction (shared-effect pleiotropic SNPs), meaning that the same allele tends to increase or decrease the risk of both SZ and HIV. This positive genetic correlation helps to explain the long-observed comorbidity of SZ and HIV, and calls for awareness that SZ patients might have higher risk for HIV partially due to the shared genetic factors acting with the same effect direction. It has been reported previously that behavioral interventions from nurses to address HIV among people with serious mental illness, including SZ (to encourage adherence to HIV and psychiatric treatment regimens), might be beneficial in terms of viral load and CD4 count (McGinty et al. 2016).
Although the overall SZ-HIV genetic correlation is positive, some associated SNPs that are shared have opposite effect directions (antagonistically pleiotropic SNPs) for these two traits, meaning that the same allele tends to increase the risk of one trait while decreasing the risk of the other. Genes identified based on the antagonistically pleiotropic SNPs of SZ and HIV are functionally enriched in the regulation of synaptic transmission and neurotransmitter transport. SZ risk scores based on SNPs having the opposite effect direction for HIV significantly predicted RSB score, while SZ risk scores based on SNPs having the same effect direction for HIV did not. These results indicate different biological mechanisms regarding the contributions to SZ-HIV pleiotropy from the two groups of SNPs, which support a genetic contribution to the complicated relationship between HIV and SZ, as well as RSB.
Risky sexual activity, as a major risk factor for HIV infection, is associated with common psychiatric disorders, including schizophrenia spectrum (Ramrakha et al. 2000). People with SZ are less likely to be sexually active and have lower rates of HIV-related risk behavior than those without SZ (Carey et al. 1999, 2001). However, individuals with SZ who are sexually active are more likely to engage in RSBs (Coverdale and Turbott 2000). Our results provide a genetic basis and a tentative biological explanation for these observations. We observed that when used to predict RSB score, SZ risk scores based on SNPs with opposite effect directions for SZ-HIV consistently outperformed (that is, showed better prediction) those based on SNPs with the same effect directions. Although our results only reached a significance level of p = 0.019, the consistent trend suggests that genetic factors that drive antagonistic pleiotropy of SZ and HIV contribute to RSBs. These genetic factors are highly enriched for biological functions such as the regulation of synaptic transmission and neurological system processes, which are likely to play a role in neural processing of risk behaviors. The low level of significance might be due to limited sample sizes, and future studies with larger sample sizes are needed to confirm our results. Nsh is a significant predictor of RSB score, which is expected as needle sharing and RSB are both risk-taking behaviors. In our polygenic risk score analyses, we observed that adjusting for DI and Nsh did not reduce the proportion of RSB variance explained by SZ risk scores, which tentatively suggests that Nsh might influence RSB through mechanisms independent of SZ. We note that this result should be interpreted with caution due to relatively small effect sizes and limited sample sizes.
Our study has several potential limitations. First, our analysis on HIV genetic factors might be underpowered. HIV risk score was not a significant predictor for RSB in any analysis. This unexpected result could be attributable to the lack of adequate statistical power for the analysis, which was based on a GWAS of 6300 cases and 7200 controls; an HIV risk score calculated from a GWAS with a larger sample could yield a different result. Second, our sample was predominantly substance dependent; behaviors that are common to individuals with substance dependence, such as NSh, are also HIV risk factors that contribute independently to HIV risk. To address this possible confounding, we controlled for DI and NSh in our analyses. Third, additional caution interpreting our results is warranted inasmuch as many environmental factors and genetic factors independent of RSB might also contribute to the comorbidity of SZ and HIV infection.
Building on current knowledge from epidemiological and clinical studies, our results have many possible interpretations. One we favor is that the shared-action SNP sets may involve a propensity to poor judgement and risky behavior, and those that act antagonistically may affect affiliative and social behaviors. We propose that investigating these SNP sets further, and, when larger samples are available, deriving genetic risk scores from them to test other phenotypes may yield important insights into several very important and varied behaviors.
Supplementary Material
Acknowledgments
This study was supported in part by Grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, P50 AA12870, R01 GM59507, and R01 DA030976 from the National Institutes of Health. Although unrelated to this work, Dr. Kranzler has been a consultant or advisory board member for Alkermes, Indivior, Lundbeck, and Otsuka. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by AbbVie, Ethypharm, Lilly, Lundbeck, and Pfizer.
Footnotes
Compliance with ethical standards
Conflict of interest Dr. Gelernter, Dr. Zhao, Dr. Farrer, Dr. Polimanti, and Qian Wang reported no biomedical financial interests or potential conflicts of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s00439-016-1737-8) contains supplementary material, which is available to authorized users.
References
- Blank MB, Mandell DS, Aiken L, Hadley TR. Co-occurrence of HIV and serious mental illness among Medicaid recipients. Psychiatric Serv (Washington, D C) 2002;53:868–873. doi: 10.1176/appi.ps.53.7.868. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JRB, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM, Consortium R, Consortium PG, Nervos GCA. An atlas of genetic correlations across human diseases and traits. Nature Genetics. 2015;47:1236. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy H, Stahl D, MacCabe JH. A systematic review and meta-analysis of the fertility of patients with schizophrenia and their unaffected relatives. Acta Psychiatr Scand. 2011;123:98–106. doi: 10.1111/j.1600-0447.2010.01623.x. [DOI] [PubMed] [Google Scholar]
- Carey MP, Carey KB, Maisto SA, Gleason JR, Gordon CM, Brewer KK. HIV-risk behavior among outpatients at a state psychiatric hospital: prevalence and risk modeling. Behav Ther. 1999;30:389–406. doi: 10.1016/S0005-7894(99)80017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey MP, Carey KB, Maisto SA, Gordon CM, Vanable PA. Prevalence and correlates of sexual activity and HIV-related risk behavior among psychiatric outpatients. J Consult Clin Psychol. 2001;69:846–850. doi: 10.1037//0022-006x.69.5.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. doi: 10.1371/journal.pgen.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournos F, Guido JR, Coomaraswamy S, Meyerbahlburg H, Sugden R, Horwath E. Sexual-activity and risk of HIV-infection among patients with schizophrenia. Am J Psychiatry. 1994;151:228–232. doi: 10.1176/ajp.151.2.228. [DOI] [PubMed] [Google Scholar]
- Coverdale JH, Turbott SH. Risk behaviors for sexually transmitted infections among men with mental disorders. Psychiatric Serv. 2000;51:234–238. doi: 10.1176/Appi.Ps.51.2.234. [DOI] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014a;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, Farrer LA. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014b;76:66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall HI, An Q, Tang T, Song R, Chen M, Green T, Kang J. Prevalence of diagnosed and undiagnosed HIV infection—United States, 2008–2012. MMWR Morb Mortal Wkly Rep. 2015;64:657–662. [PMC free article] [PubMed] [Google Scholar]
- Helleberg D, Pedersen MG, Pedersen CB, Mortensen PB, Obel N. Associations between HIV and schizophrenia and their effect on HIV treatment outcomes: a nationwide population-based cohort study in Denmark. Lancet HIV. 2015;2:e344. doi: 10.1016/S2352-3018(15)00089-2. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/Nar/Gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/Nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Hua Zhao J, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Mateo Leach I, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Ju Sung Y, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Ida Chen YD, Clarke R, Daw EW, de Craen AJ, Delgado G, Dimitriou M, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks G, Crepaz N, Senterfitt JW, Janssen RS. Meta-analysis of high-risk sexual behavior in persons aware and unaware they are infected with HIV in the United States—implications for HIV prevention programs. Jaids J Acquir Immune Defic Syndr. 2005;39:446–453. doi: 10.1097/01.Qai.0000151079.33935.79. [DOI] [PubMed] [Google Scholar]
- McGinty EE, Baller J, Azrin ST, Juliano-Bult D, Daumit GL. Interventions to address medical conditions and health-risk behaviors among persons with serious mental illness: a comprehensive review. Schizophrenia Bull. 2016;42:96–124. doi: 10.1093/schbul/sbv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren PJ, Coulonges C, Ripke S, van den Berg L, Buchbinder S, Carrington M, Cossarizza A, Dalmau J, Deeks SG, Delaneau O, De Luca A, Goedert JJ, Haas D, Herbeck JT, Kathiresan S, Kirk GD, Lambotte O, Luo M, Mallal S, van Manen D, Martinez-Picado J, Meyer L, Miro JM, Mullins JI, Obel N, O’Brien SJ, Pereyra F, Plummer FA, Poli G, Qi Y, Rucart P, Sandhu MS, Shea PR, Schuitemaker H, Theodorou I, Vannberg F, Veldink J, Walker BD, Weintrob A, Winkler CA, Wolinsky S, Telenti A, Goldstein DB, de Bakker PI, Zagury JF, Fellay J. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS Pathog. 2013;9:e1003515. doi: 10.1371/journal.ppat.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller N, Schwarz MJ. Immune System and Schizophrenia. Curr Immunol Rev. 2010;6:213–220. doi: 10.2174/157339510791823673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Polimanti R, Wang Q, Meda S, Patel K, Pearlson G, Zhao H, Farrer L, Kranzler H, Gelernter J. The interplay between risky sexual behaviors and alcohol dependence: genome-wide association and neuroimaging support for LHPP as a Risk Gene. Neuropsychopharmacology. 2016 doi: 10.1038/npp.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GCBDWG. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrakha S, Caspi A, Dickson N, Moffitt TE, Paul C. Psychiatric disorders and risky sexual behaviour in young adulthood: cross sectional study in birth cohort. BMJ. 2000;321:263–266. doi: 10.1136/bmj.321.7256.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. http://www.nature.com/nature/journal/v511/n7510/abs/nature13595.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman MV. Social skills training for schizophrenia: A step-by-step guide—Bellack AS, Mueser KT, Gingerich S, Agresta J. Journal of Psychiatry & Neuroscience. 1997;22:342–342. [Google Scholar]
- Walkup J, Blank MB, Gonzalez JS, Safren S, Schwartz R, Brown L, Wilson I, Knowlton A, Lombard F, Grossman C, Lyda K, Schumacher JE. The impact of mental health and substance abuse factors on HIV prevention and treatment. J Acquir Immune Defic Syndr. 2008;47(Suppl 1):S15–S19. doi: 10.1097/QAI.0b013e3181605b26. [DOI] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, Amin N, Buchkovich ML, Croteau-Chonka DC, Day FR, Duan Y, Fall T, Fehrmann R, Ferreira T, Jackson AU, Karjalainen J, Lo KS, Locke AE, Magi R, Mihailov E, Porcu E, Randall JC, Scherag A, Vinkhuyzen AA, Westra HJ, Winkler TW, Workalemahu T, Zhao JH, Absher D, Albrecht E, Anderson D, Baron J, Beekman M, Demirkan A, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Fraser RM, Goel A, Gong J, Justice AE, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Lui JC, Mangino M, Mateo Leach I, Medina-Gomez C, Nalls MA, Nyholt DR, Palmer CD, Pasko D, Pechlivanis S, Prokopenko I, Ried JS, Ripke S, Shungin D, Stancakova A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Afzal U, Arnlov J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Bluher M, Bolton JL, Bottcher Y, Boyd HA, Bruinenberg M, Buckley BM, Buyske S, Caspersen IH, Chines PS, Clarke R, Claudi-Boehm S, Cooper M, Daw EW, De Jong PA, Deelen J, Delgado G, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu EQ, Birnbaum HG, Shi L, Ball DE, Kessler RC, Moulis M, Aggarwal J. The economic burden of schizophrenia in the United States in 2002. J Clin Psychiatry. 2005;66:1122–1129. doi: 10.4088/jcp.v66n0906. [DOI] [PubMed] [Google Scholar]
- Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.