Abstract
Although growing numbers of oncoproteins and pro-metastatic proteins have been extensively characterized, many of these tumor-promoting proteins are not good drug targets, which represents a major barrier to curing breast cancer and other cancers. There is a need, therefore, for alternative therapeutic approaches to destroying cancer-promoting proteins. The human genome encodes approximately 100 deubiquitinating enzymes (DUBs, also called deubiquitinases), which are amenable to pharmacologic inhibition by small molecules. By removing monoubiquitin or polyubiquitin chains from the target protein, DUBs can modulate the degradation, localization, activity, trafficking, and recycling of the substrate, thereby contributing substantially to the regulation of cancer proteins and pathways. Targeting certain DUBs may lead to destabilization or functional inactivation of some key oncoproteins or prometastatic proteins, including non-druggable ones, which will provide therapeutic benefits to cancer patients. In breast cancer, growing numbers of DUBs are found to be aberrantly expressed. Depending on their substrates, specific DUBs can either promote or suppress mammary tumors. In this article, we review the role and mechanisms of action of DUBs in breast cancer, and discuss the potential of targeting DUBs for cancer treatment.
Keywords: breast cancer, deubiquitinase, ubiquitination, DUB inhibitor
1 Introduction
Protein ubiquitination is a multistep posttranslational modification process in which a highly conserved, 76-amino acid polypeptide, ubiquitin, is added to protein substrates through a cascade of reactions involving ubiquitin activation by a ubiquitin-activating enzyme (E1), followed by its transfer to a lysine residue on the substrate, which is catalyzed by ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3) (Figure 1) [1,2]. In addition, another class of polyubiquitin ligases, E4, was identified as a ubiquitin chain elongation factor family required for polyubiquitin chain assembly for certain monoubiquitinated proteins [3,4]. Ubiquitin E3 ligases can act as oncoproteins or tumor suppressors in breast cancer. For instance, cancer-predisposing mutations of BRCA1 inactivate its ubiquitin ligase activity, suggesting that the tumor suppressor role of BRCA1 is associated with its E3 ligase function [5,6]. On the other hand, SKP2, the E3 ligase of the SKP1-Cullin1-F-box protein (SCF) complex, targets the CDK inhibitor p27 for degradation and plays an oncogenic role in breast cancer [7-9].
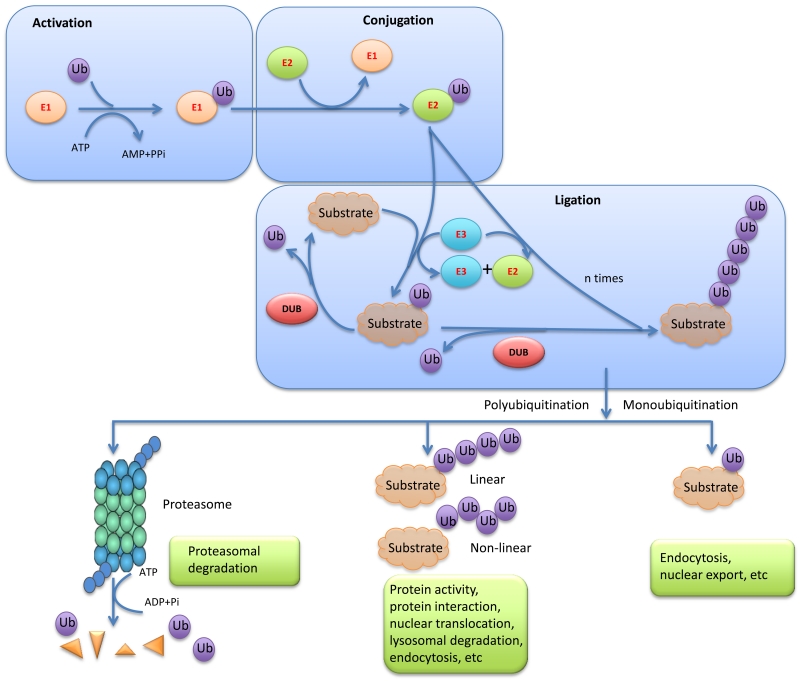
Figure 1. Ubiquitin ligases, deubiquitinating enzymes, and other components in the ubiquitination pathway.
Ubiquitin is activated by a ubiquitin-activating enzyme (E1), followed by its transfer to a lysine residue on the substrate, which is catalyzed by ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3). DUBs reverse this process by removing polyubiquitin chains or monoubiquitin from target proteins, and thus rescue proteins from proteasome-dependent degradation or modulate non-proteasomal processes.
Ubiquitination is reversed by deubiquitinases, or DUBs, a superfamily of cysteine proteases and metalloproteases that cleave ubiquitin-protein bonds (Figure 1) [10]. The human genome encodes approximately 100 DUBs, which can be classified into six families: ubiquitin-specific proteases (USPs), ubiquitin carboxy-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph disease protein domain proteases (MJDs), JAMM/MPN domain-associated metallopeptidases (JAMMs), and the monocyte chemotactic protein-induced protein (MCPIP) family [11]. The USP family is the largest and most diverse DUB family. Members of this family have a conserved catalytic domain that consists of three subdomains resembling the thumb, fingers, and palm of the right hand [12]. DUBs in the UCH family, the first structurally characterized DUB family, have six or seven β-sheets surrounded by eight α-helices, which act as a gate to preclude large substrates from getting access to the catalytic core located at the bottom of the DUB [13,14]. Thus, UCH family members can only target small peptides from the C terminus of ubiquitin. The OTU domain was initially identified in an ovarian tumor gene, which consists of five β-sheets interspersed between two helical domains [15,16]. The MJD family has four members, including the well characterized ATXN3 that is mutated in Machado-Joseph disease, and the other members are ATXN3L, JOSD1, and JOSD2 [17,18]. Unlike all other DUB families that are cysteine proteases, the JAMM family members are zinc metalloproteases [19,20]. Recent structural studies revealed that a JAMM family member, AMSH-LP (associated molecule with SH3 domain-like protease), specifically cleaves lysine 63-linked polyubiquitin from the substrate and regulates vesicle trafficking [21]. The MCPIP family has at least seven members, all of which consist of an N-terminal ubiquitin association domain, a central CCCH-type zinc-finger domain, and a C-terminal proline-rich domain [22].
DUBs regulate proteasome-dependent or lysosome-dependent degradation, localization, and recycling of substrate proteins (Figure 1), depending on the specific lysine (K) residue through which the ubiquitin chain is linked. Seven lysine residues, K6, K11, K27, K29, K33, K48, and K63, are present on the ubiquitin molecule, and K48- and K63-linked ubiquitination is best characterized. Polyubiquitin chains linked through K48, and likely K6, K11, K27, K29, and K33 mediate proteasomal degradation [23]. By removing these ubiquitin chains from target proteins, DUBs stabilize their substrates. For example, stabilization of NF-κB/RelA by USP48 [24], stabilization of MCL1 by USP9X [25], and stabilization of PTEN by OTUD3 [26] are associated with the cleavage of K48-linked polyubiquitin from the substrate. DUBs can also alter protein localization, which is usually mediated by the cleavage of K63-linked polyubiquitin from the target protein. For instance, CYLD, a USP family member involved in cylindromatosis, antagonizes K63-linked ubiquitination of BCL3 and blocks its nuclear localization [27]. In addition, DUBs can also inhibit lysosomal degradation of proteins. A recent study suggested that ubiquitinated EGFR is internalized into early endosomes, where USP2a catalyzes deubiquitination of EGFR, leading to recycling of EGFR back to the plasma membrane [28]. Since DUBs modulate protein stability, signal transduction, and other non-proteasomal functions, they contribute substantially to the regulation of key cancer proteins and pathways. In this review, we focus on DUB-mediated regulation in breast cancer (Table 1).
Table 1. DUBs involved in breast cancer.
| DUB | Substrate | Deubiquitination- induced change |
Potential role in breast cancer |
References |
|---|---|---|---|---|
|
| ||||
| USP2 | EGFR, FASN, Mdm2, RIP1, TRAF2 |
Protein stability or activity |
Promote or suppress |
28, 37-40 |
|
| ||||
| USP7 | PTEN, p53, Mdm2 | Protein stability or localization |
Promote or suppress |
91, 95-100 |
|
| ||||
| USP9X | MCL1, SMAD4, SMURF1 | Protein stability or interaction |
Promote | 25, 46, 47 |
|
| ||||
| USP10 | p53 | Protein stability and localization |
Promote or suppress |
100 |
|
| ||||
| USP13 | PTEN | Protein stability | Suppress | 26, 92 |
|
| ||||
| USP15 | Mdm2, TβR-I, R-SMADs | Protein stability or activity |
Promote | 50-52 |
|
| ||||
| USP18 | Not reported | Not reported | Promote | 41-43 |
|
| ||||
| USP28 | MYC, HIF-1α, LSD1 | Protein stability | Promote | 58-60 |
|
| ||||
| USP36 | MYC | Protein stability | Promote | 74 |
|
| ||||
| ATXN3L | KLF5 | Protein stability | Promote | 61 |
|
| ||||
| BAP1 | KLF5 | Protein stability | Promote | 62 |
|
| ||||
| BRCC36 | H2A-type histones | Protein interaction | Promote | 68-70 |
|
| ||||
| CYLD | TRAF2, TRAF6, TRAF7, TAK1, DVL, MIB2 |
Protein stability or activity |
Suppress | 77, 79-87 |
|
| ||||
| DUB3 | CDC25A | Protein stability | Promote | 73 |
|
| ||||
| OTUB1 | ERα, SMAD2/3, histones, FOXM1 |
Protein stability | Promote | 31-34 |
|
| ||||
| OTUD3 | PTEN | Protein stability | Suppress | 26 |
|
| ||||
| UCH37 | TβR-I | Protein stability | Promote | 71 |
2 Breast cancer-promoting DUBs
2.1 Deubiquitinases that regulate receptors involved in breast cancer
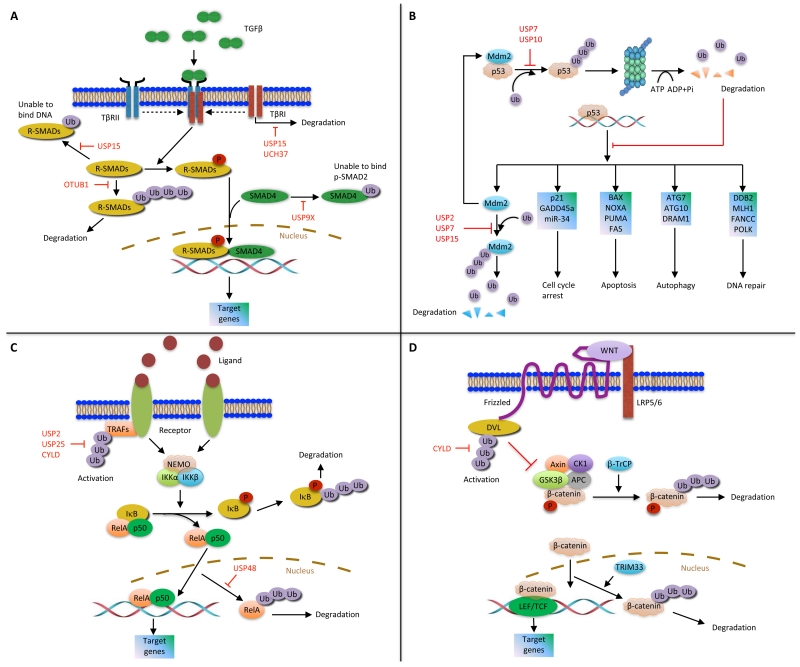
Approximately 70% of human breast tumors are positive for estrogen receptor (ER) α. Upon ligand binding, ERα translocates into the nucleus, recruits coactivator protein complexes to gene promoters, and activates the transcription of target genes [29,30]. Through mass spectrometric analysis of ERα-containing protein complexes, O’Malley and colleagues identified a member of the OTU domain family, OTUB1, as an ERα-interacting DUB. OTUB1 deubiquitinates ERα in vitro and in vivo and stabilizes ERα in the nucleus [31]. Additional substrates have also been found for this deubiquitinase. For instance, a recent study revealed that OTUB1 inhibits the ubiquitination and degradation of active SMAD2/3, thereby enhancing the activity of the TGFβ signaling pathway [32] (Figure 2A). Other studies have implicated OTUB1 in DNA damage response. It has been shown that OTUB1 inhibits DNA double-strand break-induced, RNF168-dependent polyubiquitination of histones [33]. Moreover, OTUB1 can deubiquitinate and stabilize the forkhead transcription factor FOXM1, which promotes DNA damage response and genotoxic drug resistance in breast cancer. Consistently, tissue microarray analysis demonstrated a positive correlation of OTUB1 expression with ERα expression and FOXM1 expression in human breast tumors [34].
Figure 2. DUB regulation of several signaling pathways.
(A) TGFβ pathway. (B) Mdm2-p53 pathway. (C) NF-κB pathway. (D) Wnt pathway.
Overexpression or activating mutation of EGFR, which promotes cell growth and survival, is associated with various human cancers including breast cancer [35]. A splicing isoform of USP2, USP2a, localizes to early endosomes and stabilizes internalized EGFR protein by antagonizing the ubiquitination and lysosomal degradation of EGFR [28]. Using microarray and tissue microarray analyses, Qu et al. found that USP2 was overexpressed in human breast tumors, especially in triple-negative breast cancer (TNBC), and that patients with USP2-positive breast tumors had worse clinical outcomes than patients with USP2-negative breast tumors [36]. This study also showed that USP2 upregulates MMP2 expression and induces the motility and invasiveness of TNBC cells [36]; however, it remains unknown whether MMP2 is a direct substrate of USP2. On the other hand, several USP2 substrates other than EGFR have been reported. For example, FASN (fatty acid synthase), which is overexpressed in many cancers including breast cancer and prostate cancer, is deubiquitinated and stabilized by USP2a [37]. In addition, USP2a binds and deubiquitinates Mdm2, leading to accumulation of Mdm2 and Mdm2-mediated degradation of p53 [38] (Figure 2B). Interestingly, knockdown of USP2a led to downregulation of Mdm2, upregulation of p53, and activation of p53-dependent gene transcription and cell death [38], suggesting a strategy to restore p53 in tumor cells. However, it should be noted that ectopic expression of USP2a in the breast cancer cell line MCF7 caused apoptosis, which might be mediated by reversal of K63-linked polyubiquitination of RIP1 and TRAF2 by USP2a and subsequent inhibition of NF-κB signaling [39,40] (Figure 2C). These results suggest that the function of USP2 may depend on the cellular context.
In an RNA interference screen of 106 different genes related to deubiquitination, silencing of USP18 reduced EGFR expression without affecting the levels of other receptor tyrosine kinases [41]. Mechanistic studies demonstrated that depletion of USP18 led to upregulation of microRNA miR-7, which in turn suppressed EGFR mRNA translation and induced apoptosis [41,42]. The USP18 substrate responsible for regulating miR-7 remains to be identified. Another RNA interference screen of 53 DUBs revealed that loss of USP18 enhanced apoptosis triggered by bortezomib or etoposide [43]. This was further corroborated by the finding that overexpression of USP18 suppressed IFN-α-, TRAIL-, or bortezomib-induced apoptotic signaling in MCF7 breast cancer cells; surprisingly, a catalytically inactive mutant of USP18 exhibited similar effects [43]. Taken together, the anti-apoptotic functions of USP18 may depend on its catalytic and non-catalytic activity.
2.2 USP9X
USP9X, an X-linked USP family member, was found to be overexpressed in human breast tumors relative to adjacent normal tissues through restriction fragment differential display polymerase chain reaction (RFDD-PCR) and proteomic analyses [44]. The Drosophila homolog of USP9X, FAM, interacts with and stabilizes β-catenin, an oncogenic protein in colon cancer, breast cancer, and other cancers [45]; however, whether FAM/USP9X directly deubiquitinates β-catenin has not been determined. By screening a DUB siRNA library, Piccolo and colleagues identified FAM/USP9X as a positive regulator of TGFβ signaling. FAM/USP9X is required for TGFβ-induced migration of MDA-MB-231 breast cancer cells; mechanistically, FAM/USP9X inhibits the monoubiquitination of SMAD4, a modification that blocks the association of SMAD4 with phospho-SMAD2 [46] (Figure 2A). USP9X has also been shown to antagonize polyubiquitination. For example, by removing K48-linked polyubiquitin chains from MCL1, USP9X inhibits the proteasomal degradation of MCL1, a pro-survival BCL2 family member that is overexpressed in multiple cancer types and contributes to chemoresistance and tumor recurrence [25]. Moreover, USP9X deubiquitinates and stabilizes SMURF1, a member of the NEDD4 family of ubiquitin ligases; silencing USP9X expression in MDA-MB-231 breast cancer cells destabilized SMURF1 and inhibited SMURF1-dependent cell migration [47]. Collectively, USP9X may promote breast cancer through deubiquitination of multiple substrates.
2.3 USP15
USP15 is a DUB associated with the COP9 signalosome (CSN), a conserved protein complex implicated in DNA damage response. Specific CSN subunits are upregulated in human breast cancer, positively correlate with Mdm2 expression, and antagonize p53-mediated tumor suppression [48]. Owing to the homology between CSN and the 19S lid complex of the proteasome, CSN is hypothesized to regulate the degradation of polyubiquitinated proteins [49]. Indeed, USP15 has been found to directly deubiquitinate and stabilize Mdm2, which in turn inhibits the expression of p53 target genes, promotes tumor cell survival, and suppresses antitumor T cell responses [50] (Figure 2B). Moreover, Seoane and colleagues reported that the USP15 gene is amplified in breast cancer, ovarian cancer, and glioblastoma, and that USP15 deubiquitinates and stabilizes type I TGFβ receptor, thereby enhancing TGFβ signaling and tumor growth [51] (Figure 2A). USP15 can also inhibit monoubiquitination. Recently, Piccolo and colleagues identified USP15 as a DUB for receptor-activated SMADs (R-SMADs) [52]. Upon ligand binding, TGFβ and BMP receptors phosphorylate R-SMADs, which then bind SMAD4 to form a transcriptional complex and regulate target gene transcription. USP15 is required for TGFβ and BMP responses, such as TGFβ-induced breast cancer cell migration. Mechanistically, USP15 reverses R-SMAD monoubiquitination, a posttranslational modification that blocks the DNA-binding ability of R-SMADs [52] (Figure 2A). Taken together, USP15 can enhance TGFβ signaling by opposing both TGFβ receptor polyubiquitination and R-SMAD monoubiquitination, which may contribute to tumor progression. Oncomine data revealed upregulation of USP15 mRNA levels in human breast cancer, suggesting that USP15 may play a breast tumor-promoting role [53].
2.4 USP28
MYC is an oncogenic transcription factor that regulates cell growth, proliferation, and apoptosis. Amplifications or activating mutations of the MYC gene are associated with multiple types of human cancer [54]. MYC protein is normally subjected to degradation by the ubiquitin-proteasome pathway; however, the stability of MYC is elevated in human tumors as a result of mutations of the MYC gene or alterations in the regulators of MYC protein degradation [55-57]. Identified in a retroviral shRNA library screen, USP28 has been shown to decrease MYC polyubiquitination and increase MYC stability by antagonizing the activity of the SCFFBW7 ubiquitin ligase complex [58]. Silencing USP28 expression in breast, colon, lung, and cervical cancer cells resulted in downregulation of MYC protein and inhibition of cell proliferation, which suggested a cancer-promoting role of USP28 [58]. USP28 also opposes the ubiquitination and degradation of other FBW7 substrates, such as HIF-1α, which in turn promotes angiogenesis [59]. In addition, USP28 confers stem cell-like traits on breast cancer cells by deubiquitinating and stabilizing LSD1, a chromatin modulator that demethylates H3K4me1/2 and is overexpressed in breast, bladder, lung, and colon tumors [60]. Knockdown of USP28 in human breast cancer cells inhibited tumorigenicity and cancer stem cell-like properties. In tumors from patients with breast cancer, USP28 protein is highly expressed and correlates with the level of LSD1 protein [58,60].
2.5 ATXN3L and BAP1: the KLF5 deubiquitinases
KLF5 (Krüppel-like factor 5) is a zinc finger transcription factor that is highly expressed in basal-like breast cancer and promotes cell proliferation, survival, migration, and tumorigenesis [61]. A recent study identified ATXN3L, a member of the MJD family, as a KLF5 DUB by siRNA screening [61]. ATXN3L directly binds, deubiquitinates, and stabilizes KLF5. Knockdown of ATXN3L decreased KLF5 protein levels and inhibited the proliferation of breast cancer cells [61]. Another KLF5 DUB that stood out in siRNA screening was BAP1 (BRCA1-associated protein 1), which also directly binds KLF5 and stabilizes KLF5 through deubiquitination [62]. Knockdown of BAP1 in TNBC cells inhibited tumorigenesis and metastasis, which could be partially rescued by restoration of KLF5 expression, suggesting that KLF5 mediates, at least in part, the breast cancer-promoting function of BAP1 [62]. These studies shed light on DUBs that are potential antitumor targets in basal-like breast cancer.
2.6 Other DUBs upregulated in breast cancer
Some of the deubiquitinases involved in regulating immune responses are aberrantly expressed in breast cancer. USP25 was first identified as a deubiquitinase gene located in the chromosomal region 21q11.2 [63]. Using RFDD-PCR and proteomic analyses, Deng et al. observed overexpression of USP25 in human breast tumors [44]; however, the function of USP25 in breast cancer remains unclear. Mutational analysis revealed that the ubiquitin-associated domain and ubiquitin-interacting motifs of USP25 regulate its ubiquitination state, and that the conjugation of ubiquitin or SUMO to the K99 residue of USP25 modulates substrate recognition [64,65]. A recent study showed that USP25 suppresses IL-17 signaling by reversing K63-linked ubiquitination of TRAF6 and TRAF5, and that IL-17-mediated inflammation was enhanced in Usp25-/- mice [66] (Figure 2C). Whether USP25 plays a functional role in breast cancer warrants further investigation.
Ubiquitination and deubiquitination play a pivotal role in DNA damage response. The BRCA1 ubiquitin ligase is implicated in many cellular processes, including DNA repair, cell cycle, and chromatin remodeling. Germline mutations in BRCA1 and BRCA2 predispose women to breast cancer and ovarian cancer [67]. BRCC36 (BRCA1/BRCA2-containing complex subunit 36), a member of the JAMM family of DUBs, is highly expressed in breast tumors [68]. Depletion of BRCC36 abrogated BRCA1 phosphorylation and nuclear foci formation in breast cancer cells exposed to ionizing radiation (IR), thereby sensitizing these tumor cells to IR-induced apoptosis [69]. On the other hand, BRCC36 is recruited to DNA lesions as a component of the RAP80 complex upon IR exposure and counteracts RNF8-UBC13-mediated ubiquitination of H2A-type histones [70]. The precise role of BRCC36 in BRCA1-associated breast and ovarian cancer risk remains to be determined.
Targeting specific DUBs may provide a strategy to overcome therapy resistance. UCH37 (also known as UCHL5) is a UCH family member that is upregulated in breast cancer [53]. Wicks et al. reported that UCH37 can deubiquitinate and stabilize type I TGFβ receptor and augment TGFβ signaling [71] (Figure 2A). b-AP15, an inhibitor of USP14 and UCH37, reverses resistance of tumor cells to the proteasome inhibitor bortezomib [72].
Cell cycle proteins are regulated by ubiquitination and deubiquitination. USP17L2 (also known as DUB3) is a deubiquitinase responsible for the stabilization of the Tyr/Thr phosphatase CDC25A, which activates cell-cycle progression and is overexpressed in many human cancers [73]. Overexpression of DUB3 induced oncogenic transformation, while knockdown of DUB3 promoted the ubiquitination and degradation of CDC25A, leading to the arrest of cells at G1/S and G2/M phases. In a subset of patients with breast cancer, DUB3 and CDC25A were found to be co-expressed at high levels in the tumors [73].
In addition, high expression levels of USP36 (a MYC DUB) [74], USP32 (a membrane-bound DUB) [75], and USP9Y (a Y-linked DUB) [44] have also been observed in human breast tumors. Their roles and mechanisms of action in breast cancer have yet to be elucidated.
3 Breast cancer-suppressing DUBs
3.1 CYLD
CYLD is a unique K63 linkage-specific deubiquitinase in the USP family that lacks the zinc finger domain responsible for distal ubiquitin interaction. The CYLD gene was first identified as a candidate tumor suppressor gene mutated in cylindromatosis, which is characterized by the development of multiple benign skin tumors [76]. A recent study revealed that CYLD is downregulated in human breast cancer and correlates with clinical outcomes [77].
CYLD regulates multiple pathways by deubiquitinating key signaling components, which has been best characterized for the NF-κB pathway. In canonical NF-κB signaling, TRAF2 and TAK1 are activated in response to ligand stimulation, leading to activation of the IKK complex consisting of two catalytic subunits IKKα and IKKβ and the regulatory protein IKKγ (also known as NEMO). Subsequently, IκB is phosphorylated by the IKK complex and degraded by the proteasome, releasing the p50-p65 dimer that enters the nucleus to activate gene transcription [78]. By screening an shRNA library targeting 50 human DUBs, Bernards and colleagues identified CYLD as a negative regulator of NF-κB signaling [79]. Mechanistically, CYLD binds to NEMO and TRAF2 and reverses non-K48-linked polyubiquitination of TRAF2, thereby blocking TRAF2-mediated activation of the IKK complex [79-81] (Figure 2C). Additional components of the NF-κB pathway, such as TRAF6, TRAF7, and TAK1, have also been reported to be regulated by CYLD [82]. Moreover, IKKε, a noncanonical IKK family member, phosphorylates CYLD at serine 418 and reduces its deubiquitinase activity, leading to induction of oncogenic transformation [83]. This implicates CYLD in breast cancer, since IKKε is over-expressed in more than 30% of breast tumors [84]. Notably, Cyld-deficient mice exhibited enhanced NF-κB activity and increased susceptibility to colonic inflammation and colitis-associated cancer [85], providing in vivo proof that CYLD is a bona fide suppressor of inflammation and tumorigenesis.
CYLD also has NF-κB-independent functions. For instance, CYLD interacts with Dishevelled (DVL) and counteracts its K63-linked ubiquitination, leading to inhibition of this cytoplasmic effector in the Wnt/β-catenin pathway (Figure 2D). Silencing CYLD expression resulted in accumulation of β-catenin and activation of its target genes [86]. Moreover, MIB2, an E3 ligase for the Notch ligand JAG2, is deubiquitinated and stabilized by CYLD. Depletion of CYLD led to upregulation of JAG2 expression and activation of Notch signaling, and CYLD-deficient tumor cells were sensitive to γ-secretase inhibitors that target the Notch pathway [87]. Therefore, targeting the DUB downstream pathways could be a useful therapeutic strategy for cancers that have lost a tumor-suppressing DUB.
3.2 The PTEN deubiquitinases
The tumor suppressor PTEN, a lipid phosphatase that antagonizes the PI3K-AKT pathway, is frequently lost in human cancer [88]. Whereas approximately 5% of sporadic breast tumors harbor PTEN mutations [89], loss of PTEN protein is found in a much higher percentage of tumors [90], suggesting that posttranscriptional and posttranslational regulation may contribute substantially to the absence of PTEN. Several E3 ligases target PTEN for proteasomal degradation. On the other hand, reversal of PTEN monoubiquitination by USP7 (also known as HAUSP) alters PTEN subcellular localization without affecting its protein level [91]. Unlike the great majority of DUB screening studies that rely on siRNA libraries, our laboratory used two alternative screening approaches to identify the PTEN DUB. In the first approach, we co-transfected a panel of triple epitope-tagged DUBs with MYC-tagged PTEN into 293T cells, and then pulled down the DUB with S-protein beads and used immunoblotting to detect PTEN. In the second approach, we performed tandem affinity purification and mass spectrometric analysis of PTEN-containing protein complexes. These two approaches identified the same set of five deubiquitinases that physically associate with PTEN, and only one of them, USP13, stabilizes PTEN protein via direct binding and deubiquitination of PTEN [92]. Depletion of USP13 in breast cancer cells promoted AKT phosphorylation, cell proliferation, glycolysis, and tumor growth through downregulation of PTEN. Conversely, overexpression of USP13 suppressed tumorigenesis in PTEN-positive but not PTEN-null breast cancer cells. Importantly, USP13 protein is downregulated in human breast cancer and correlates with PTEN protein levels [92]. This work identified USP13 as the first DUB that regulates PTEN polyubiquitination and protein stability, which has been independently confirmed by several recent studies [26,93]. Intriguingly, USP13 and a newly identified PTEN deubiquitinase, OTUD3, have a synergistic effect in regulating PTEN, phospho-AKT, and tumor growth in breast cancer [26].
3.3 The p53 deubiquitinases
The tumor suppressor protein p53 is regulated by Mdm2-mediated ubiquitination and degradation and has a short half-life (5-20 minutes) [94]. The first DUB for p53, USP7 (HAUSP), was identified by Gu and colleagues through mass spectrometric analysis of affinity-purified p53-interacting proteins. Wild-type USP7, but not its catalytically inactive mutant, deubiquitinates and stabilizes p53 [95] (Figure 2B). Interestingly, a biomarker of poor clinical outcome in breast cancer, TSPYL5, which is encoded by a gene located in a frequently amplified chromosomal region, binds to USP7 and suppresses its ability to deubiquitinate and stabilize p53, leading to increased p53 ubiquitination and oncogenic transformation [96]. Surprisingly, both the Gu group and the Vogelstein group reported that complete ablation of USP7 resulted in stabilization of p53; this is because USP7 deubiquitinates and stabilizes Mdm2 more potently under physiologic conditions [97,98] (Figure 2B). However, under genotoxic stress conditions, binding of Mdm2 to USP7 is impaired as a result of ATM-mediated phosphorylation of Mdm2, shifting the balance towards stabilization of p53 [99,100]. Collectively, these findings reveal a dynamic role of USP7 (HAUSP) in regulating the Mdm2-p53 pathway.
Lou and colleagues then added another deubiquitinase, USP10, to the loop [101]. Unlike USP7, USP10 is a p53 DUB (Figure 2B), but not an Mdm2 DUB. In unstressed cells, USP10 is predominantly localized in the cytoplasm, which deubiquitinates cytoplasmic p53 and inhibits Mdm2-mediated p53 nuclear export and degradation. Following DNA damage, phosphorylation of USP10 by ATM leads to translocation of USP10 into the nucleus, where USP10 deubiquitinates and stabilizes nuclear p53 [101]. Because USP10 may stabilize both wild-type p53 and mutant p53, this deubiquitinase can potentially act as a tumor suppressor or an oncoprotein depending on p53 status, which warrants future investigation. It should be noted that somatic mutations in only three genes (TP53, PIK3CA, and GATA3) occurred at >10% incidence across all breast cancers [102], and that USP10 is overexpressed in human breast tumors compared with adjacent normal tissues [44].
4 Development of DUB inhibitors for cancer therapy
FDA approval of the proteasome inhibitor bortezomib for treating multiple myeloma validated the concept of targeting the proteasome for cancer treatment [103]. However, extended treatment with bortezomib is associated with drug resistance and toxicity [104]. Therapeutic strategies targeting specific DUBs, instead of the entire ubiquitin-proteasome system, might be better tolerated [105]. The rationale for targeting DUBs is that therapeutic inhibition of certain DUBs will inactivate some key oncoproteins or pro-metastatic proteins (including non-druggable ones), either by destabilizing these proteins or by changing their subcellular localization or activity; this will provide therapeutic benefits to patients whose cancers are driven by these tumor-promoting proteins. Although no DUB inhibitors have yet entered clinical trials, progress is being made in developing them as therapeutic strategies.
Similar to kinase inhibitors, DUB inhibitors range from broad pan-DUB inhibitors to specific inhibitors of single DUBs. Cyclopentenone prostaglandins, the first DUB active-site inhibitors, induced accumulation of polyubiquitinated proteins and caused apoptosis in colon cancer cells [106,107]. WP1130, a partially selective DUB inhibitor that inhibits the DUB activity of USP9X, USP5, USP14, and UCH37, triggered rapid accumulation of polyubiquitinated proteins in aggresomes and induced tumor cell apoptosis [108]. P5091, an inhibitor of USP7, induced apoptosis in multiple myeloma cells [109], while inhibitors of the USP1-UAF1 deubiquitinase complex, pimozide and GW7647, reversed the resistance of non-small cell lung cancer cells to cisplatin treatment [110]. Moreover, concurrent inhibition of DUBs (via b-AP15, an inhibitor of USP14 and UCH37) and autophagy (via vorinostat or chloroquine) led to synergistic killing of triple-negative breast cancer cells, providing a rationale for combination treatment of DUB-targeting agents with other drugs [111].
Several strategies have been developed to enable high-throughput screening (HTS) of DUB inhibitors. Ub-AMC (ubiquitin-7-amino-4-methylcoumarin), a fluorogenic substrate for a wide range of DUBs, is commonly used to measure DUB activity. Once the AMC fluorophore is cleaved from Ub-AMC by a DUB, fluorescence can be detected. Using HTS with Ub-AMC as the substrate, multiple compounds including HBX 41,108 were identified from a chemical library to inhibit the DUB activity of USP7 [112]. Similarly, 215 out of 63,052 compounds were found to inhibit USP14, and three of them showed selectivity for USP14. Further tests demonstrated that at least one of those three, IU1, inhibited USP14’s function and accelerated the degradation of several substrates that have been implicated in neurodegenerative disease [113]. However, screening for DUB inhibitors using Ub-AMC has two disadvantages. First, false positives are common, since AMC has an excitation wavelength in the UV range. Second, Ub-AMC is not hydrolyzed efficiently by the UBP/USP class of DUBs, weakening its application to some extent [114,115]. The FRET (fluorescence resonance energy transfer) assay, which selectively incorporates the FRET fluorescence donor (terbium) or acceptor (fluorochrome) onto ubiquitin, has been used to identify novel small-molecule inhibitors of AMSH [116]. Finally, the Ub-PLA2 assay developed by Progenra consists of ubiquitin fused to the N-terminus of the reporter enzyme phospholipase A2 (PLA2). The basis of this assay is that PLA2 requires a free N-terminus for its catalytic activity to cleave phospholipids into lysophospholipids and non-esterified fatty acids [117]. Therefore, when PLA2 is fused to ubiquitin, it is catalytically inactive. When a DUB is present, it cleaves ubiquitin from PLA2, freeing PLA2 to act on its substrate. There are multiple commercial PLA2 substrates, including the fluorescent phospholipid NBD C6-HPC [115,118]. The Ub-PLA2 assay has been used to identify P5091 and P22077 as USP7 inhibitors [109,119]. Compared with other strategies, the Ub-PLA2 fusion proteins represent more physiologically relevant substrates while not as sensitive for detecting the activity of the UCH family [115].
In summary, while development of DUB inhibitors are still in the early phase, recent studies have provided the proof of principle that selective small-molecule inhibitors of cancer-promoting DUBs (e.g., USP7) can induce cancer cell death. DUB inhibitors with improved efficacy, specificity, and safety may emerge as new agents for the treatment of cancer.
Acknowledgments
We thank Richard R. Behringer, Rachel K. Miller, Eric C. Swindell, and Sarah Lawhon for critical reading of the manuscript. The authors’ research is supported by US National Institutes of Health grants R01CA166051 and R01CA181029, a Cancer Prevention and Research Institute of Texas grant RP150319, a Stand Up To Cancer Innovative Research Grant, and an R. Lee Clark Fellows Award.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258(13):8206–8214. [PubMed] [Google Scholar]
- 2.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. doi:70/1/503 [pii] 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96(5):635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 4.Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci U S A. 2009;106(38):16275–16280. doi: 10.1073/pnas.0904305106. doi:10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. The Breast Cancer Linkage Consortium Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62(3):676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci U S A. 2001;98(9):5134–5139. doi: 10.1073/pnas.081068398. doi:10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1(4):193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 8.Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, et al. Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci U S A. 2001;98(9):5043–5048. doi: 10.1073/pnas.081474898. doi:10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, et al. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Invest. 2002;110(5):633–641. doi: 10.1172/JCI15795. doi:10.1172/JCI15795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. [Research Support, U.S. Gov’t, P.H.S. Review] FASEB J. 1997;11(14):1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 11.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31(19):2373–2388. doi: 10.1038/onc.2011.443. doi:10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 12.Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111(7):1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 13.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18(14):3877–3887. doi: 10.1093/emboj/18.14.3877. doi:10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280(2):1512–1520. doi: 10.1074/jbc.M410770200. doi:10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 15.Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25(2):50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- 16.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4(5):517–522. doi: 10.1038/sj.embor.embor824. doi:10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhlbrodt K, Janiesch PC, Kevei E, Segref A, Barikbin R, Hoppe T. The Machado-Joseph disease deubiquitylase ATX-3 couples longevity and proteostasis. Nat Cell Biol. 2011;13(3):273–281. doi: 10.1038/ncb2200. doi:10.1038/ncb2200. [DOI] [PubMed] [Google Scholar]
- 18.Nicastro G, Menon RP, Masino L, Knowles PP, McDonald NQ, Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci U S A. 2005;102(30):10493–10498. doi: 10.1073/pnas.0501732102. doi:10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. doi:10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. doi:10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 21.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455(7211):358–362. doi: 10.1038/nature07254. doi:10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 22.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med. 2010;207(13):2959–2973. doi: 10.1084/jem.20092641. doi:10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286(12):10457–10465. doi: 10.1074/jbc.M110.149633. doi:10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweitzer K, Naumann M. CSN-associated USP48 confers stability to nuclear NF-kappaB/RelA by trimming K48-linked Ub-chains. Biochim Biophys Acta. 2015;1853(2):453–469. doi: 10.1016/j.bbamcr.2014.11.028. doi:10.1016/j.bbamcr.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463(7277):103–107. doi: 10.1038/nature08646. doi:10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 26.Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17(9):1169–1181. doi: 10.1038/ncb3218. doi:10.1038/ncb3218. [DOI] [PubMed] [Google Scholar]
- 27.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125(4):665–677. doi: 10.1016/j.cell.2006.03.041. doi:10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Zanata SM, Kim J, Peterson MA, Di Vizio D, Chirieac LR, et al. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene. 2013;32(13):1660–1669. doi: 10.1038/onc.2012.188. doi:10.1038/onc.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14(2):121–141. [PubMed] [Google Scholar]
- 30.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296(5573):1642–1644. doi: 10.1126/science.1071884. doi:10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 31.Stanisic V, Malovannaya A, Qin J, Lonard DM, O’Malley BW. OTU Domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) alpha and affects ERalpha transcriptional activity. J Biol Chem. 2009;284(24):16135–16145. doi: 10.1074/jbc.M109.007484. doi:10.1074/jbc.M109.007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herhaus L, Al-Salihi M, Macartney T, Weidlich S, Sapkota GP. OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun. 2013;4:2519. doi: 10.1038/ncomms3519. doi:10.1038/ncomms3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466(7309):941–946. doi: 10.1038/nature09297. doi:10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 34.Karunarathna U, Kongsema M, Zona S, Gong C, Cabrera E, Gomes AR, et al. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene. 2015 doi: 10.1038/onc.2015.208. doi:10.1038/onc.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shostak K, Chariot A. EGFR and NF-kappaB: partners in cancer. Trends Mol Med. 2015;21(6):385–393. doi: 10.1016/j.molmed.2015.04.001. doi:10.1016/j.molmed.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Qu Q, Mao Y, Xiao G, Fei X, Wang J, Zhang Y, et al. USP2 promotes cell migration and invasion in triple negative breast cancer cell lines. Tumour Biol. 2015;36(7):5415–5423. doi: 10.1007/s13277-015-3207-7. doi:10.1007/s13277-015-3207-7. [DOI] [PubMed] [Google Scholar]
- 37.Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5(3):253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26(4):976–986. doi: 10.1038/sj.emboj.7601567. doi:10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahul-Mellier AL, Datler C, Pazarentzos E, Lin B, Chaisaklert W, Abuali G, et al. De-ubiquitinating proteases USP2a and USP2c cause apoptosis by stabilising RIP1. Biochim Biophys Acta. 2012;1823(8):1353–1365. doi: 10.1016/j.bbamcr.2012.05.022. doi:10.1016/j.bbamcr.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Mahul-Mellier AL, Pazarentzos E, Datler C, Iwasawa R, AbuAli G, Lin B, et al. De-ubiquitinating protease USP2a targets RIP1 and TRAF2 to mediate cell death by TNF. Cell Death Differ. 2012;19(5):891–899. doi: 10.1038/cdd.2011.185. doi:10.1038/cdd.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duex JE, Sorkin A. RNA interference screen identifies Usp18 as a regulator of epidermal growth factor receptor synthesis. Mol Biol Cell. 2009;20(6):1833–1844. doi: 10.1091/mbc.E08-08-0880. doi:10.1091/mbc.E08-08-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duex JE, Comeau L, Sorkin A, Purow B, Kefas B. Usp18 regulates epidermal growth factor (EGF) receptor expression and cancer cell survival via microRNA-7. J Biol Chem. 2011;286(28):25377–25386. doi: 10.1074/jbc.M111.222760. doi:10.1074/jbc.M111.222760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 2010;70(2):655–665. doi: 10.1158/0008-5472.CAN-09-1942. doi:10.1158/0008-5472.CAN-09-1942. [DOI] [PubMed] [Google Scholar]
- 44.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, et al. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104(1):21–30. doi: 10.1007/s10549-006-9393-7. doi:10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- 45.Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999;4(12):757–767. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- 46.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123–135. doi: 10.1016/j.cell.2008.10.051. doi:10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 47.Xie Y, Avello M, Schirle M, McWhinnie E, Feng Y, Bric-Furlong E, et al. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 2013;288(5):2976–2985. doi: 10.1074/jbc.M112.430066. doi:10.1074/jbc.M112.430066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121(3):851–865. doi: 10.1172/JCI44111. doi:10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee M-H, Zhao R, Phan L, Yeung S-CJ. Roles of COP9 signalosome in cancer. Cell cycle. 2011;10(18):3057–3066. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol. 2014;15(6):562–570. doi: 10.1038/ni.2885. doi:10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med. 2012;18(3):429–435. doi: 10.1038/nm.2619. doi:10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 52.Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13(11):1368–1375. doi: 10.1038/ncb2346. doi:10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 53.Sacco JJ, Coulson JM, Clague MJ, Urbe S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62(2):140–157. doi: 10.1002/iub.300. doi:10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 55.Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95(6):2104–2110. [PubMed] [Google Scholar]
- 56.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20(7):2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, et al. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia. 2006;20(9):1572–1581. doi: 10.1038/sj.leu.2404317. doi:10.1038/sj.leu.2404317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9(7):765–774. doi: 10.1038/ncb1601. doi:10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 59.Flugel D, Gorlach A, Kietzmann T. GSK-3beta regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1alpha. Blood. 2012;119(5):1292–1301. doi: 10.1182/blood-2011-08-375014. doi:10.1182/blood-2011-08-375014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, et al. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013;5(1):224–236. doi: 10.1016/j.celrep.2013.08.030. doi:10.1016/j.celrep.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ge F, Chen W, Qin J, Zhou Z, Liu R, Liu L, et al. Ataxin-3 like (ATXN3L), a member of the Josephin family of deubiquitinating enzymes, promotes breast cancer proliferation by deubiquitinating Kruppel-like factor 5 (KLF5) Oncotarget. 2015;6(25):21369–21378. doi: 10.18632/oncotarget.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. doi: 10.1038/ncomms9471. doi:10.1038/ncomms9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valero R, Marfany G, Gonzalez-Angulo O, Gonzalez-Gonzalez G, Puelles L, Gonzalez-Duarte R. USP25, a novel gene encoding a deubiquitinating enzyme, is located in the gene-poor region 21q11.2. Genomics. 1999;62(3):395–405. doi: 10.1006/geno.1999.6025. doi:10.1006/geno.1999.6025. [DOI] [PubMed] [Google Scholar]
- 64.Denuc A, Bosch-Comas A, Gonzalez-Duarte R, Marfany G. The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS One. 2009;4(5):e5571. doi: 10.1371/journal.pone.0005571. doi:10.1371/journal.pone.0005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohideen F, Lima CD. SUMO takes control of a ubiquitin-specific protease. Mol Cell. 2008;30(5):539–540. doi: 10.1016/j.molcel.2008.05.010. doi:10.1016/j.molcel.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong B, Liu X, Wang X, Chang SH, Liu X, Wang A, et al. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat Immunol. 2012;13(11):1110–1117. doi: 10.1038/ni.2427. doi:10.1038/ni.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.King MC, Marks JH, Mandell JB, New York Breast Cancer Study, G. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. doi:10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 68.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12(5):1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 69.Chen X, Arciero CA, Wang C, Broccoli D, Godwin AK. BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res. 2006;66(10):5039–5046. doi: 10.1158/0008-5472.CAN-05-4194. doi:10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- 70.Al-Hakim A, Escribano-Diaz C, Landry MC, O’Donnell L, Panier S, Szilard RK, et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 2010;9(12):1229–1240. doi: 10.1016/j.dnarep.2010.09.011. doi:10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, et al. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24(54):8080–8084. doi: 10.1038/sj.onc.1208944. doi:10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- 72.Tian Z, D’Arcy P, Wang X, Ray A, Tai YT, Hu Y, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123(5):706–716. doi: 10.1182/blood-2013-05-500033. doi:10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pereg Y, Liu BY, O’Rourke KM, Sagolla M, Dey A, Komuves L, et al. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell Biol. 2010;12(4):400–406. doi: 10.1038/ncb2041. doi:10.1038/ncb2041. [DOI] [PubMed] [Google Scholar]
- 74.Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci U S A. 2015;112(12):3734–3739. doi: 10.1073/pnas.1411713112. doi:10.1073/pnas.1411713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akhavantabasi S, Akman HB, Sapmaz A, Keller J, Petty EM, Erson AE. USP32 is an active, membrane-bound ubiquitin protease overexpressed in breast cancers. Mamm Genome. 2010;21(7-8):388–397. doi: 10.1007/s00335-010-9268-4. doi:10.1007/s00335-010-9268-4. [DOI] [PubMed] [Google Scholar]
- 76.Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25(2):160–165. doi: 10.1038/76006. doi:10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 77.Hayashi M, Jono H, Shinriki S, Nakamura T, Guo J, Sueta A, et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res Treat. 2014;143(3):447–457. doi: 10.1007/s10549-013-2824-3. doi:10.1007/s10549-013-2824-3. [DOI] [PubMed] [Google Scholar]
- 78.Karin M, Yamamoto Y, Wang QM. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3(1):17–26. doi: 10.1038/nrd1279. doi:10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 79.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424(6950):797–801. doi: 10.1038/nature01811. doi:10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 80.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424(6950):801–805. doi: 10.1038/nature01802. doi:10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 81.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–796. doi: 10.1038/nature01803. doi:10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 82.Massoumi R. CYLD: a deubiquitination enzyme with multiple roles in cancer. Future Oncol. 2011;7(2):285–297. doi: 10.2217/fon.10.187. doi:10.2217/fon.10.187. [DOI] [PubMed] [Google Scholar]
- 83.Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol Cell. 2009;34(4):461–472. doi: 10.1016/j.molcel.2009.04.031. doi:10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–1079. doi: 10.1016/j.cell.2007.03.052. doi:10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 85.Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JM, et al. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Invest. 2006;116(11):3042–3049. doi: 10.1172/JCI28746. doi:10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tauriello DV, Haegebarth A, Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, et al. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37(5):607–619. doi: 10.1016/j.molcel.2010.01.035. doi:10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 87.Rajan N, Elliott RJ, Smith A, Sinclair N, Swift S, Lord CJ, et al. The cylindromatosis gene product, CYLD, interacts with MIB2 to regulate notch signalling. Oncotarget. 2014;5(23):12126–12140. doi: 10.18632/oncotarget.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13(5):283–296. doi: 10.1038/nrm3330. doi:10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 89.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11(4):289–301. doi: 10.1038/nrc3037. doi:10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjold B, Rutqvist LE, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13(12):3577–3584. doi: 10.1158/1078-0432.CCR-06-1609. doi:10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 91.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] Nature. 2008;455(7214):813–817. doi: 10.1038/nature07290. doi:10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15(12):1486–1494. doi: 10.1038/ncb2874. doi:10.1038/ncb2874 ncb2874 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiang S, Fang J, Wang S, Deng B, Zhu L. MicroRNA135b regulates the stability of PTEN and promotes glycolysis by targeting USP13 in human colorectal cancers. Oncol Rep. 2015;33(3):1342–1348. doi: 10.3892/or.2014.3694. doi:10.3892/or.2014.3694. [DOI] [PubMed] [Google Scholar]
- 94.Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12(19):2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 95.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416(6881):648–653. doi: 10.1038/nature737. doi:10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 96.Epping MT, Meijer LA, Krijgsman O, Bos JL, Pandolfi PP, Bernards R. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat Cell Biol. 2011;13(1):102–108. doi: 10.1038/ncb2142. doi:10.1038/ncb2142. [DOI] [PubMed] [Google Scholar]
- 97.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428(6982) doi: 10.1038/nature02501. 1 p following 486, doi:10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 98.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13(6):879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 99.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18(5):565–576. doi: 10.1016/j.molcel.2005.04.024. doi:10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 100.Brooks CL, Li M, Hu M, Shi Y, Gu W. The p53--Mdm2--HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene. 2007;26(51):7262–7266. doi: 10.1038/sj.onc.1210531. doi:10.1038/sj.onc.1210531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140(3):384–396. doi: 10.1016/j.cell.2009.12.032. doi:10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. doi:10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kane RC, Farrell AT, Sridhara R, Pazdur R. United States Food and Drug Administration approval summary: bortezomib for the treatment of progressive multiple myeloma after one prior therapy. Clin Cancer Res. 2006;12(10):2955–2960. doi: 10.1158/1078-0432.CCR-06-0170. doi:12/10/2955 [pii] 10.1158/1078-0432.CCR-06-0170. [DOI] [PubMed] [Google Scholar]
- 104.Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;8(5):407–419. doi: 10.1016/j.ccr.2005.10.013. doi:S1535-6108(05)00334-X [pii] 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 105.Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3(2):191–199. doi: 10.2217/14796694.3.2.191. doi:10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mullally JE, Fitzpatrick FA. Pharmacophore model for novel inhibitors of ubiquitin isopeptidases that induce p53-independent cell death. Mol Pharmacol. 2002;62(2):351–358. doi: 10.1124/mol.62.2.351. [DOI] [PubMed] [Google Scholar]
- 107.Mullally JE, Moos PJ, Edes K, Fitzpatrick FA. Cyclopentenone prostaglandins of the J series inhibit the ubiquitin isopeptidase activity of the proteasome pathway. J Biol Chem. 2001;276(32):30366–30373. doi: 10.1074/jbc.M102198200. doi:10.1074/jbc.M102198200. [DOI] [PubMed] [Google Scholar]
- 108.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70(22):9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. doi:10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 109.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. doi:10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, et al. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol. 2011;18(11):1390–1400. doi: 10.1016/j.chembiol.2011.08.014. doi:10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vogel RI, Coughlin K, Scotti A, Iizuka Y, Anchoori R, Roden RB, et al. Simultaneous inhibition of deubiquitinating enzymes (DUBs) and autophagy synergistically kills breast cancer cells. Oncotarget. 2015;6(6):4159–4170. doi: 10.18632/oncotarget.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8(8):2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. doi:10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 113.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–184. doi: 10.1038/nature09299. doi:10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tirat A, Schilb A, Riou V, Leder L, Gerhartz B, Zimmermann J, et al. Synthesis and characterization of fluorescent ubiquitin derivatives as highly sensitive substrates for the deubiquitinating enzymes UCH-L3 and USP-2. Anal Biochem. 2005;343(2):244–255. doi: 10.1016/j.ab.2005.04.023. doi:10.1016/j.ab.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 115.Goldenberg SJ, McDermott JL, Butt TR, Mattern MR, Nicholson B. Strategies for the identification of novel inhibitors of deubiquitinating enzymes. Biochem Soc Trans. 2008;36(Pt 5):828–832. doi: 10.1042/BST0360828. doi:10.1042/BST0360828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Arnst JL, Davies CW, Raja SM, Das C, Natarajan A. High-throughput compatible fluorescence resonance energy transfer-based assay to identify small molecule inhibitors of AMSH deubiquitinase activity. Anal Biochem. 2013;440(1):71–77. doi: 10.1016/j.ab.2013.05.017. doi:10.1016/j.ab.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dijkstra BW, Drenth J, Kalk KH. Active site and catalytic mechanism of phospholipase A2. Nature. 1981;289(5798):604–606. doi: 10.1038/289604a0. [DOI] [PubMed] [Google Scholar]
- 118.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, et al. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17(6):1035–1043. doi: 10.1110/ps.083450408. doi:10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18(11):1401–1412. doi: 10.1016/j.chembiol.2011.08.018. doi:10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]