Abstract
Expansions of (CAG)n•(CTG)n trinucleotide repeats are responsible for over a dozen neuromuscular and neurodegenerative disorders. Large-scale expansions are typical for human pedigrees and may be explained by iterative small-scale events such as strand slippage during replication or repair DNA synthesis. Alternatively, a distinct mechanism could lead to a large-scale repeat expansion at a step. To distinguish between these possibilities, we developed a novel experimental system specifically tuned to analyze large-scale expansions of (CAG)n•(CTG)n repeats in Saccharomyces cerevisiae. The median size of repeat expansions was ~60 triplets, though additions in excess of 150 triplets were also observed. Genetic analysis revealed that Rad51, Rad52, Mre11, Pol32, Pif1, and Mus81 and/or Yen1 proteins are required for large-scale expansions, whereas proteins previously implicated in small-scale expansions are not involved. Based on these results, we propose a new model for large-scale expansions based on recovery of replication forks broken at (CAG)n•(CTG)n repeats via break-induced replication.
Introduction
Expansions of (CAG)n•(CTG)n repeats are responsible for over a dozen neuromuscular and neurodegenerative disorders in humans, including Huntington’s disease (HD), myotonic dystrophy (DM1), and numerous forms of spinocerebellar ataxia (SCA)1,2. Individuals with adult-onset HD typically have 40–80 (CAG)n repeats in the coding region of the HTT gene. Longer CAG tracts do occur but are rare and associated with juvenile onset3. In contrast, individuals with DM1 commonly have hundreds of (CTG)n repeats in the 3’UTR of the DMPK gene, reaching up to 4000 copies in severe cases4. The molecular mechanisms of (CAG)n•(CTG)n (hereafter abbreviated CAG) repeat expansions have been intensively studied in model organisms and human cells, recapitulating many properties observed in human patients and pedigrees such as length-dependent increase in repeat instability2,5,6. CAG sequences were shown to form stable hairpins and slipped-strand DNA structures both in vitro and in vivo, which stall replication forks, promote replication fork reversal, and cause chromosomal breakage in a length-dependent manner7–9.
All models of CAG repeat expansions implicate the deleterious impact of their secondary structures on DNA replication, transcription, and repair processes10,11. DNA polymerase slippage followed by hairpin formation on the nascent DNA strand can lead to small-scale expansions if the hairpin persists to the next round of replication6,11. Strand slippage and hairpin formation can also occur during repair DNA synthesis in the course of base excision repair (BER)12,13, nucleotide excision repair (NER)14, and transcription-coupled repair (TCR)15. In all the above scenarios, expansion size is limited by slippage events that are normally small-scale. Thus, these models can explain large-scale expansions by the iterative succession of independent small-scale events. For example, oxidized DNA bases can lead to subsequent BER where strand displacement creates a DNA hairpin that is refractory to cleavage by flap endonuclease. This hairpin would then result in a single expansion, and many rounds of oxidation, repair, and expansions would create a “toxic oxidation cycle” to generate large-scale expansions13.
Most experimental systems to study CAG repeat expansions deal with relatively small-scale events16–20, which we define as an increase up to 20 repeats. The first selectable system in budding yeast deliberately looked at the instability of a short (i.e. (CAG)25) starting tract to simulate the change from normal to pre-mutation length alleles, as in HD16; ~10 repeats were added at a rate of ~105. Yeast studies for longer CAG repeats (45-to-155 units) consistently detected small-scale expansions that occurred at a percentile level (~1%)17. Altogether, these yeast systems enabled powerful genetics analysis of small-scale repeat expansion, establishing the importance of replication fork integrity, chromatin remodeling, specialized helicases, and nuclear localization of the repeat8,21–25.
In a Drosophila experimental system, the scale of repeat expansions was even smaller: the majority of events were additions of just one or two repeats to a long (CAG)270 tract18. In mice, much longer CAG repeats were required to show disease phenotypes than in humans. Similarly to yeast, mice predominantly displayed small-scale expansions during both intergenerational transmissions and in somatic tissues13,26. An exception is the curiously small-sized humanized DM1 mice carrying 430 to >1000 CAG repeats, which exhibit jumps in excess of hundreds of repeats during intergenerational transmission27. Hairpin formation and the role of replication on CAG repeat instability has been confirmed in human cells28,29. Additionally, large-scale expansions were recovered from a very long starting tract of 800 repeats30. Yet in such experimental systems, extensive genetic analyses remain challenging.
Given the lack of experimental systems to detect large-scale CAG expansions, it is impossible to ascertain whether they occur via a distinct mechanism, or result from the sequential accumulation of small-scale expansions. Previous studies of large-scale expansions of (GAA)n•(TTC)n and (ATTCT)n•(AGAAT)n repeats in a yeast system led us to propose a template-switching model for large-scale expansion during DNA replication31,32. Genetic analysis of these large-scale events revealed dramatic differences with small-scale CAG repeat expansions studied by others. It remained unclear whether differences in scale of expansions, repeat sequences, or experimental model system accounted for these differences.
To address this problem, we established a new system to detect and analyze large-scale CAG expansions in S. cerevisiae. The median size of repeat expansions in this system was ~60 triplets, while additions in excess of 150 triplets were also observed. Our genetic analysis revealed that Rad51, Rad52, Mre11, Pol32, Pif1 and Mus81 or Yen1 proteins are required for large-scale expansions, whereas proteins previously implicated in small-scale expansions are not involved. These results point to a mechanism that is distinct from small-scale CAG expansions, which is based on the recovery of replication forks broken at (CAG)n•(CTG)n repeats by the interplay between break-induced replication and broken fork repair. Thus, large-scale CAG expansions could a represent a striking example of genome instability arising from break-induced replication machinery.
Results
Large-scale CAG repeat expansions can be recovered in budding yeast
To investigate large-scale CAG expansions, we capitalized on a system previously developed in the lab to study GAA repeats33. This system relies on the well-characterized GAL1 promoter, where the distance (i.e. spacer) between the upstream activating sequence (UASGAL) and TATA box (PGAL) is constrained such that transcriptional activation no longer occurs when the spacer is too long34. We cloned CAG repeats into this spacer, upstream of the forward-selection marker CAN1. These constructs were then integrated into chromosome III, ~1 kb away from the replication origin ARS306 (Fig. 1A, S1, S2).
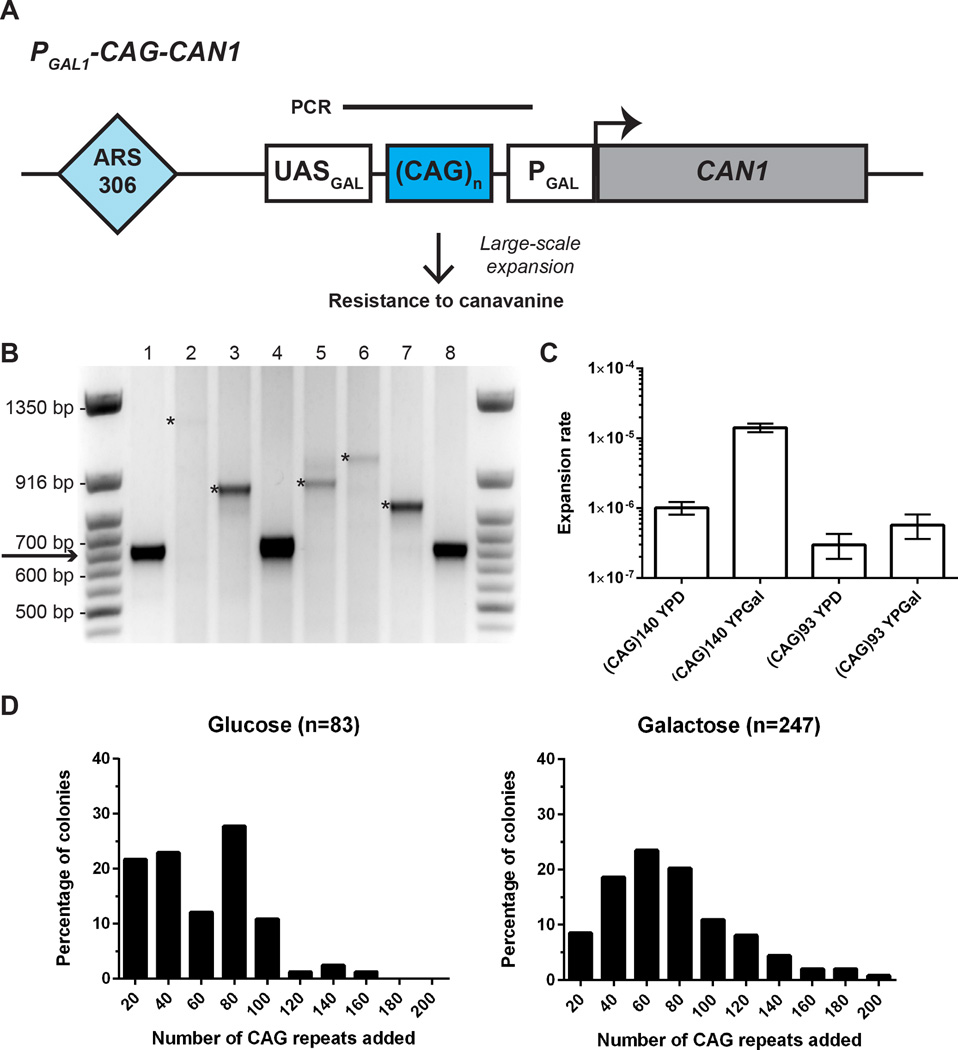
Figure 1. Large-scale CAG repeat expansions can be recovered and analyzed in budding yeast.
(A) Experimental system to study large-scale CAG repeat expansions. In the starting strain, the distance between the upstream activating sequence (UASGAL) and TATA box promoter (PGAL) allows transcription of the forward selection marker CAN1, which encodes an arginine permease. Large-scale expansions will result in CanR clones. Bar above the CAG repeats indicate product of single colony PCR used for determination of repeat length. (B) Agarose gel showing PCR amplification of CAG repeat length for CanR clones. Arrow points to the initial length of (CAG)140. Only expanded clones are used to calculate expansion rates. Asterisks in lanes 2, 3, 5, 6, and 7 indicate expanded clones. (C) Rate of expansion (per cell per division) for (CAG)140 and (CAG)93, for non-selective growth on glucose and galactose. Rates and 95% confidence intervals (error bars) were calculated based on distribution of expanded clones in at least 10 independent cultures using the Ma-Sandri-Sarkar maximum likelihood estimator with a correction for sampling and plating efficiency. (D) Distribution of repeats added to (CAG)140 for non-selective growth on glucose and galactose. The median number of repeats added is 61.7 repeats (interquartile range 30.7–85.3) for glucose and 68.3 repeats (interquartile range 48.0–95.7) for galactose.
Our starting strain contained a selectable cassette with 140 (CAG)n repeats, corresponding to a mild disease-size repeat in myotonic dystrophy (DM1), which undergoes large-scale expansions during intergenerational transmission. The CAG sequence was positioned on the lagging strand template for DNA replication because the CTG orientation is known to be deletion-prone35,36. We reasoned that if a large-scale expansion (which we designate as >20 repeats) occurred during non-selective growth, the increased spacer distance would preclude expression of CAN1. This would permit colony formation on plates containing canavanine, a toxic analog of arginine, in the presence of galactose (Figure 1A).
The rate of large-scale CAG expansions was determined in fluctuation test experiments, with non-selective growth occurring on either glucose or galactose. The length of CAG repeats in individual CanR clones was then determined by single colony PCR (Fig. 1B, S3). The rate of large-scale (CAG)140 expansion corresponded to 1.4 × 10−5 per replication when cells were pre-grown on galactose and 10-fold lower (1.0 × 10−6) when pre-grown on glucose (Fig. 1C).
The number of added (CAG)n tracts was determined and plotted for each CanR clone (Fig. 1D). For pre-growth on both galactose and glucose, the median size of large-scale expansions corresponded to ~60 repeats. Remarkably, for both growth conditions, we observed multiple clones that had added in excess of 150 repeats, which more than doubles the starting length of CAG repeats. We sequenced the expanded repeats of 21 CanR clones and found that the CAG tracts were pure and did not contain any large insertions that could account for the increased PCR product size (Table S1). Additionally, 18 out of 19 sequenced CanR clones with (CAG)n expansions did not contain any point mutations in CAN1, while the remaining clone had a silent mutation in the CAN1 ORF. We also constructed a strain carrying a cassette with a shorter (CAG)93 repeat tract. The expansion rate for the (CAG)93 repeat was an order of magnitude lower (3.0 × 10−7 on glucose and 5.7 × 10−7 on galactose) than for (CAG)140 (Fig. 1C), highlighting that the likelihood of large-scale expansions in yeast increases with starting tract length and mimicking the genetic anticipation phenomenon observed in human pedigrees.
Notably, the rate for large-scale expansions of the (CAG)140 run was considerably lower than the rate of small-scale expansions previously determined for similar size CAG tracts17. To determine how frequently small-scale expansions occurred in our experimental system, we grew our strain with (CAG)140 repeats under non-selective conditions followed by analyzing the repeat’s length by single colony PCR. Similar to the previous data, we observed relatively high frequencies of small-scale expansions in strains grown on either glucose or galactose (~1.0%), and even higher frequencies of contractions, particularly on galactose (17.5% versus 5.3% on glucose) (Table S2).
Genetic control of large-scale CAG repeat expansions is distinct from small-scale expansion
Using our selectable system, we took a candidate gene approach to identify genes involved in large-scale CAG expansions. Srs2 is a DNA helicase that has been shown to unwind CAG repeats in vitro37. Also, eliminating Srs2 function resulted in an increased rate of small-scale expansions8,21. We reasoned that if large-scale expansions observed by us result from multiple small-scale events, Srs2 deletion would similarly show an increased expansion rate in our experimental system. However, we saw no effect of srs2Δ on large-scale CAG expansions (Fig. 2A).
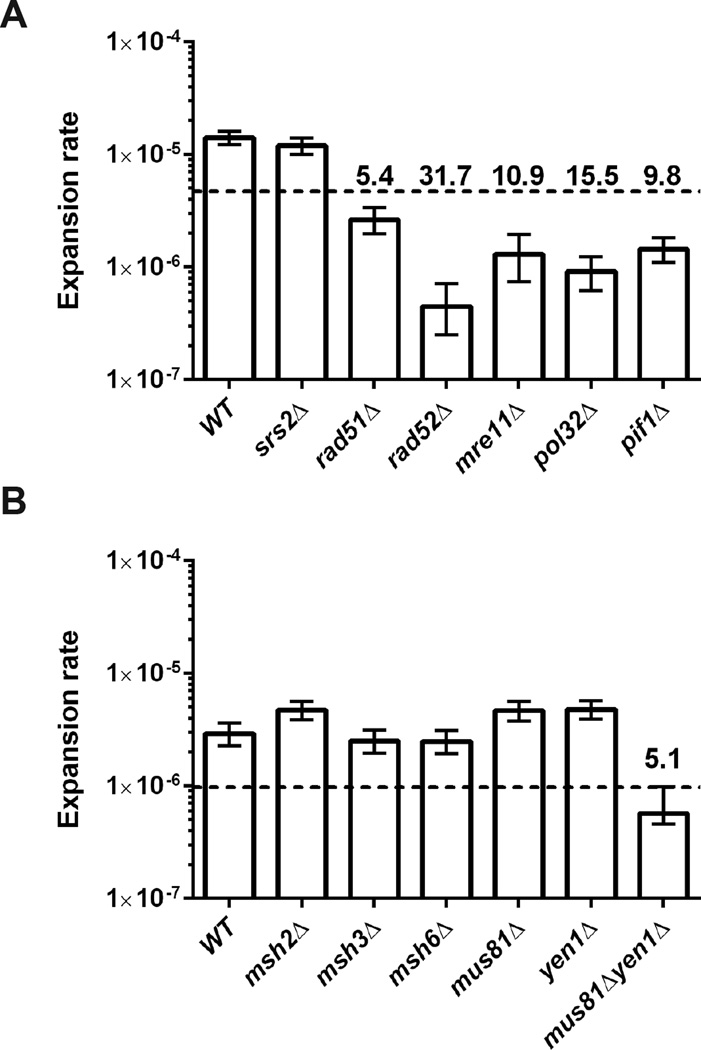
Figure 2. Genetic control of large-scale CAG repeat expansions.
Effect of different gene knockouts on the large-scale expansion rate of (CAG)140 pre-grown on galactose. (A) 60 µg/mL canavanine concentration and (B) 200 µg/mL canavanine concentration. Rates of expansion and 95% confidence intervals (error bars) were calculated based on distribution of expanded clones in at least 9 independent cultures using the Ma-Sandri-Sarkar maximum likelihood estimator with a correction for sampling and plating efficiency. Dashed line designates 3-fold decrease from wild-type. Numbers above the dashed line show fold decrease compared to wild-type.
Mismatch repair (MMR) proteins have been shown to affect CAG expansions in several experimental systems38. Msh2–Msh3 (MutSβ) typically binds to long insertion or deletion loops to promote repair, but in the context of CAG repeats, Msh2–Msh3 promotes expansions. Analysis of MMR proteins using a yeast system for detecting small-scale expansions found that msh3Δ reduced, whereas msh6Δ increased, CAG expansion rate39. In contrast, we observed no difference in the rates of large-scale expansions between various MMR deficient strains as compared to wild-type (Fig. 2B).
Genetic control of large-scale CAG repeat expansions implicates genes required for homologous recombination and specifically the break-induced replication pathway
The role of homologous recombination (HR) in CAG repeat expansion has been studied in several yeast systems, indicating differing results that may reflect distinct aspects of CAG repeat instability. Eliminating Rad51 and Rad52 proteins had no effect on expansion rate of short tracts of (CAG)13 or (CAG)25 repeats16,21. Chromosomal fragility and expansions of (CAG)70 run were enhanced by loss of these recombination proteins40. However, in mutant backgrounds where (CAG)70 expansions were elevated, this increase was dependent on HR proteins8,40. In contrast, large-scale expansions of GAA and ATTCT repeats were not affected by loss of Rad51 or Rad52. Strikingly, we found that the rate of large-scale CAG expansion was reduced 32-fold in the rad52Δ mutant and 5-fold in rad51Δ compared to the wild type strain (Fig. 2A), implicating a role for HR in this process.
HR encompasses double-strand break repair (DSBR), synthesis-dependent strand annealing (SDSA), and break-induced replication (BIR), all of which require end-resection by the MRX (Mre11-Rad50-Xrs2) complex41. Inactivating Mre11 diminishes large-scale expansions of CAG repeats 10-fold (Fig. 2A). DSBR and BIR repair two-ended or one-ended DNA breaks, formation of which may require resolution of a double or single Holliday junction, respectively. We tested single and double knockouts of the resolvase genes MUS81 and YEN1 and found that only the double mutant showed a decrease in expansion rate (Fig. 2B), thus indicating an overlapping role for these proteins in resolving Holliday junctions associated with the formation of large-scale CAG repeat expansions.
To discriminate between the distinct pathways of HR, we looked at the role of Pol32 - a non-essential subunit of polymerase δ that is required for BIR42, and much less for other branches of HR. In BIR, DNA synthesis occurs in the context of a displacement loop (D-loop), potentially to the end of the chromosome. Remarkably, eliminating POL32 reduced the expansion rate 15-fold (Fig. 2A). Following end resection and strand invasion during BIR, Pif1 helicase stimulates DNA synthesis. We observed a strong (10-fold) reduction of expansion rate for pif1Δ and a more modest decrease for the pif1-m2 mutant43, which eliminates nuclear activity while maintaining function in the mitochondria. (Fig. 2A, S1). The smaller effect of the pif1-m2 allele may be due to residual Pif1 activity in the nucleus, which has been reported previously for other BIR assays44,45. Knock out of REV3 gene, encoding for DNA polymerase zeta, showed only a 2-fold decrease in CAG expansion rate, indicating that the role of Pol32 and Pif1 in large-scale CAG expansion is not primarily dependent on translesion synthesis (Fig. S4).
Large-scale CAG repeat expansions are a replication-dependent event associated with replication fork stalling
In eukaryotes, BIR was most extensively characterized in budding yeast in the context of an irreparable one-ended double strand break (DSB) generated by HO endonuclease41,46 or chromosome fragmentation47. Because these events were almost exclusively repaired in G2 phase, the D-loop could be involved in DNA synthesis to the end of the chromosome. Though a remarkable feat, repair of the broken chromosome comes at a cost given the high mutagenicity of BIR synthesis48. BIR was proposed to repair one-ended DSBs resulting from replication fork breakage, as well. However, recent work investigating this subject concluded that while repair of the one-ended DSB uses error-prone Pol32-dependent synthesis initially, its scope is limited owing to the arrival of a converging replication fork followed by Mus81 or Yen1-dependent D-loop cleavage, referred as Broken Fork Repair (BFR)49. Thus, the trade-off between BIR and BFR repair pathways depends on the proximity or activity of a convergent replication origin.
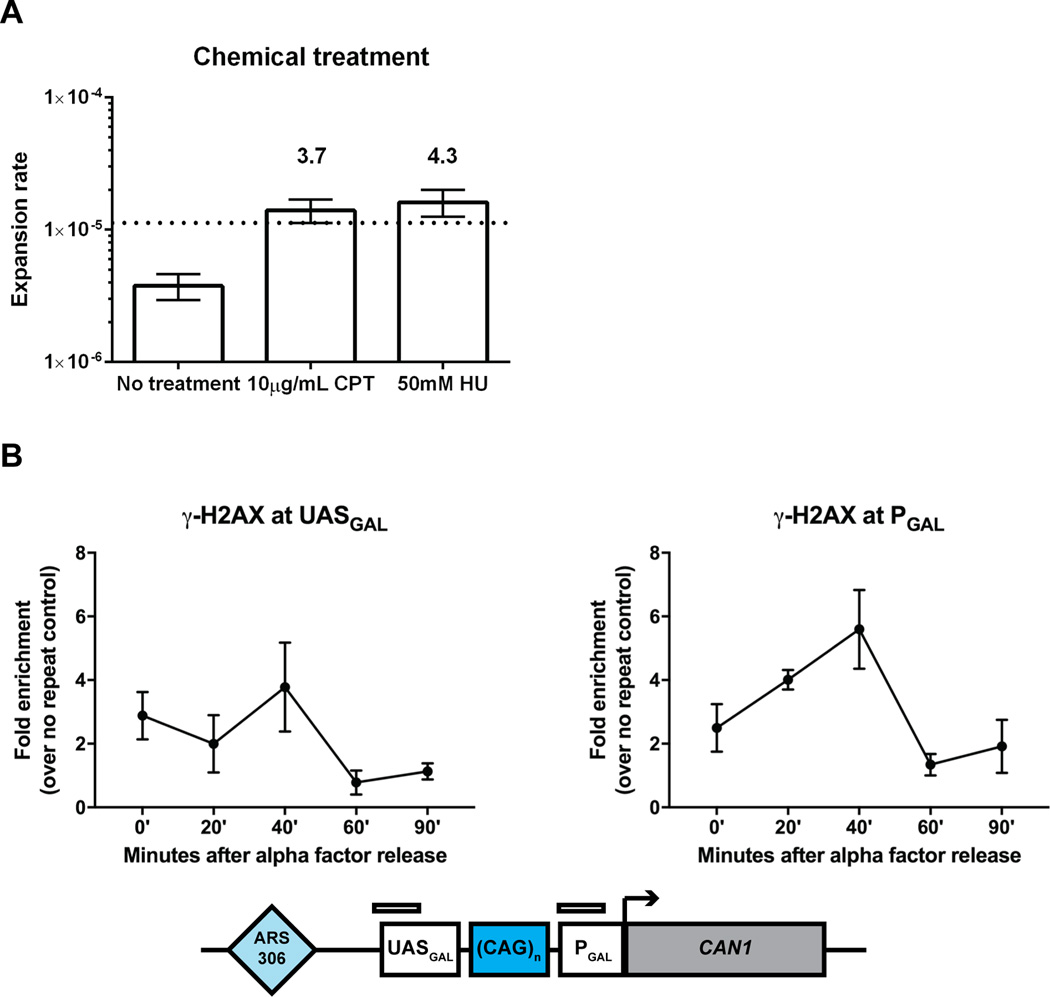
To test whether large-scale CAG repeat expansions occur during S-phase in our system, we treated cells with low doses of camptothecin, a topoisomerase I inhibitor, which triggers replication fork breakage in S-phase, as well as hydroxyurea to increase replication fork stalling and collapse. We found that both treatments increased (over 3-fold) the rate of large-scale CAG expansion (Figure 3A).
Figure 3. Large-scale CAG repeat expansions are a replication-dependent event associated with replication fork stalling and collapse.
(A) Effect of 10 µg/mL camptothecin and 50 mM hydroxyurea treatment on large-scale expansion rate of (CAG)140 pre-grown on galactose. Rates of expansion and 95% confidence intervals (error bars) were calculated based on distribution of expanded clones in at least 9 independent cultures using the Ma-Sandri-Sarkar maximum likelihood estimator with a correction for sampling and plating efficiency. Numbers above the confidence intervals reflect fold increase over the wild-type. Canavanine concentration was used at 120 µg/mL. (B) Enrichment of γ-H2AX at the (CAG)140 locus following release from alpha factor arrest by chromatin immunoprecipitation and quantitative PCR. The mean and range are plotted for each time point. P = 0.054 by one-way ANOVA comparison of all time points for γ-H2AX at PGAL.
Previous studies have convincingly demonstrated that long CAG repeats promote replication fork stalling, breakage, and the formation of joint molecules consistent with recombination intermediates8,9. To confirm whether replication fork stalling and breakage occur in our CAG system, we looked for accumulation of the γ-H2AX histone variant – a marker for both fork stalling and DSB repair50,51 - at our repetitive run during S-phase. Using ChIP analysis, we indeed saw enrichment of γ-H2AX at the (CAG)140 repeat, which peaked 40 minutes following release into S-phase from alpha factor arrest (Figure 3B).
Discussion
Our results clearly show that the genetic control of large-scale CAG expansions is different from the genetic control of small-scale CAG expansions. Most importantly, the role of principal players in break-induced replication (BIR), the Pol32 subunit of DNA polymerase delta and Pif1 helicase in large-scale CAG expansions, was never before observed for CAG or other expandable repeats.
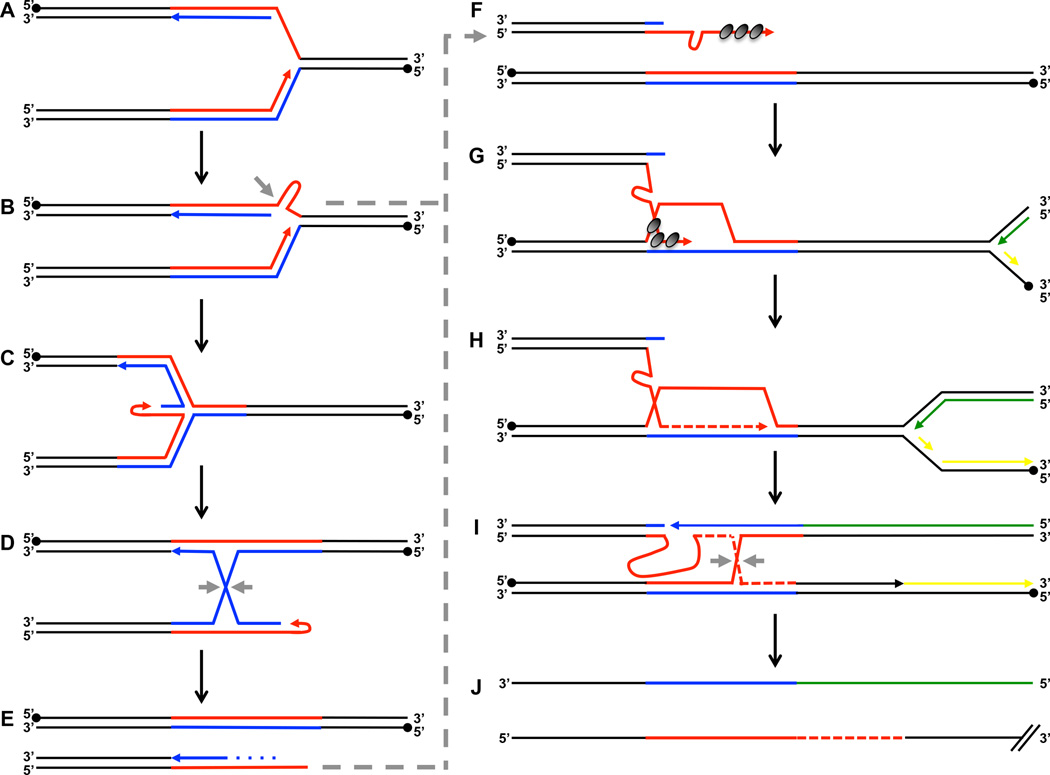
Based on these results, we propose a comprehensive model of large-scale CAG repeat expansions. We believe that large-scale expansions of long CAG tracts are rooted in their ability to form stable hairpin structures during DNA synthesis, which ultimately leads to replication fork stalling8,24 (Fig. 4A,B). Such stalling, including at various triplet repeats, was previously shown to cause fork reversal52,53 (Fig. 4C). Isomerization of the resulting four-way junction (chicken-foot structure) will lead to the formation of a Holliday junction (Fig. 4D), whose resolution by proteins such as Mus81 or Yen1 would result in a one-ended DSB (Fig. 4E). Alternatively, endonucleases could act directly on the hairpin structure at the stalled fork to create a one-ended DSB (Fig. 4B).
Figure 4. Model of large-scale CAG repeat expansions.
(A) Replication fork progression proceeds through the repetitive region. In this experimental system, CAG is on the lagging strand template (red) and CTG is on the leading strand template (blue). Closed circles denote 5’ end of template strand, and closed arrows denote 3’ end of nascent strand. (B) Stable hairpin formation on exposed single-stranded DNA. Gray arrow denotes potential site of endonuclease cleavage, which could then directly proceed to F. (C) Replication fork reversal. (D) Four-way junction from C isomerizes to form a Holliday junction, which can be cleaved by resolvase proteins (gray arrows). (E) A one-ended double strand break is subject to 5’ to 3’ resection (dotted line), resulting in single-stranded repetitive sequence. (F) The single-stranded repetitive sequence is coated by RPA, then Rad51 (dark gray circles) and forms stable hairpin structure(s). (G) Out-of-register invasion results in formation of a D-loop. A left-ward moving convergent replication fork shows the leading strand (green) and lagging strand (yellow) emanating from the adjacent replication origin. (H) Pol32-dependent DNA synthesis continues, further extending D-loop progression. (I) The D-loop converges with the left-ward moving replication fork resulting in a Holliday junction, which needs to be resolved (gray arrows). (J) The nascent DNA strand (bottom) would have accumulated extra CAG repeats equivalent to the out-of-register invasion step resulting in a large-scale expansion.
The one-ended DSB would be subject to end resection by the MRX complex, creating a 3’-single stranded DNA (ssDNA) tail stabilized both by proteins (RPA and Rad51) as well as hairpin formation of the repetitive sequence (Fig. 4F). To restart the replication fork, the one-ended DSB will invade the sister chromatid and create a D-loop (Fig 4G). Since this one-ended DSB occurs within a long repetitive run, it would tend to invade its repetitive counterpart out-of-register. Indeed, out-of-register invasion has been previously proposed as a mechanism of CAG repeat instability. However, these studies involved artificially induced two-ended DSBs54 or DSBs generated during meiotic recombination55. Notably, in our model, hairpin formation on the ssDNA portion of the repeat tract would exacerbate this out-of-register invasion potentially explaining the bias for repeat expansions observed during intergenerational transmissions in human pedigrees. The convergent replication origin (ARS307) is ~30 kb away in our experimental system. Thus, it would take just ~10 minutes for the converging fork to reach the stall site. Consequently, BIR would only progress over a relatively short distance past the break (Fig. 4H). After its collision with the converging fork, a single Holliday junction would need to be resolved to separate the newly synthesized DNA molecule, which would have accumulated extra CAG repeats in the nascent DNA strand equivalent to the out-of-register invasion step. Mus81, Yen1, or the two proteins together could act at this step in addition to its earlier role in Holliday junction resolution of the isomerized four-way junction. Thus, the genetic control of large-scale expansions of CAG repeats has characteristics of both BIR and BFR.
Intriguingly, the proposed mechanism of large-scale CAG repeat expansion is distinct from mechanisms described for small-scale expansions of short CAG tracts. In those cases, loss of RAD51 or RAD52 genes had little, if any, effect on the rate of (CAG)25 expansions16,21. A likely explanation for this difference is that long CAG tracts are more susceptible to fork stalling and DNA breakage than the shorter ones. Supporting this reasoning, it was recently found that expanded CAG repeats are more likely than unexpanded repeats to localize to the nuclear periphery during S-phase of the cell cycle22. This observation could point to the repair of one-ended DSB, as proposed by us, at a nuclear pore.
Additionally, our genetic analysis of large-scale expansions of a different trinucleotide repeat, (GAA)n, was inconsistent with HR, BIR, and BFR pathways, but implied template-switching during DNA replication as a mechanism for expansions31. While fork stalling at long GAA runs, similarly to CAG runs, results in DSB formation56, we believe that the difference in expansion pathways could be due to the triplex-forming potential of the GAA repeat, which would hide ssDNA formed upon end-resection from the HR machinery.
Note that the proposed mechanism has underlying similarities with microhomology-mediated break-induced replication (MMBIR) pathway, which was brought forth to explain copy number variation and chromosomal rearrangements in humans57 and budding yeast58. Our data add large-scale expansions of CAG repeats as another striking example of genome instability arising from BIR and BFR, highlighting the fundamental importance of these processes in DNA damage repair, albeit at the expense of repeat instability and mutagenesis.
Could BIR account for CAG repeat expansions observed in human patients? While it is too early to say, we believe that it is an attractive model for large-scale expansions in dividing cells. In contrast, somatic instability was reported to result from cumulative small mutations59, making mechanisms such as toxic oxidation cycles more likely. Though the timing of expansion in human cases is unclear, it is possible that CAG-induced replication fork stalling and subsequent repair might occur during cell divisions of early embryonic development or during division of spermatogonial stem cells, which are exciting areas of future investigation. Thus, the price of BIR to repair DNA damage at long CAG repeats may contribute to the development and severity of human genetic disease.
Online Methods
Plasmids
CAG repeats were obtained from pGL2-CTG14060. The repeats were cut out with AvrII and SfoI and initially cloned into a pYES3-TET644 derivative (described as (GAA)061, which had been cut with BsaBI and an engineered AvrII site. This plasmid was used as template for generating a PCR product (primers JK109 and JK110) containing CAG repeats with NcoI and NotI handles. The PCR product was cloned into pYes3-G4G1C1-T150-GAA10033, which had been cut with NcoI and NotI to remove the GAA repeats. As this construct did not allow selection of large-scale CAG repeat expansions, the length of the “spacer” was increased using PCR products with NcoI handles generated from the bacterial tetracycline resistance gene (Forward JK161 with reverse JK162, JK163, or JK164). The plasmid used in the present study for analyzing large-scale expansion of (CAG)140 is pYes3-G4G1C1-Fori-CAG140-tetbal1-rev (799 bp). For analyzing (CAG)93, a plasmid containing contracted CAG repeats and a longer fragment from the tetracycline resistance gene was used, pYes3-G4G1C1-Fori-CAG93-tetbal2-rev (708 bp). The repeat’s integrity in these constructs was verified by plasmid sequencing using primers FlankL and CanF. A no repeat control plasmid was constructed by cloning a PCR fragment from the tetracycline resistance gene (primers JK161 and JK165) into the NcoI and SphI sites of pYes3-G4G1C1-T150-GAA100, which is called pYes3-G4G1C1_Fori_tet340. All bacterial cloning steps were carried out in the Escherichia coli SURE2 strain (Agilent). Primers are available in Table S3.
Yeast strains
All strains in this study are isogenic to Saccharomyces cerevisiae wild-type (WT) strain CH1585 (MATa, leu2-Δ1, trp1-Δ63, ura3–52, his3–200), an S288c-related haploid strain used in our previous studies31. For transformation into the CH1585 can1::KanMX strain33, the construct was digested from the plasmid using SwaI. Transformants were selected on synthetic complete media lacking tryptophan. The cassette is positioned ~1 kb downstream of ARS306 replacing SGD coordinates 75594–75641 on chromosome III. Correct integration was verified by PCR (primers TrpsF and 36aR or A36bF and CanF). Integrity of CAG repeat length was verified by PCR and Sanger sequencing using primers FlankL and CanF.
Gene knockouts were constructed using a PCR-based method for direct gene replacement with pAG32 (HphMX4) or pAG25 (NatMX4)62 used as template for PCR. Yeast strains are available in Table S4.
Fluctuation assay and calculation of rates
Colonies were grown on rich media containing glucose (YPD) or galactose (YPGal) for 72 hours. Individual colonies were suspended in 0.2–1 mL sterile water, serially diluted, and plated on synthetic complete media containing galactose and canavanine [2% galactose, 0.67% yeast nitrogen base, 0.2% drop-out mix synthetic minus arginine (US Biological D9518), 2% agar, and canavanine sulfate (Sigma C9758)] as well as on YPD media for determination of total cell number. Non-selective growth was on galactose for all knockout and mutant strains unless noted otherwise. The standard concentration of canavanine used was 60 µg/mL. 120 and 200 µg/mL concentrations were also used to minimize background colony growth for some strains/conditions as noted in the text. For each experiment, at least 12 independent colonies were grown, though colonies that had an initial contraction or small expansion were excluded from the analysis. All rates are determined from at least 9 independently grown cultures with verified ((CAG)140 or (CAG)93) starting length. Colonies on YPD was counted on Day 3. CanR colonies were counted on Day 4 for 60 or 120 µg/mL and Day 5 for 200 µg/mL canavanine concentration. PCR was performed on all or at least 8 CanR clones from each plate to determine CAG repeat length. The total number of expanded clones by PCR or the number of CanR clones multiplied by the total frequency of expanded clones by PCR was used to determine expansion rates. Rates and 95% confidence intervals were calculated using the Ma-Sandri-Sarkar maximum likelihood estimator (MSS-MLE) method with correction for plating efficiency determined as z-1/zln(z), where z is the fraction of the culture analyzed63. The average number of viable cells grown on YPD (Nt) was used in all calculations.
18 out of 19 sequenced CanR clones with (CAG)n expansions did not contain any point mutations in CAN1.
Single colony PCR to determine expansion size and generate products for sequencing
Genomic DNA was isolated from CanR clones using a previously described method64. Cells were resuspended in 1.5 µL of 0.5 mg/mL lyticase solution [0.9 M Sorbitol, 0.1 M EDTA (pH 7.4)] in microplates, incubated at 37°C for 15 min, then resuspended in 50 µL of water. The samples were incubated at 100°C for 5 min and centrifuged at 2,500 × g for at least 2 min. For PCR analysis of CAG repeat length, reactions included 1× Green GoTaq reaction buffer (M7911, Promega), 0.16 mM dNTP mix, 0.8 µM of each primer, 0.5 units of Taq DNA polymerase (SibEnzyme or Thermo Scientific), and 1 µL of the DNA supernatant in a 12.5 µL total reaction volume. Primers JK316 and JK317 result in a 544 bp product for (CAG)140. Primers JK318 and JK319 result in a 625 bp product for (CAG)140 (Figure 1B). Primers JK153 and JK171 result in a 462 bp product for (CAG)140 and 321 bp (CAG)93. 10 µL PCR products were run on 1.5% agarose in 0.5× TBE alongside 50 bp and 100 bp DNA ladders (NEB). PCR products were sized using TotalLab Quant software for 1D DNA gels.
The same PCR method was used to generate products for sequencing the CAG repeats (FlankL and CanF) or the CAN1 gene (JK167 and JK168, JK169 and JK170). PCR products were Exo-SAP treated (Affymetrix) and sequenced by Eton Bioscience or University of Chicago Sequencing Core).
Chromatin immunoprecipitation
Strain YJK146 containing (CAG)140 and the no repeat control YJK154 was analyzed by chromatin immunoprecipitation (ChIP) using antibodies recognizing phosphorylated H2A Serine129 (ab15083, Abcam). This modification has been referred to as γ-H2AX in yeast65. Yeast were grown to an O.D. of ~0.8 in YPGal, arrested in G1 using alpha factor, then washed twice with water and released into YPGal. 50 mL cultures of 0, 20, 40, 60, and 90 minute time points were cross-linked with 1% formaldehyde for 15 minutes at room temperature, and quenched with glycine. Cells were lysed mechanically with 0.5 mm glass beads. The chromatin-containing cell suspension was sonicated to yield sheared DNA in the range of approximately 100–1500 bp. Antibody bound DNA was immunoprecipitated (IP) using Protein G Dynabeads (Invitrogen). Samples were analyzed by quantitative PCR using QuantStudio 6 (Applied Biosystems). Relative quantities of IP and input DNA were determined using a standard curve for primers at the CAG locus as well as non-enriched control, ACT1. IP/input values for the CAG locus is normalized to ACT1. The graph shows the fold-enrichment of the CAG strain compared to the no repeat control strain.
Statistical methods
Rates of expansion and 95% confidence intervals (error bars) were calculated based on distribution of expanded clones in at least 9 independent cultures (number depends on clones found to have the correct starting repeat length) using the Ma-Sandri-Sarkar maximum likelihood estimator with a correction for sampling and plating efficiency. Median and interquartile values are reported in Figure 1D. ChIP timepoints were analyzed by one-way ANOVA.
Supplementary Material
Acknowledgments
Research in the lab of S.M.M. is supported by NIH grants R01GM60987 and P01GM105473 and generous contribution from the White family. J.C.K. was supported by the NIH Training in Education and Critical Research Skills postdoctoral program (K12GM074869), and by Tufts University. We thank C. Freudenreich for valuable comments to the manuscript, G. Ira and S. Jinks-Robertson for helpful discussions, K. Spivakovsky-Gonzalez for her help analyzing MSH mutants and D. Padmanhaban for helping with strain construction.
Footnotes
Accession Codes
Not applicable
Data Availability Statement
Source data for Figures in the main text are provided online. All other data is available upon request.
Author Contributions
J.C.K. and S.M.M. designed the study; J.C.K., S.T.H., and T.D. performed experiments; J.C.K., S.T.H., T.D., and S.M.M. analyzed data; K.A.S. provided reagents; J.C.K. and S.M.M. wrote the manuscript.
Competing Financial Interests Statement
None
Data availability
Source data for Figures 1, 2, and 3 are available with the paper online. All other data is available upon request.
References
- 1.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature Reviews Genetics. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nature Reviews Genetics. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nance MA, Myers RH. Juvenile onset Huntington's disease--clinical and research perspectives. Mental Retardation and Developmental Disabilities Research Reviews. 2001;7:153–157. doi: 10.1002/mrdd.1022. [DOI] [PubMed] [Google Scholar]
- 4.Thornton CA. Myotonic dystrophy. Neurologic Clinics. 2014;32:705–719. viii. doi: 10.1016/j.ncl.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JC, Mirkin SM. The balancing act of DNA repeat expansions. Current Opinion in Genetics & Development. 2013;23:280–288. doi: 10.1016/j.gde.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Usdin K, House NC, Freudenreich CH. Repeat instability during DNA repair: Insights from model systems. Critical Reviews in Biochemistry and Molecular Biology. 2015;50:142–167. doi: 10.3109/10409238.2014.999192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 8.Kerrest A, et al. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat Struct Molec Biol. 2009;16:159–167. doi: 10.1038/nsmb.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 10.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends in Biochemical Sciences. 2012;37:162–172. doi: 10.1016/j.tibs.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Concannon C, Lahue RS. Nucleotide excision repair and the 26S proteasome function together to promote trinucleotide repeat expansions. DNA Repair. 2014;13:42–49. doi: 10.1016/j.dnarep.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miret JJ, Pessoa-Brandao L, Lahue RS. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan JL, Andrews KJ, Zakian VA, Freudenreich CH. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol Cell Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung J, van Jaarsveld MT, Shieh SY, Xu K, Bonini NM. Defining genetic factors that modulate intergenerational CAG repeat instability in Drosophila melanogaster. Genetics. 2011;187:61–71. doi: 10.1534/genetics.110.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Human Molecular Genetics. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya S, Lahue RS. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Molecular and Cellular Biology. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su XA, Dion V, Gasser SM, Freudenreich CH. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes & Development. 2015;29:1006–1017. doi: 10.1101/gad.256404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debacker K, et al. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biology. 2012;10:e1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viterbo D, Michoud G, Mosbach V, Dujon B, Richard GF. Replication stalling and heteroduplex formation within CAG/CTG trinucleotide repeats by mismatch repair. DNA Repair. 2016;42:94–106. doi: 10.1016/j.dnarep.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nature Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 26.Mollersen L, Rowe AD, Larsen E, Rognes T, Klungland A. Continuous and periodic expansion of CAG repeats in Huntington's disease R6/1 mice. PLoS Genetics. 2010;6:e1001242. doi: 10.1371/journal.pgen.1001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomes-Pereira M, et al. CTG trinucleotide repeat "big jumps": large expansions, small mice. PLoS Genetics. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, et al. Altered replication in human cells promotes DMPK (CTG)(n). (CAG)(n) repeat instability. Mol Cell Biol. 2012;32:1618–1632. doi: 10.1128/MCB.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) × (CAG) repeat hairpins in human cells. Nature Chemical Biology. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakatani R, Nakamori M, Fujimura H, Mochizuki H, Takahashi MP. Large expansion of CTG*CAG repeats is exacerbated by MutSbeta in human cells. Scientific Reports. 2015;5:11020. doi: 10.1038/srep11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shishkin AA, et al. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Molecular Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherng N, et al. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah KA, McGinty RJ, Egorova VI, Mirkin SM. Coupling transcriptional state to large-scale repeat expansions in yeast. Cell Rep. 2014;9:1594–1602. doi: 10.1016/j.celrep.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobi KC, Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Molecular and Cellular Biology. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E.coli. Nat Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 36.Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Molecular and Cellular Biology. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharyya S, Lahue RS. Srs2 helicase of Saccharomyces cerevisiae selectively unwinds triplet repeat DNA. The Journal of Biological Chemistry. 2005;280:33311–33317. doi: 10.1074/jbc.M503325200. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt MH, Pearson CE. Disease-associated repeat instability and mismatch repair. DNA Repair. 2016;38:117–126. doi: 10.1016/j.dnarep.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Kantartzis A, et al. Msh2–Msh3 interferes with Okazaki fragment processing to promote trinucleotide repeat expansions. Cell Rep. 2012;2:216–222. doi: 10.1016/j.celrep.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundararajan R, Gellon L, Zunder RM, Freudenreich CH. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 43.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 44.Wilson MA, et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakofsky CJ, et al. Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Molecular Cell. 2015;60:860–872. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harbor Perspectives in Biology. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrow DM, Connelly C, Hieter P. "Break copy" duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biology. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayle R, et al. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee CS, Lee K, Legube G, Haber JE. Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nature Structural & Molecular Biology. 2014;21:103–109. doi: 10.1038/nsmb.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szilard RK, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nature Structural & Molecular Biology. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297(5581):599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 53.Follonier C, Oehler J, Herrador R, Lopes M. Friedreich's ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nature Structural & Molecular Biology. 2013;20:486–494. doi: 10.1038/nsmb.2520. [DOI] [PubMed] [Google Scholar]
- 54.Richard G-F, Goellner GM, McMurray CT, Haber JE. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. Embo J. 2000;19:2381–2390. doi: 10.1093/emboj/19.10.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jankowski C, Nasar F, Nag DK. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2134–2139. doi: 10.1073/pnas.040460297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H-M, et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. Embo J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genetics. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genetics. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higham CF, Morales F, Cobbold CA, Haydon DT, Monckton DG. High levels of somatic DNA diversity at the myotonic dystrophy type 1 locus are driven by ultra-frequent expansion and contraction mutations. Human Molecular Genetics. 2012;21:2450–2463. doi: 10.1093/hmg/dds059. [DOI] [PubMed] [Google Scholar]
Methods-only References
- 60.Raca G, Siyanova EY, McMurray CT, Mirkin SM. Expansion of the (CTG)(n) repeat in the 5'-UTR of a reporter gene impedes translation. Nucleic Acids Research. 2000;28:3943–3949. doi: 10.1093/nar/28.20.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah KA, et al. Role of DNA Polymerases in Repeat-Mediated Genome Instability. Cell Reports. 2012;2:1088–1095. doi: 10.1016/j.celrep.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 63.Rosche WA, Foster PL. Determining Mutation Rates in Bacterial Populations. Methods. 2000;20:1–14. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aksenova AY, et al. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19866–19871. doi: 10.1073/pnas.1319313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee C-S, Lee K, Legube G, Haber JE. Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nat Struct Mol Biol. 2013;21:103–109. doi: 10.1038/nsmb.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.