Abstract
Autism spectrum disorders (ASD) are common, complex and heterogeneous neurodevelopmental disorders. Cellular and molecular mechanisms responsible for ASD pathogenesis have been proposed based on genetic studies, brain pathology, and imaging, but a major impediment to testing ASD hypotheses is the lack of human cell models. Here, we reprogrammed fibroblasts to generate induced pluripotent stem cells (iPSCs), neural progenitor cells (NPCs) and neurons from ASD individuals with early brain overgrowth and non-ASD controls with normal brain size. ASD-derived NPCs display increased cell proliferation due to dysregulation of a β-catenin/BRN2 transcriptional cascade. ASD-derived neurons display abnormal neurogenesis and reduced synaptogenesis leading to functional defects in neuronal networks. Interestingly, defects in neuronal networks could be rescued by IGF-1, a drug that is currently in clinical trials for ASD. This work demonstrates that selection of ASD subjects based on endophenotypes unraveled biologically relevant pathway disruption and revealed a potential cellular mechanism for the therapeutic effect of IGF-1.
Keywords: Autism spectrum disorders, induced pluripotent stem cells, macrencephaly, disease modeling, cell proliferation, neuronal networks, multi-electrode arrays, IGF-1, personalized medicine
INTRODUCTION
Autism spectrum disorders (ASD) comprise a group of complex neurodevelopmental disorders that affect more than 1% of children in the United States. ASD is characterized by impaired social interaction and limited and repetitive interests and behavior at its core. ASDs are phenotypically and etiologically heterogeneous, making it challenging to uncover the underlying genetic, pathologic and cellular pathophysiology. Family history and twin studies suggest that, at least in some cases, these disorders share genetic roots1, 2. Mounting evidence proposes that heritable and de novo genetic variation plays a significant role, but these studies also demonstrate striking genetic heterogeneity3-5. Neuropathological imaging and gene expression studies of postmortem brains from ASD patients have revealed disruption of developmental and proliferation gene networks6, 7. Recent studies integrating ASD candidate genes with spatiotemporal coexpression networks demonstrate that gene expression converge on the transcriptional regulation in pyramidal, glutamatergic cortical neurons during mid-fetal human development8, 9. One relevant observation in ASD pathophysiology has been the occurrence of macrencephaly and altered growth trajectory with early overgrowth and later normalization in a subset of affected individuals. An increase in brain size in autistic individuals in the first three years of life precedes the first clinical signs10-15, and excess neuron numbers are reported for abnormally enlarged young ASD brains16. Excess cortical neuron numbers and patches of abnormal cortical organization and cell migration are pathologies that also implicate mid-fetal development as being crucial in ASD pathophysiology16, 17. Gene expression changes in postmortem brain overlap with developmentally regulated genes involved in cortical patterning as well as in cell cycle, proliferation and neural differentiation6, 7. Taken together, these observations highlight the relevance of early fetal brain development factors in the pathophysiology of ASD.
One of the major impediments to ASD research is the genetic and brain pathological heterogeneity that makes it difficult to produce relevant animal and cell models. Reprogramming of somatic cells to a pluripotent state by over-expression of specific genes has been accomplished using human cells18, 19. Induced pluripotent stem cells (iPSCs) are attractive models for understanding complex diseases and disorders with heritable and sporadic conditions20. Although iPSCs have been generated for monogenetic ASD diseases4, 21, 22, the demonstration of disease-specific pathogenesis in complex and heterogeneous disease such as sporadic ASD is a current challenge in the field23. Nonetheless, extending the iPSC modeling technology beyond monogenetic ASD to the study of non-syndromic forms of autism could uncover molecular and cellular pathways that overlap among many forms of autism, leading to a better understanding of the disease and potentially developing novel ASD biomarkers and targets for therapeutics24.
We reasoned that ASD patients sharing a common phenotype, early developmental brain enlargement ranging from mild to extreme macrencephaly, might also share underlying molecular and cellular pathway dysregulation. We therefore pre-selected ASD infants and toddlers who displayed this phenotype, including pre-selection that provided a range from mild to extreme that enabled generalization of results to ASD beyond those with pure and extreme macrencephaly. We took advantage of reprogramming technologies to generate iPSCs from a cohort of ASD patients who displayed brain overgrowth early in life. Neural progenitor cells (NPCs) derived from ASD-iPSCs displayed altered proliferation resulting from dysregulation of a β-catenin/BRN2 transcriptional cascade. As a consequence, we observed that ASD-derived neurons formed fewer excitatory synapses and matured into defective neuronal networks with less bursting. Importantly, all ASD patients showed improved network strength after treatment with IGF1 (a drug that is currently in clinical trial for ASD), but the levels of improvement were unique to the patients, revealing a potential novel assay to pre-screen patients for future clinical trials. Together, our results suggest that, when stratified into measurable endophenotypes, idiopathic ASD can be modeled using iPSC technology to reveal novel cellular and molecular mechanisms underlying brain abnormalities.
MATERIALS AND METHODS
Patient ascertainment
Subjects were recruited through the UCSD Autism Center of Excellence from a pool of volunteers formerly included in previous brain imaging studies. Control subjects were selected randomly from lists of typically developing individuals who had had an MRI scan when they were toddlers. ASD subjects were selected from lists of ASD subjects who had been identified and diagnosed with ASD and MRI scanned when they were toddlers; from among these potential ASD subjects, we selected those with larger than normal average total brain volume as compared to typically developing toddlers. The ASD subjects demonstrated a behavioral presentation consistent with autism as defined by the criteria set forth in the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV; APA)25. Assessment of the history and presence of the disorder was achieved via standardized behavioral, cognitive, and functional assessments, including the appropriate Wechsler Intelligence Scale, the Autism Diagnostic Observation Schedule (ADOS), the Autism Diagnostic Interview, Revised (ADI-R), and Vineland Adaptive Behavior Scales (VABS). Participants in the control group had no history of psychological, genetic, or other disorder. This project was approved by the Ethics Committees of the institutes at which the study was conducted. After a complete description of the study, written informed consent was provided by the parents.
CNV (Copy-Number Variation) analyses
High-confidence CNV calls were made using the CNVision pipeline as previously described 7, 26. In brief, patients and controls were genotyped using the Illumina 660W array. Probe intensity signals were analyzed to make CNV calls using e types of software PennCNV, QuantiSNP and GNOSIS. High-confidence calls were made by merging the prediction output of these 3 different algorithms. Rare events were defined based on the database of genomic variants (DGV) (http://dgv.tcag.ca/).
Exome sequencing
Fibroblasts derived from the 8 ASD individuals were cultured. DNA was extracted and exome libraries were generated for each line using Nextera Rapid Capture Exome kit from Illumina. Illumina Hi-Seq 2500 Pair-end sequencing was conducted (read length=100bp), resulting in 308,082,362 reads in average for each library. Short reads were aligned to the human genome (hg19) using BWA 27 and Samtools 28. Mutations were first filtered based on the reading depth (D>=10), alignment quality (q>=20) and base quality (Q>=30), and 4712088 variants passed filtering. Next, we used two different software programs, ANNOVAR31 and IlluminaVariantStudio (https://www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/datasheet_illumina_variantstudio_software.pdf) to annotate the variants. The results from ANNOVAR 29 and IlluminaVariantStudio were very similar (Supplementary Table S1A), so we further analyzed variants found in the IlluminaVariantStudio only. There were a total of 485 stop-gains and 103 stop losses found in all ASD lines, but several variants were found in multiple lines. Therefore, a total of 275 unique stop gain/loss mutations were detected, of which 222 were listed in dbSNP while 53 rare mutations were not (Supplementary Table S1B). We further filtered the 58278 missense variants found in IlluminaVariantStudio by PolyPhen and SIFT scores. Of these, 3403 variants were considered both “deleterious” in SIFT and “probably damaging” in PolyPhen. Again, several of these variants were found in multiple patients, so the total number of unique “deleterious” and “probably damaging” SNVs was 1829. Of these, 1564 were found in dbSNP while 265 rare variants were not (Supplementary Table S1C).
Cellular reprogramming
IPSCs were obtained from skin fibroblasts of ASD patients and controls, collected by the UC San Diego Autism Center of Excellence. Briefly, fibroblasts were transduced with retroviruses containing OCT4, SOX2, KFL4, and MYC to induce overexpression of these genes 30. Two days after transduction, the cells were transferred to a co-culture system with murine embryonic fibroblasts (mEFs) maintained with DMEM/F12 (Invitrogen, CA), 20% Knockout Serum Replacement (Invitrogen, CA), 1% non-essential amino acids, and 100 μM beta-mercaptoethanol and treated with 1 mM valproic acid (Sigma) for 5 days. The iPSC colonies were identified after approximately 2 weeks in this culture system, transferred to Matrigel (BD Biosciences)-coated plates, and maintained in mTeSR media (Stem Cell Technologies). To avoid potential variability caused by retroviral insertions in the genome, the experiments were performed with at least 2 independent iPSCs clones.
Karyotype
Standard G-banding karyotype analyses were performed by Cell Line Genetics (Madison, WI).
Neural differentiation
The iPSC colonies were plated on Matrigel (BD Biosciences)-coated plates and maintained for 5 days in mTSeR medium (Stem Cell Technologies). On the fifth day, the medium was changed to N2 media [DMEM/F12 medium supplemented with 1× N2 supplement (Invitrogen) and 1× B27 supplement (Invitrogen). After 2 days, the colonies were removed from the plate and cultured in suspension as EBs for 2 weeks using NPC medium with dorsomorphin during the entire procedure. The EBs were then gently dissociated with accutase (Gibco), plated on Matrigel-coated dishes, and maintained in NBF medium (DMEM/F12 medium supplemented with 1× N2, 1× B7 supplements, 20 ng/mL FGF2 and 1% penicillin/streptomycin). The rosettes that emerged after 3 or 4 days were manually selected, gently dissociated with accutase, and plated in dishes coated with 10 μg/mL poly-ornithine and 5 μg/mL laminin. This NPC population was expanded using NBF medium. To differentiate the NPCs into neurons, the cells were re-plated in the absence of FGF2, with regular medium changes every 3 or 4 days.
Immunofluorescence
Cells were washed once with PBS and then permeabilized with 0.5% Triton X-100 in PBS for 5 minutes. After washing with PBS and blocking with 10% Donkey serum, 0.5% Triton X-100 in PBS (blocking solution), cells were incubated with the primary antibodies diluted in blocking solution for 16 hours at 4°C. Primary antibodies used were: anti-Pax6 (Covance PRB-278P); anti-Sox1 (R&D, AF3369); anti-Otx2 (Milipore, AB9566); anti-Vimentin (abcam, RV202) anti-phospho-Histone H3 (Sigma, H0412); anti-Ki67 (Novocastra NCL-Ki67p); Anti-Brn2 (Santa Cruz, cs6029) anti-Brn2 (2R2) 31; anti-Ngn2 (Santa-cruz, sc-19233); anti-Mash1/Ascl1 (BD 556604 and Chemicon AB5696s); anti-NKX2.1 (Novacastra, TTF-1-L-CE); anti-Olig2 (Millipore, AB9610); anti-Tuj1 (Covance, MMS-435P) and anti-Doublecortin (Santa Cruz, cs-8066); anti-GABA (Sigma, A2052), anti-Map2AB (Abcam, ab5392). After PBS washes, cells were incubated with labeled secondary antibodies for 60 minutes. Nuclei were stained after incubation with 1 μg/ml DAPI for 5 minutes. The slides were mounted using prolong-gold (Invitrogen, P36930).
Proliferation and LiCl treatment of NPC lines
After NPCs grew close to confluence (4~9 days), the cell number in each well was counted using a cell counter (Invitrogen countess automated cell counter). The population doubling time was inferred for each well based on the culture time and the ratio between input and harvest. Differences in doubling time between LiCl-treated and untreated conditions in each NPC line were compared using 2-tail t test. Also, doubling time differences between LiCl-treated and untreated conditions were tested for all 5 autistic lines (treated=27, non-treated=28) and all 5 control lines (treated=11, non-treated=10) using the two-tail t test.
Cell cycle analysis
Monolayer-adherent NPCs were harvested, washed with PBS, resuspended in PBS and fixed for 1 hour in 4°C in 100% ethanol (−20°C). Cells were collected, resuspended in PBS containing 100 μg/ml RNase A for 30 minutes in 37°C, and stained with 1 mg/ml propidium iodide solution (10 mg / 1×10^6 cells stained) for 2 hours at 4°C. For each sample, 50,000 cells were collected in the appropriate gate for analysis on FACSCalibur (BD Biosciences, Mountain View, CA) using FlowJo software (Tree Star, Ashland, OR).
Western blotting
NPCs cultured as monolayers were washed with cold PBS. Protein was extracted with 50 mM Tris pH 7.6, 500 mM NaCl, 1 mM EDTA, 1% NP-40, 1% deoxycholate, 0.1% SDS and protease inhibitor cocktail (Roche, # 1 836 153). Lysates were centrifuged for 25 minutes at 15,000 rpm at 4°C. The resulting supernatant was collected and the total protein content of the samples was determined by the BCA Protein Assay Kit (Pierce Biotechnology, #23225). SDS gel electrophoresis was performed utilizing 15 μ5 protein per lane. Gels were transferred to nitrocellulose membranes (BioRad, 9004-70-0). Proteins were detected with anti-Pax6 (Covance PRB-278P), anti-Sox1 (R&D, AF3369), anti-Otx2 (Milipore, AB9566), anti-Vimentin (abcam, RV202), anti-Brn2 (2R2) 31, anti-β-III-tubulin (Covance, MMS-435P) and anti-GAPDH (Acris, ACR001P). All membranes were visualized using ECL (Thermo Scientific, 34080) and exposure to autoradiography film (Denville Scientific Inc. E3018).
RT-PCR (Reverse Transcription Polymerase Chain Reaction)
Total cellular RNA was extracted with TRIzol (Invitrogen) and purified on a column using the RNeasy Mini Kit. DNAase treatment was performed on the column. Extracted RNA (1 μg) samples were reverse transcribed into cDNA according to the manufacturer’s protocol for RT-PCR (BioRad, 170-8891). RT-qPCR was performed by using SsoFast EvaGreen Supermix (Biorad) and run on the Eppendorf Mastercycler EP Realplex 2 thermal cycler. The specific primers used for RT-qPCR were: HPRT1 realtime primers.com, VHPS-4263, Pax6 realtime primers.com, VHPS-6650, Otx2 realtime primers.com, VHPS-6546, Sox1 Left primer: GGGAAAACGGGCAAAATAAT; Right primer: TTTTGCGTTCACATCGGTTA. Brn2 Left primer: TGACGATCTCCACGCAGTAG; Right primer: GAGGGTGTGGGACCCTAAAT.
Neurotransmitter Receptors quantitative polymerase chain reaction (qPCR) Array
For neurotransmitter receptors analysis, iPSC-derived neurons from ASD and related controls were rapidly frozen and placed in TRIzol (Life Technologies) for subsequent RNA isolation and purification. One microgram of RNA of each individual/clone were converted into cDNA using the RT2 First Strand Kit (SABiosciences). Quantitative RNA expression analysis of 84 neurotransmitter receptors-related genes was simultaneously analyzed using human RT2 Profiler PCR Array (#PAHS-060Z; SABiosciences) on a Bio-Rad CFX96 following the manufacturer’s instructions. Data normalization was based on correcting all Ct values for the average Ct values of five constantly expressed housekeeping genes present on the array, and reported as fold changes over similar values from controls samples.
NPC transfection
Transfection was performed in NPC medium, which was replaced 24 hours following transfection. Transfection efficiency was measured by GFP expression and was ~45%. Super Top-Flashx8 (50 ng), super Fop-Flashx8 (50 ng) and Renilla (10 ng) plasmids were transiently transfected into NPCs (40,000 cells per well of a 96-well plate) using Lipofectamine LTX+ in NPC medium, which was replaced 24 hours following transfection. After 48 hours, cells were lysed in the 96-well plates with passive lysis buffer (Promega). Firefly luciferase and Renilla luciferase activity in the lysates were measured on a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT) in replicates of 3, using the Dual-Luciferase reporter assay system (Promega). The ratios for firefly luciferase:Renilla luciferase were determined and expressed as relative luciferase activity.
Quantification of neuronal synaptic puncta
Neuronal tracing was performed on neurons in which the shortest dendrite was at least 3 times longer than the cell soma diameter, as determined using a semi-automatic ImageJ plug-in (NeuroJ). Spines and VGLUT1 puncta were quantified after 3-dimensional reconstruction of z-stack confocal images. The same density of neurons was plated in each condition. Final cell density was confirmed by DAPI. Only Tuj1- or Map2-positive neurons were scored. Images were taken randomly for each individual and from 2 different experiments, using at least 2 different clones. Quantification was performed blind to the cell genotype. No distinction was made between different types of spines due to the unviability of this assessment using the present method. All experiments were performed with independent clones and different controls. All analyses were performed double blinded to avoid bias.
Image acquisition and analysis
The immunofluorescent slides were viewed and photographed with 40× or 60× objectives using a Nikon C1si microscope. Analysis of the thickness and/or cell counts of immunohistochemical staining were captured and quantified across the entire image taken by the Image-Pro Plus system (version 5.1; Media Cybernetics).
Whole-cell patch clamp and electrophysiology analyses
Whole-cell patch clamp recordings were performed on neurons co-cultured with rat hippocampal astrocytes for 8 weeks. The same density of neurons was plated in each condition. Neurons were labeled with a synapsin-GFP lentiviral marker. Cells were perfused with ACSF solution containing (in mM): 119 NaCl, 2.5 KCl, 1 NaH2PO4, 26 NaHCO3, 2 CaCl2, 1.3 MgSO4, 11 glucose, 0.1 picrotoxin. 2-5 MΩ pipettes were pulled on a horizontal micropipette puller (Sutter P-97). For sEPSPs and Na+/K+ currents, pipettes were filled with a cesium-gluconate internal solution that consisted of (in mM): 130 D-Gluconic Acid, 130 CsOH, 5 NaCl, 12 Phosphocreatine, 10 HEPES, 10 EGTA, 3 adenosine triphosphate, 0.2 guanosine triphosphate (pH 7.3). For spontaneous and evoked action potentials, cells were recorded with a potassium-gluconate internal solution that consisted of (in mM): 120 K-gluconate, 15 KCl, 4 MgCl2, 0.1 EGTA, 10 Hepes, 4 MgATP, 0.3 Na2GTP, 7 phosphocreatine (pH, 7.2). The mean access resistance was 7.5 ± 1.0 MΩ. Recordings were collected using an Axoclamp-200B amplifier (Axon Instruments) and analyzed with Clampfit 10 software (Molecular Devices). Signals were low-pass filtered at 2kHz and digitized at 5kHz. Na+/K+ currents were recorded by stepping cells in +5-mV increments. For evoked action potentials, current injections were applied to hold cells at approximately −50 mV. A series of 300-ms current steps were injected into cells, starting at −0.2 nA, and increasing by +0.1 nA increments. Statistical comparisons of control and autism cells were made using two-tailed Student t-tests with a significance threshold of p = 0.05.
Multi-electrode arrays (MEA)
Using the 12-well MEA plate from Axion Biosystems, we plated cells derived from 5 control individuals and 7 ASD patients in triplicate. Each well was seeded with 10,000 NPCs that were induced into neuronal differentiation as described above. Each well was coated with poly-ornithine and laminin prior to cell seeding. Cells were fed once a week and measurements were taken before media changes. Recordings were performed in a Maestro MEA system and AxIS software (Axion Biosystems) using a bandwidth with a filter for 10Hz to 2.5kHz cutoff frequencies. Spike detection was performed using an adaptive threshold set to 5.5 times the standard deviation of the estimated noise on each electrode. Each plate rested for 5 minutes in the Maestro Instrument, and then was recorded for 10 minutes to calculate the spike rate per well. Multi-electrode data analysis was performed using the Axion Biosystems Neural Metrics Tool, and an active electrode was considered once 5 spikes occurred over the length of 1 minute (5 spikes/min). Bursts were identified in the data recorded from each individual electrode using an adaptive Poisson surprise algorithm. Network bursts were identified for each well; using a non-adaptive algorithm requiring a minimum of 10 spikes with a maximum inter-spike interval of 100ms. Only the wells that exhibited bursting activity were included in this analysis. For the rescue experiments, 10ng/ml of recombinant human IGF1 (PeproTech) was added to the neuronal cultures on day 35 of differentiation. Media was changed every week (with or without the drug for the blank wells) for the duration of the experiment.
RNA sequencing
RNAs were isolated from 88 cell lines (31 iPSCs, 29 NPCs and 28 neurons) derived from idiopathic ASD patients with macrencephaly and non-autistic controls using the RNeasy Mini kit (QIAGEN). A total of 1,000 ng of RNA was used for library preparation using the Illumina TruSeq RNA Sample Preparation Kit. The RNAs were sequenced on Illumina HiSeq2000 with 50bp paired-end reads, generating 50 million high quality sequencing fragments per sample on average. We first utilized TopHat2 aligner to align the reads against the human reference genome from the 1000 Genomes Project (i.e., human_g1k_v37), and then summarized the read counts to Genecode11 union-exon gene models using htseq-count (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html). Of the reads, 85% could be uniquely mapped, and 74% of those uniquely aligned reads fell into exonic and UTR regions. Next, we processed the expression data for each experimental stage separately. At every differentiation stage, outlier samples were defined by low inter-sample correlations as previously described 32 and subsequently excluded in the follow-up expression analysis. Eighty-three samples were retained for further analysis, including 28 iPSCs, 30 NPCs, and 25 neuronal samples. Gene-based read counts were further median-normalized and log2-transformed to bring their distribution to normality. Batch effects were corrected afterwards using R ComBat package with the non-parametric model option. Lastly, we filtered expressed genes requiring count per million (cpm) values larger than 1 in at least 1 group of the samples (ASD or controls). Consequently, 14,291 genes in iPSCs, 14,413 in NPCs, and 14,974 in neurons were identified as expressed and were retained for further analysis. All gene expression data is deposited at GEO (GSE67528).
Differential expression (DE) analysis
DE analysis at static experimental stage was performed using the linear mixed effect model from R limma package. Samples derived from the same subject were considered as replicates, where their variations were accounted for the random effects in the model. RNA qualities may greatly influence the DE analysis. Therefore, we included RNA integrity numbers (RIN) as a covariate in our regression model. DE genes were detected based on the significance threshold of p-value <0.005, unless we specified otherwise. To identify the genes showing differentiation-induced expression changes, we first paired progenitors with neurons if they were from the same iPSC clone and plated for differentiation in 1 experiment. Next, we computed the gene-based ratios of progenitor expression over neuronal expression for every differentiation pair and we used the log2-transformed ratios as the input for the follow-up regression model. The comparison was conducted using the linear mixed model via the lme function provided by R package nlme. Ratios computed for the same subject were considered as replicates. Significant events were detected if p-value <0.005.
Functional enrichment analysis
Gene ontology (GO) enrichment analysis was assessed using the GO-Elite Pathway Analysis tool. GO-Elite performs permutations to obtain an over-representation Z score and enrichment p-value for each GO term. We used the default permutations settings in GO-Elite package (see supplementary materials for details).
Weighted gene correlation network analysis (WGCNA)
We conducted signed co-expression network analysis on 25 neuronal samples using R WGCNA package, as previously described 6, 8. Briefly, 14,974 expressed protein-coding genes were used in our analysis, and the soft thresholding power was chosen to be 16 based on the approximate scale-free topology criterion. We set the minimum module size to 100 genes and required the dissimilarities among modules to be at least 15%. Each identified module was summarized by the module eigengene (first component of the standardized expression patterns) and was thereafter correlated with ASD disease status. The disease association significance was evaluated by a linear mixed effect model via R lme function. Four modules (brown, tan, magenta, and purple) were considered significantly associated with ASD status with an association of FDR<0.1. Furthermore, genes were prioritized based on their correlations with the module eigengene, named kME. The top 200 connected genes ranked by kME values were used to generate the network plots via the R igraph package.
Statistical Analyses
All statistical analyses were performed using SPSS version 12. The control and ASD groups were compared by Student’s t-test and, for the doubling time, we used the repeated measurement analysis. For the pCAG-Brn2 over-expression and LiCl experiments two-way ANOVA was used. Where appropriate, Student’s t-test post-hoc analysis was performed with Bonferroni correction for multiple comparisons, and the p-values presented are those of the corrected values.
A summary Table containing number of patients and control cell lines used of each experiment can be found on Supplementary Table S7.
RESULTS
Identification of genetic variants in macrencephaly ASD individuals and controls
We recruited 8 ASD Individuals with quantitative MRI-validated early brain enlargement ranging from mild brain enlargement to macrencephaly and 5 age/gender-matched control individuals for skin biopsies and phenotypic characterization (Supplementary Table S2). The ASD donors displayed larger brain size compared to the normal average brain size of typically developing control subjects at any given age (Figure 1A, B). Copy Number Variation (CNV) analysis, using DNA extracted from the donors’ whole blood did not show the presence of any rare structural variant known to be associated with ASD (Supplementary Table S3). A small duplication involving intron 2 of the NLGN1 gene was detected in one ASD case; however, this event has also been reported in typically developing controls (http://dgv.tcag.ca/). Exome sequencing was performed on DNA from the 8 ASD fibroblasts. Variants were called by two pipelines (lluminaVariantStudio and ANNOVAR) that allowed for the detection of variant alleles with higher frequency, since we reasoned that both rare and common variants may contribute to the ASD phenotypes. Using these criteria, more than 50,000 total variants (58866 and 53101 from each of the two pipelines) in all 8 libraries were found (Supplementary Table S1A). Since both pipelines gave similar results, we focused our analysis on stop-gain, stop-loss and non-synonymous variants identified by the lluminaVariantStudio pipeline. A total of 275 unique stop gain/loss mutations were detected, of which 222 were listed in dbSNP while 53 rare mutations were not (Supplementary Table S1B). Filtering the non-synonymous variants that were considered both “deleterious” in SIFT and “probably damaging” in PolyPhen, a total of 1829 unique variants were detected (Supplementary Table S1C). Of these, 1564 were found in dbSNP (common) while 265 rare variants were not in dbSNP (Supplementary Table S1C).
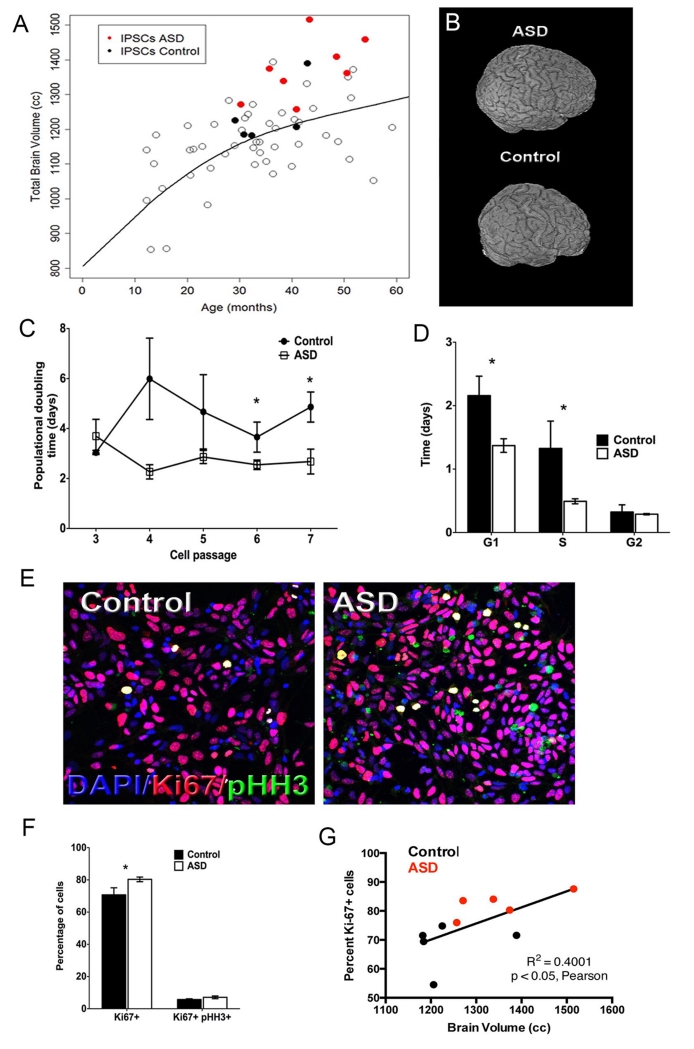
Figure 1.
Altered ASD iPSC-derived NPCs proliferation. (A) Left panel, scatterplot of Total Brain Volume (TBV) across ages. Open black circles indicate brain size of typically developing subjects. Black solid dots represent control donors. Red solid dots represent ASD donors. (B) Three-dimensional reconstructions of the brain from one control donor and one ASD donor. (C) iPSCs from ASD and control were differentiated to NPCs. From passages 2 to 6 cells were plated at the same density and population-doubling time at each passage was calculated. Results of all lines (2 clones per line) are presented as mean ± SEM (*repeated measurements p=0.02, post-hoc p<0.04). (D) Adherent monolayer NPCs from control and ASD iPSCs were dissociated, counted for calculation of population doubling time and prepared for cell cycle analysis. Results are presented as the time spent in each cell cycle stage (n≥4, mean ± SEM, ANOVA p<0.04, post-hoc p<0.04 for comparing the time spent in G1 phase in the ASD NPCs with those of the control NPCs, respectively). (E) Control and ASD NPCs were immunostained with DAPI (Blue), anti-pHH3 (Green) and anti-ki67 (Red) (Scale bar: 200 μm). Representative images of the staining are shown. (F) Quantification of the percentage of Ki67+ and Ki67+pHH3+ labeled cells are presented as mean ± SEM (n≥5; *p<0.03 for comparing the results of the ASD with those of the control NPCs). (G) Pairwise correlation between individual brain size (volume) and respective NPC cell line proliferation rates (% of Ki67 positive cells).
Notably, novel stop-gains were found in both pipelines in the canonical Wnt pathway genes CTNNB1, coding for β-catenin, and FZD6, coding for Frizzled 6 (Table 1A). Additionally, two more common missense variants listed in dbSNP in FRZB (rs288326), coding for a secreted Frizzled antagonist, and WNT10B (rs121908120), coding for a WNT ligand, were also detected (Table 1A). These three missense variants were predicted to be damaging by both SIFT and PolyPhen algorithms. No mutations were found in 2 alleles of a single gene in any patient (i.e., all variants were heterozygous). In addition, stop-gain or damaging mutations by both SIFT and PolyPhen algorithms of 97 known autistic genes found in SFARI Gene (https://sfari.org/resources/sfari-gene) were detected (Supplementary Table S4), including one mutation found in PTEN (rs181113165), previously implicated in ASD brain growth trajectory33. Of the mutations found in these 97 genes, 78 were found in dbSNP and 19 were novel. Gene ontogeny (GO) analysis revealed that 11 GO terms were significantly represented among the variants detected in these ASD samples after correction for multiple testing (Table 1B, Supplementary Table S5). These GO terms included sensory perception, cognition, neural systems, adhesion terms, as well as G-protein and cell surface receptor signaling terms. Pathway analysis using DAVID revealed that two pathways, olfactory transduction and ABC transporters, were significantly represented among the variants detected in these ASD samples after correction for multiple testing (Table 1C, Supplementary Table S6).
Table 1.
All variants were both deleterious (SIFT) and “probably damaging” (PolyPhen).
| A. WNT pathway genes with deleterious variants in ASD subjects. Rows in bold indicate that the variant resulted in a stop gain, and the asterisk (*) represents a stop codon. | |||||||
|---|---|---|---|---|---|---|---|
| WNT pathway related genes | Sample | coordiant | Variant | dbSNP ID | Allele Freq | Amino Acids | Codons |
| CTNNB1 | arch | chr3:41266229 | C>C/T | 0 | Q/* | Caa/Taa | |
| FZD6 | acai | chr8:104330986 | C>C/T | 0 | R/* | Cga/Tga | |
| FRZB | acai | chr2:183703336 | G>G/A | rs288326 | 5 | R/W | Cgg/Tgg |
| WNT10A | arch | chr2:219755011 | T>T/A | rs121908120 | 1 | F/I | Ttt/Att |
| B. Significant GO terms for genes with deleterious variants in ASD subjects. | |||||||
|---|---|---|---|---|---|---|---|
| Term | Count | % | PValue | Fold Enrichment |
Bonferroni | Benjamini | FDR |
| Sensory Perception of Smell | 111 | 7.43 | 4.62E-30 | 3.28 | 1.52E-26 | 1.52E-26 | 8.4E-27 |
| Sensory Perception of Chemical Stimulus |
113 | 7.56 | 4.02E-27 | 3.01 | 1.32E-23 | 6.62E-24 | 7.31E-24 |
| Sensory Perception | 148 | 9.91 | 3.37E-23 | 2.33 | 1.11E-19 | 3.7E-20 | 6.13E-20 |
| Cognition | 152 | 10.17 | 5.88E-20 | 2.13 | 1.94E-16 | 4.85E-17 | 1.07E-16 |
| Neurological System Process | 173 | 11.58 | 1.85E-15 | 1.82 | 6.22E-12 | 1.24E-12 | 3.43E-12 |
| G-protein Coupled Receptor Protein Signaling Pathway |
159 | 10.64 | 7.75E-14 | 1.80 | 2.55E-10 | 4.26E-11 | 1.41E-10 |
| Biological Adhesion | 99 | 6.63 | 9.57E-09 | 1.80 | 3.16E-05 | 4.51E-06 | 1.74E-05 |
| Cell Surface Receptor Linked Signal Transduction |
210 | 14.06 | 1.26E-08 | 1.44 | 4.14E-05 | 5.17E-06 | 2.28E-05 |
| Cell Adhesion | 98 | 6.56 | 1.81E-08 | 1.78 | 5.97E-05 | 6.64E-06 | 3.3E-05 |
| Cell-Cell Adhesion | 47 | 3.15 | 8.11E-07 | 2.17 | 0.002671 | 0.000267 | 0.001476 |
| Homophilic Cell Adhesion | 25 | 1.67 | 7.49E-05 | 2.43 | 0.218923 | 0.022212 | 0.13627 |
| C. Significant pathways for genes with deleterious variants in ASD subjects. | |||||||
|---|---|---|---|---|---|---|---|
| Term | Count | % | PValue | Fold Enrichment | Bonferroni | Benjamini | FDR |
| Olfactory Transduction | 106 | 7.10 | 4.02E-29 | 3.12 | 7.36E-27 | 7.36E-27 | 4.97E-26 |
| ABC Transporters | 14 | 0.94 | 7.74E-05 | 3.55 | 0.014069 | 0.007059 | 0.095583 |
Generation of iPSCs from macrencephaly ASD individuals and controls
ASD and non-affected control fibroblasts were transduced with 4 retroviral reprogramming vectors (Sox2, Oct4, c-Myc and Klf4), as described elsewhere 30. Following 2 to 3 weeks of culture in human embryonic stem cells (hESC)-supporting conditions, compact refractile ESC-like colonies emerged from a background of fibroblasts. iPSC colonies were then manually picked and cultured under feeder-free conditions. Cells were mechanically expanded for at least 10 passages and tested for the expression of pluripotent markers. We obtained several clones that continuously expressed pluripotent markers, such as Nanog, Lin28, Tra-1-81 and Sox2, from each control wild-type (WT)-iPSCs and ASD-iPSCs. We excluded all karyotypically unstable clones from further experiments (Figure S1).
iPSC-derived NPCs from ASD Individuals with early brain overgrowth display rapid proliferation
Our recent analyses showed that iPSC-derived neural cell types could recapitulate human fetal cortical development and thus could be used to model this developmental stage in ASD34. We used our previous published protocol to generate NPCs from iPSCs4 in the presence of Noggin. Briefly, we initiated neural differentiation by plating 1-week-old embryoid bodies (EBs) treated with Noggin. After a week in culture, EB-derived rosettes became apparent in the dish. Rosettes were then manually collected, dissociated and re-plated. The NPCs derived from rosettes formed a homogeneous population after a few passages from ASD individuals and controls, and continued to proliferate in the presence of FGF2 as adherent monolayers.
We hypothesized that an alteration of the rates of NPC proliferation could result in early brain overgrowth. Proliferation was measured in (2 clones per line) by calculating the population doubling time from plating at passages 3 to 7 (P3-7) in continuous culture, and the doubling times of all ASD and control lines were pooled to calculate averages. From P4, the population doubling time decreased in ASD NPCs from all 8 Individuals compared to NPCs from all 5 controls, reaching statistical significance at P6 (Figure 1C and Figure S2A). Surprisingly, all ASD NPC clones proliferated faster than all control NPC clones. Cell cycle analysis at P6 revealed that shortening of G1 and S phases was the main reason for the decrease in the population doubling time, with no change of G2-M phase length (Figure 1D and Figure S2B). Double labeling for Ki67 and pHH3 revealed an increased percentage of Ki67+ (cycling cells) in ASD relative to control NPCs, whereas the percentage of pHH3+Ki67+ (G2-M phase mitotic cells) was unaffected in autistic NPCs (Figure 1E, F and Figure S2C). After patient and control age-matching correction, we performed pair-wise correlations between brain size (volume) and NPC proliferation and doubling time. We observed a positive correlation with NPC proliferation and an inverse correlation with NPC doubling time across all samples (ASD and controls) (Figure 1G and Figure S2D). Strikingly, proliferation (represented by percentage of Ki67 positive cells) is robustly significant in its correlation with brain volume, suggesting that it may be proliferation that specifically contributes to the large brain early on in ASD. These findings demonstrate that iPSC-derived NPCs from ASD Individuals with macrencephaly proliferated faster than those derived from controls and NPC proliferation is correlated increase in brain volume. To our knowledge, this is the first time that clinical data is significantly correlated to an iPSC-neural phenotype and open the doors for potential use of vitro assays as diagnostic tools for complex multigenic disorders such ASD.
β-catenin/BRN2 transcriptional activity regulates proliferation of NPCs from ASD individuals
Based on the genomic analyses of the ASD individuals, we hypothesized that the alterations in the canonical Wnt pathway might govern the aberrant proliferation in ASD-derived NPCs. To test our hypothesis, we examined Wnt/β-catenin transcriptional activity using TOP-flash assays and found that β-catenin transcriptional activity was indeed reduced in ASD NPCs compared to control NPCs (Figure 2A and Figure S2E). Activation of canonical Wnt signaling with 5 mM LiCl, which inhibits GSK3β-mediated degradation of β-catenin, elevated β-catenin transcriptional activity in both ASD and control NPCs. However, β-catenin transcriptional activity was significantly reduced in LiCl-treated ASD NPCs compared to control NPCs, suggesting that the cause of reduced β-catenin transcriptional activity is downstream of GSK3β. While the elevation of β-catenin transcriptional activity with LiCl treatment in ASD NPCs is modest, LiCl treatment was able to rescue the overproliferation defects on ASD NPCs. Importantly, since the level of Wnt transcriptional activity required for normal proliferation is currently unknown, it is conceivable that a moderate increase of Wnt is sufficient to rescue abnormal over proliferation in ASD. In our experiments, LiCl treatment increased Wnt transcriptional activity by over 2 fold on ASD NPCs and that is potentially sufficient to elicit normal proliferation. Activation of canonical Wnt signaling with Wnt3A resulted in a marked but non-significant elevation of β-catenin transcriptional activity in both control and ASD NPCs, with a similar trend of reduced β-catenin transcriptional activity in the ASD NPCs (Figure 2A and Figure S2E). Notably, NPCs derived from the patient with the heterozygous stop-gain mutation in CTNNB1, Arch, displayed the weakest responses to LiCl and Wnt3a stimulation (Figure S2E). To determine if this reduced β-catenin transcriptional activity was responsible for the growth defects displayed, we repeated the proliferation experiment in ASD-derived and control NPCs in the presence and absence of LiCl. In the absence of LiCl, all ASD-derived NPC lines displayed faster growth (reduced doubling times) than all control NPC lines. LiCl treatment slowed the growth of ASD-derived NPCs close to doubling times of control NPCs, whereas the growth of control NPCs was unaffected (Figure 2B and Figure S2F). These findings indicate that the reduced β-catenin transcriptional activity played a functional role in the accelerated proliferation found in ASD-derived NPCs.
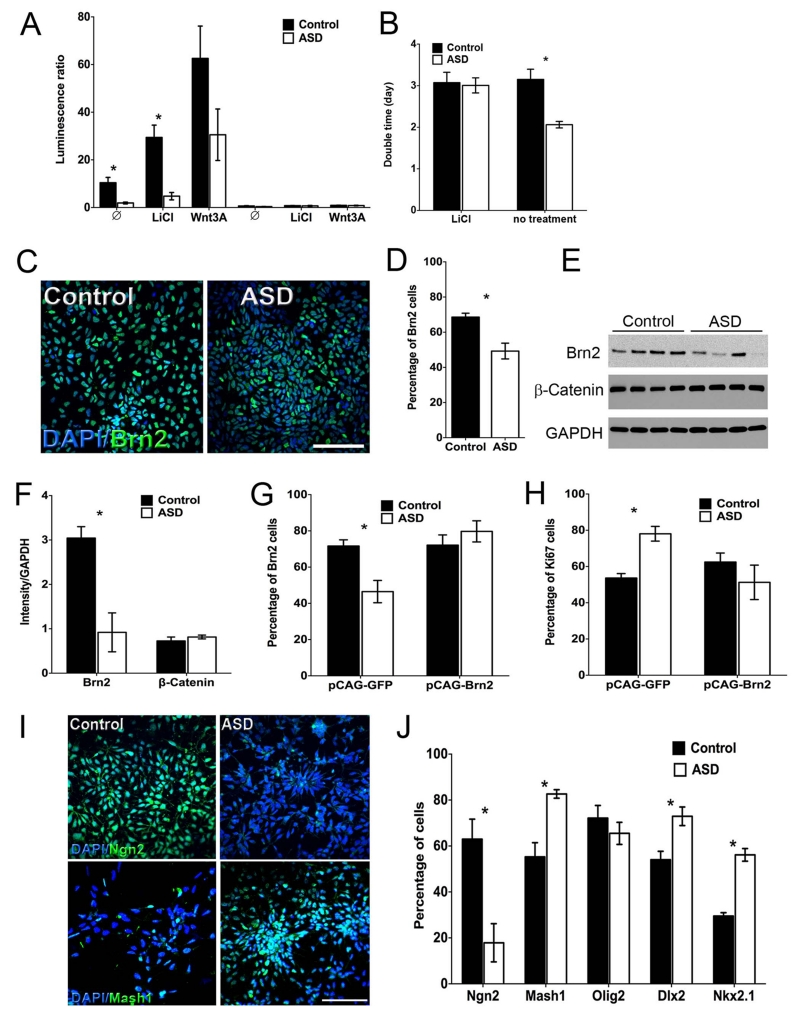
Figure 2.
Regulation of NPC proliferation by the Wnt pathway and differential expression of early fate determinants. (A) Control and ASD NPCs were transfected with TOP-Flash and firefly renilla reporters and treated with either 5 mM LiCl or 100 ng/ml Wnt3A. Results are presented as mean ± SEM (n≥5, *p<0.04 for comparing the luminescence ratio in the ASD NPCs with those of the control NPCs). (B) Doubling time of control and ASD NPCs, treated or not with 5 mM LiCl. Results are presented as mean ± SEM (n≥5, ANOVA<0.05 *p<0.05 for comparing the ASD NPCs doubling time with control NPCs). (C) Control and ASD NPCs were fixed and immunostained for Brn2 and representative images of the staining are shown (Scale bar: 200 μm). (D) Quantification of the percentage of Brn2+-labeled cells is presented as mean±SEM (n≥5; *p<0.001 for comparing the results of the ASD with those of the control NPCs). (E) Representative immunoblot of control and ASD-derived NPCs that were lysed and immunoblot for Brn2, β-catenin and GAPDH. (F) The levels of Brn2 and β-catenin, which were normalized to GAPDH levels were quantified and results are presented as mean ± SEM (n≥5; *p<0.03 for comparing the results of the ASD with those of the control NPCs). (G) The percentage of Brn2+ and (H) Ki67+ cells were measured. Results are presented as mean ± SEM (n≥4, ANOVA<0.02 *p<0.05, comparing the results of the GFP-transfected ASD NPCs with those of the control and the Brn2 transfected ASD NPCs). (I) Representative images of the control and ASD NPCs immunostained for Ngn2 and Mash1. (Scale bar: 200um) (G) Quantification of the percentage of Ngn2+, Mash1+, Olig2, Dlx2+ and Nkx2.1+ labeled cells is presented as mean ± SEM (n≥5; *p<0.03 for comparing the results of the ASD with those of the control NPCs).
Dvl1−/−3+/− mutant mice display reduced social interaction and transient embryonic brain overgrowth, the result of reduced β-catenin transcriptional activity and reduced levels of the transcription factor Brn2 or Pou3f235. To test if BRN2 levels were similarly affected in NPCs from ASD patients with early brain overgrowth, we performed immunocytochemistry for BRN2 on ASD and control NPCs. The percentage of BRN2+ cells was in fact reduced in ASD NPCs compared to controls (Figure 2C, D and Figure S2G), Western blot analysis confirmed that BRN2 protein levels were reduced in ASD NPCs compared to controls (Figure 2E, F and Figure S2H). To examine whether exogenous BRN2 rescued the increased rate of proliferation observed in ASD NPCs, we transfected CAG-BRN2 into control and ASD NPCs and performed immunocytochemistry for BRN2 and Ki67. Transfection efficiency was similar between control and ASD NPCs (around 45%). The number of DAPI+ cells (as an index of proliferation) in ASD NPCs was similar to controls after BRN2 overexpression (Figure 2G, H and Figure S2I, J). The expression pattern of BRN2 and Ki67 in the BRN2 transfected ASD NPCs resembled that of control NPCs, providing additional support for the direct regulation of proliferation of autistic NPCs by BRN2. These data also suggest that partially conserved pathways regulate proliferation of NPCs in a mouse model of social behavior and ASD-derived NPCs from patients with early brain overgrowth (Figure S2K).
ASD-derived neurons display reduced synaptogenesis
Evidence from the literature suggests an imbalance in excitatory versus inhibitory signals in developing ASD36, 37. In addition, analysis of Brn1/Brn2 double mutant mice31, 38 demonstrated that these genes regulate neurogenesis and proliferation in the cerebral cortex of glutamatergic precursor fields. Therefore, we tested whether glutamatergic and GABAergic cell fate determination were affected in ASD NPCs. Indeed, a reduction in the percentage of glutamatergic NGN2+ NPCs in ASD NPCs compared to controls was observed (Figure 2I, J and Figure S2L). In contrast, markers of GABAergic inhibitory precursors present in the subpallium (MASH1, DLX2 and NKX2.1) were up-regulated in ASD compared to control NPCs, whereas OLIG2, another subpallium marker, was unchanged between control and ASD NPCs (Figure 2J and Figure S2L). In light of these findings, we tested whether this change in fate determination had an impact on neurogenesis and synaptogenesis. We did not detect a significant alteration in ASD neuronal survival after differentiation when compared to controls, as measured by MAP2 staining.
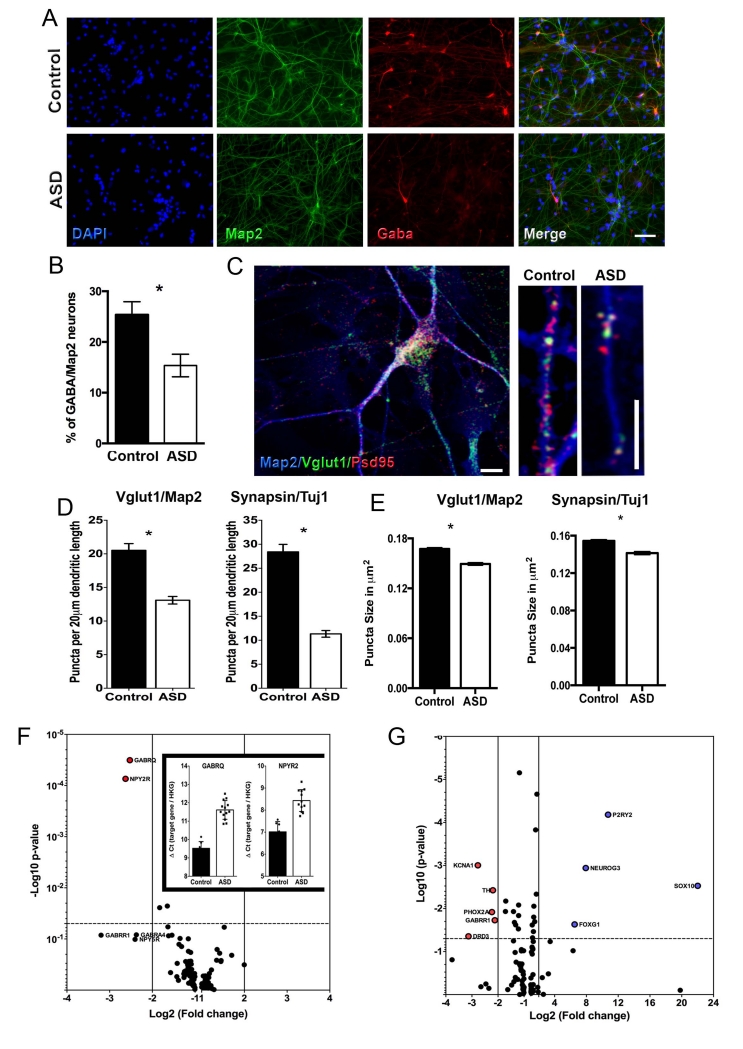
Upon neuronal differentiation of these NPCs, we detected a significant reduction in inhibitory excitatory neurotransmitters GABA (γ-amino butyric acid) ASD neurons compared to controls (Figure 3A, B and S3A) while the percentage of neurons (assessed by Map2 staining) and astrocytes (assessed by GFAP staining) was unchanged (Figure S3 B, C). We also found a clear reduction in the density of both Synapsin and VGLUT1 (vesicular glutamate transpoter-1) puncta from ASD neurons, suggesting a specific defect in vesicular glutamate transport in ASD cultures (Figure 3C-E and Figure S3D-G). To confirm the specificity of glutamatergic neurons in our cultures, we demonstrated that VGLUT1 puncta were mostly adjacent to the postsynaptic density-95 (PSD95) protein, a postsynaptic glutamatergic marker39 (Figure 3C). The distribution of puncta densities for both Synapsin and VGLUT1 displayed a distinctly increased number of ASD neurites with lower densities and an increased number of control-derived neurites with higher densities, corroborating the defect in glutamatergic synapses. We also found significant differences in the area size of synaptic puncta between the 2 groups (Figure S3H, I).
Figure 3.
Synaptic alterations in ASD neuronal networks. (A) Representative images of cells after neuronal differentiation. iPSC-derived neurons express a marker for inhibitory neurons, GABA. Bar = 20 μm. (B) The graph shows the quantification of GABA-positive neurons in all ASD-derived neurons compared to all controls (*p<0.001). (C) Representative images of cells after neuronal differentiation. iPSC-derived neurons express markers for excitatory neurons, such as postsynaptic density protein 95 (Psd95) and vesicular glutamate transporter 1 (VGlut1). (Scale bar: 5 μm) (D) Bar graphs show synaptic density and (E) synaptic puncta size in ASD and control neurons. (**p<0.001 for comparing the results of the ASD with those of the control neurons for (D) and *p<0.05 for (E)). (F and G) Volcano plots of qPCR array data. Plot illustrates differences in expression patterns of neurotransmitters receptor-related genes of iPSC-derived neurons from ASD (n = 13) and related controls (n = 7). The blue-gray plots represent more than or equal to 2.0-fold differentially expressed genes between the groups at P<0.05 (unpaired two-sample Student’s t-test). Insert: Bar graphs show the distribution of ΔCt values (inverse to gene expression) of each individual for GABRQ and NYR2 genes in control and ASD.
A decrease in the number of excitatory synapses may directly affect other neurotransmission systems in ASD cultures40. To investigate whether the expression of neurotransmitters was affected in ASD-derived neurons, we performed a qPCR array of 84 neurotransmitter receptors-related genes in both ASD and control neurons. Interestingly, we detected a tendency for GABA-related and neuropeptide Y receptors to be down-regulated in ASD-derived neurons (Figure 3F and Supplementary Table S8). In this experiment, only the subunits GABRQ and NPY2R were significantly down-regulated in ASD compared to control neurons. We then investigated the expression of 86 different gene indicators for different developmental stages of neural differentiation, neurotransmitters maturation and cortical layer formation using the Fluidigm Biomark qPCR Dynamic Array (Figure 3G and Supplementary Table S8). We detected down-regulation of the GABA receptor GABRR1, plus other relevant neurotransmitters and synapse-related genes, such as KCNA1, TH, PHOX2A and DRD3, in ASD neurons. In contrast, genes related to early stages of neural differentiation, such as NEUROG3, FOXG1, SOX10 and P2RY2, were significantly up-regulated in ASD. Together the results described above indicate that, while ASD neurons initiate an earlier differentiation program, ASD neurons do not mature as extensively as controls.
Network defects in ASD-derived neurons
Next, electrophysiological recordings using single-cell patch clamp were used to investigate the functional properties of iPSC-derived neurons 6 weeks after differentiation. Spontaneous action potentials were observed in control and ASD-derived neurons. However, no difference was observed in the frequency of spontaneous action potentials between iPSC-derived neurons from control (4.3 ± 1.2Hz, n = 21) and ASD (3.0 ± 0.7Hz, n = 27, p = 0.3) cultures. iPSC-derived neurons further exhibited spontaneous excitatory postsynaptic currents (sEPSCs), with comparable amplitudes (p = 0.7) between control (26.5 ± 2.6pA, n = 20) and ASD (27.7 ± 2.6pA, n = 32) cultures. Likewise, no significant difference was found in terms of mean sEPSC frequency (control: 7.6 ± 1.4Hz, ASD: 5.3 ± 1.0Hz, p = 0.2, Figure S4A-D). Despite the lack of significant sEPSC frequency differences between control and ASD neurons, there was a trend of lower frequencies in ASD cultures (Figure S4E). Both control and ASD-derived cells exhibited characteristics of functional neurons, including transient voltage-gated sodium and potassium currents evoked by voltage-step depolarizations and action-potentials generated following depolarizing current injections.
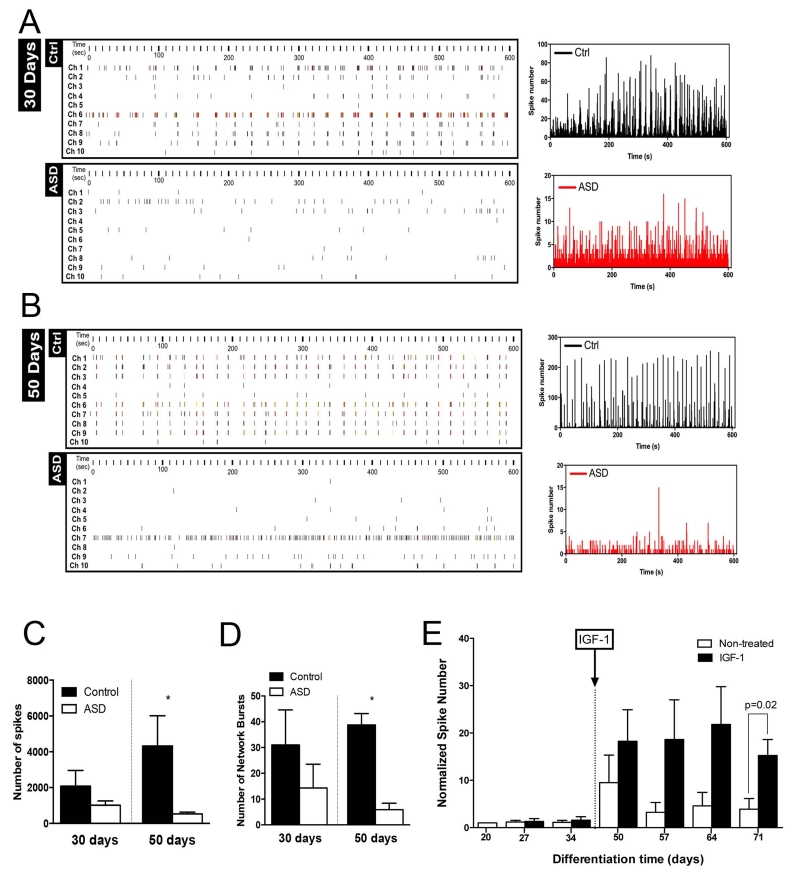
Next, we analyzed spike activity within the neuronal networks by multielectrode arrays (MEA). Connectivity defects have been commonly observed in postmortem ASD brains, but connectivity cannot be studied during early stages of embryogenesis41, 42. Unlike single-cell patch clamp recordings, MEA allow for the dynamic interrogation of how human-derived neurons behave in circuits. We plated paired control and ASD NPCs to mature in MEA dishes and recorded their activity over time. After 30 days, the total number of spikes in control and ASD neuronal networks was similar. As the cultures matured, the number of spikes increased in control neurons but the numbers of spikes in ASD did not, leading to a significant difference between ASD and control networks (Figure 4A-C). After 50 days, we also detected a reduction in the number of synchronized bursts, i.e., spikes that were not singular or random (sequential 10 spikes over 100 ms) in the ASD cultures (Figure 4D). While the number of network bursts was variable at 30 days of maturation, with a tendency toward more synchronized events in control neurons, within 50 days of maturation this difference was 6 times higher in control compared to ASD neurons and highly significant (Figure 4C, D).
Figure 4.
Functional defects in ASD-derived neuronal networks shown by Multielectrode array activity (MEA). Representative spike raster plots of 10 channels from one well plated with control and ASD cultures over (A) 30 days and (B) 50 days. XY graphs on the left show overlay of a representative generated from the raw data of a spike raster plot, using the number of spikes recorded over 10 minutes at 30 days and 50 days of culture maturation (n=3 wells per cell type). (C) Total number of spikes from data obtained from controls (n=6) and ASD (n=10) clonal lines differentiating over 30 days and controls (n=4) and ASD (n=9) clonal lines at 50 days after differentiation over 10 minutes of recording. Results are presented as mean ± SEM (*p=0.0046 for comparing the results of the ASD with control networks). (D) Number of network bursts from wells that were able to generate bursts (10 spikes over 100 ms). Results are presented as mean ± SEM (*p<0.0001 for comparing the results of the ASD with control networks). (E) ASD neuronal cultures were treated with IGF1 as indicated in the graph (arrow) and was kept in the cultures for the duration of the experiment.
We then treated the cells with IGF1, a drug that has been previously shown to rescue synaptic deficits in other ASD monogenetic syndromes such as Rett and Phelan-McDermid Syndromes, and is currently in clinical trials for idiopathic ASD4, 21. When IGF1 was used during the differentiation process, we observed an increase in the number of GABAergic neurons in all ASD-derived cultures, but no effect in controls (Figure S4 F-I). We next used IGF1 to treat mature neuronal cultures after differentiation, starting on day 35 of differentiation. Collectively, ASD patients displayed a tendency to improve neuronal spike number that became significant at day 72 of differentiation (Figure 4E). Interestingly, when we looked at individual patients’ response to the drug, while most patients improved the neuronal activity, Apex did not show any response (Figure S4J). Further investigation should clarify the mechanistic reasons for such drug response variation, but nonetheless, the assay may have the sensitivity to pre-screen patients for future clinical trials. To address the involvement of stabilization of Wnt-β-cat pathway in NPCs and the consequence effect on neuronal function, we treated NPCs with LiCl and assessed the differentiation by quantifying the number of GABAergic neurons after 4 weeks of differentiation and also analyzed neuronal functional maturation using Multi Electrode Array (MEA) technology. Interestingly, treatment of NPCs with LiCl did not improve the percentage of GABA positive cells 4 weeks after differentiation or improved MEA activity on ASD neurons (Figure S4I and S4K). Hence, contrary to IGF1 treatment, LiCl treatment (and consequent stabilization Wnt-β-catenin pathway and rescue of NPC proliferation) does not affect neuronal function significantly, suggesting that there are multiple pathways involved in the ASD phenotypical deficit or that stabilization of the Wnt-β-catenin pathway is also required at the early stages of differentiation35. These results demonstrate an impairment followed by a partial rescue with IGF1, but not LiCl of network formation in ASD-derived neuronal cultures.
RNA sequencing in neural cells identifies altered gene expression in macrencephaly ASD individuals
Strong evidence implicates a convergence of pathways at the transcriptome level in postmortem brains from ASD patients8, 32. Therefore, we performed RNA sequencing analysis in iPSCs and in iPSC-derived NPCs and neurons to evaluate whether any convergent transcriptional alterations could be identified to be associated with early brain overgrowth in ASD during development in this in vitro model. At the transcriptome level, samples were explicitly separated into 3 distinct clusters by cell-type differences based on the top 2 principal components, strongly supporting the reproducibility of our iPSC generation and neural differentiation procedures (Figure S5A, B).
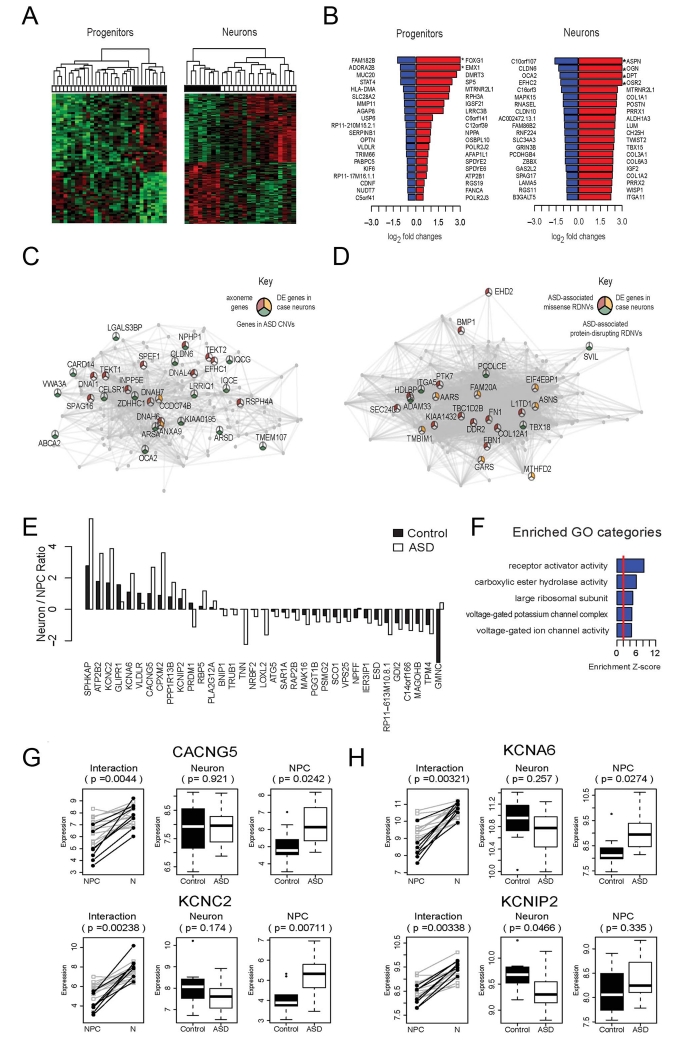
We first conducted a standard differential expression (DE) analysis to evaluate individual gene expression pattern changes at the NPC and neuronal stages. We identified 71 genes that were significantly differentially expressed in ASD compared to controls at the NPC stage and 154 genes at the neuronal stage, which by hierarchical clustering clearly distinguished(the ASD cell lines from controls (Figure 5A and Supplementary Table S9). The expression fold changes of the top dysregulated genes are shown in Figure 5B. Of note, while the expressions of both CTNNB1 (coding for β-catenin) or POU3F2 (coding for BRN2) mRNAs were reduced, neither was significantly altered in ASD-derived NPCs, although β-catenin activity in the canonical Wnt pathway is primarily regulated at the post-transcriptional level. To reveal the biological pathways associated with the differentially expressed genes, we performed GO enrichment analysis. We found that the up-regulated genes in ASD progenitors were significantly enriched for GO category of brain development, which agrees with the observations of the high NPC proliferation rate and differentiation defects in ASD cultures (Figure S5C, D). At the neuronal stage, the up-regulated genes in ASD were enriched for the GO categories related to extracellular matrix, whereas the down-regulated genes were significantly enriched for the GO categories of cilium and axoneme, consistent with the observed synaptic dysregulation (Figure S5E, F).
Figure 5.
Gene expression changes in neurons and NPCs derived from idiopathic ASD Individuals with macrencephaly. (A) Hierarchical clustering using the differentially expressed (DE) genes identified in NPCs (left) and neurons (right). The heatmap shows color-coded scaled expression values, up-regulation in red and down-regulation in green. Each column represents a line from an iPSC clone. The horizontal bar on the top shows disease status: white cell lines are derived from ASD cases, and black represents cell lines from controls. (B) Log2 transformed fold changes of the top 20 down-regulated (blue) and top 20 up-regulated (red) genes in patient neural progenitors (left) and neurons (right) as compared to controls. Genes highlighted with * have log2 fold change > 3. (C) Visualization of the top connections in the brown module, which is down-regulated in ASD neurons. Genes are connected if their pairwise correlation is larger than 0.8. Pie chart: genes in GO category “axoneme” (red); genes in ASD CNVs (yellow); differentially expressed genes (p<0.005)(green). (D) Visualization of the top connections in the tan module, which shows up-regulation in ASD neurons. Genes are connected if their pairwise correlation is larger than 0.8. Pie chart: genes that are previously identified to be affected by ASD-associated missense (red) and protein disrupting (green) rare de novo variation (RDNVs), and differentially expressed genes (p<0.005) (yellow). (E) Barplot showing the ratios of neuronal/progenitor expression of the DE genes in the progenitor to neuron transition. Black bars represent the ratios in control samples, and white bars represent the ratios in ASD samples. (F) The top 5 enriched GO categories among the genes showing differentiation-dependent expression changes in ASD Individuals vs. controls. (G and H) Dynamic expression patterns (log2 transformed read counts) of the genes that show significant differentiation-dependent expression alterations during neuronal differentiation (p<0.005) and that are in the GO category “voltage gated ion channel activity.” White: cell samples derived from ASD individuals; black: cell samples from controls. P-value in the interaction plot shows the significance of the interaction effect between cell type and disease status. P-values in the neuronal and progenitor plots represent the significance of the difference between ASD cases vs. controls.
To explore the network organization of the transcriptome6, 43, 44, we next applied a Weighted Gene Co-expression Network Analysis (WGCNA) to provide a higher order view of the biological processes altered in the patient cells. Given the substantial transcriptomic alterations in the patient-derived neurons, we mainly focused on determining the gene co-expression organizations at the neuronal stage. We identified 17 gene co-expression modules in the assigned co-expression network (each labeled in a different color). We also observed a striking clustering pattern of the case-associated genes and known ASD susceptibility genes (curated from SFARI database), respectively, upon the identified co-expression structure by plotting the module assignment under the gene clustering dendrogram (Figure 5C, D and Figure S5E, F).
Comparing the expression patterns of the modules between ASD and controls (see Methods), we found 4 modules that were significantly associated with ASD status (FDR <0.1): brown, tan, purple, and magenta (Supplementary Table S10). Remarkably, known autism candidate genes were significantly enriched in these autism-correlated modules (Supplementary Table S11). Genes that were commonly affected by ASD-associated CNVs were overrepresented in the brown module (OR=1.5, enrichment p-value=0.02), whereas the tan module was enriched for genes known to be affected by ASD-associated protein-disrupting and missense rare de novo variations (RDNVs) (OR=1.6, p-value=0.02). Because co-expressed genes imply co-regulation, our results provide further evidence for the suggested convergent transcriptional regulation in ASD8.
Since ASD NPCs exhibited abnormal differentiation phenotypes, we evaluated whether there were dynamic expression changes in patient cells upon differentiation. Our analysis identified 35 genes showing significant differences in their expression trajectories between ASD NPCs and controls in the progenitor to neuron transition (p<0.005) (Figure 5E). These genes were significantly enriched for voltage-gated cation channels (Figure 5F), including CACNG5, KCNA6, KCNC2, and KCNIP2. As shown in Figure 5G and H, those channel genes were up-regulated in control cells, but this increase was attenuated in the ASD cells. These data further implicate voltage-gated cation channel dysregulation in ASD neural differentiation, consistent with decreased excitatory glutamatergic synapses, likely affecting synaptic transmission.
DISCUSSION
The use of iPSCs to study genetic disorders is a powerful tool to dissect molecular and cellular pathways implicated in disease pathology during early stages of human neurodevelopment. However, modeling highly complex idiopathic disorders such as ASD is challenging due to a high level of heterogeneity in the patient population. Here, we took advantage of iPSCs derived from a carefully clinically characterized cohort of ASD patients who have an anatomical phenotypic trait that occurs in about 20-30% of idiopathic ASD: an early developmental enlargement of brain volume, including macrencephaly, that is frequently associated with poor prognosis. Recent evidence suggests that abnormal brain growth and organization in ASD are underway by mid-fetal life16, 17 and continue during the first few years of life10, 13, 15, 45-47. The ASD patients recruited here had their brain scans between the ages of 2 and 5 years, confirming the anatomical phenotype. Their brain volumes ranged between 3.3% and 23.5% (median=12%, mean=12.5%) above the normal average for age when compared with large samples of age-matched control subjects10.
We hypothesized that the increased brain volume and neuron numbers found in ASD might result from increased rates of proliferation in neural progenitors. Consistent with this hypothesis, NPCs derived from iPSCs of such autistic patients displayed rapid rates of proliferation when compared to NPCs derived from non-affected typically developing individuals, a finding that also supports the cell cycle dysregulation/proliferation theory proposed to explain early brain overgrowth in some ASD13, 16, 48. Interestingly, all ASD-derived NPCs displayed faster proliferation than all control-derived NPCs. This cellular phenotype results from alterations in a canonical Wnt-β-catenin/BRN2 transcriptional cascade. In further support of this iPSC-derived NPC phenotype, we found that Dvl mutants displayed adult social behavior abnormalities and transient brain overgrowth during embryonic deep layer cortical neuron formation mediated by a similar β-catenin/Brn2/Tbr2 transcriptional cascade35. Transient activation of the canonical Wnt pathway in utero normalized the adult social behavior deficits in Dvl mutant mice. Thus, common transcriptional mechanisms that include β-catenin and BRN2 appear to regulate NPC proliferation in a mouse model with social and repetitive behavioral deficits and a cellular model of human autism. Cross-talk between the Wnt and insulin signaling pathways has been demonstrated in different disorders such as diabetes and cancer49-53. IGF-1 has been shown to stimulate the β-catenin pathway through signaling cascades involving the activation of phosphatidylinositol 3-kinase (PI 3-K)/Akt and the inhibition of glycogen synthase kinase-3 (GSK-3)53. Since LiCl treatment of ASD NPCs stabilized cell proliferation via inhibition of GSK-3 and subsequent increase in β-catenin levels, it is feasible to hypothesize that stimulation of IGF-1 pathway could potentially rescue proliferation deficits in developing ASD NPCs. Future studies will elucidate the mechanistic nature of the relationship between WNT and IGF1 pathways in neurodevelopmental disorders.
Finally, stop-gain mutations were found in the canonical Wnt pathway genes CTNNB1 and FZD6, while a novel missense mutation was found in DVL2, and two more common missense variants listed in dbSNP in FRZB and WNT10B were also detected, consistent with an important role for the canonical Wnt pathway in the pathogenesis of brain enlargement in ASD. It is known that Wnt signaling controls NPCs proliferation and differentiation in a temporal and context-dependent fashion during brain development. Early in neurogenesis, Wnt stimulation results in NPC proliferation, whereas later in development Wnts induce neuronal differentiation by promoting terminal neurogenesis 54-56. Moreover, the same transcription factors may have opposing effects in different cell types. Finally, a missense mutation was found in PTEN (rs181113165) in one patient, consistent with a role for this gene in ASD brain growth trajectory and Wnt signaling33. Taken together, these findings indicate that increased proliferation rates found in NPCs are the likely cause of abnormal prenatal numbers of cortical neurons in the genesis of autism 16.
These proliferation abnormalities were associated with abnormalities in differentiation. The percentage of excitatory NGN2+ NPCs was reduced, whereas markers of inhibitory precursors present in the subpallium (MASH1, DLX2 and NKX2.1) were up-regulated in ASD compared to control NPCs. Interestingly, a decrease of NGN2+ NPC did not resulted in an increase in GABAergic neurons, but ASD neuronal cultures have a significant reduction in inhibitory neurons. When examining excitatory synapses, ASD-derived neurons also displayed decreased Vglut1 puncta density. This imbalance of excitatory/inhibitory neurons could also explain our network results during neuronal maturation in the MEAs, where we detected a significant reduction in the amount of spontaneous activity in ASD neurons, leading to a pronounced deficiency in network connectivity that was ameliorated with addition of IGF1. IGF-1 is a naturally occurring neurotrophic factor essential for brain development and plasticity57, 58. In the CNS, IGF-1 binds to its receptor (IGF1R), and activates MAP Kinase and PI3 kinase pathways, which are both involved in synaptic function and plasticity59, 60. IGF1 treatment of rodent neurons in vitro resulted in an increase in neuronal outgrowth, as well as increased expression of Synapsin1, a pre-synaptic marker responsible for regulating neurotransmitter release, and post-synaptic density protein-95 (PSD-95), a scaffolding protein responsible for proper synapse and receptor formation61, 62. While the precise mechanisms through which IGF1 induces changes in synaptic proteins and improve neuronal function in ASD cultures remain to be elucidated, one possibility is that it acts indirectly on neurons through the activation of IGF1R and its many neurotrophic downstream cascades. Another possibility is that IGF1 contributes to the increase in synaptic markers by acting on glial cells and improving maintenance of synapses. Future studies will reveal and dissect precise pathways downstream of IGF1 during different stages of neuronal development and neuronal circuit maturation in ASD.
The proliferation and maturation defects displayed by ASD patient-derived NPCs and neurons are consistent with early brain overgrowth, but the dramatic in vitro phenotypes are perhaps greater than one would expect based on the pathological and MRI studies of ASD brains so far reported. In support of our data, a recent study with macrocephaly ASD also revealed high level of NPC proliferation in another four ASD individuals, which may be due to a deficiency in FOXG1-mediated pathways63. Thus, it is conceivable that the phenotypes are amplified in vitro due to the lack of tissue architectural constraints that occur during in vivo development. However, it is unclear if these proliferation and differentiation defects result from macrencephaly, ASD or both. The consistency in proliferation and differentiation phenotypes shared among the ASD-patient derived cells in these two studies suggests that these findings are relevant to ASD pathology. Future studies will be needed to clearly distinguish these possibilities; using normocephalic ASD-patient derived NPCs and neurons as well as iPSC models of macrencephaly without ASD.
In summary, we have used a selected group of individuals with mild to extreme macrencephaly to address a specific hypothesis about the origin of ASD: namely, that a specific, early developmental brain enlargement might be generated from an underlying molecular and cellular pathway dysregulation. We employed patient-derived iPSCs to investigate this hypothesis and found that NPCs derived from this cohort displayed increased proliferation and abnormal differentiation, consistent with this hypothesis. Our data support the hypothesis that proliferation of NPCs is tightly regulated during mammalian brain development and that abnormalities of proliferation can lead to a long lasting differentiation abnormalities and defects seen in ASD.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the California Institute for Regenerative Medicine (CIRM) TR2-01814 and TR4-06747, the National Institutes of Health through the NIH Director’s New Innovator Award Program (1-DP2-OD006495-01), an R01 MH100175-01 and U19MH107367 from NIMH, the International Rett Syndrome Foundation (IRSF grant # 2915); a NARSAD Independent Investigator Award to A.R.M., and NIMH Autism Center of Excellence Program Project grant (to E.C., K.P., A.W.-B., and F.H.G.); the work was supported by the Helmsley Trust, the JPB Foundation, the Engmann Foundation, a grant from the CDMRP Autism Research Program (to A.W.-B. and F.H.G.); a KL2 CTRI (KL2TR00099) to T.P and Postdoctoral Translational Fellowship from Autism Speaks to H.B.
Footnotes
The authors declare no conflict of interest.
Supplementary information is available at Molecular Psychiatry’s website.
REFERENCES
- 1.Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. The American journal of psychiatry. 1997;154(2):185–190. doi: 10.1176/ajp.154.2.185. [DOI] [PubMed] [Google Scholar]
- 2.Ronald A, Happe F, Bolton P, Butcher LM, Price TS, Wheelwright S, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45(6):691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 3.Garber K. Neuroscience. Autism’s cause may reside in abnormalities at the synapse. Science. 2007;317(5835):190–191. doi: 10.1126/science.317.5835.190. [DOI] [PubMed] [Google Scholar]
- 4.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow ML, Pramparo T, Winn ME, Barnes CC, Li HR, Weiss L, et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS genetics. 2012;8(3):e1002592. doi: 10.1371/journal.pgen.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155(5):997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courchesne E, Campbell K, Solso S. Brain growth across the life span in autism: age-specific changes in anatomical pathology. Brain Res. 2011;1380:138–145. doi: 10.1016/j.brainres.2010.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courchesne E, Redcay E, Morgan JT, Kennedy DP. Autism at the beginning: microstructural and growth abnormalities underlying the cognitive and behavioral phenotype of autism. Dev Psychopathol. 2005;17(3):577–597. doi: 10.1017/S0954579405050285. [DOI] [PubMed] [Google Scholar]
- 12.Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, et al. Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry. 2011;68(5):467–476. doi: 10.1001/archgenpsychiatry.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57(2):245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- 14.Courchesne E, Carper R, Akshoomoff N. Evidence of brain overgrowth in the first year of life in autism. JAMA : the journal of the American Medical Association. 2003;290(3):337–344. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- 15.Shen MD, Nordahl CW, Young GS, Wootton-Gorges SL, Lee A, Liston SE, et al. Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain : a journal of neurology. 2013;136(Pt 9):2825–2835. doi: 10.1093/brain/awt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306(18):2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 17.Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, et al. Patches of disorganization in the neocortex of children with autism. The New England journal of medicine. 2014;370(13):1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 20.Freitas BC, Trujillo CA, Carromeu C, Yusupova M, Herai RH, Muotri AR. Stem cells and modeling of autism spectrum disorders. Exp Neurol. 2012 doi: 10.1016/j.expneurol.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503(7475):267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17(12):1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltrao-Braga PC, Pignatari GC, Russo FB, Fernandes IR, Muotri AR. In-a-dish: induced pluripotent stem cells as a novel model for human diseases. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2013;83(1):11–17. doi: 10.1002/cyto.a.22231. [DOI] [PubMed] [Google Scholar]
- 24.Griesi-Oliveira K, Acab A, Gupta AR, Sunaga DY, Chailangkarn T, Nicol X, et al. Modeling non-syndromic autism and the impact of TRPC6 disruption in human neurons. Molecular psychiatry. 2014 doi: 10.1038/mp.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gmitrowicz A, Kucharska A. [Developmental disorders in the fourth edition of the American classification: diagnostic and statistical manual of mental disorders (DSM IV -- optional book)] Psychiatria polska. 1994;28(5):509–521. [PubMed] [Google Scholar]
- 26.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 31.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295(5559):1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 32.Voineagu I. Gene expression studies in autism: moving from the genome to the transcriptome and beyond. Neurobiol Dis. 2012;45(1):69–75. doi: 10.1016/j.nbd.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Huang WC, Sejourne J, Clipperton-Allen AE, Page DT. Pten Mutations Alter Brain Growth Trajectory and Allocation of Cell Types through Elevated beta-Catenin Signaling. J Neurosci. 2015;35(28):10252–10267. doi: 10.1523/JNEUROSCI.5272-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernandez IA, Marchetto MC, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83(1):69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belinson H, Nakatani J, Babineau BA, Birnbaum RY, Ellegood J, Bershteyn M, et al. Prenatal beta-catenin/Brn2/Tbr2 transcriptional cascade regulates adult social and stereotypic behaviors. Mol Psychiatry. 2016 doi: 10.1038/mp.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubenstein JL. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr Opin Neurol. 2010;23(2):118–123. doi: 10.1097/WCO.0b013e328336eb13. [DOI] [PubMed] [Google Scholar]
- 37.Zikopoulos B, Barbas H. Altered neural connectivity in excitatory and inhibitory cortical circuits in autism. Frontiers in human neuroscience. 2013;7:609. doi: 10.3389/fnhum.2013.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, et al. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16(14):1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16(7):2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000;407(6801):189–194. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- 41.Hahamy A, Behrmann M, Malach R. The idiosyncratic brain: distortion of spontaneous connectivity patterns in autism spectrum disorder. Nat Neurosci. 2015;18(2):302–309. doi: 10.1038/nn.3919. [DOI] [PubMed] [Google Scholar]
- 42.Davis G, Plaisted-Grant K. Low endogenous neural noise in autism. Autism : the international journal of research and practice. 2014 doi: 10.1177/1362361314552198. [DOI] [PubMed] [Google Scholar]
- 43.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462(7270):213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carper RA, Moses P, Tigue ZD, Courchesne E. Cerebral lobes in autism: early hyperplasia and abnormal age effects. NeuroImage. 2002;16(4):1038–1051. doi: 10.1006/nimg.2002.1099. [DOI] [PubMed] [Google Scholar]
- 46.Sparks BF, Friedman SD, Shaw DW, Aylward EH, Echelard D, Artru AA, et al. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59(2):184–192. doi: 10.1212/wnl.59.2.184. [DOI] [PubMed] [Google Scholar]
- 47.Hazlett HC, Poe M, Gerig G, Smith RG, Provenzale J, Ross A, et al. Magnetic resonance imaging and head circumference study of brain size in autism: birth through age 2 years. Arch Gen Psychiatry. 2005;62(12):1366–1376. doi: 10.1001/archpsyc.62.12.1366. [DOI] [PubMed] [Google Scholar]
- 48.Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, et al. Mapping early brain development in autism. Neuron. 2007;56(2):399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Jimenez C, Garcia-Martinez JM, Chocarro-Calvo A, De la Vieja A. A new link between diabetes and cancer: enhanced WNT/beta-catenin signaling by high glucose. Journal of molecular endocrinology. 2014;52(1):R51–66. doi: 10.1530/JME-13-0152. [DOI] [PubMed] [Google Scholar]
- 50.Palsgaard J, Emanuelli B, Winnay JN, Sumara G, Karsenty G, Kahn CR. Cross-talk between insulin and Wnt signaling in preadipocytes: role of Wnt co-receptor low density lipoprotein receptor-related protein-5 (LRP5) J Biol Chem. 2012;287(15):12016–12026. doi: 10.1074/jbc.M111.337048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116(Pt 7):1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desbois-Mouthon C, Cadoret A, Blivet-Van Eggelpoel MJ, Bertrand F, Cherqui G, Perret C, et al. Insulin and IGF-1 stimulate the beta-catenin pathway through two signalling cascades involving GSK-3beta inhibition and Ras activation. Oncogene. 2001;20(2):252–259. doi: 10.1038/sj.onc.1204064. [DOI] [PubMed] [Google Scholar]
- 53.Siddle K. Signalling by insulin and IGF receptors: supporting acts and new players. Journal of molecular endocrinology. 2011;47(1):R1–10. doi: 10.1530/JME-11-0022. [DOI] [PubMed] [Google Scholar]
- 54.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297(5580):365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 55.Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131(12):2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 56.Munji RN, Choe Y, Li G, Siegenthaler JA, Pleasure SJ. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J Neurosci. 2011;31(5):1676–1687. doi: 10.1523/JNEUROSCI.5404-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng CM, Reinhardt RR, Lee WH, Joncas G, Patel SC, Bondy CA. Insulin-like growth factor 1 regulates developing brain glucose metabolism. Proc Natl Acad Sci U S A. 2000;97(18):10236–10241. doi: 10.1073/pnas.170008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciucci F, Putignano E, Baroncelli L, Landi S, Berardi N, Maffei L. Insulin-like growth factor 1 (IGF-1) mediates the effects of enriched environment (EE) on visual cortical development. PLoS One. 2007;2(5):e475. doi: 10.1371/journal.pone.0000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshii A, Constantine-Paton M. BDNF induces transport of PSD-95 to dendrites through PI3K-AKT signaling after NMDA receptor activation. Nat Neurosci. 2007;10(6):702–711. doi: 10.1038/nn1903. [DOI] [PubMed] [Google Scholar]
- 60.Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. J Neurochem. 2001;76(1):1–10. doi: 10.1046/j.1471-4159.2001.00054.x. [DOI] [PubMed] [Google Scholar]
- 61.Ozdinler PH, Macklis JD. IGF-I specifically enhances axon outgrowth of corticospinal motor neurons. Nat Neurosci. 2006;9(11):1371–1381. doi: 10.1038/nn1789. [DOI] [PubMed] [Google Scholar]
- 62.Corvin AP, Molinos I, Little G, Donohoe G, Gill M, Morris DW, et al. Insulin-like growth factor 1 (IGF1) and its active peptide (1-3)IGF1 enhance the expression of synaptic markers in neuronal circuits through different cellular mechanisms. Neurosci Lett. 2012;520(1):51–56. doi: 10.1016/j.neulet.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 63.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162(2):375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.