Abstract
This paper focuses on the problem of feature extraction and the classification of microvascular morphological types to aid esophageal cancer detection. We present a patch-based system with a hybrid SVM model with data augmentation for intraepithelial papillary capillary loop recognition. A greedy patch-generating algorithm and a specialized CNN named NBI-Net are designed to extract hierarchical features from patches. We investigate a series of data augmentation techniques to progressively improve the prediction invariance of image scaling and rotation. For classifier boosting, SVM is used as an alternative to softmax to enhance generalization ability. The effectiveness of CNN feature representation ability is discussed for a set of widely used CNN models, including AlexNet, VGG-16, and GoogLeNet. Experiments are conducted on the NBI-ME dataset. The recognition rate is up to 92.74% on the patch level with data augmentation and classifier boosting. The results show that the combined CNN-SVM model beats models of traditional features with SVM as well as the original CNN with softmax. The synthesis results indicate that our system is able to assist clinical diagnosis to a certain extent.
Keywords: Microvascular type classification, Feature representation, Convolutional neural network, Support vector machine (SVM), Data augmentation
Introduction
Feature design for image recognition has been studied for decades. Powerful features, such as local binary pattern (LBP) [1], scale-invariant feature transform (SIFT) [2], speeded up robust features (SURF) [3], and histograms of oriented gradients (HOG) [4], have been proposed to promote the development of classical computer vision and pattern recognition tasks. However, these traditional handcrafted features are unsatisfactory for distinctive tasks, especially medical image processing.
Recently, deep learning using convolution neural networks (CNNs) has gained much success in visual recognition tasks such as image classification and object detection [5–8]. Since the descriptors acquired from these neural networks (e.g., AlexNet [5] and OverFeat [6]) are quite powerful, it is popular to treat these CNNs, trained on large natural image datasets (e.g., ImageNet [9]), as generic feature extractors. By reusing the knowledge gained from past related tasks [10–12], it is now much easier to tackle more challenging tasks such as image retrieval [5], semantic segmentation [13], fine grained recognition [14], and emotion recognition [15].
Support vector machine (SVM) is popular for classification, particularly for medical signal processing [16–18]. For recognition, great attention has been paid to the fusion of neural networks and SVM. The benefits of their combination have been confirmed by prior works on pedestrian detection [19], face recognition [20], and handwritten digit recognition [21]. Razavian et al. [14] use an off-the-shelf CNN representation with linear SVM to address recognition tasks. The results suggest it to be a strong competitor to more sophisticated and highly tuned state-of-the-art methods on various datasets.
Another way to advance recognition is called data augmentation [22], where transformations such as deformation and translation [5] have led to significant improvements in prediction accuracy and system robustness.
This paper focuses on the recognition of intraepithelial papillary capillary loops (IPCLs), a kind of esophageal microvessel, whose types are closely related to the depth of tumor invasion of esophageal squamous cell carcinoma. While the recognition results are meaningful for cancer detection and treatment [23], the task suffers from challenges such as inter-class similarity, intra-class variety, and data imbalance. Hence, efficient feature representations, strong classifiers are desired.
In this paper, we propose a CNN-SVM model for the recognition of IPCLs. The model is tested on the NBI-ME dataset (Sect. 2.2). The key idea of our method is to train a specialized CNN called NBI-Net to extract robust hierarchical features from image patches and provide them to SVM classifiers. We also investigate the feature representation ability of deep models and examine the characteristics of narrowband imaging (NBI) images through data augmentation. Comparisons with traditional feature extractors and a plain CNN model show that the proposed model outperforms them.
Related Work
With poor prognosis when diagnosed at an advanced stage, esophageal cancer ranks as the sixth most common cause of cancer-related death [23].
NBI is a technology that enhances vessel imaging based on the spectral absorption of hemoglobin. Recent developments in narrowband imaging with magnified endoscopy (NBI-ME) and medical image processing technologies allow clear visualization of the esophageal microvascular structure, facilitating cancer detection in the early stage [24, 25].
IPCL Type Definition
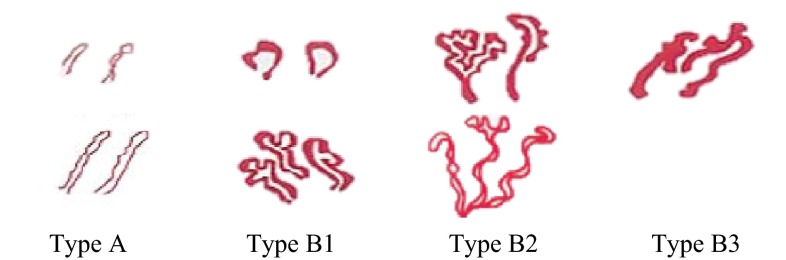
In clinical practice, IPCLs are observed as brown loops on NBI-ME images. Their types demonstrate characteristic morphological changes (Fig. 1) according to the cancer infiltration. The microvessel types are classified into four classes according to the magnified endoscopy diagnostic criteria for esophageal cancer proposed by the Japan Esophageal Society [26] and researchers [24]. The types, illustrated in Fig. 2, are defined as follows:
Type A normal vessels, or vessels with slight dilation and tortuosity
Type B1 dilated and tortuous vessels of various diameters and shapes and with intact loop formation
Type B2 irregularly and dendritically branched vessels with no loop formation
Type B3 obviously thicker vessels than surrounding ones
Fig. 1.
Morphological changes of vessels [23]
Fig. 2.
Illustration of typical IPCL types [26]
Image Acquisition and Annotation
Our NBI-ME dataset contains 261 full-size images of 67 patient cases captured from January 2013 to February 2015 at the Department of Gastroenterology, the First Affiliated Hospital of Anhui Medical University. Confirmed by biopsy of esophagectomy specimens, image regions were manually annotated after collection. Besides giving a scalar label type, label curves were carefully drawn on each original full-size image.
Task Challenges
Data problems are sometimes the bottleneck in a pattern recognition system. For NBI image acquisition, non-uniform illumination and camera noise result in a reduction of image quality. In addition, the magnification of NBI images changes with the distance from the tissue to the camera lens. Thus, parts of raw images must be discarded due to image distortion.
The classification task suffers from inter-class similarity and intra-class variety (Fig. 3). Medically, tumors progress gradually and continuously from low to high grade. However, IPCLs are factitiously classified into four discrete types so that it is sometimes difficult to distinguish two adjacent types. The texture pattern of IPCLs varies from case to case, leading to highly intra-class variety.
Fig. 3.
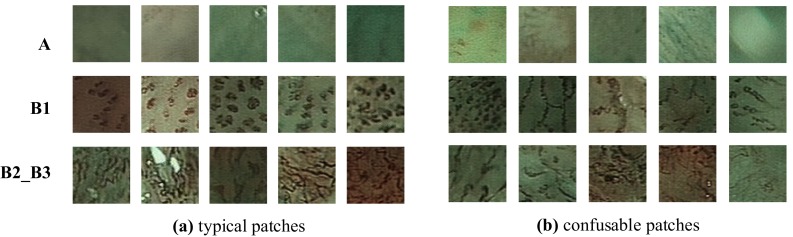
a Typical and b confusable patches
In addition, data imbalance is a problem for model optimization. For instance, a class with fewer training samples is easily belittled when the optimal target is minimizing the training error of the whole dataset. As lesions for B2 and B3 are much fewer than those for A and B1, to mitigate data imbalance, we simplified the problem to a three-class (A, B1, and B2_B3) classification task, on condition that types B2 and B3 have much in common.
Conventional Features
According to clinical experience, texture and shape play crucial roles in microvascular morphological type recognition. Conventional features, namely pyramid histogram of words (PHOW) [27], LBP, and pyramid histogram of oriented gradient (PHOG), are used in our experiments.
PHOW, an effective texture feature, is a combination of SIFT and the bag of words model. Variants of dense SIFT descriptors, extracted at multiple scales, are clustered into visual words and the histograms of these words are treated as descriptions of images. PHOG is similar but describes shape. LBP is a set of local descriptors that capture the appearance of an image cell (a small neighborhood around a pixel), recording local pixel intensity difference.
Support Vector Machine
SVM was originally proposed for binary classification. Supposing a training set , where feature vector and label scalar , the soft margin SVM tries to find a hyperplane that satisfies the following constrained optimization:
| 1 |
| 2 |
where is the weight vector for , is the intercept of the hyperplane, vector contains the slack variables, and is the adjustable penalty parameter controlling the trade-off between the maximization of the margin and the minimization of the classification error.
By importing a kernel function, SVM is able to solve a nonlinear separable problem by transforming the feature vector into a high-dimensional space.
Architecture
We want to design a data-adaptive and customer-friendly system to aid clinical diagnosis. For real-time recognition, a batch processing program is required to train and test images parallelly at a constrained time cost. Advanced CNNs, sped up by a GPU, are an excellent match for this job.
The flow diagram of our system is shown in Fig. 4. In our system, image patches are generated from marked regions for the CNN by a greedy patch-generating algorithm (GPGA) and the locations of all patches are recorded for further synthesis via Gaussian-weighted voting. The CNN functions as a trainable feature extractor and the SVM acts as a predictor.
Fig. 4.
Flow diagram of proposed system
Greedy Patch-Generating Algorithm
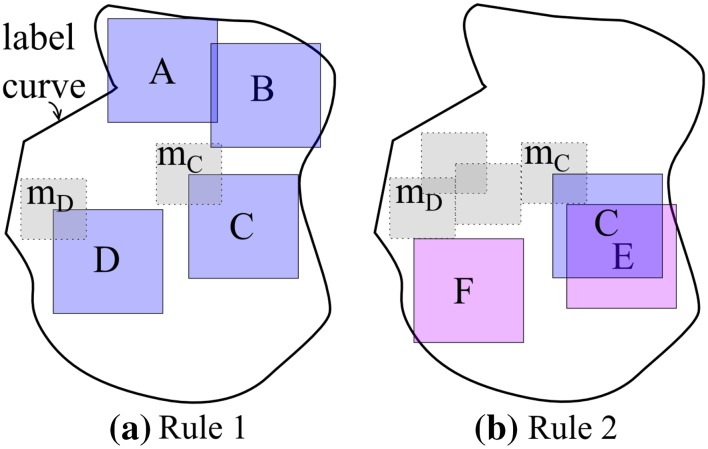
Since fixed-size image patches are required by the CNN, we employ a GPGA to take advantage of marked regions following these two rules:
Rule 1, most (≥95%) of a patch must be bounded within the label curve, e.g., patches A and B in Fig. 5a
Rule 2, the area-overlapping-percentage of any two patches should be exempted from too high (≤75%), e.g., patches C and E in Fig. 5b.
Fig. 5.
Schematic of greedy patch-generating algorithm
Rule 2 can be achieved by invalidating a smaller gray area (e.g., m C for patch C, m D for patch D) around the upper-left point of previously generated patches. These gray areas are then marked unreachable for new patches.
Convolutional Neural Network
We build our network following the popular three-stage designs (i.e., convolutional layers, pooling layers, and fully connected layers) with modifications, including rectified linear units (ReLU), local response normalization (LRN), and dropout, to prevent overfitting.
The input layer size is one of the most important parameters when building a CNN. For example, the typical 5-layer LeNet-5 [28] handles 28 × 28 images and the 8-layer AlexNet takes images of size 256 × 256. A deeper and wider network is expected to learn richer hierarchical features, but needs a larger number of training samples and more iterations for fine-tuning. Limited by the amount of data,1 it is difficult to train and tune a large network.
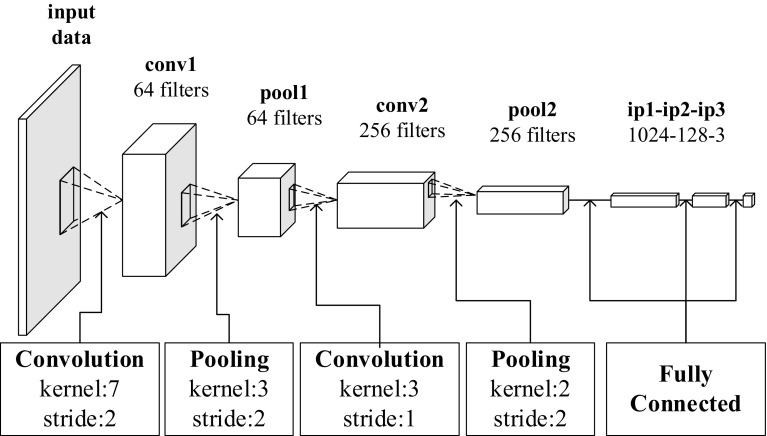
By taking account of our patch size and sample magnitude (see Sect. 4 for details), we started from a five-layer model. Inspired by AlexNet and GoogLeNet [8], we increased the kernel size of the first convolutional layer (conv1) from 5 to 7 to widen the filter “sight” and placed a max pooling layer (pool1) with a size of = and stride = 2. A larger kernel also could speed up training and decrease CNN depth. We set and obtained an overlapping pooling layer to reduce overfitting. Smaller convolutional and pooling kernel sizes were chosen, as features of higher abstraction would be captured by the second convolutional layer (conv2). The last three layers were 1024-d, 128-d, and 3-d fc layers (fc1 to fc3), with softmax as the output function. The width of each layer was tuned according to the system demand analysis. The architecture of our CNN and the blob shape before fc layers are summarized in Fig. 6 and Table 1.
Fig. 6.
Architecture of proposed NBI-Net
Table 1.
Blob shapes of proposed NBI-Net model
| Input data | conv1 | pool1 | conv2 | pool2 | |
|---|---|---|---|---|---|
| Default | 3 × 64 × 64 | 64 × 29 × 29 | 64 × 14 × 14 | 256 × 12 × 12 | 256 × 6 × 6 |
| Cropped | 3 × 56 × 56 | 64 × 25 × 25 | 64 × 12 × 12 | 256 × 10 × 10 | 256 × 5 × 5 |
Layers conv1, conv2, ip1, and ip2 were equipped with ReLU [29] to avoid gradient vanishing and speed up convergence. In addition, we used dropout in the fully connected layers, with a dropout probability of 0.5, to help prevent units from co-adapting and generate more robust features by learning on different random subsets [5, 30].
Our system was built with Caffe [31], a powerful deep learning framework developed by the Berkeley Vision and Learning Center (BVLC), with the NVIDIA CUDA cuDNN [32] library and trained on a single NVIDIA GPU.
Experiments
We selected a patch width of 64 as a trade-off between accuracy and data proportion. About 6.5 k patch samples were generated using GPGA. The performance was measured using average precision (AP) on the patch level.
In addition to CNN, we implemented extractors of PHOW, LBP, and PHOG. Raw RGB patches were directly fed to each feature extractor. No pre-processing method was applied except mean component removal in CNN.
When applying the cross-validation strategy for testing model performance, we sliced the dataset on the case level rather than the patch level because images from the same patient case were similar in texture pattern and patches were generated with an overlap.2 The classification accuracy in table was the aggregated accuracy of all folds.
The time cost depended on the dataset and model complexity, and could be sensitive to programming and hardware. In this work, we trained our models on NVIDIA GPUs (GTX 980 and GTX 970). During the CNN training stage, we used an initial learning rate of 0.01 or 0.005 and scheduled two tenfold decreases for fine-tuning. The models were iterated with 50 epochs (about 30,000 gradient steps). Most models finished in several minutes, but the largest one took about 2 h due to data augmentation.
A GPU-accelerated SVM [33, 34] has been suggested to deal with high-dimensional feature vectors. Principal component analysis (PCA) is optional and should be carefully used for dimensionality reduction.
In Sect. 4.1, we examine the characteristics and distribution of the NBI dataset and improve the precision of NBI-Net via data augmentation. The CNN representation ability analysis and performance comparison of models are respectively presented in Sects. 4.2 and 4.3.
Data Augmentation
To be robust, a model for pattern recognition should make predictions that are invariant to various inputs of a given label. A straightforward approach is to collect a large number of training samples with abundant variation, regardless of the difficulties in data collection and labeling.
Another way to deal with this problem is data augmentation, which is achieved by adding sample replicas with label preservation. Various kinds of affine transform may take effect depending on the characteristics and distribution of a dataset. We apply rescaling, rotation, and flipping on the full-size images and used Caffe embedded cropping for patches. We prioritized operation on the image level over that on the patch level because operation on the image level could afford more randomness of data augmentation benefited from our GPGA.
Rescaling
The NBI images were acquired on different scale factors. The non-normalized scale factor may confuse our model and weaken generalization ability. For rescaling, as shown in Fig. 7a, images were rescaled on a spatial pyramid using:
| 3 |
where is the length of the side for mode . The factor was set to from experience.
Fig. 7.
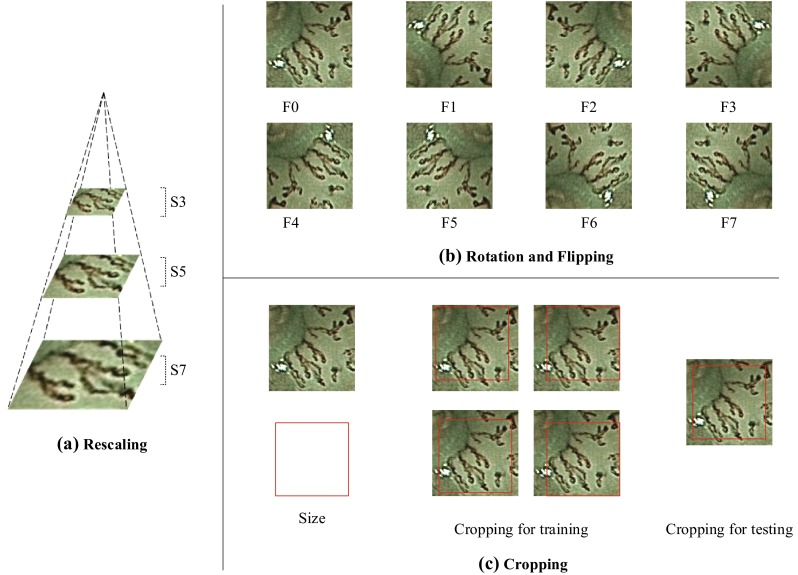
Illustration of data augmentation with a rescaling, b rotation and flipping, and c cropping
Rotation and Flipping
Unlike natural images (e.g., images of human faces or buildings), there is no clear principal direction of NBI images. Rotation and flipping were introduced to strengthen rotational invariance. In this work, the dataset was augmented via roughly rotating each image by every 90 degree and doubled by flipping. This produced eight modes, as shown in Fig. 7b.
Cropping
Cropping was implemented by randomly shifting on both horizontal and vertical lines in the training stage. For testing, patches were cropped at the center, as shown in Fig. 7c. The length proportion of a cropping was 56/64 for NBI-Net, 227/256 for AlexNet, and 224/265 for VGG-16 or GoogLeNet.
Table 2 shows the configurations and relations of models A-I. Model A, NBI-Net without any data augmentation, acted as the baseline in this test. The steady increase in the recognition rates of models A, G, and H shows that rotation and flipping operations were effective. Unlike rotation and flipping, which have high priority, the scaling operations had to be picked carefully, as models A-F show. While the robustness of CNN was improved with rescaling modes S4 and S6, modes S3 and S7 seem to introduce a lot of noise, which confused models E and F, resulting in a decrease in accuracy. The intensity of the rescaling should be gradually increased. For all NBI-Net models, cropping did not improve accuracy, as it did for models trained on ImageNet, such as AlexNet (see Table 3). Although cropping reduces precision, it may prevent overfitting in the visualization of model losses.
Table 2.
Configurations of datasets under augmentation and CNN average precision
| NBI-Net model | Rescaling mode | Rotation/flipping mode | Data complexity | AP w/o cropping | Compared to base | AP w/cropping | Compared to base |
|---|---|---|---|---|---|---|---|
| S5 | F0 | 1 | 90.03 | (Base) | 89.42 | −0.61 | |
| B | S4–S5 | F0 | ×1.3 | 90.28 | 0.25 | 89.63 | −0.40 |
| C | S5–S6 | F0 | ×3.6 | 91.45 | 1.42 | 89.45 | −0.58 |
| D | S4–S6 | F0 | ×3.9 | 91.87 | 1.84 | 90.62 | 0.59 |
| E | S4–S7 | F0 | ×10.2 | 90.43 | 0.40 | 88.69 | −1.34 |
| F | S3–S6 | F0 | ×3.9 | 91.65 | 1.62 | 90.13 | 0.10 |
| G | S5 | [F0–F7]/2 | ×4 | 91.42 | 1.39 | 90.11 | 0.08 |
| H | S5 | F0–F7 | ×8 | 91.80 | 1.77 | 90.00 | −0.03 |
| I | S4–S6 | F0–F7 | ×3.9 × 8 | 92.31 | 2.28 | 91.24 | 1.21 |
“[]/2” represents a random 50% downsampling
The top results have been styled with bold and italic
The best results of comparative group are styled with italic
Table 3.
Average precision of CNN models with linear SVMs
| Model | Feature layer | Avg | ||
|---|---|---|---|---|
| fc1 | fc2 | fc3 | ||
| AlexNet | 87.14 | 88.48 | 87.29 | 87.64 |
| AlexNet Cropping | 87.91 | 88.93 | 87.99 | 88.28 |
| VGG-16 | 89.20 | 90.02 | 89.65 | 89.62 |
| VGG-16 Cropping | 89.88 | 91.67 | 90.68 | 90.74 |
| GoogLeNet | 85.62 | 86.20 | – | 85.91 |
| GoogLeNet Cropping | 87.57 | 86.83 | – | 87.20 |
| NBI-Net (model A) | 90.93 | 90.64 | 91.21 | 90.93 |
| NBI-Net Aug (model I) | 92.87 | 92.38 | 92.97 | 92.74 |
Layer “fc1” is first fully connected layer before softmax or layer before softmax1 in GoogLeNet, or named “fc6” in AlexNet
Principal component analysis (PCA) here is optional and should be carefully used for dimensionality reduction
The top results have been styled with bold and italic
The best results of comparative group are styled with italic
Model I without cropping (NBI-Net Aug), which is a combination of models D and H, was best, improving accuracy by 2.28% against the baseline.
CNN Feature Descriptor Analysis
The ImageNet Large Scale Visual Recognition Challenge (ILSVRC) has become the standard benchmark for object recognition. It contains millions of images belonging to thousands of object categories. State-of-the-art models AlexNet, VGG-16, and GoogLeNet are all top winners of this challenge. While models trained on ImageNet can be transferred well to natural image sets such as Caltech-101, there is currently no clear understanding of how they will do on medical image sets.
The ImageNet dataset consists of thousands of object classes, with only a small number resembling an NBI scene. A comparison was made between pre-trained models and our NBI-Net trained on the NBI image set. For testing, NBI patches were fed to CNN feature extractors. Since features from fully connected layers are more discriminative than those from convolutional layers [12], linear SVMs were applied after fully connected layers.
According to Table 3, all CNN models have a recognition rate of at least 85%. This indicates that stacked convolutional networks show obvious adaptability to feature descriptions.
The accuracy of VGG-16 Cropping, the best performing generic model trained on ImageNet, is close to that of NBI-Net trained on the NBI dataset. The results imply a similarity between natural and medical images in terms of basic feature representation. Starting from a pre-trained model for IPCL recognition is thus worthy of consideration.
Model Comparison
Here, we only compare models; feature fusion and model ensembles are beyond the scope of this study.
Some key parameters of the experiments are as follows. The PHOW feature was extracted from the 2-level spatial pyramid with 200 visual words. The PHOG feature was obtained from the 4-level pyramid with 8 angle bins. Uniform-58 LBP was applied to each cell of size . For the CNN, we took the output of fully connected layers ip1, ip2, and ip3. Both linear SVM and SVM with a radial basis function (RBF) kernel were used in the experiments except for the original CNN (CNN-softmax group). For multiclass problems, we adopted the one-against-one strategy for SVMs. The class that received the most votes won.
In Table 4, the results show that CNN models with linear SVM are equivalent to CNN models with SVM with the RBF kernel, which means that CNN features are almost linearly separable whereas conventional features are not. Linear SVM was used for further study in consideration of algorithm complexity.
Table 4.
Average precision of models using SVM and dimension of features
| Feature extractor | Average precision | Average precision | Feature dimension |
|---|---|---|---|
| (Linear SVM) | (SVM with RBF kernel) | ||
| PHOW | 80.24 | 85.16 | 4000 |
| LBP | 82.39 | 82.90 | 928 |
| PHOG | 49.04 | 62.60 | 680 |
| AlexNet Cropping | 88.28 | 88.12 | 4096/4096/3 |
| VGG-16 Cropping | 90.74 | 90.49 | 4096/4096/3 |
| GoogLeNet Cropping | 87.20 | 87.33 | 1000/1000 |
| NBI-Net Aug | 92.74 | 92.70 | 1024/128/3 |
The top results have been styled with bold and italic
The best results of comparative group are styled with italic
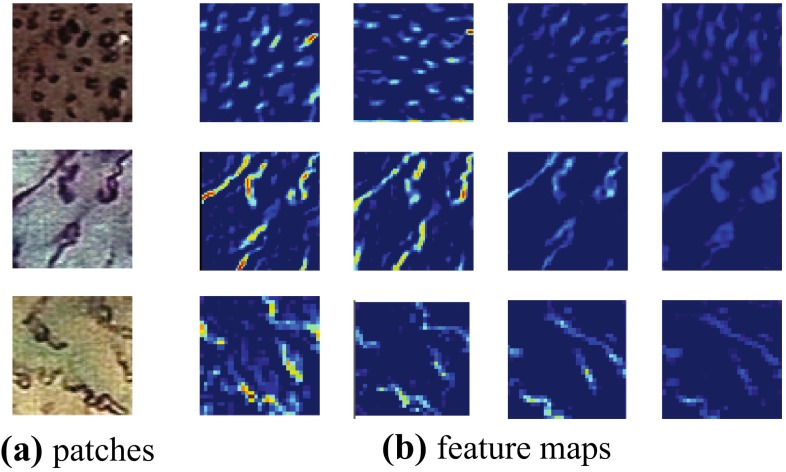
A comparison among rows shows that CNN features significantly outperform conventional handcrafted features. It can also be verified from the visualization of middle-layer feature maps (shown in Fig. 8) that CNN features fit the “disorganized” IPCL pattern very well, which is difficult to achieve using manual design.
Fig. 8.
a Patches and b their feature maps from conv-1 layer of NBI-Net
A comparison between hybrid CNN-SVM (NBI-Net with linear SVM) and plain CNN (NBI-Net with softmax) is shown in Table 5. The fusion of CNN and SVM slightly boosted accuracy by 0.90% (0.43% with Aug). The gain is mainly due to the use of a different optimization criterion. The learning algorithm of softmax is based on empirical risk minimization, which attempts to minimize the prediction loss on the training set. In contrast, SVM aims to minimize the generalization error by using structural risk minimization principles for the testing set. As a result of a maximized margin, the generalization ability of SVM is greater than that of softmax.
Table 5.
Accuracy of NBI-Net models with softmax and linear SVM
| Model | Softmax | Linear SVM | Classifier boosting |
|---|---|---|---|
| NBI-Net | 90.03 | 90.93 | 0.90 |
| NBI-Net Aug | 92.31 | 92.74 | 0.43 |
CNN = NBI-Net with softmax
CNN-SVM = NBI-Net with linear SVM
Aug (Augmentation) is optional for both CNN and CNN-SVM
The top results have been styled with bold and italic
A combination of data augmentation and classifier boosting improved accuracy by 2.71% (=92.74% − 90.03%) and led to a high recognition rate of 92.74%. Figure 9 shows the synthesis results and the original mark curves; our system offers correct prediction in most regions. Thus, our system may be able to assist clinical judgement to a certain extent.
Fig. 9.
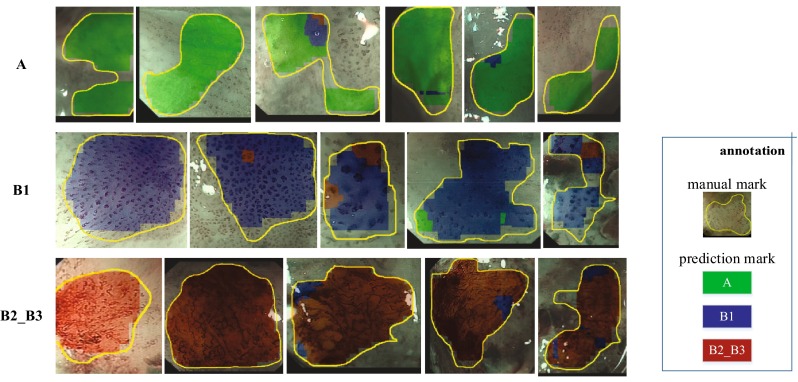
Synthesis results for full-size images. Representative images of labels A, B1, and B2_B3 are placed in first, second, and third rows, respectively. Recognized IPCL regions are colored in green/blue/red for types A/B1/B2_B3
Conclusion and Future Work
In this paper, a patch-based system with a hybrid CNN-SVM model was proposed for IPCL recognition to aid clinical diagnosis. The performance of the CNN model was improved by data augmentation and classifier boosting. Experimental results show that features learned by the CNN beat manually designed features in terms of efficiency and linear separability. A switch from softmax to SVM appears to be beneficial for generalization ability.
For future work, the IPCL recognition precision will be further increased by using a larger dataset, better understanding of IPCLs types, and the fine-tuning of pre-trained CNN models.
Footnotes
Although a data augmentation can help increase the number of patches, the data distribution is still limited.
It may be cheating to distribute patches from the same case or the same full-size image to the training set and testing set concurrently.
References
- 1.Ojala T, Pietikäinen M, Harwood D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognition. 1996;29(1):51–59. doi: 10.1016/0031-3203(95)00067-4. [DOI] [Google Scholar]
- 2.Lowe DG. Distinctive image features from scale-invariant keypoints. International Journal of Computer Vision. 2004;60(2):91–110. doi: 10.1023/B:VISI.0000029664.99615.94. [DOI] [Google Scholar]
- 3.Bay H, Ess A, Tuytelaars T, Van Gool L. Speeded-up robust features (SURF) Computer Vision and Image Understanding. 2008;110(3):346–359. doi: 10.1016/j.cviu.2007.09.014. [DOI] [Google Scholar]
- 4.Dalal, N., & Triggs, B. (2005). Histograms of oriented gradients for human detection. In 2005 IEEE computer society conference on computer vision and pattern recognition, vol. 1, proceedings, pp. 886–893.
- 5.Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems, 2012, pp. 1097–1105.
- 6.Sermanet, P., Eigen, D., Zhang, X., Mathieu, M., Fergus, R., & LeCun, Y. (2013). Overfeat: Integrated recognition, localization and detection using convolutional networks. arXiv preprint arXiv:1312.6229.
- 7.Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
- 8.Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., et al. Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 1–9.
- 9.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. Imagenet large scale visual recognition challenge. International Journal of Computer Vision. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 10.Pan SJ, Yang Q. A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering. 2010;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 11.Yang L, Hanneke S, Carbonell J. A theory of transfer learning with applications to active learning. Machine Learning. 2013;90(2):161–189. doi: 10.1007/s10994-012-5310-y. [DOI] [Google Scholar]
- 12.Zeiler, M. D., & Fergus, R. (2014). Visualizing and understanding convolutional networks. In European Conference on Computer Vision, 2014 (pp. 818–833). Springer.
- 13.Girshick, R., Donahue, J., Darrell, T., & Malik, J. (2014). Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2014, pp. 580–587.
- 14.Razavian, A., Azizpour, H., Sullivan, J., & Carlsson, S. CNN features off-the-shelf: An astounding baseline for recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition workshops, 2014, pp. 806–813.
- 15.Ng, H.-W., Nguyen, V. D., Vonikakis, V., & Winkler, S. (2015) Deep learning for emotion recognition on small datasets using transfer learning. In Proceedings of the 2015 ACM on international conference on multimodal interaction, 2015, ACM, pp. 443–449.
- 16.Kecman V. Learning and soft computing: Support vector machines, neural networks, and fuzzy logic models. Cambridge: MIT Press; 2001. [Google Scholar]
- 17.Chu F, Wang L. Applications of support vector machines to cancer classification with microarray data. International Journal of Neural Systems. 2005;15(06):475–484. doi: 10.1142/S0129065705000396. [DOI] [PubMed] [Google Scholar]
- 18.Khandoker AH, Palaniswami M, Karmakar CK. Support vector machines for automated recognition of obstructive sleep apnea syndrome from ECG recordings. IEEE Transactions on Information Technology in Biomedicine. 2009;13(1):37–48. doi: 10.1109/TITB.2008.2004495. [DOI] [PubMed] [Google Scholar]
- 19.Szarvas, M., Yoshizawa, A., Yamamoto, M., & Ogata, J. (2005). Pedestrian detection with convolutional neural networks. In Intelligent vehicles symposium, 2005. Proceedings. IEEE, 2005. IEEE, pp. 224–229.
- 20.Mori, K., Matsugu, M., & Suzuki, T. (2005) Face Recognition using SVM fed with intermediate output of CNN for face detection. In MVA, 2005, pp. 410–413.
- 21.Niu X-X, Suen CY. A novel hybrid CNN–SVM classifier for recognizing handwritten digits. Pattern Recognition. 2012;45(4):1318–1325. doi: 10.1016/j.patcog.2011.09.021. [DOI] [Google Scholar]
- 22.Chatfield, K., Simonyan, K., Vedaldi, A., & Zisserman, A. (2014). Return of the devil in the details: Delving deep into convolutional nets. arXiv preprint arXiv:1405.3531.
- 23.Uedo N, Fujishiro M, Goda K, Hirasawa D, Kawahara Y, Lee J, et al. Role of narrow band imaging for diagnosis of early-stage esophagogastric cancer: current consensus of experienced endoscopists in Asia-Pacific region. Digestive endoscopy: Official Journal of the Japan Gastroenterological Endoscopy Society. 2011;23:58–71. doi: 10.1111/j.1443-1661.2011.01119.x. [DOI] [PubMed] [Google Scholar]
- 24.Inoue H, Kaga M, Ikeda H, Sato C, Sato H, Minami H, et al. Magnification endoscopy in esophageal squamous cell carcinoma: A review of the intrapapillary capillary loop classification. Annals of Gastroenterology. 2014;28(1):41. [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuchi D, Iizuka T, Yamada A, Furuhata T, Yamashita S, Nomura K, et al. Utility of magnifying endoscopy with narrow band imaging in determining the invasion depth of superficial pharyngeal cancer. Head and Neck. 2015;37(6):846–850. doi: 10.1002/hed.23683. [DOI] [PubMed] [Google Scholar]
- 26.Takubo K, Makuuchi H, Fujita H, Isono K. Japanese classification of esophageal cancer, tenth edition: Part I. Esophagus. 2009;6(1):1–25. doi: 10.1007/978-4-431-68616-3_1. [DOI] [Google Scholar]
- 27.Bosch, A., Zisserman, A., & Munoz, X. (2007). Image classification using random forests and ferns. In IEEE 11th international conference on computer vision, 2007. ICCV 2007. IEEE, pp. 1–8.
- 28.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 29.Nair, V., & Hinton, G. E. Rectified linear units improve restricted boltzmann machines. In Proceedings of the 27th international conference on machine learning (ICML-10), 2010, pp. 807–814.
- 30.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: A simple way to prevent neural networks from overfitting. The Journal of Machine Learning Research. 2014;15(1):1929–1958. [Google Scholar]
- 31.Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J., Girshick, R., et al. (2014). Caffe: Convolutional architecture for fast feature embedding. In Proceedings of the ACM international conference on multimedia, 2014.ACM, pp. 675–678.
- 32.Chetlur, S., Woolley, C., Vandermersch, P., Cohen, J., Tran, J., Catanzaro, B., et al. (2014). cuDNN: Efficient primitives for deep learning. arXiv preprint arXiv:1410.0759.
- 33.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2(3):27. [Google Scholar]
- 34.Athanasopoulos, A., Dimou, A., Mezaris, V., & Kompatsiaris, I. (2011). GPU acceleration for support vector machines. In WIAMIS 2011: 12th international workshop on image analysis for multimedia interactive services, Delft, The Netherlands, April 13–15, 2011, 2011: TU Delft; EWI; MM; PRB.