Abstract
Ovo, which encodes a transcription factor with Zn-finger domains, is evolutionarily conserved among animals. In Drosophila, in addition to its zygotic function for egg production, maternal ovo activity is required in primordial germ cells (PGCs) for expression of germline genes such as vasa and nanos. In this study, we found that maternal Ovo accumulates in PGC nuclei during embryogenesis. In these cells, ovo serves a dual function: activation of genes expressed predominantly in PGCs, and conversely suppression of somatic genes. Reduction of ovo activity in PGCs makes them unable to develop normally into germ cells of both sexes. In mice, knockout of the ovo ortholog, Ovol2, which is expressed in PGCs, decreases the number of PGCs during early embryogenesis. These data strongly suggest that ovo acts as part of an evolutionarily conserved mechanism that regulates germline development in animals.
The germline is the only cell lineage destined to produce the next generation, while the soma gives rise to the body tissues. In animals, two distinct modes of germline establishment have been reported, “preformation” and “epigenesis“1,2. In the animal species with the preformation mode of germline establishment, maternal factors directing germline fate are localized in a specialized ooplasm, or germ plasm, that is partitioned into primordial germ cells (PGCs)3. By contrast, in certain species with the epigenesis mode, PGCs are specified by inductive signals secreted from the surrounding tissues. For example, in mice, bone morphogenetic proteins (BMPs) and Wnt signaling are both essential for PGC induction in the epiblast4,5,6,7. Irrespective of the modes of the germline establishment, zygotic expression of vasa (vas), nanos (nos), and piwi is evident in the PGCs of a variety of animal groups1,8. Therefore, it is possible that the mechanism underlying the activation of germline genes is conserved throughout the animal species.
To elucidate how germline gene expression is activated in PGCs, we have identified the pertinent maternal transcription factors in Drosophila, which uses the preformation mode of germline formation9. In this species, the germ plasm is localized in the posterior pole of cleavage embryos (stage 1–2) and is subsequently partitioned into PGCs, also called pole cells (stage 3–4). PGCs pass through the midgut epithelium to associate with the somatic components of the gonads (stage 9–12), and then aggregate with each other to form the embryonic gonads (stage 14). Following embryonic development, PGCs differentiate into germ cells (i.e., mature oocytes or sperm) within the gonads. Expression of vas begins in PGCs at around stage 9, and continues in germline cells10,11. By contrast, piwi and nos transcripts are both maternally supplied in PGCs, and their zygotic expression is evident in PGCs around stage 159 (BDGP: http://www.fruitfly.org). The germline-specific expression of these genes is thought to be activated by maternal factors localized in the germ plasm. Accordingly, we identified maternal mRNAs that are enriched in the germ plasm and encode transcription factors9, and then performed RNA interference (RNAi)-dependent knockdown experiments in order to determine their contribution to germline-specific vas and nos expression in PGCs. The results revealed six transcripts required for vas and/or nos gene expression9.
Ovo, one of these six maternal transcripts, belongs to a family of genes encoding DNA-binding proteins with C2H2 zinc-finger domains9,12. Ovo is evolutionarily conserved across a wide range of animals13. In Drosophila, ovo produces three alternate isoforms: Ovo-A and Ovo-B function as a negative and positive transcriptional regulator in the germline during oogenesis, respectively14, whereas Svb is required for epidermal differentiation15,16. During oogenesis, Ovo-B activates transcription of ovo itself and ovarian tumor (otu) by binding their promoter regions, and this transcriptional activation is required for production of mature oocytes17,18. Thus, in the absence of zygotic ovo activity, female adults never produce progeny; consequently, genetic approaches have failed to clarify the role of maternal ovo in progeny. To overcome this problem, ovo-targeting dsRNA was injected into early embryos; however, this approach caused embryonic lethality due to the lack of epidermal differentiation, which also requires zygotic ovo function9. Thus, it remains unclear whether maternal ovo function is required for the normal development of the germline.
Irrespective of the two modes of germline formation, ovo orthologs are widely conserved among animal species13. For example, three ovo orthologs have been identified in the mouse nuclear genome13. The zinc-finger domains of Ovol1, Ovol2/MOVO, and Ovol3 share 74%, 68%, and 58% amino-acid identity, respectively, with Drosophila ovo19,20. Previous work demonstrated that Ovol2 only partially rescues the defect in Drosophila oogenesis caused by ovo mutation20, suggesting that the ovo gene family may exert similar functions in the germline in different animal species. However, the role of ovo orthologs in germline development in mouse embryos remains unclear.
Here we show that, in Drosophila, maternal Ovo-B protein is predominantly expressed in PGC nuclei. Its function is required in PGCs to activate the gene expression highly enriched in the germline, and conversely to suppress the somatic genes. Furthermore, Ovo-B must be intact for proper development of the germline both in females and males. Moreover, in mice, ovol2 is zygotically expressed in PGCs, and its knockdown decreases the number of PGCs at early embryogenesis. Therefore, we propose that ovo plays an evolutionarily conserved role in the regulation of germline development in both the fruit fly and mouse.
Results and Discussion
Ovo-B is the major isoform in PGCs of Drosophila embryos
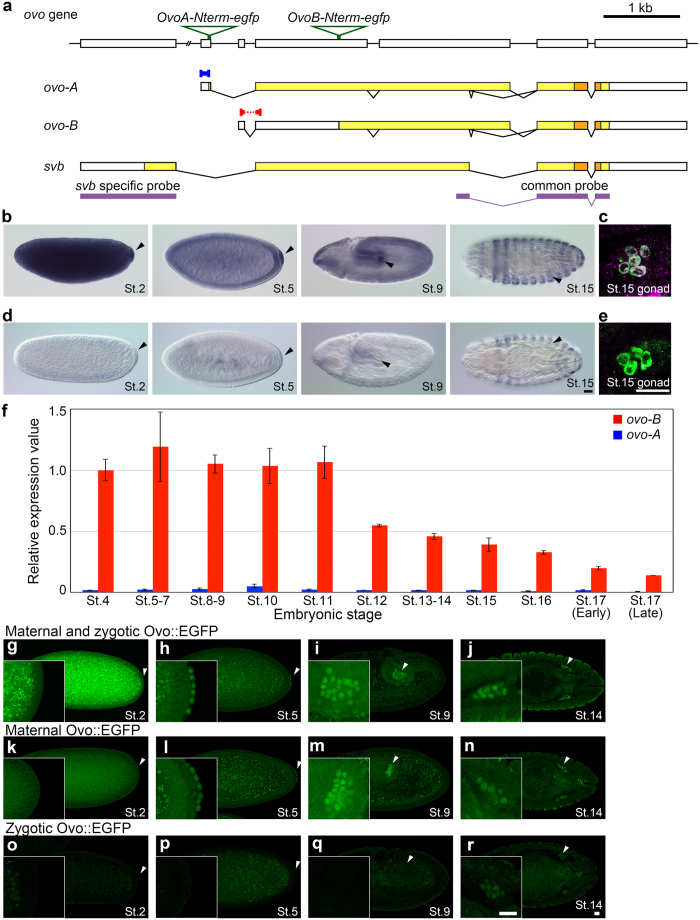
The ovo locus encodes three proteins, Ovo-A, Ovo-B, and Svb, which share a common C-terminal region containing Zn-finger DNA-binding domains (Fig. 1a). The signal detected by common probes for the three transcripts encoding Ovo-A, Ovo-B, and Svb is distributed almost uniformly in cleavage and syncytial blastodermal embryos from stage 2 to stage 4 (Fig. 1b). Subsequently, the signal decreases in the somatic region and is enriched in PGCs of the cellular blastodermal embryos at stage 5 (Fig. 1b). The signal remains detectable in PGCs until the end of embryogenesis (Fig. 1b,c). This spatio-temporal expression of ovo RNA is compatible with observations reported previously9,21,22. To determine which isoform is expressed in PGCs, we performed whole-mount in situ hybridization (WISH) and quantitative RT-PCR (qRT-PCR). WISH analysis using an svb-specific probe detected signal in the epidermis, but not in early embryos or PGCs (Fig. 1d,e). This observation strongly suggests that the ovo-A and/or ovo-B transcripts, but not svb, are present in PGCs. However, we could not confirm this by WISH using probes specific to ovo-A and ovo-B, presumably because the probes were too short (127 and 78 bases, respectively) to detect signals in embryos. qRT-PCR analysis detected both ovo-A and ovo-B transcripts in PGCs (Fig. 1f). In these experiments, we generated cDNA pools from PGCs of embryos at 11 different stages23,24, and amplified cDNAs using primer sets specific to ovo-A and ovo-B (Fig. 1a) (see Supplementary Materials and Methods). We detected ovo-B transcript in PGCs throughout the embryonic stages. Expression levels of ovo-B remained constant from stage 4 to stage 11, but decreased from stage 12 and thereafter to approximately 10% of peak levels at stage 17. By contrast, the level of ovo-A transcript was less than 5% of that of ovo-B throughout the embryonic stages. Thus, ovo-B is the major isoform of ovo transcripts in PGCs.
Figure 1. Structure of the ovo gene and its expression during embryogenesis.
(a) Genomic organization of the ovo locus, which encodes three transcripts: ovo-A, ovo-B, and svb. Exons (boxes), introns (straight lines), protein coding regions (yellow boxes), and zinc-finger DNA-binding domains (orange boxes) are shown. Regions corresponding to the probes for in situ hybridization (purple bars) and the ovo-A– and ovo-B–specific region amplified by qRT-PCR analysis (blue and red bars, respectively) are also indicated. Sites where Egfp was inserted are indicated by green triangles. The ovoA-Nterm-egfp and ovoB-Nterm-egfp knock-in alleles contain the gene in the N-terminal regions of the Ovo-A and Ovo-B proteins, respectively. (b–e) Expression of ovo transcripts during embryogenesis. Probes used for in situ hybridization are indicated in (a). Embryos (b and d) and gonads (c and e) were hybridized with a common probe (b and c) and an svb-specific probe (d and e). Gonads (c and e) were double-stained for ovo or svb transcript (magenta) and Vasa protein (green), a marker for germ plasm and PGCs. The developmental stage of each embryo is shown in lower right. Arrowheads show germ plasm and PGCs. Scale bars: 20 μm. (f) The levels of ovo-A (blue) and ovo-B transcript (red) relative to the expression level of ovo-B in st.4 PGCs were determine by qRT-PCR, and plotted against the developmental stages of embryogenesis. Means ± SEM of three biological replicates are shown. (g–r) Expression of maternal and/or zygotic Ovo protein during embryogenesis. The Ovo-EGFP fusion protein encoded by ovoB-Nterm-egfp was detected using an anti-GFP antibody (green). Developmental stages are shown at lower right. Insets show close-up images of germ plasm and PGCs. Maternal and zygotic Ovo-EGFP protein was detected in embryos derived from females homozygous for ovoB-Nterm-egfp mated with males hemizygous for ovoB-Nterm-egfp (g–j). Maternal Ovo-EGFP was detected in embryos with YFP staining, derived from ovoB-Nterm-egfp/FM7c, Dfd-GMR-nvYFP females mated with y w males (k–n). Zygotic expression of Ovo-EGFP was detected in only female embryos produced from y w females mated with ovoB-Nterm-egfp hemizygous males, since the ovo gene is on X chromosome. Scale bars: 20 μm.
Next, we examined the expression of Ovo protein in PGCs. Because we were unable to raise an antibody against Ovo protein, we inserted a tag sequence encoding an enhanced green fluorescent protein (EGFP) into the N-terminus of the Ovo-B coding region (ovoB-Nterm-egfp) (Fig. 1a), enabling us to detect Ovo-B protein using an anti-GFP antibody. Knock-in of GFP at this site produced a GFP fusion of Ovo-A and Svb as well as Ovo-B. Given that ovo-B is the major isoform of ovo transcript in PGCs, it is reasonable to assume that the vast majority of the GFP signal detected in PGCs represents Ovo-B expression. This idea is further supported by our observation that GFP fused to the Ovo-A–specific region was undetectable in PGCs throughout embryogenesis (Supplementary Fig. S1).
In embryos produced from females homozygous for ovoB-Nterm-egfp, the GFP signal was ubiquitously distributed throughout embryos at stage 2 (Fig. 1g). Subsequently, the signal started to accumulate in nuclei of PGCs after stage 4, and this accumulation was maintained in the nuclei of PGCs until at least stage 16 (Fig. 1h–j). By contrast, the GFP signal was decreased in the soma, and barely detectable in their nuclei except for a subset of epidermal cells, where Svb was zygotically expressed from stage 12 onward16 (Fig. 1j).
The ovo-B transcript is supplied maternally to PGCs, and its levels decreased during embryogenesis (Fig. 1f), suggesting that zygotically expressed ovo-B transcript makes little, if any, contribution to the production of Ovo protein in PGCs. To confirm this, we examined zygotic expression of GFP in PGCs from the paternally transmitted ovoB-Nterm-egfp allele, which produced only a very weak GFP signal in female PGCs after stage 14 (Fig. 1o–r). This expression pattern is compatible with the previous finding that zygotic ovo expression is detectable in PGCs at stage 17 in a female specific manner25. By contrast, GFP expression from the maternal ovoB-Nterm-egfp allele was detectable in PGCs from stage 4 until at least stage 16 (Fig. 1k–n). Taken together, our observations show that Ovo-B protein produced from the maternal ovo-B transcript is dominant in PGCs during embryogenesis.
Function of maternal Ovo-B is required for both female and male germline development in Drosophila
Next, we examined the role of maternal Ovo-B in PGCs. Because oogenesis is completely arrested in the ovaries in the absence of ovo-B function14,26, we were unable to obtain embryos lacking maternal ovo-B activity. Furthermore, knockdown of maternal ovo-B by injection of ovo-targeting dsRNA caused embryonic lethality due to the lack of epidermal differentiation9. Consequently, we were unable to examine the post-embryonic phenotypes caused by knockdown of maternal Ovo-B function. To overcome these problems, we used Ovo-A to specifically reduce Ovo-B activity in PGCs; this strategy was based on the prior observation that the Ovo-A isoform antagonizes Ovo-B and represses the transcription induced by Ovo-B during oogenesis14.
We conducted two types of Ovo-B knockdown experiments, each involving a different duration of ovo-A over-expression. ovo-A expression was induced using the nos-Gal4-VP16 (nos-Gal4) transgene, which expresses the transcriptional activator Gal4-VP16 under the control of the germline-specific nos promoter11. In embryos derived from females with nos-Gal4, the transcript is maternally supplied and partitioned into PGCs via the nos 3′UTR11. In PGCs, Gal4-VP16 produced from the maternal transcript (maternal nos-Gal4) activated UAS-dependent gene expression from stage 11 until at least the end of embryogenesis (Supplementary Fig. S2a). By contrast, zygotic expression of nos-Gal4 (zygotic nos-Gal4) activated UAS-dependent gene expression in the germline from stage 15 to adulthood (Supplementary Fig. S2b). We induced UASp-Ovo-A expression in the germline by maternal and zygotic nos-Gal4 from stage 11 onward, and by maternal nos-Gal4 from stage 11 to at least the end of embryogenesis.
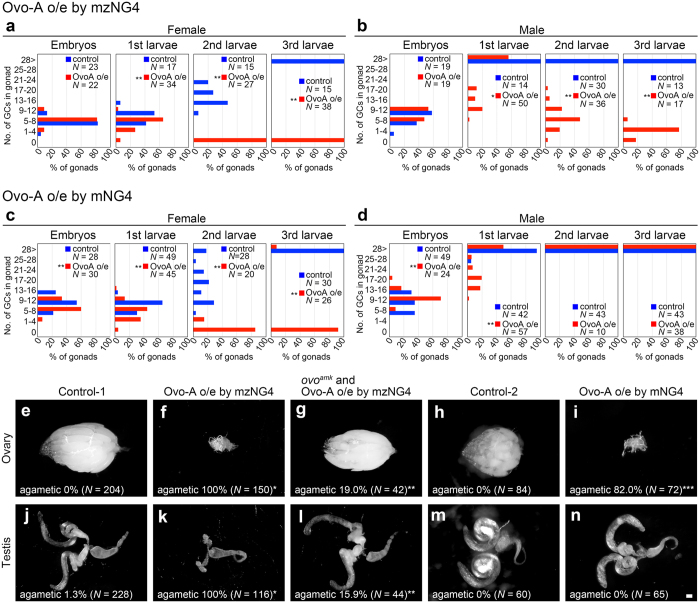
Using this system, we first examined germline phenotypes resulting from Ovo-A expression under the control of maternal and zygotic nos-Gal4 (mzNG4). Ovo-A–expressing PGCs normally migrated into the gonads, and the number of these cells was almost identical to that observed in control embryos (Fig. 2a,b, Supplementary Fig. S3a, b). However, the number of Ovo-A–expressing germline cells during larval development was significantly reduced relative to controls (Fig. 2a,b, Supplementary Fig. S3a,b); consequently, larvae of both sexes developed into agametic adults (Fig. 2f,k, Supplementary Fig. S3e,f). Since maternal Ovo-A activity is required for germline development14, we cannot exclude the possibility that the above mentioned germline phenotype is caused by the upregulation of Ovo-A rather than the reduction of Ovo-B activity by Ovo-A overexpression. However, we found that Ovo-B expression from the ovoamk transgene14 was able to rescue the agametic phenotype caused by Ovo-A expression (Fig. 2g,l, Supplementary Fig. S3e,f). This clearly indicates that this phenotype is caused by the reduction of Ovo-B activity. Therefore, Ovo-B activity is essential for germline development.
Figure 2. Germline-specific expression of Ovo-A causes germline loss in males and females.
(a–d) Distribution of the number of Vasa-positive germline cells per gonad of Ovo-A–expressing (red) and control (blue) females (a and c) and males (b and d) at embryonic stage 15–16, the first, second, and third instar larval stages. Ovo-A was expressed under the control of mzNG4 in progeny derived from nos-Gal4 homozygous females mated with UASp-Ovo-A (line#4-2) homozygous males (red bars in a and b). Progeny derived from nos-Gal4 homozygous females mated with y w males were used as controls (blue bars in a and b). Furthermore, Ovo-A was overexpressed under the control of mNG4 in YFP-positive progeny derived from nos-Gal4/TM6B, Dfd-GMR-nvYFP females mated with UASp-Ovo-A (line#4-2) homozygous males (red bars in c and d). YFP-positive progeny derived from nos-Gal4/TM6B, Dfd-GMR-nvYFP females mated with y w males were used as controls (blue bars in c and d). N: the number of gonads observed. Significance of differences relative to controls was calculated using Fisher’s exact test (*P < 0.05, **P < 0.01). (e–n) Ovaries (e–i) and testes (j–n) of adult progeny derived from nos-Gal4 homozygous females mated with y w males (control-1) (e and j), nos-Gal4 homozygous females mated with UASp-Ovo-A (line#4-2) homozygous males (f and k), or ovoamk and nos-Gal4 homozygous females mated with UASp-Ovo-A (line#4-2) homozygous males (g and l); and YFP-positive adult progenies derived from nos-Gal4/TM6B, Dfd-GMR-nvYFP females mated with y w males (control-2) (h and m) or nos-Gal4/TM6B, Dfd-GMR-nvYFP females mated with UASp-Ovo-A (line#4-2) homozygous males (i and n). The ovoamk transgene expresses only Ovo-B under the control of the ovo-B promoter14. Ovaries and testes were stained with an anti-Vasa antibody, and the gonads without Vasa signal were categorized as agametic. The percentage of agametic gonads is shown at the bottom of each panel. N: the number of gonads examined. Significance of difference relative to control-1 (*), Ovo-A o/e by mzNG4 (**), or control-2 (***) was calculated by Fischer’s exact test (P < 0.01). Scale bar: 100 μm.
A mutation that depletes zygotic Ovo-B activity results in female-specific germline loss after the third instar larval stage27,28. This defect obviously differs from that caused by Ovo-A induction by mzNG4, which induces germline loss as early as the first instar larval stage in both sexes (Fig. 2a,b, Supplementary Fig. S3a,b). Thus, it is reasonable to speculate that maternal, rather than zygotic, Ovo-B is required in germline cells for their development in both males and females. To confirm this, we decreased maternal Ovo-B activity by inducing Ovo-A in PGCs under the control of maternal nos-Gal4 (mNG4). In females, Ovo-A expression decreased the number of germline cells during larval development (Fig. 2c, Supplementary Fig. S3c), resulting in adult sterility (Fig. 2i, Supplementary Fig. S3e). This phenotype is consistent with that caused by Ovo-A expression under the control of mzNG4 (Fig. 2a,f, Supplementary Fig. S3a,e). By contrast, in males, a subtle but statistically significant decrease in the number of germline cells was evident in first-instar larvae (Fig. 2d). However, the number of germline cells was restored to control levels after the second instar larval stage (Fig. 2d). These weak phenotypes may result from poor expression of Ovo-A in PGCs due to the half dose of maternal nos-Gal4 driver (see legend of Fig. 2). Together, these observations suggest that maternal Ovo-B is required in PGCs for normal development in females and males.
Maternal Ovo-B activates expression of genes highly enriched in PGCs and suppresses somatic gene expression in PGCs
Because Ovo-B protein acts as a transcriptional activator14 and is present in nuclei of PGCs during embryogenesis (Fig. 1h–j), we speculated that maternal Ovo-B plays an important role in regulating gene expression in PGCs. To investigate this possibility, we compared transcriptomes from Ovo-B knockdown (Ovo-B KD) and control PGCs, which were isolated at stage 16 by fluorescence-activated cell sorting (FACS) from embryos expressing Ovo-A under the regulation of mzNG4 and control embryos, respectively. In our microarray analysis (see Supplementary Materials and Methods), a total of 13,167 genes were expressed in either control PGCs, Ovo-B KD PGCs, or both. Among them, 401 genes were down-regulated and 510 genes were up-regulated in Ovo-B KD PGCs (Supplementary Tables S1 and S2).
We performed Gene Ontology (GO) enrichment analysis of the genes down- or up-regulated in Ovo-B KD PGCs. Among the genes down-regulated in Ovo-B KD PGCs, no GO category was statistically significantly enriched (Supplementary Table S3). By contrast, the genes up-regulated in Ovo-B KD PGCs were enriched for GO terms associated with development of somatic tissues and organs (q-value < 0.05) (Supplementary Table S4), suggesting that Ovo-B represses expression of genes involved in somatic development in PGCs.
This is further supported by our observation that genes expressed mainly in somatic cells (soma-enriched genes) were up-regulated in Ovo-B KD PGCs, whereas genes expressed predominantly in PGCs (PGC-enriched genes) were down-regulated. We first selected 347 soma-enriched and 145 PGC-enriched genes by comparing the transcriptomes of stage-16 PGCs and whole embryos (see Supplementary Materials and Methods). Among 347 soma-enriched genes, 240 were up-regulated in Ovo-B KD (Fig. 3, Supplementary Table S5). By contrast, among 145 PGC-enriched genes, 98 were down-regulated in Ovo-B KD PGCs (Fig. 3, Supplementary Table S6).
Figure 3. Gene expression significantly changed both in the comparison between control and Ovo-B KD PGCs, and between PGCs and whole embryos.
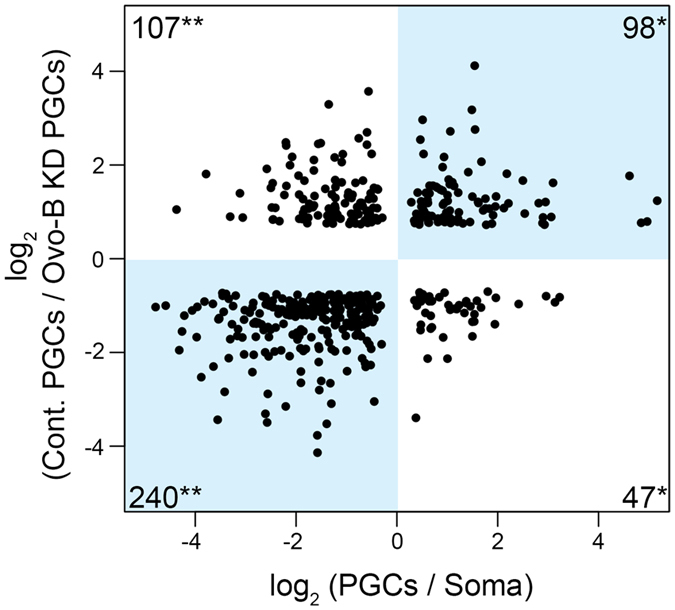
The x-axis shows the log2 fold change in gene expression between PGCs and whole embryos. The y-axis shows the log2 fold change in gene expression between control PGCs and Ovo-B KD PGCs. Spots represent genes whose expression levels differed significantly between controls and Ovo-B KD PGCs (q-value < 0.05), and between PGCs and whole embryos (q-value < 0.05). When multiple microarray probes mapped to a single gene, the average value of the log2 fold change was used. The numbers of genes plotted are shown in the corners of each quadrant. Among 145 PGC-enriched genes, which are plotted in the right half, more genes were down-regulated than up-regulated by Ovo-B KD [*P < 0.01 (Fisher’s exact test)]. By contrast, among 347 soma-enriched genes, plotted in the left half, more genes were up-regulated than down-regulated by Ovo-B KD [**P < 0.01 (Fisher’s exact test)].
The above data show that maternal Ovo-B plays an important role in activating expression of PGC-enriched genes, as well as in repressing somatic gene expression in PGCs. At least five genes (CG13741, CG14838, cup, fal, and piwi), whose expression patterns are annotated only with the anatomical ontology terms “germ cells and/or gonads” at stage 13–16 in the BDGP WISH database, were down-regulated in Ovo-B KD PGCs (Supplementary Table S1). According to the BDGP database, the four genes other than fal are specifically expressed in the germline at stage 13–16, and cup and piwi are both involved in germline development29,30, suggesting that Ovo-B is required to activate the “germline genes” in PGCs.
Three genes, vas, nos, and piwi, are regarded as the germline genes whose expression is commonly observed in the germline in multiple animal species1,8. Because probes for detecting endogenous vas and nos transcripts were unavailable in the microarrays we used (see Supplementary Materials and Methods), we monitored the expression of these genes by qRT-PCR. Our analysis revealed no significant difference in the relative expression level of vas between control PGCs [mean ± SEM in log2 (the number of biological replicates: N): 0 ± 0.27 (N = 3)] and Ovo-B KD PGCs [0.11 ± 0.14 (N = 3); P > 0.05 (Student’s t-test)]. Similarly, we observed no significant difference in the relative expression of nos between control PGCs [0 ± 0.08 (N = 3)] and Ovo-B KD PGCs [0.28 ± 0.22 (N = 3); P > 0.05]. However, injection of dsRNA against ovo into early embryos, which causes more severe defect in germline development during embryogenesis than Ovo-A overexpression does31, is able to suppress zygotic expression of vas and nos in PGCs9. Furthermore, our present microarray data shows that piwi expression in PGCs was also affected by Ovo-B KD (Supplementary Table S1). Based on these findings, we conclude that in PGCs, maternal Ovo-B acts as a transcriptional activator for vas, nos, and piwi, which are known to be expressed predominantly in the germline of a wide range of animal species. This led us to speculate that ovo plays an evolutionarily conserved role in germline development.
Mouse Ovol2 is required for normal development of PGCs
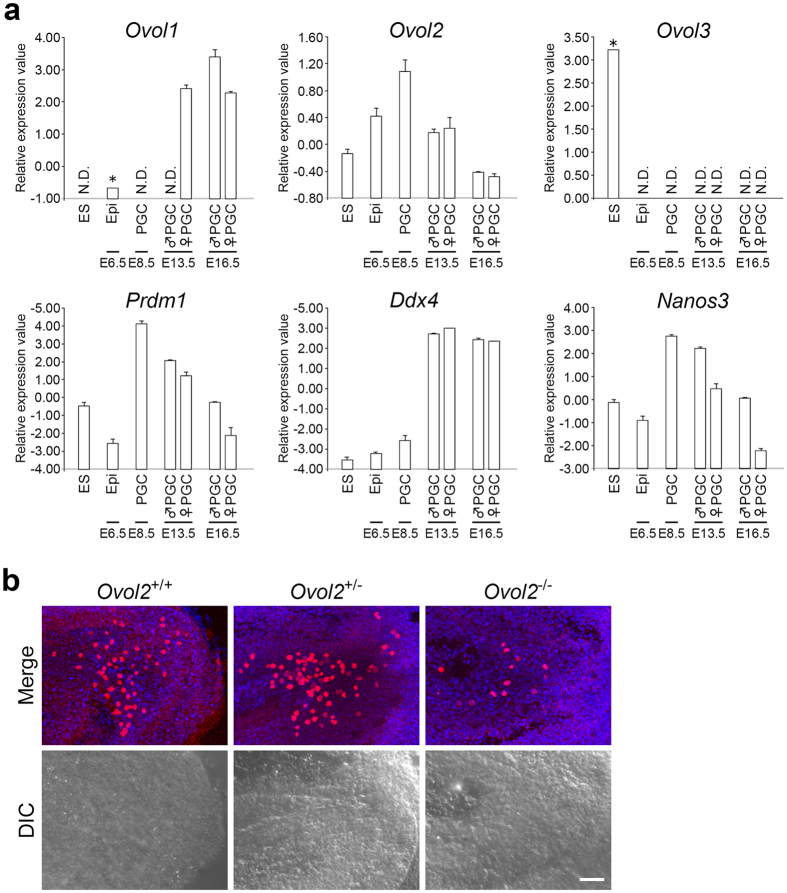
Mouse has three ovo orthologs, Ovol1, Ovol2, and Ovol3; the zinc-finger domains of these genes exhibit significant sequence similarity with Drosophila ovo19,20. To investigate whether an ovo ortholog is required for germline development in mouse embryos, we focused on Ovol2, which was clearly expressed in early PGCs (Fig. 4a). Ovol2 was expressed in epiblasts of E6.5 embryos, as described previously32, and in PGCs isolated from E8.5 embryos; its expression level declined thereafter (Fig. 4a). Later, in spermatocytes, expression of Ovol2 protein is detected again33. By contrast, Ovol3 expression was undetectable in the PGCs at E8.5, E13.5, and E16.5 (Fig. 4a). We detected Ovol1 expression in PGCs only after E13.5 (Fig. 4a), suggesting that this gene plays a minimal role, if any, in the formation and early development of PGCs. In males, Ovol1 is required in the germline at later stages for pachytene progression of meiotic spermatogenic cells34. In females, deletion of Ovol1 gene causes reduced fertility, but this is due to somatic defects, i.e., structural abnormalities in the urogenital tract34.
Figure 4. Ovol2 is expressed in mouse PGCs and is required for their development.
(a) Relative expression levels (log2 scale) of three mouse ovo-like genes (Ovol1, Ovol2, and Ovol3) and three germline markers (Prdm1, Ddx4, and Nanos3) were plotted against cell type (ES cells, epiblast, and PGCs) at the indicated embryonic stages. Data represent means ± SEM of two biological replicates. *Indicates that the microarray signal was detected in only one of two replicates. N.D. Indicates that no signal was detected in either replicate. Successful isolation of PGCs using FACS was confirmed by expression of the early PGC maker Prdm1 (E8.5) and the late PGC makers Ddx4 and Nanos3 (E13.5 and E16.5). (b) PGCs stained with an anti-Oct4 antibody (red) and the nuclei stained with TO-PRO-3 (blue) in mouse E8.0 wild type (Ovol2+/+), heterozygous (Ovol2+/−) or homozygous (Ovol2−/−) embryos for Ovol2. Upper panels show fluorescence images, and lower panels show the corresponding differential interference contrast (DIC) images. Scale bar: 50 μm.
To determine whether Ovol2 mutation affects early germline development in mouse embryos, we counted PGCs in E8.0–E8.5 embryos, in which Ovol2 is highly expressed (Fig. 4a). Even in the absence of Ovol2 activity, somatic development proceeded normally, and the morphology of entire embryo and the neural and mesodermal tissues seemed to be intact at E8.032,35. However, significantly fewer PGCs were detected by immunostaining for OCT4, a marker for PGCs, in Ovol2 null embryos than in controls (Fig. 4b). For example, we examined embryos with three to eight pairs of somites in both control (+/−) and null mutant (−/−) embryos, and found that the null mutants contained far fewer OCT4-positive PGCs [number of PGCs ± SEM (number of embryos examined: N): 25.0 ± 7.5 (N = 6)] than controls [104.5 ± 20.5 (N = 4); P < 0.01 (Student t-test)]. Subsequently, the mutant embryos exhibited lethality by E10.5 due to defects in neural, gut, and heart development, and vascular angiogenesis32,35. Consequently, we were unable to follow the developmental fate of PGCs in later-stage mutant embryos.
Evolutionarily conserved role of ovo in germline development in Drosophila and mice
The observations described above indicate that, in the absence of Ovol2 activity, the number of PGCs was reduced in the embryos with three to eight somites (Fig. 4b). This defect resembled the phenotype observed in Drosophila expressing Ovo-A in PGCs (Fig. 2 and Supplementary Fig. S3). Accordingly, we propose that ovo plays an evolutionarily conserved role in germline development in these two animal species.
In mice, BMPs and Wnt are both required to induce PGCs in the epiblast of early embryos4,5,6,7. Ovol2 acts downstream of the BMP signaling pathway to regulate the cell fate decision between neuroectoderm and mesendoderm36. In addition, the Wnt signaling pathway is required to induce Ovol2 in the embryonic stem cells (ESC) and the colorectal cancer cells36,37. Therefore, it is reasonable to speculate that Ovol2 is a downstream target for BMP and Wnt signaling pathways during PGC induction in epiblast. This is compatible with our data that Ovol2 expression is upregulated in E8.5 PGCs compared with those in epiblasts and ESCs (Fig. 4a). The transcription factors (TFs), Blimp1, Prdm14, and Tfap2c, direct epiblast-like-cells (EpiLCs) into a PGC state and are also key downstream components of the BMP and Wnt pathways in the process of PGC induction6,38; therefore, Ovol2 may act synergistically with these TFs to facilitate PGC formation.
During mouse PGC formation, these TFs activate germline and pluripotent genes, repress somatic genes, and reconstitute epigenetic programs38. However, the genetic network regulating PGC formation appears to be diverse even among mammals39. For example, reduction of Prdm14 does not affect human PGC formation40, but is sufficient to induce PGC fate in mice38. Furthermore, Prdm14 is not conserved in Drosophila genome (OrthoDB ver.9: http://www.orthodb.org), and neither expression nor function of Blimp1 and Tfap2c orthologs has been reported in fly PGCs (FlyBase: http://flybase.org). These observations suggest that Blimp1, Prdm14, and Tfap2c do not act as a conserved set of TFs directing germline gene expression. By contrast, we show here that in Drosophila, maternal Ovo-B activates expression of genes highly enriched in PGCs and conversely suppresses somatic gene expression, thereby contributing to determination of “germness”. The ovo gene family is conserved across animal species, and encodes transcription factors regulating gene expression in various differentiation processes13. In light of the fact that an ovo ortholog is required for PGC formation in mice, we propose that Ovo acts as part of an evolutionarily conserved mechanism regulating the genetic network of germline development. In the future, it would be of particular interest to identify genes downstream of Ovol2 in mouse PGCs, and to determine whether these genes also act downstream of Ovo-B in the Drosophila germline. Our data provide an important first step toward elucidation of a mechanism of germline gene regulation common to a wide range of animal species.
Materials and Methods
Drosophila stocks
Nos-Gal4-VP16 (nos-Gal4)11 was used to induce expression of UASp-Ovo-A (line#4-2, line#7, line#8) in the germline. The ovoamk transgene was used to rescue the agametic phenotype caused by Ovo-A overexpression. ovoamk carries the ovo genomic fragment expressing only ovo-B mRNA under the control of ovo-B promoter14. To examine the duration of UASp-Ovo-A expression driven by nos-Gal4, P{UASp-GFPS65C-aTub84B}3 (UASp-GFP) (Bloomington Stock center; #7373) was used. To determine the genotypes of embryos derived from females hemizygous for nos-Gal4 and females hemizygous for ovoB-Nterm-egfp (see EGFP knock-in mediated by CRISPR/Cas9), nos-Gal4/TM6B, P{Dfd-GMR-nvYFP}4 and ovoB-Nterm-egfp/FM7c, P{Dfd-GMR-nvYFP}1 were used, respectively. For EGFP knock-in mediated by CRISPR/Cas9, y2 cho2 v1; Sp/CyO, P{nos-Cas9}2 A (NIG-Fly; CAS-0004) and P{nos-phiC31\int.NLS}X; attP40(II) (NIG-Fly; TBX-0002) were used. Flies were maintained on a standard Drosophila medium at 25 °C.
Transgenes
The 5′- and 3′-fragments of ovo-A cDNA were amplified from an embryonic cDNA library41. Primer pairs ovoA5′-Fw/ovoA5′-Rv and ovoA3′-Fw/ovoA3′-Rv (Supplementary Table S7) were used for amplifying the 5′- and 3′-fragments, respectively. The amplified fragments were ligated using the endogenous Not I site and inserted into the KpnI/HindIII sites of pBS-KS nos3′UTR42. Next, the Kpn I/Xba I fragment containing the ovo-A-nos3′UTR hybrid gene was cloned into pUASp43. Germline transformation was performed using y w embryos as recipients. Three independent w+ transformants were mated with y w females to establish a homozygous stock of UASp-Ovo-A.
EGFP knock-in mediated by CRISPR/Cas9
See Supplementary Materials and Methods for details of the EGFP knock-in procedure.
mRNA and protein expression analysis
See Supplementary Materials and Methods for details of the procedures used for in situ hybridization, quantitative RT-PCR to detect the ovo-A and ovo-B isoforms, immunostaining, microarray analysis of Ovo-B KD and control PGCs, microarray analysis of PGCs and whole embryos, and quantitative RT-PCR to detect vas and nos.
Mouse ovo-like gene analysis
Microarray analysis of purified mouse PGCs was performed as described44. Expression levels of ovo-like genes and other germline markers were extracted from the microarray data. Microarray data were deposited in GEO under Accession No. GSE82020.
To count PGCs in mouse embryos, we used null-mutant mice lacking Ovol2 exon 3, which encodes the first and second of four zinc-finger domains35. Oct4-positive PGCs in mouse embryos were counted as follows. Embryos of early somite stage (E8.0–8.5) were dissected and immunostained with anti-OCT4 antibody (Santa Cruz Biotechnology) as described45. Z-stack confocal images were taken from each embryo using a Zeiss 510, and optical slices were analyzed using the ImageJ software. OCT4-positive PGCs were detected, and their X-Y positions in each section were recorded. PGCs with the same (or almost the same) X-Y positions were considered to be identical, and total PGCs were counted by manual inspection of all optical sections.
All mouse experiments conformed to the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation of RIKEN BioResource Center.
Additional Information
How to cite this article: Hayashi, M. et al. Conserved role of Ovo in germline development in mouse and Drosophila. Sci. Rep. 7, 40056; doi: 10.1038/srep40056 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. B. Oliver for ovoamk flies, and Dr. A. Nakamura for advice on EGFP knock-in mediated by CRISPR/Cas9. We also thank the Functional Genomics Facility of the NIBB Core Research Facilities for technical support, and the NIG stock center (Japan) and the Bloomington Drosophila Stock Center for providing us with fly strains and Developmental Studies Hybridoma Bank for antibodies. This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (KAKENHI Grant Numbers: 25114002 and 24247011).
Footnotes
Author Contributions M.H., Y.S. and S.K. designed the experiments using fruit fly. M.H., Y.S., M.S. (Hokkaido University) and S.S. performed the fly experiments and bioinformatic analysis. M.S. (RIKEN BioResource Center), K.A. and S.I. designed and performed the experiments using mouse. M.H., Y.S. and S.K. wrote the paper. All authors reviewed manuscript.
References
- Extavour C. G. & Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884 (2003). [DOI] [PubMed] [Google Scholar]
- Extavour C. G. M. Evolution of the bilaterian germ line: lineage origin and modulation of specification mechanisms. Integr. Comp. Biol. 47, 770–785 (2007). [DOI] [PubMed] [Google Scholar]
- Ikenishi K. Germ plasm in Caenorhabditis elegans, Drosophila and Xenopus. Development, growth & differentiation 40, 1–10 (1998). [DOI] [PubMed]
- Lawson K. A. et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424–436 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y., Liu X. M., Marble A., Lawson K. A. & Zhao G.-Q. Requirement of Bmp8b for the Generation of Primordial Germ Cells in the Mouse. Molecular Endocrinology (2009). [DOI] [PubMed] [Google Scholar]
- Aramaki S. et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell 27, 516–529 (2013). [DOI] [PubMed] [Google Scholar]
- Cantú A. V. & Laird D. J. Wnt and Bmp fit germ cells to a T. Dev. Cell 27, 485–487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C. E., Swartz S. Z. & Wessel G. M. A conserved germline multipotency program. Development 137, 4113–4126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu J. et al. Maternal RNAs encoding transcription factors for germline-specific gene expression in Drosophila embryos. Int. J. Dev. Biol. 52, 913–923 (2008). [DOI] [PubMed] [Google Scholar]
- Lasko P. F. & Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 4, 905–921 (1990). [DOI] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L. & Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8, 243–246 (1998). [DOI] [PubMed] [Google Scholar]
- Mével-Ninio M., Terracol R. & Kafatos F. C. The ovo gene of Drosophila encodes a zinc finger protein required for female germ line development. EMBO J. 10, 2259–2266 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. et al. Molecular phylogeny of OVOL genes illustrates a conserved C2H2 zinc finger domain coupled by hypervariable unstructured regions. PLoS One 7, e39399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J. et al. OVO transcription factors function antagonistically in the Drosophila female germline. Development 127, 881–892 (2000). [DOI] [PubMed] [Google Scholar]
- Chanut-Delalande H., Fernandes I., Roch F., Payre F. & Plaza S. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 4, 1549–1561 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T. et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329, 336–9 (2010). [DOI] [PubMed] [Google Scholar]
- Lü J. & Oliver B. Drosophila OVO regulates ovarian tumor transcription by binding unusually near the transcription start site. Development 128, 1671–1686 (2001). [DOI] [PubMed] [Google Scholar]
- Bielinska B., Lü J., Sturgill D. & Oliver B. Core promoter sequences contribute to ovo-B regulation in the Drosophila melanogaster germline. Genetics 169, 161–172 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. et al. Ovol2, a mammalian homolog of Drosophila ovo: gene structure, chromosomal mapping, and aberrant expression in blind-sterile mice. Genomics 80, 319–325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unezaki S. et al. Characterization of the isoforms of MOVO zinc finger protein, a mouse homologue of Drosophila Ovo, as transcription factors. Gene 336, 47–58 (2004). [DOI] [PubMed] [Google Scholar]
- Mevel-Ninio M., Terracol R., Salles C., Vincent A. & Payre F. ovo, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech. Dev. 49, 83–95 (1995). [DOI] [PubMed] [Google Scholar]
- Salles C., Mével-Ninio M., Vincent A. & Payre F. A germline-specific splicing generates an extended ovo protein isoform required for Drosophila oogenesis. Dev. Biol. 246, 366–376 (2002). [DOI] [PubMed] [Google Scholar]
- Shigenobu S., Arita K., Kitadate Y., Noda C. & Kobayashi S. Isolation of germline cells from Drosophila embryos by flow cytometry. Dev. Growth Differ. 48, 49–57 (2006). [DOI] [PubMed] [Google Scholar]
- Hashiyama K., Hayashi Y. & Kobayashi S. Drosophila Sex lethal gene initiates female development in germline progenitors. Science 333, 885–888 (2011). [DOI] [PubMed] [Google Scholar]
- Casper A. L. & Van Doren M. The establishment of sexual identity in the Drosophila germline. Development 136, 3821–3830 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B., Perrimon N. & Mahowald A. P. The ovo locus is required for sex-specific germ line maintenance in Drosophila. Genes Dev. 1, 913–923 (1987). [DOI] [PubMed] [Google Scholar]
- Rodesch C., Geyer P. K., Patton J. S., Bae E. & Nagoshi R. N. Developmental analysis of the ovarian tumor gene during Drosophila oogenesis. Genetics 141, 191–202 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab S. & Steinmann-Swicky M. Female germ cells of Drosophila require zygotic ovo and otu product for survival in larvae and pupae respectively. Mech. Dev. 54, 205–210 (1996). [DOI] [PubMed] [Google Scholar]
- Keyes L. N. & Spradling A. C. The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development 124, 1419–1431 (1997). [DOI] [PubMed] [Google Scholar]
- Cox D. N., Chao A. & Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514 (2000). [DOI] [PubMed] [Google Scholar]
- Jankovics F. et al. Functional analysis of the Drosophila embryonic germ cell transcriptome by RNA interference. PLoS One 9, e98579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay D. R., Hu M., Li B., Rhéaume C. & Dai X. The mouse Ovol2 gene is required for cranial neural tube development. Dev. Biol. 291, 38–52 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizaki R., Yao I., Katano T., Matsuda T. & Ito S. Restricted expression of Ovol2/MOVO in XY body of mouse spermatocytes at the late pachytene stage. J. Androl. (2011). [DOI] [PubMed] [Google Scholar]
- Li B. et al. Ovol1 regulates meiotic pachytene progression during spermatogenesis by repressing Id2 expression. Development 132, 1463–1473 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unezaki S., Horai R., Sudo K., Iwakura Y. & Ito S. Ovol2/Movo, a homologue of Drosophila ovo, is required for angiogenesis, heart formation and placental development in mice. Genes to Cells 12, 773–785 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang T. et al. The zinc finger transcription factor Ovol2 acts downstream of the bone morphogenetic protein pathway to regulate the cell fate decision between neuroectoderm and mesendoderm. J. Biol. Chem. 288, 6166–6177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G. D. et al. OVOL2, an inhibitor of WNT signaling, reduces invasive activities of human and mouse cancer cells and is down-regulated in human colorectal tumors. Gastroenterology 150, 659–671.e16 (2016). [DOI] [PubMed] [Google Scholar]
- Nakaki F. et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature 501, 222–226 (2013). [DOI] [PubMed] [Google Scholar]
- Tang W. W. C. et al. A unique gene regulatory network resets the human germline epigenome for development. Cell 161, 1453–1467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawa F. et al. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 34, 1009–1024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. H. & Kafatos F. C. Functional cDNA libraries from Drosophila embryos. J. Mol. Biol. 203, 425–437 (1988). [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K. & Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1, 431–437 (1999). [DOI] [PubMed] [Google Scholar]
- Rørth P. Gal4 in the Drosophila female germline. Mech. Dev. 78, 113–118 (1998). [DOI] [PubMed] [Google Scholar]
- Ikeda R. et al. Large, male germ cell-specific hypomethylated DNA domains with unique genomic and epigenomic features on the mouse X chromosome. DNA Res. 20, 549–565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M. et al. Molecular identification of tw5: Vps52 promotes pluripotential cell differentiation through cell-cell interactions. Cell Rep. 2, 1363–1374 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.