Abstract
The genome is burdened with repetitive sequences that are generally embedded in silenced chromatin. We have previously demonstrated that Lsh (lymphoid-specific helicase) is crucial for the control of heterochromatin at pericentromeric regions consisting of satellite repeats. In this study, we searched for additional genomic targets of Lsh by examining the effects of Lsh deletion on repeat regions and single copy gene sequences. We found that the absence of Lsh resulted in an increased association of acetylated histones with repeat sequences and transcriptional reactivation of their silenced state. In contrast, selected single copy genes displayed no change in histone acetylation levels, and their transcriptional rate was indistinguishable compared to Lsh-deficient cells and wild-type controls. Microarray analysis of total RNA derived from brain and liver tissues revealed that <0.4% of the 15 247 examined loci were abnormally expressed in Lsh−/−embryos and almost two-thirds of these deregulated sequences contained repeats, mainly retroviral LTR (long terminal repeat) elements. Chromatin immunoprecipitation analysis demonstrated a direct interaction of Lsh with repetitive sites in the genome. These data suggest that the repetitive sites are direct targets of Lsh action and that Lsh plays an important role as ‘epigenetic guardian’ of the genome to protect against deregulation of parasitic retroviral elements.
INTRODUCTION
Packaging of DNA into chromatin organizes the genome into ‘active’ zones of euchromatin and ‘repressed’ zones of heterochomatin (1–4). These opposing states of chromatin compaction ultimately control the access of DNA binding factors to their specific target sites. The chromatin-dependent organization of the genome plays an important role in the control of fundamental cellular processes such as tissue-specific gene expression, gene imprinting, X-chromosome inactivation and mitosis (5–9).
Chromatin states can be inherited by the next cell generation, thus preserving specific gene expression pattern, a phenomena known as epigenesis. Epigenetic modifications of chromatin are largely determined by specific posttranslational modifications of histone tails or covalent modification of cytosine residue (10–14). The transcriptionally active euchromatin is frequently characterized by hypomethylation of DNA, hyperacetylation of histone 3 and histone 4 tails, and methylation of the histone 3 tails at Lys-4. The majority of single copy genes are contained in euchromatin. In contrast, heterochromatin represents a condensed form of chromatin and is generally marked by DNA hypermethylation, and hypoacetylation of histones 3 and 4. A specific methylation of histone 3 at Lys-9 serves as a recognition signal for binding of the major heterochromatin protein 1 (HP1). Heterochromatin contains largely repetitive sequences derived from transposable elements. These repeats include LTR (long terminal repeat) containing retroviral-derived sequences, long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs), simple repeats, and major and minor satellite sequences. Although, the suppression of these parasitic repeat elements in the genome is considered to be important, the regulation and establishment of silencing chromatin around repeat regions is not yet understood.
Lsh (lymphoid-specific helicase) is a member of the SNF2 family of chromatin remodeling proteins (15). SNF2 homologues are important components of SNF/SWI chromatin remodeling complexes that require ATP to disrupt histone–DNA interactions, and to allow for sliding of the nucleosomes along the DNA (16,17). Lsh has been previously cloned from a lymphoid precursor library and characterized in proliferating lymphoid tissues (15,18,19). Lsh is ubiquitously expressed during embryogenesis and targeted deletion of Lsh leads to early death of the newborn with signs of renal and lymphoid defects (18,20). Embryonal fibroblasts derived from Lsh−/− embryos display mitotic defects and embryos show reduced birth weight (19,21). Thus, the chromatin structure shaped by Lsh plays a crucial role during normal embryonic development.
We have previously characterized Lsh as a major epigenetic regulator in mice. Targeted deletion of Lsh in mice leads to a global loss of CpG methylation (22). This reduction in CpG methylation has been recently confirmed in an independently generated Lsh-deficient mouse strain (PASG) (23). Similarly, a mutation of the DDM1 gene (decrease in DNA methylation 1), a close homologue of Lsh in Arabidopsis thaliana, results in genomic hypomethylation (24). Since Lsh and DDM1 both belong to the SNF2 family of chromatin remodeling proteins, it suggests that chromatin structure plays a crucial role in determining DNA methylation patterns.
Lsh shows a preferential association with pericentric heterochromatin, and the loss of CpG methylation is most substantial around pericentromeric heterochromatin sequences (22,25). Moreover, Lsh−/− cells show an abnormal pattern of histone methylation at pericentromeric heterochromatin (26). This suggests that Lsh plays an important role in controlling epigenetic modifications at pericentromeric heterochromatin. In this study, we attempted to search for additional genomic targets of Lsh. By screening epigenetic modifications and transcription at repeat sites that are usually embedded in silenced chromatin, and at single copy genes that are frequently embedded in euchromatin, we report here that, in particular, repetitive elements are effected by Lsh. Our study supports the idea that Lsh is preferentially recruited to repeat sites in the genome and that Lsh contributes to silencing of repetitive elements to preserve genomic integrity.
MATERIALS AND METHODS
Mice tissue and cell culture
Embryonic tissues from crosses between Lsh+/− mice were obtained at the indicated day of gestation, and the genotype was performed by PCR analysis as previously described (18). Primary murine embryonal fibroblast (MEF) cells were isolated and cultured in growth medium as previously described (21). Embryonal carcinoma cell line P19 was purchased from ATCC and cultured according to the manufacturer's protocol.
Southern blotting of PCR products
Amplified DNA was separated by agarose gel electrophoresis and transferred to NYTRAN nylon transfer membranes (Schleicher & Schuell). After 1 h pre-hybridization with Rapid-hyb buffer (Amersham Pharmacia Biotech), hybridization was carried out for 1 h at 42°C with the radiolabeled oligonucleotide probes. The probes were end-labeled with [γ-32P]ATP using the RTS T4 Kinase Labeling System (Gibco BRL) and purified with ProbeQuant G-50 Micro Columns (Amersham Pharmacia Biotech). Blots were washed as described in the manufacturer's protocol and visualized by autoradiography.
Western analysis
Samples derived from P19 and MEF cells were dispersed into single cell suspensions in phosphate-buffered saline (PBS). Nuclei were extracted by using Nuclei EZ prep nuclei isolation kit (Sigma). The nuclei pellets were lysed into half volume of low salt buffer [10 mM HEPES, 25% Glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.5 mM DTT, 0.2 μM EDTA and 25 μM p-nitrophenyl-p′-guanidinobenzoate (PNPGB)] and half volume of high salt buffer (10 mM HEPES, 25% Glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.5 mM DTT, 0.2 μM EDTA and 25 μM PNPGB) The samples were mixed and then extracted on ice for 30 min. After spinning at 15 000 g for 15 min at 4°C, the supernatants were collected and protein concentrations determined with the Bradford assay. Western blot was performed as described previously (26).
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIPs) were performed as described in detail elsewhere (27). In brief, cells were cross-linked with 1% formaldehyde, lysed and sonicated on ice to generate DNA fragments with an average length of ∼200–800 bp. After pre-cleaning, 1% of each sample was saved as input fraction. Immunoprecipitation was performed using specific antibodies against the indicated peptides or immunoglobulin G (IgG) used as control. The eluted complex was directly used for western blotting analysis, or the cross-linking was reversed for nucleic acids preparation and PCR analysis as described in (27). Amplification conditions were as follows: 94°C for 3 min; 94°C for 1 min; 62°C for 2 min; 72°C for 3 min (10 cycles for major satellite sequences, 18 cycles for minor satellite sequences and intracisternal A particle (IAP), 25 cycles for single copy genes) and 72°C for 7 min. The following primer pairs were used for sequence detection: IAP (accession no. NM_010490), 5′-(463–484), 3′-(741–762 bp), probe (689–709); major satellite (accession no. X06899), 5′-(9–30), 3′-(113–132), probe (53–72); minor satellite (accession no. Z22164), 5′-(113–132), 3′-(220–242), probe (166–189); IgF2R (accession no. L06446), 5′-(2021–2040) 3′-(2144–2163), probe (2049–2068); APRT (accession no. M11310), 5′-(2486–2506), 3′-(2568–2589), probe (2520–2539); H19 (accession no. U19619), 5′-(2609–2630), 3′-(2745–2766), probe (2658–2681); β-globin (accession no. J00413), 5′-(4885–4904), 3′-(4966–4995), probe (2658–2681); Pgk-2 (accession no. M17299) 5′-(1564–1585), 3′-(1694–1715), probe (1586–1609). For ChIPs using anti-Lsh antibodies (18), PCRs were performed at the following conditions: 94°C for 3 min; 94°C for 1 min; 62°C for 2 min; 72°C for 3 min (12 cycles for major satellite sequences, 20 cycles for minor satellite sequences, 22 cycles for IAP and LINE, 25 cycles for SINE) and 72°C for 7 min. The following primer pairs were used for LINE and SINE detection: LINE (accession no. D84391), 5′-(5557–5576), 3′-(5648–5656), SINE (accession no. AC002121), 5′-(13374–13393), 3′-(13354–13374). Quantitative determination of single copy genes by real-time PCR was performed as previously described (22).
RT–PCR analysis
Samples from primary MEF cells and the indicated embryonic tissues were homogenized with the QIAshredder spin column and total RNA was isolated using RNAeasy Mini kit (Qiagen). An aliquot of 1 μg of total RNA from each sample was subjected to first strand cDNA synthesis using an Omniscript RT kit (Qiagen) with or without reverse transcriptase (+RT or −RT). Additional controls were performed for major satellite sequences analysis by treating total RNA with Rnase (Qiagen). PCR analysis was performed in serial dilutions (1:10) for the detection of transcripts. The β-actin gene served as control. The following primer sets were used: IAP (accession no. NM_010490), 5′-(691–702), 3′-(779–798); PGK-1 (accession no. NM 008828.1), 5′-(1364–1385), 3′-(1507–1528); PGK-2 (accession no. M17299), 5′-(1715–1737), 3′-(1918–1939); IgF2R (accession no. NM-010515), 5′-(8495–8514), 3′-(8713–8732); APRT (accession no. M 11310), 5′-(2845–2866), 3′-(2990–3011); H19 (accession no. NM-023123), 5′-(1268–1280), 3′-(1652–1672); β-globin (accession no. M10524), 5′-(108–127), 3′-(268–299). For major and minor satellite sequences analysis, total RNA was extracted with Trizol (Invitrogen). Any genomic DNA present was eliminated with TURBO DNA-freeTM Kit (Ambion). The first strand cDNA synthesis reaction was performed from 50 ng of purified total RNA using Super ScriptTM Kit (Invitrogen) with oligo(dT) or strand-specific oligos for cDNA synthesis of major satellite sequences (Forward: 5′-GTCGTCAAGTGGATGTTTATCAT-3′; Reverse: 5′-ATAA ACATCCACTTGACGACTTG-3′). PCRs were performed with the following conditions: 95°C for 15 min; 94°C for 30 min; 64°C for 1 min; 72°C for 1 min (28 cycles) and 72°C for 3 min. The primers for major satellite sequences were: 5′-GCGAGAAAACTGAAAATCACG, 3′-CTTGCCATATTCCACGTCCT.
Microarray analysis
Total RNA was prepared from liver or brain tissue of Lsh−/− embryos of day 17.5 of gestation and wild-type littermate controls. The NIA mouse cDNA array is composed of 15 247 unique clones derived from 11 embryonic libraries. Details of the library composition, hybridization protocol and data analysis are available online (http://lgsun.grc.nia.nih.gov/cDNA/), and as supplemental Materials and Methods section. The average clone size is about 1500 bp in length. All clones in the library have been sequence verified [(28); D. Munroe unpublished data]. Pair-wise comparison of Lsh−/− and Lsh+/+ tissues was performed. Three arrays were performed for each pair (including inversion of the dye labeling). Every hybridization experiment was performed in triplicates, and the data was normalized by background subtraction. Only sequences that had a more than 2-fold rise in at least two arrays were scored. For analysis of repetitive elements, first, the parental sequences of the clones were determined using Unigene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=unigene) and then the sequences were examined by the Repeatmasker program (http://www.repeatmasker.org).
RESULTS
Lsh deficiency alters histone acteylation levels at repetitive elements
We have previously demonstrated that the absence of Lsh results in alterations of histone methylation levels that occur specifically at repetitive sequences (26). ChIP analysis revealed a raise in methylation at Lys-4 of histone 3 at major and minor satellite repeats, LTR elements and LINEs. In contrast, a number of selected single copy genes failed to reveal any changes in histone methylation levels. Thus, we tested the idea that Lsh specifically controlled histone modifications at repeat sites that are usually embedded in silenced chromatin versus single copy genes frequently residing in euchromatin.
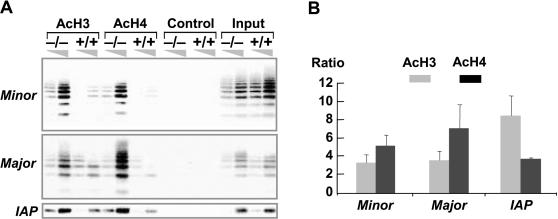
First, we examined the histone acetylation levels at repeat elements that are usually low in acetylation using ChIP. Chromatin from MEFs derived from Lsh wild-type or Lsh−/− embryos were precipitated using anti-acetyl-histone 3 (Lys-9 and Lys-14) or anti-acetyl-histone 4 (Lys-5/8/12/16) antibodies. As shown in Figure 1A, repetitive sequences were greatly enriched in the chromatin precipitated material derived from Lsh-deficient samples using antibodies against acetylated histone 3 and histone 4. This increase in acetylated chromatin precipitates was prevalent at all repetitive elements tested, such as minor and major satellite sequences, that are present at pericentromeric heterochromatin and LTR elements (such as the IAP), that are located at silenced chromatin throughout the genome. As a control, wild-type chromatin displayed very low levels of histone 3 and histone 4 acetylation at repetitive elements. On average, the association of acetylated histone 3 and histone 4 with repeat sequences was about 3- to 8-fold higher in Lsh-deficient chromatin as compared to that of wild-type controls (Figure 1B). This increased association of acetylated chromatin with repeats could not be explained by a general increase in acetylated histones, since neither western blot analysis nor immunefluorescence techniques revealed a general increase in histone acetylation levels (data not shown).
Figure 1.
Increased association of acetylated histones with repetitive elements in Lsh-deficient cells. (A) ChIP assay was performed from Lsh-deficient and wild-type chromatin extracts using indicated antibodies (normal rabbit IgG as control) to detect acetylation of histone 3 (Lys-9/14) or histone 4 (Lys-5/8/12/16). Precipitated DNA or chromatin input was diluted (1:3) for the detection of linear PCR amplification followed by Southern analysis. (B) Results of ChIPs were quantified using a PhosporImager, and the ratio of Lsh−/− to wild-type controls was calculated. The average values of four to five experiments are plotted in the bar graph.
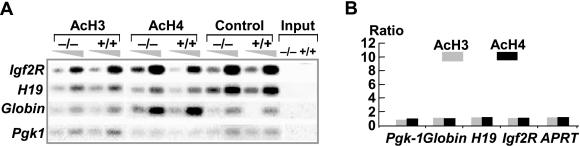
Next, we investigated the effect of Lsh on histone acetylation levels at a number of selected single copy sequences: such as the X-linked phosphoglycerate kinase 1 gene (Pgk-1), tissue-specific genes such as β-globin and Pgk-2, the imprinting genes H19 and Igf2R and the house keeping gene adenine phosphoribosyl transferase (APRT). Since the acetylation level was indistinguishable when compared with the wild-type and Lsh−/− samples using conventional PCR technique (Figure 2A), real-time PCR was employed for greater sensitivity. However, as shown in Figure 2B, no changes in the degree of either histone 3 or histone 4 acetylation levels were detectable. (Note: The acetylation level of the Pgk-2 gene was too low to be measurable in wild-type embryonic body tissue or MEFs, and the absence of Lsh could not raise Pgk-2 histone acetylation to detectable levels, data not shown).
Figure 2.
Unchanged histone acetylation levels at single copy sequences. ChIP assay was performed using antibodies as indicated to detect acetylation of histone 3 (Lys-9/14) or histone 4 (Lys-5/8/12/16) in nuclear extracts derived from Lsh−/−, Lsh+/+ MEFs followed by PCR analysis. (A) Southern analysis. (B) Taqman-PCR analysis. The average values were calculated from four experiments.
Thus, changes in histone acetylation levels after Lsh deletion closely corresponded to previously observed alterations in histone methylation levels (26) and were only detectable at repetitive sites as opposed to single copy sites.
Gene transcription is only elevated at retroviral elements and not at selected single copy genes
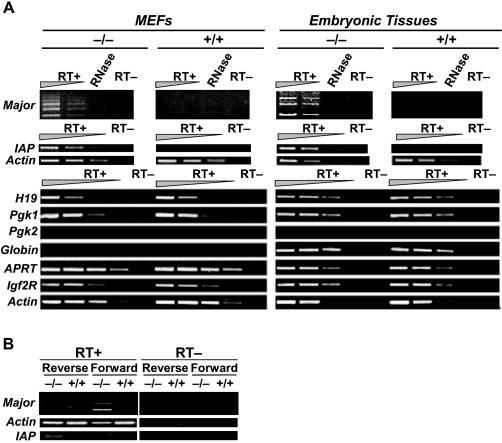
In order to further test the effect of Lsh deletion on repetitive sites versus single copy genes and to evaluate also functional consequences of altered histone acetylation levels, we measured gene expression levels by RT–PCR analysis. First, the expression levels of repetitive sequences were examined using RNA derived from Lsh−/− MEFs or Lsh−/− embryos (Figure 3A). Repeat sequences are generally silenced in heterochromatin and transcripts are hardly detectable or even undetectable in wild-type tissues. However, the disturbance of heterochromatin in the absence of Lsh as previously reported and the raise of histone acetylation levels as reported in this study resulted in the reactivation of the suppressed repeats. As shown in Figure 3A, major satellite transcripts were increased in Lsh−/− MEFs and embryonic tissue. Primarily, the forward strand was detectable in Lsh-deficient cells (Figure 3B). Similarly, preferential expression of forward strands for centromeric repeats has been reported in yeast cells with a mutation of the HP1 homologue Swi6 (29). Possibly, transcripts of the reverse strand are less stable. Furthermore, the repetitive LTR element (such as the IAP, also known as ERV class II) was enhanced in embryonal fibroblasts or embryonic tissue, as we have previously reported (26). In contrast, transcripts for minor satellite repeats were completely undetectable in either Lsh−/− or Lsh+/+ tissues (data not shown). The difference between the transcriptional rates of major versus minor satellite sequences may be due to the different distribution of retrotransposable elements, which contain regulatory sequences for transcription. These elements are dispersed in pericentromeric heterochromatin where major satellites are embedded and are not present in centromeric regions, which contain minor satellites. In order to determine the size of the major satellite transcripts, northern analysis was performed (data not shown). Lsh-deficient cells gave varying degrees of smear ranging from 4 kb to 200 bp, which may be expected based on the variability of promoter positions and transcriptional start sites (see above). However, a cross-hybridization with traces of genomic DNA was also observed. RNase treatment as well as DNase treatment abolished the signal leaving the true source of the smear undetermined.
Figure 3.
Deregulation of transcripts with repetitive elements in the absence of Lsh. (A) Total RNA derived from Lsh−/− or Lsh+/+ MEF cells or Lsh−/− and Lsh+/+ embryos of day 13.5 of gestation was subjected to RT–PCR analysis. Repeat sequences were analyzed in dilutions of 1:3, single copy genes in dilutions of 1:10. Omission of reverse transcriptase (−RT) or treatment with RNase served as further controls. (B) Analysis of strand-specific transcripts using forward and reverse primers during reverse transcription. Oligo(dt) primed cDNA served as control for detection of β-actin and IAP transcripts.
Next, several selected single copy genes were assayed using RNA derived from either Lsh−/− MEF cells or Lsh−/− embryos. None of the widely expressed genes, Igf2R, APRT, β-actin, H19 or Pgk-1 revealed any differences in gene expression levels comparing the wild-type and Lsh−/− samples (Figure 3A). β-globin, a tissue-specific gene, is usually not expressed in embryonal fibroblasts and the tissue-specific barrier of gene expression was not released upon Lsh deletion and genomic hypomethylation. Pgk-2 is specifically expressed in testis and was undetectable in MEFs or embryonic tissue, and its silenced state in most tissues was apparently not reactivated upon Lsh deletion.
Thus, Lsh deficiency appears to specifically alter gene expression levels at repetitive sites but not at selected single copy genes. The transcriptional change corresponds to alterations in histone modifications that preferentially occurred at repeat sites. These results support the hypothesis that Lsh plays a specific role in protecting the chromatin state and function at repetitive elements in the genome.
Microarray analysis of Lsh-deficient brain and liver tissues
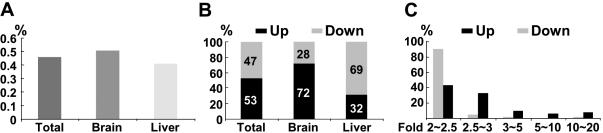
The previous RT–PCR analysis allowed only the investigation of a few genomic loci affected by Lsh deficiency. In order to carry out a genome-wide screening for other targets of Lsh effects, we obtained a gene expression profile using the NIA embryonic cDNA microarray. Total RNA was prepared from Lsh−/− brain or liver tissues (day 17.5 of gestation) and from wild-type littermate controls. Of the 15 247 available sequences on the array, about 12 616 were analyzed with brain RNA and 13 026 with liver RNA. Of these, about 0.5% was altered more than 2-fold in the Lsh−/− brain and about 0.4% was altered in the Lsh−/−liver in comparison with wild-type controls (Figure 4A). Since only moderate changes in gene expression levels were detected, these results suggest that Lsh is not a major transcriptional regulator such as SNF2, a chromatin remodeling protein that transcriptionally controls about 6% of genes in Saccharomyces cerevisiae (30).
Figure 4.
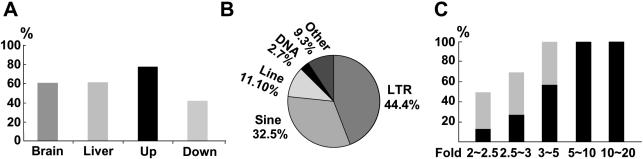
Microarray analysis of gene expression after Lsh deletion. Total RNA derived from Lsh−/− or Lsh+/+ liver or brain tissue of day 17.5 gestation embryos were subjected to Microarray analysis. Results represent the average of three hybridizations performed for each tissue type. (A) Proportion of clones from the whole microarray that are abnormally up- or downregulated more than 2-fold above wild-type controls. (B) Proportion of deregulated genes that are either up- or downregulated in the brain or liver tissues. (C) Proportion of deregulated genes that are up- or downregulated and their degree of deregulation expressed as a fold above wild-type control.
The majority of sequences in the brain were increased in the absence of Lsh, suggesting that Lsh mainly silences specific transcripts in the brain (Figure 4B). Surprisingly, the majority of clones in the liver were decreased; however, the magnitude was marginal, since more than 97% of downregulated sequences in the liver showed a change of <2.5-fold (Figure 4C). Thus, based on the degree of deregulation, Lsh deletion mainly resulted in upregulation of selected clones suggesting an overall suppressive effect of Lsh on gene transcription.
Many of the aberrantly expressed sequences were of unknown function (Table 1). Since Lsh deficiency or CpG hypomethylation can lead to reactivation of LTR elements (Figure 3), we screened the parental sequences for the presence of repetitive sequences using the Repeatmasker Program. As shown in Figure 5A, about 60% of all altered brain or liver clones contained repeats in the parental sequences, among the upregulated sequences the percentage was even ∼80%. The following repeat families were identified by the Repeatmasker Program (Figure 5B): 45% of the repeat containing clones belonged to ‘LTR elements’ (such as endogenous retroviral-derived elements ERV class I and ERV class II), about 33% belonged to the SINE family (such as Alu/B1, B2-B4 and MIR elements) and 11% revealed LINEs (such as L1 and L2 elements) and only 3% had sequences derived from ‘DNA elements’ (such as the MER1 and MER2 type). Low complexity sequences or poly (A) tails were not included in the repeat analysis. The detection of clones containing repeat sequences was possible despite the presence of COT1 DNA in the hybridization protocol. COT1 DNA is commonly used to suppress non-specific hybridization since it is enriched in repetitive sequences, such as SINEs B1, B2 or LINE L1. Thus, the hybridization to LINE or SINE sequence containing clones was probably mediated by the unique sequence portion of identified clones. A few clones on the array consisted mainly of LTR elements and served quasi as positive controls. For example, the clones H3020E05 or H3053D11, that consist 85–90% of LTR elements. Their hybridization was probably due to the high homology that LTR elements frequently share with each other. For example, LTR containing clones H3020E05 and H3020E06 are 99% homologous in their repeats, and H3081A04 and H3010A11 are 95% homologous. The extent of abnormal gene expression correlated with the presence of repeats and in particular with the presence of retroviral-derived LTR elements (Figure 5C) supporting the idea that Lsh plays an important part in silencing parasitic elements in the genome.
Table 1. Microarray analysis of Lsh-deficient brain and liver tissues.
| Brain | Liver | ||||||
|---|---|---|---|---|---|---|---|
| Fold | FeatureID | UniGene | Gene | Fold | FeatureID | UniGene | Gene |
| 20.9 | H3020E05 | −11.4 | H3095E04 | ||||
| 20.7 | H3020E06 | 10.3 | H3025G09 | Mm.4533 | Apoa4 | ||
| 11.9 | H3073B03 | Mm.173779 | 4 | C0027A11 | Mm.277406 | Stat1 | |
| 11.1 | H3081A04 | 3.9 | H3157D02 | Mm.33902 | Igtp | ||
| 10.4 | H3053D11 | 3.7 | H3028D07 | Mm.192826 | |||
| 9.3 | H3085H12 | 3.6 | H3144G06 | Mm.273195 | Car1 | ||
| 9.3 | H3010A11 | 3.3 | H3020E05 | ||||
| 8.6 | H3066G11 | Mm.351303 | 3.1 | H3020E06 | |||
| 6.9 | H3081A03 | 2.9 | H3081A04 | ||||
| −3.6 | L0223A07 | Mm.272226 | Msh4 | 2.8 | H3034E11 | Mm.210336 | Lgals4 |
| 3.1 | H3039F05 | Mm.13694 | Oat | 2.9 | H3047D01 | Mm.252145 | Ckmt1 |
| 3 | H3155H01 | Mm.343840 | 2.4 | H3013E04 | Mm.8687 | Cap1 | |
| 2.8 | H3139H05 | Mm.330987 | Rps23 | −2.4 | H30096D08 | ||
| 2.7 | H3154G09 | Mm.218457 | −2.4 | H3097D08 | |||
| 2.7 | L0216C09 | Mm.28251 | Tulp4 | −2.3 | H3104G01 | Mm.14526 | Mylpf |
| 2.7 | H3103F06 | Mm.1387 | Rab11a | −2.3 | H3108F03 | Mm.329866 | |
| 2.6 | L0266A11 | −2.3 | H3043F12 | Mm.1070 | Fech | ||
| 2.6 | H3111D06 | 2.3 | H3093H10 | Mm.339215 | |||
| 2.6 | H3124H09 | Mm.331 | Ubc | 2.3 | H3095F01 | ||
| 2.5 | L0238A07 | Mm.143813 | Midn | 2.2 | H3149G10 | Mm.41506 | |
| 2.5 | J1043E09 | Mm.41637 | Arfip2 | −2.2 | H3045A12 | Mm.196110 | Hba-a1 |
| 2.5 | L0203H07 | Mm.195020 | 2.1 | H3053D11 | |||
| 2.5 | H3139B10 | Mm.103092 | 2.1 | H3010A11 | |||
| −2.5 | H3118A03 | Mm.288567 | Hbb-y | 2.1 | H3109A08 | Mm.327565 | |
| −2.5 | J0073B10 | Mm.355624 | −2.1 | H3024H06 | Mm.273053 | Srprb | |
| −2.5 | H3045A12 | Mm.196110 | Hba-a1 | −2.1 | H3026F02 | Mm.1090 | Gpx1 |
| 2.4 | H3007D02 | Mm.2344 | Gnb1 | −2.1 | H3108H11 | Mm.162497 | |
| 2.4 | H3016D05 | Mm.302037 | −2.1 | H3083F09 | Mm.292049 | Phtf | |
| 2.4 | H3131B09 | Mm.43871 | Trim35 | −2.1 | J0073B10 | Mm.355624 | |
| 2.4 | H3119F02 | Mm.133919 | Tcfl1 | −2.1 | H3117B08 | ||
| 2.4 | H3127D01 | Mm.88216 | Snrpb | −2.1 | H3003H11 | Mm.247824 | Ubadc1 |
| 2.4 | H3147H06 | Mm.10702 | Cacybp | −2.1 | H3122H11 | Mm.196110 | Hba-a1 |
| 2.4 | H3147D06 | Mm.6839 | Cdk4 | −2.1 | H3003F02 | Mm.99 | Rrm2 |
| 2.4 | H3095D10 | Mm.300697 | −2 | H3129C03 | Mm.7248 | Slc4a1 | |
| −2.3 | H3118C11 | Mm.196110 | Hba-a1 | −2 | H3125H07 | Mm.196110 | Hba-a1 |
| −2.3 | H3118A06 | Mm.288567 | Hbb-y | −2 | L0065G04 | ||
| 2.3 | H3052D09 | Mm.333388 | Chd4 | −2 | H3032A06 | Mm.12267 | Pabpc4 |
| 2.3 | H3041A06 | Mm.218316 | −2 | H3027G12 | Mm.4048 | Myd116 | |
| 2.3 | C0174D09 | −2 | H3048G11 | Mm.24021 | Blrrb7 | ||
| 2.3 | H3087F10 | Mm.3823 | Elavl2 | −2 | J0738B12 | Mm.39704 | |
| 2.3 | H3060F05 | Mm.33908 | −2 | H3024E12 | Mm.28632 | MCT4 | |
| 2.3 | H3008B09 | Mm.243024 | −2 | C0108B10 | |||
| 2.3 | H3097C09 | Mm.113724 | Nfia | −2 | H3155C06 | Mm.29742 | Cd24 |
| 2.3 | H3115F09 | Mm.8739 | Sgce | −2 | H3122H10 | Mm.196110 | Hba-a1 |
| 2.2 | H3111D05 | Mm.10853 | −2 | L0060H12 | Mm.196110 | Hba-a1 | |
| 2.2 | H3104H06 | Mm.784 | Nbr1 | −2 | H3024D04 | Mm.212066 | Abi2 |
| −2.2 | H3122H08 | Mm.30702 | −2 | H3021F12 | Mm.7141 | Pcna | |
| −2.2 | L0034C10 | Mm.233825 | Hbb-b1 | −2 | H3118C11 | Mm.196110 | Hba-a1 |
| −2.2 | L0072C01 | Mm.218679 | −2 | H3028F05 | Mm.34102 | Odc | |
| −2.2 | H3117B08 | −2 | H3013H07 | Mm.7884 | |||
| −2.2 | H3122H12 | Mm.196110 | Hba-a1 | −2 | L0058G12 | ||
| −2.2 | H3118A04 | Mm.288567 | Hbb-y | −2 | H3001F07 | Mm.24043 | Ermap |
| −2.1 | L0035C02 | Mm.233825 | Hbb-b1 | −2 | H3124D02 | Mm.1070 | Fech |
| −2.1 | H3057C12 | Mm.349140 | −2 | H3088C10 | Mm.222228 | Cks2 | |
| −2.1 | H3154A11 | Mm.336481 | |||||
| 2 | H3076H01 | Mm.240490 | |||||
| 2 | H3013E04 | Mm.8687 | Cap1 | ||||
| 2 | H3147D01 | Mm.195310 | |||||
| 2 | H3016F05 | Mm.260064 | Eif3s4 | ||||
| 2 | L0241C09 | Mm.24591 | |||||
| 2 | H3120F01 | Mm.200365 | Sv2a | ||||
| −2 | H3118A05 | Mm.288567 | Hbb-y | ||||
| −2 | H3122H11 | Mm.196110 | Hba-a1 | ||||
| −2 | H3118D11 | Mm.182630 |
Figure 5.
Presence of repeat elements in deregulated sequences. (A) Proportion of deregulated clones in brain or liver tissues that contain a repeat element. (B) Proportion of clones that contain different types of repeat elements. Note that a few clones contain more than one type of repeat elements. About 9% of clones did contain neither LTR elements, SINE, LINE nor DNA elements, and only simple repeats were present. (C) Whole bars indicate the proportion of deregulated sequences that contain repeats in relation to their magnitude of deregulation. Black bars indicate the proportion of deregulated sequences containing an LTR.
Among 14 upregulated sequences that did not contain repeats, six were uncharacterized, three were related to ribosomal proteins and five remaining genes appear functionally not related to each other (Table 1). Possibly their transcriptional increase is due to close proximity to retroviral-derived sequences. Among 32 sequences that were downregulated and did not contain repeat sequences, 8 were not characterized and 16 were sequences derived from the globin locus. The 8 remaining genes appear functionally unrelated. Since most of the downregulated sequences were found in the liver and were only altered 2- to 2.5-fold (see above), it is possible that their decrease is partly due to a defect in hematopoietic cell numbers in the liver as discussed below.
Lsh targets directly to repetitive sequences
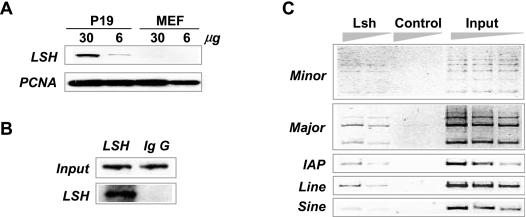
The Lsh protein shows preferential localization to pericentromeric heterochromatin, composed of major and minor satellite repeats (25). Furthermore, Lsh deletion primarily affects repetitive sequences with respect to transcription and posttranslational modification of histone tails. Thus, we hypothesized that Lsh may be directly recruited to repetitive sequences in the genome. To test this hypothesis, we performed ChIP analysis to examine the association of Lsh with specific DNA regions. The embryonic cell line P19 was selected for ChIP analysis, since western analysis revealed a more than 5-fold higher amount of endogenous Lsh protein in comparison with embryonal fibroblasts (Figure 6A). ChIPs were performed using a C-terminal antibody against Lsh to precipitate the endogenous Lsh protein after cross-linking to chromatin. Western analysis demonstrated a substantial enrichment of Lsh protein in the specifically precipitated DNA–protein complex in comparison with the IgG control (Figure 6B). PCR analysis of the immunoprecipitated DNA revealed an increase in repetitive sequences such as major and minor satellites, LINE, SINE and LTR elements (such as IAP) in the anti-Lsh precipitated chromatin fraction in comparison with the IgG control (Figure 6C). In contrast, the amount of single copy genes (such as APRT and actin) was not enriched in anti-Lsh precipitated chromatin compared to IgG controls (data not shown). Thus, Lsh is directly recruited to repetitive elements in the genome, making parasitic elements a primary target of Lsh action.
Figure 6.
Lsh targets to repetitive sequences. (A) Western analysis of Lsh expression in the embryonal carcinoma cell-line P19 and embryonal fibroblasts (MEFs) by using a specific antibody raised against the C-terminal portion of Lsh. (B) ChIPs were performed on P19 cells using the C-terminal antibody against Lsh or an IgG control. The precipitated material was examined by western analysis using the C-terminal anti-Lsh antibody for detection. (C) ChIPs were performed from P19 chromatin extracts using the C-terminal antibody against Lsh or an IgG control. Precipitated DNA or chromatin input was analyzed for the presence of the indicated repeat sequences. DNA was diluted (1:3) for the detection of linear PCR amplification.
DISCUSSION
Lsh is a guardian of heterochromatin
Repeat sequences derived from transposable elements are spread throughout the genome and may impose a threat to the genome integrity (31). Here, we demonstrate a specific role for Lsh on epigenetic regulation of repetitive elements that are frequently embedded within heterochromatin. Lsh deletion causes an increased association of acetylated histones with repetitive elements and this elevation closely correlates with an altered transcriptional rate. In contrast, no effect of histone acetylation or transcription was observed at several selected single copy genes. A similar pattern of histone methylation changes occurring only at repeat sequences as opposed to single copy sites in Lsh deleted tissue had been previously reported (26). Using microarray analysis in the present study, we show that Lsh primarily affects sequences that contain repeat elements. Furthermore, we provide evidence that Lsh is directly recruited to repetitive sites in the genome. These data suggest that Lsh plays an important role as an epigenetic regulator at repetitive parasitic elements, and we propose that Lsh specifically acts as guardian of heterochromatin organization and function.
How is Lsh targeted specifically to repeat sites?
Recently, several reports suggest that small RNA molecules are involved in the establishment of heterochromatin in lower organisms (32–34). For example, in fission yeast transcripts derived from centromeric regions can be detected and lead to the formation of double-stranded RNA (29,35–37). The long RNA transcripts are then targeted by components of the RNA interference machinery and cleaved into small 21–23 bp long RNA molecules. These small molecules then return to sites of their transcription and are crucial for the recruitment of H3–K9 methyltransferase activity and establishment of heterochromatin. Recent evidence has suggested that in mammalian cells also, heterochromatin formation depends on the RNA interference machinery (38). The recruitment of Suv39 proteins (H3–K9 methyltransferases) and HP1 may depend on an RNA component (39). Suv39 methyltransferases are important for the recruitment of DNA methyltransferases, DNA methylation at major satellite repeats and transcriptional regulation of satellite regions (39,40). Since the presence of Lsh is also crucial for DNA methylation at satellite repeats and the regulation of satellite transcripts, targeting of Lsh may similarly involve an RNA component. Some repetitive elements, to which Lsh is recruited, may be transcribed on both strands or contain an intrinsic repeat that facilitates the formation of double-stranded RNA, leading to small RNA molecules by the RNA interference pathway. These small RNA molecules may be instructional for the recruitment of Suv39h1/h2, HP1, and other critical components of heterochromatin formation such as Lsh. Deletion of Suv39 proteins in contrast to deletion of DNA methyltransferases moderately increases gene expression levels of major satellite repeats (40). Thus, DNA methylation alone may not sufficiently regulate the transcription of satellite sequences and Lsh may affect heterochromatin structure beyond DNA methylation. Alternatively, DNA methylation may have different effects in distinct cell types, being only critical in somatic cells (as used in this study) and not in stem cells (40). The observation that Lsh controls transcription and heterochromatin formation at major satellite sequences is consistent with a model in which small RNA molecules are instructional for recruitment of Lsh. However, the presence of small RNA molecules derived from major satellites has not been reported as yet in mammalian cells.
Does Lsh affect single copy sequences?
Currently, though we cannot exclude a direct impact of Lsh on selected single copy genes, it is possible that some of the abnormally expressed single copy sequences are indirectly affected by Lsh deletion. For example, the downregulation of single copy sequences (such as globin sequences, MyD116 or CD24) may be in part due to different cellular compositions in the analyzed tissue since Lsh deletion impairs lymphoid development (18) and hematopoiesis in general (T. Fan and K. Muegge, unpublished data). The liver is the site of hematopoiesis during embryogenesis and Lsh deletion may lead, for example, to reduced numbers of erythrocyte precursors in liver tissue, thus causing a moderate decrease in globin gene expression. Similarly, reduced PCNA (Table 1) expression may reflect mitotic problems as we have recently reported for MEF cells (21) rather than direct suppression of the PCNA promoter. As an alternate possibility to explain effects on some single copy genes, the spreading of chromatin states may be responsible. In fission yeast, it has been recently reported that the close proximity to LTR elements spreads heterochromatin formation and leads to silencing of genes (37). Upon deletion of Lsh, repeat elements may change their chromatin state and since repetitive elements can be located in introns, in promoter regions or in close proximity of coding sequences, these altered heterochromatin configuration may then spread into a few single copy sequences. This model may also explain the observation of moderate CpG methylation loss of Lsh−/− cells at selected single copy genes as judged by Southern analysis contrasting the more dramatic reduction in DNA methylation at repeat elements (22). Similarly, DDM1 mutations (a homologue of Lsh in A.thaliana) cause depletion of DNA methylation at repeats and only after consecutive rounds of breeding can an effect on single copy sites be observed (24).
What is the functional consequence of demethylated repeats?
Analysis of the whole genome revealed that the bulk of the mammalian genome is burdened with mobile elements (30). Up to 40–50% of the murine or human genome contain repeat sequences derived from transposable elements, including retrotransposon with LTR elements or without LTRs (such as LINE or SINE) that are able to amplify and transfer to new locations in the genome utilizing RNA intermediates or DNA transposons (41). More recent reviews consider a role for repeat elements in shaping the genome during evolution (42). For example, repetitive elements may have been incorporated into promoter regions or transposable elements may have been ‘tamed’ and used for endogenous gene rearrangement during V(D)J recombination. However, repeats are generally considered as selfish, parasitic elements whose mobility poses the threat of genomic mutations (41–43). DNA methylation may play a role in the transcriptional silencing and thus repression of transposable elements. In line with this idea, it had been reported that the transcription of the IAP is enhanced in the absence of dnmt1 and CpG methylation (44). In DDM1 mutants (a close homologue of Lsh in A.thaliana), reactivation and transposition of endogenous mobile elements was observed causing insertional mutagenesis (45). It is estimated that in the mouse genome up to 10% of all mutations are a consequence of retroviral insertion (41). The insertions may promote recombination or may disrupt coding sequences. Insertions may also lead to aberrant gene expression patterns by spreading of heterochromatin leading to silencing of adjacent tumor suppressor genes. Whether the reactivation of transposable elements in the absence of Lsh can indeed threaten genome integrity and promote tumorigenesis awaits further investigation.
In brief, normal heterochromatin organization is important for gene silencing, imprinting and centromere function. The burden of transposable elements in the mammalian genome demands additional strict control to suppress the activation and mobility of parasitic repeat elements. We propose that Lsh participates in normal heterochromatin organization and function and may be crucial in taming these parasitic elements to ensure genomic integrity.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Joost Oppenheim, Nancy Colburn and Howard Young for critical review of the manuscript. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, and Alternative Medicine Program, National Institutes of Health, under Contract No. NO1-CO-12400.
REFERENCES
- 1.Felsenfeld G. and Groudine,M. (2003) Controlling the double helix. Nature, 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 2.Khorasanizadeh S. (2004) The nucleosome: from genomic organization to genomic regulation. Cell, 116, 259–272. [DOI] [PubMed] [Google Scholar]
- 3.Fahrner J.A. and Baylin,S.B. (2003) Heterochromatin: stable and unstable invasions at home and abroad. Genes Dev., 17, 1805–1812. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff S. (2000) Heterochromatin function in complex genomes. Biochim. Biophys. Acta, 1470, O1–O8. [DOI] [PubMed] [Google Scholar]
- 5.Hashimshony T., Zhang,J., Keshet,I., Bustin,M. and Cedar,H. (2003) The role of DNA methylation in setting up chromatin structure during development. Nature Genet., 34, 187–192. [DOI] [PubMed] [Google Scholar]
- 6.Belotserkovskaya R., Saunders,A., Lis,J.T. and Reinberg,D. (2004) Transcription through chromatin: understanding a complex FACT. Biochim. Biophys. Acta, 1677, 87–99. [DOI] [PubMed] [Google Scholar]
- 7.Li E. (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nature Rev. Genet., 3, 662–673. [DOI] [PubMed] [Google Scholar]
- 8.Verona R.I., Mann,M.R. and Bartolomei,M.S. (2003) Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol., 19, 237–259. [DOI] [PubMed] [Google Scholar]
- 9.Sims R.J. III, Nishioka,K. and Reinberg,D. (2003) Histone lysine methylation: a signature for chromatin function. Trends Genet., 19, 629–639. [DOI] [PubMed] [Google Scholar]
- 10.Richards E.J. and Elgin,S.C. (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell, 108, 489–500. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 12.Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 13.Goll M.G. and Bestor,T.H. (2002) Histone modification and replacement in chromatin activation. Genes Dev., 16, 1739–1742. [DOI] [PubMed] [Google Scholar]
- 14.Eberharter A. and Becker,P.B. (2002) Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep., 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis C.D., Geiman,T., Vila-Storm,M.P., Osipovich,O., Akella,U., Candeias,S., Nathan,I., Durum,S.K. and Muegge,K. (1996) A novel putative helicase produced in early murine lymphocytes. Gene, 169, 203–207. [DOI] [PubMed] [Google Scholar]
- 16.Becker P.B. and Horz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. [DOI] [PubMed] [Google Scholar]
- 17.Peterson C.L. (2002) Chromatin remodeling enzymes: taming the machines. Third in review series on chromatin dynamics. EMBO Rep., 3, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiman T.M. and Muegge,K. (2000) Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc. Natl Acad. Sci. USA, 97, 4772–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiman T.M., Tessarollo,L., Anver,M.R., Kopp,J.B., Ward,J.M. and Muegge,K. (2001) Lsh, a SNF2 family member, is required for normal murine development. Biochim. Biophys. Acta, 1526, 211–220. [DOI] [PubMed] [Google Scholar]
- 20.Geiman T.M., Durum,S.K. and Muegge,K. (1998) Characterization of gene expression, genomic structure, and chromosomal localization of Hells (Lsh). Genomics, 54, 477–483. [DOI] [PubMed] [Google Scholar]
- 21.Fan T., Yan,Q., Huang,J., Austin,S., Cho,E., Ferris,D. and Muegge,K. (2003) Lsh-deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res., 63, 4677–4683. [PubMed] [Google Scholar]
- 22.Dennis K., Fan,T., Geiman,T., Yan,Q. and Muegge,K. (2001) Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev., 15, 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L.Q., Lee,D.W., Zhang,Q., Xiao,W., Raabe,E.H., Meeker,A., Miao,D., Huso,D.L. and Arceci,R.J. (2004) Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev., 18, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeddeloh J.A., Stokes,T.L. and Richards,E.J. (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet., 22, 94–97. [DOI] [PubMed] [Google Scholar]
- 25.Yan Q., Cho,E., Lockett,S. and Muegge,K. (2003) Association of lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol. Cell. Biol., 23, 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan Q., Huang,J., Fan,T., Zhu,H. and Muegge,K. (2003) Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J., 22, 5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J., Durum,S.K. and Muegge,K. (2001) Cutting edge: histone acetylation and recombination at the TCR gamma locus follows IL-7 induction. J. Immunol., 167, 6073–6077. [DOI] [PubMed] [Google Scholar]
- 28.Kargul G.J., Dudekula,D.B., Qian,Y., Lim,M.K., Jaradat,S.A., Tanaka,T.S., Carter,M.G. and Ko,M.S. (2001) Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nature Genet., 28, 17–18. [DOI] [PubMed] [Google Scholar]
- 29.Volpe T.A., Kidner,C., Hall,I.M., Teng,G., Grewal,S.I. and Martienssen,R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science, 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- 30.Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 31.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 32.Schramke V. and Allshire,R. (2004) Those interfering little RNAs! Silencing and eliminating chromatin. Curr. Opin. Genet. Dev., 14, 174–180. [DOI] [PubMed] [Google Scholar]
- 33.Ekwall K. (2004) The RITS complex-A direct link between small RNA and heterochromatin. Mol. Cell, 13, 304–305. [DOI] [PubMed] [Google Scholar]
- 34.Elgin S.C. and Grewal,S.I. (2003) Heterochromatin: silence is golden. Curr. Biol., 13, R895–R898. [DOI] [PubMed] [Google Scholar]
- 35.Hall I.M., Shankaranarayana,G.D., Noma,K., Ayoub,N., Cohen,A. and Grewal,S.I. (2002) Establishment and maintenance of a heterochromatin domain. Science, 297, 2232–2237. [DOI] [PubMed] [Google Scholar]
- 36.Reinhart B.J. and Bartel,D.P. (2002) Small RNAs correspond to centromere heterochromatic repeats. Science, 297, 1831. [DOI] [PubMed] [Google Scholar]
- 37.Schramke V. and Allshire,R. (2003) Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science, 301, 1069–1074. [DOI] [PubMed] [Google Scholar]
- 38.Fukagawa T., Nogami,M., Yoshikawa,M., Ikeno,M., Okazaki,T., Takami,Y., Nakayama,T. and Oshimura,M. (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nature Cell Biol., 6, 784–791. [DOI] [PubMed] [Google Scholar]
- 39.Maison C., Bailly,D., Peters,A.H., Quivy,J.P., Roche,D., Taddei,A., Lachner,M., Jenuwein,T. and Almouzni,G. (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nature Genet., 30, 329–334. [DOI] [PubMed] [Google Scholar]
- 40.Lehnertz B., Ueda,Y., Derijck,A.A., Braunschweig,U., Perez-Burgos,L., Kubicek,S., Chen,T., Li,E., Jenuwein,T. and Peters,A.H. (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol., 13, 1192–1200. [DOI] [PubMed] [Google Scholar]
- 41.Deininger P.L. and Batzer,M.A. (2002) Mammalian retroelements. Genome Res., 12, 1455–1465. [DOI] [PubMed] [Google Scholar]
- 42.van de Lagemaat L.N., Landry,J.R., Mager,D.L. and Medstrand,P. (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet., 19, 530–536. [DOI] [PubMed] [Google Scholar]
- 43.Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 44.Walsh C.P., Chaillet,J.R. and Bestor,T.H. (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet., 20, 116–117. [DOI] [PubMed] [Google Scholar]
- 45.Miura A., Yonebayashi,S., Watanabe,K., Toyama,T., Shimada,H. and Kakutani,T. (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature, 411, 212–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.