Abstract
Aedes aegypti is a major vector for arboviruses such as dengue, chikungunya, and Zika viruses. During acquisition of a viremic bloodmeal, an arbovirus infects mosquito midgut cells before disseminating to secondary tissues, including the salivary glands. Once virus is released into the salivary ducts it can be transmitted to another vertebrate host. The midgut is surrounded by a basal lamina (BL) in the extracellular matrix, consisting of a proteinaceous mesh composed of collagen IV and laminin. BL pore size exclusion limit prevents virions from passing through. Thus, the BL likely requires remodelling via enzymatic activity to enable efficient virus dissemination. Matrix metalloproteinases (MMPs) are extracellular endopeptidases that are involved in remodelling of the extracellular matrix. Here, we describe and characterize the nine Ae. aegypti encoded MMPs, AeMMPs 1-9, which share common features with other invertebrate and vertebrate MMPs. Expression profiling in Ae. aegypti revealed that Aemmp4 and Aemmp6 were upregulated during metamorphosis, whereas expression of Aemmp1 and Aemmp2 increased during bloodmeal digestion. Aemmp1 expression was also upregulated in the presence of a bloodmeal containing chikungunya virus. Using polyclonal antibodies, AeMMP1 and AeMMP2 were specifically detected in tissues associated with the mosquito midgut.
Keywords: Aedes aegypti, midgut, bloodmeal, matrix metalloproteinase, basal lamina, virus dissemination, catalytic domain, phylogeny
Introduction
The yellow fever mosquito, Aedes aegypti, is a major vector for flaviviruses (family: Flaviviridae; genus: Flavivirus) such as dengue and Zika viruses and alphaviruses (family: Togaviridae; genus: Alphavirus) such as chikungunya virus (CHIKV). These viruses are currently the most prevalent arboviruses that infect humans and are co-circulating in many regions of the tropical world (Weaver and Reisen, 2010; Weaver, 2013).
The arboviral disease cycle requires persistent infection of an arthropod vector before virus can be transmitted to a new vertebrate host (reviewed in: Franz et al., 2015). A mosquito acquires virus from a viremic host along with the bloodmeal. The bloodmeal enters the midgut and within several hours, virions will infect the epithelial cells lining the midgut. Hours later, virions disseminate from midgut epithelial cells to infect secondary tissues or cells, such as hemocytes, fat body, nerve tissue, and eventually the salivary glands. Once these are infected, virus is released along with saliva when the mosquito is feeding on a vertebrate host. Like all epithelia, the midgut epithelium is lined with a basal lamina (BL), a sheet-like network of extracellular matrix (ECM) components including collagen IV, laminin, and proteoglycans (Yurchenco and O'Rear, 1994). The typical BL pore size exclusion limit in mosquitoes is only 8-9 nm, which is too small for flavivirus or alphavirus virions, which are 50-80 nm in diameter, to pass through (Houk et al., 1981). Thus, it has been postulated that the BL needs to be remodelled, thereby increasing its pore size exclusion limit before virions can disseminate from the midgut to secondary tissues. This hypothesis is supported by observations with another model system, the lepidopteran insect Trichoplusia ni and the baculovirus Autographa californica M nucleopolyhedrovirus (AcMNPV), which also must overcome a midgut escape barrier in the caterpillar in order to establish a systemic infection (Means and Passarelli, 2010; Passarelli, 2011). A midgut escape mechanism for AcMNPV has been proposed, which involves the activity of a viral fibroblast growth factor, MMPs, and caspases, resulting in BL remodelling to enhance virus dissemination from the midgut.
In vertebrates, metalloproteinases are the most important enzymes responsible for ECM remodelling (Cawston and Young, 2010). Metalloproteinases are grouped into two major families of zinc-dependent endopeptidases: MMPs and a disintegrin and metalloproteinase (ADAM) / ADAM with thrombospondin motifs (ADAMTS) proteinases. In humans, endopeptidases of the ADAMs family have been implicated in the control of membrane fusion, cytokine and growth factor shedding, and cell migration, as well as processes such as muscle development, fertilization, and cell fate determination (Seals and Courtneidge, 2003). MMPs cleave a wide range of ECM proteins (including collagenase IV and laminin), degrade intercellular junctions, generate cleavage products that act as novel signalling molecules, and/or modify the action of latent or active signalling molecules thereby changing cell and tissue physiology (Page-McCaw et al., 2007). Specifically, MMPs are essential for connective tissue remodelling during embryonic development, angiogenesis, bone growth, and healing of wounds (Nagase and Woessner, 1999; Vu and Werb, 2000; Rowe and Weiss, 2008). Vertebrate MMPs are grouped according to substrate specificity into collagenases, matrilysins, stromelysins, and gelatinases, the latter two degrading laminin and/or collagen IV (Brinckerhoff and Matrisian, 2002). Another classification scheme distinguishes between "membrane-type" MMPs (MT-MMPs) and "other" MMPs (Browner et al., 1995).
MMPs are expressed as zymogens with a characteristic domain structure consisting of a propeptide domain, a catalytic domain, and a hemopexin domain (Massova et al., 1998). The propeptide domain is located at the N-terminus and contains a conserved cysteine residue containing amino acid (aa) sequence motif, PRCGXXD, known as the cysteine switch (CS), which helps to maintain an inactive MMP conformation. Upon activation of the enzyme, the propeptide domain is cleaved releasing the cysteine from a Zn2+ ion, allowing access to the catalytic domain (Lopez-Pelegrin et al., 2015). The catalytic domain is located downstream of the propeptide domain and is identified by the highly conserved amino acid sequence motif HEXXGHXXGXXH, containing the Zn-binding site (Bode et al., 1993). The hemopexin domain is located at the C-terminus of the protein and is connected to the catalytic domain via a flexible linker region (Fasciglione et al., 2012). The hemopexin domain plays a major role in substrate specificity (Overall, 2002; Brew et al., 2000). To date, there have been at least 23 human MMPs (HuMMPs) identified and characterized (Brinckerhoff and Matrisian, 2002; Mott and Werb, 2004; Jackson et al., 2010). Several HuMMPs have diverse functional domain structures, especially at their C-termini. Some of them do not contain a hemopexin domain (Uria and Lopez-Otin, 2000; Velasco et al., 1999), while the HuMT-MMPs have either a transmembrane domain or a glycosylphosphatidylinositol (GPI) anchor associated with their hemopexin domain (Sato et al., 1994).
MMPs are not restricted to vertebrates; they also have been described and analysed in plants such as Arabidopsis thaliana (Marino et al., 2014) and tomato (Li et al., 2015), in the nematode Caenorhabditis elegans (Wada et al., 1998) and in insects such as Drosophila melanogaster, Manduca sexta, Tribolium castaneum, and the malaria vector, Anopheles gambiae (Llano et al., 2000, 2002; Page-McCaw et al., 2003; Page-McCaw, 2008; Mitten et al., 2012, Vishnuvardan et al., 2013; Knorr et al., 2009; Goulielmaki et al., 2014). In insects, MMP activity is required for wound healing and tissue rearrangements during moults and metamorphosis involving ECM degradation (Page-McCaw et al., 2003; Knorr et al., 2009; Bond et al., 2011; Miller et al., 2008; Mitten et al., 2012). Furthermore, insect MMPs have been shown to be involved in innate immunity responses to tissue invasion by pathogens (Knorr et al., 2009; Schmidt et al., 2011; Goulielmaki et al., 2014). For Ae. aegypti, MMPs and their possible roles in biological processes have not been described so far.
Here, we examine nine putative MMPs found in the genome of Ae. aegypti. We analyse the phylogenetic relationships of these Ae. aegypti MMPs (AeMMPs) with those of other arthropods and the well-described HuMMPs. We use qRT-PCR to conduct expression profiling of Aemmp genes during metamorphosis, bloodmeal digestion and arbovirus infection using CHIKV as a model. Antibodies to two AeMMPs, which are overexpressed during bloodmeal digestion were produced to analyse their expression at the protein level in Western blots and in situ.
Results
Identification of MMPs in the genome of Ae. aegypti
Searches of the genome sequence of Ae. aegypti in VectorBase (AaegL3) revealed the presence of nine putative AeMMP genes, which we are designating AeMMP1-9 according to homologies with other arthropod MMPs and the order of their gene accession numbers in VectorBase (Table 1). AeMMP1and AeMMP2 are orthologs of MMP1 and MMP2 of Ixodes scapularis, D. melanogaster, Bombyx mori, T. castaneum, An. gambiae, Culex quinquefasciatus, and Ae. albopictus, respectively. AeMMP3 is an ortholog of MMP3 of T. castaneum, An. gambiae, Cx. quinquefasciatus, and Ae. albopictus. With the exception of AeMMP2 (chromosome 3) and AeMMP3 (chromosome 1), all AeMMP genes are encoded on chromosome 2. For AeMMP9, a chromosome location could not be identified. AeMMPs predict similar lengths as HuMMPs (261-707 aa) with AeMMP3 (605 aa) and AeMMP9 (273 aa) having the longest and shortest amino acid sequences, respectively (Table 1). Five of the nine AeMMPs contain the signature propeptide, catalytic, and hemopexin domains, while AeMMP9 is a truncated gene copy (paralog) of AeMMP1, lacking the hemopexin domain (Fig. 1A). Generally, the catalytic domains of MMPs are highly conserved, whereas propeptide and hemopexin domains typically show high levels of divergence (Massova et al., 1998; Overall, 2002). Alignment of the propeptide domains of the AeMMPs showed the presence of a CS motif (Fig. 1B). Even though the CS motifs of AeMMPs vary slightly from the canonical mammalian CS motif, PRCGXXD, they all contain the conserved cysteine residue at or nearby amino acid position 80. Other MMPs such as HuMMP28 also encode a variation of the canonical CS motif without compromising their function (Lohi et al., 2001). In addition, AeMMP1, AeMMP3, AeMMP6, AeMMP7, and AeMMP9 contain a putative RXXR furin cleavage site. Alignment of the catalytic domains revealed that the MMP hallmark amino acid sequence motif HEXGHXXGXXH was present in all nine AeMMPs (Fig. 1C). All AeMMPs, except AeMMP9, possess a flexible linker region that links the catalytic domain to the C-terminal hemopexin domain. Further, AeMMP1 is the only AeMMP that appears to be membrane-associated as it is predicted to possess a GPI anchor (based on in silico prediction using the web-based tool GPI-Som: http://genomics.unibe.ch/cgi-bin/gpi.cgi?ref=1&id). We also noticed that AeMMP2 had been incorrectly annotated in VectorBase. We performed 5' RACE for Aemmp2 and discovered that the translational start site is located at the end of exon 4 (Fig. S1). Also, when aligning the amino acid sequence of AeMMP2 with its homolog from Ae. albopictus (AaMMP2), it became obvious that according to its annotation in VectorBase, AeMMP2 but not AaMMP2, contains 120 additional N-terminal amino acids. Regardless, AeMMP2, similar to AeMMP5 and AeMMP8, lacks a propeptide domain. In summary, five of the nine AeMMPs that we have annotated possess the typical domains found in most MMPs (propeptide, catalytic, and hemopexin domains), whereas three of them lack the propeptide, and one lacks the hemopexin domain, demonstrating a high level of diversity within this gene family in mosquitoes.
Table 1.
Identification and designation of putative MMP genes in the genome of Aedes aegypti based on VectorBase searches.
| gene ID | supercontig | chromosome | designation | homology to | aac | transcript |
|---|---|---|---|---|---|---|
| AAEL005666 | 1.169 | 2pa | Aemmp1 e | Aa, Ag, B, C, D, I, Tb | 573 | 3050 |
| AAEL000788 | 1.16 | 3q | Aemmp2 e | Aa, Ag, B, C, D, I, T | 526d | 1581d |
| AAEL002982 | 1.74 | 1q | Aemmp3 | Aa, Ag, C, T | 605 | 2214 |
| AAEL002655 | 1.63 | 2q | Aemmp4 e | Aa, C | 481 | 2249 |
| AAEL002661 | 1.63 | 2q | Aemmp5 | Aa, C | 430 | 1615 |
| AAEL002665 | 1.63 | 2q | Aemmp6 e | Aa, C | 492 | 1507 |
| AAEL002672 | 1.63 | 2q | Aemmp7 | Aa, C | 493 | 2347 |
| AAEL002677 | 1.63 | 2q | Aemmp8 | Aa, C | 479 | 1440 |
| AAEL017046 | 1.1354 | ? | Aemmp9 | - | 273 | 822 |
Based on supercontig mapping data (Timoshevsky et al., 2014; Juneja et al., 2014)
Aa = Aedes albopictus, Ag = Anopheles gambiae, B = Bombyx mori, C = Culex quinquefasciatus, D = Drosophila melanogaster, I = Ixodes scapularis, T = Tribolium castaneum
aa = amino acids
aa sequence determination and transcript length are based on 5' RACE data shown in Supplemental Figure S1
mRNA transcript was detectable by RT-PCR
Figure 1.
Characterization of the functional domains of AeMMPs. (A) Mapping of key domains for AeMMPs1-9; CS = cysteine switch; RXXR = furin cleavage site; GPI = glycosylphosphatidylinositol anchor. (B) Amino acid sequence alignment of the propeptide domains of AeMMPs. CS motifs are highlighted in red; a canonical CS motif from a vertebrate MMP is shown for comparison. (C) Amino acid sequence alignment of the catalytic domains of AeMMPs. Catalytic domain amino acid sequences of DmMMP1 and HuMMP9 are included for comparison.
Similarity of AeMMPs with MMPs of humans and other arthropods
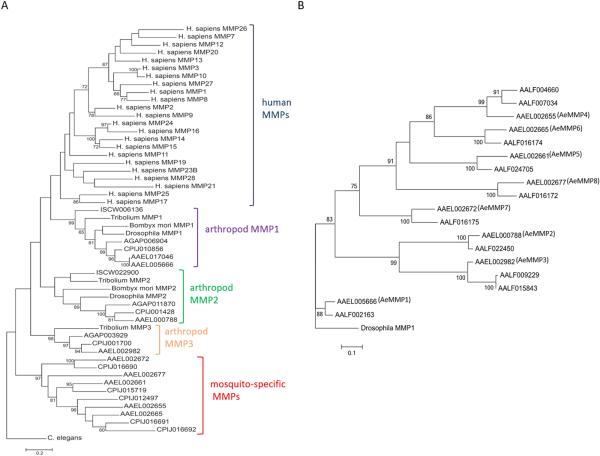
Maximum likelihood phylogenetic analyses based on nucleotide sequences revealed that arthropods such as D. melanogaster, B. mori, T. castaneum, I. scapularis, and mosquitoes (An. gambiae, Cx. quinquefasciatus, and Ae. albopictus) all possess orthologues of AeMMP1 and AeMMP2 (Fig. 2A). T. castaneum and mosquitoes also share orthologues of AeMMP3. Apparently, the Culex and Aedes mosquito genomes underwent a further MMP gene expansion with Culex having five additional MMPs, three of which are orthologous between Ae. aegypti and Cx. quinquefasciatus, and Aedes sp. possessing up to seven additional MMPs (Fig. 2A, B). All arthropod MMPs cluster together but separately from the mammalian MMPs, with the arthropod MMP1 being most similar to HuMMPs. We performed a second maximum likelihood phylogenetic study comparing the MMP nucleotide sequences between Ae. aegypti and Ae. albopictus (Fig. 2B). It became apparent that all AeMMPs are homologs to those of Ae. albopictus (AaMMPs). AeMMP9 is an exception as it is a gene duplication of AeMMP1 but lacking the hemopexin domain. In Ae. albopictus, MMP genes underwent further expansion as there are two AaMMPs homologous to AeMMP4 and two homologous to AeMMP3. The Ae. albopictus genome contains another nucleotide sequence, which is homologous to that of AeMMP1; however, the sequence only encodes the hemopexin domain (lacking catalytic and propeptide domains) and cannot be considered a functional MMP. Those MMPs of Cx. quinquefasciatus and Ae. albopictus that are homologous to AeMMP2, AeMMP5, and AeMMP8 also lack a propeptide domain.
Figure 2.
Phylogenetic relationships of AeMMPs with human and other arthropod MMPs. Maximum likelihood phylogeny for (A) arthropod and human AeMMPs and (B) for AeMMPs and Ae. albopictus (Aa)MMPs based on nucleotide sequences from which the linker encoding sequences (between catalytic and hemopexin motifs) have been removed. Bootstrap values greater than 59 are shown and indicate support as a percentage of 1000 replicates. Scale bars represent 0.2 (A) and 0.1 (B) substitutions per site. Species used for analysis: I. scapularis (ISCW), An. gambiae (AGAP), Cx. quinquefasciatus (CPIJ), Ae. aegypti (AAEL), Ae. albopictus (AALF).
Expression profiles of AeMMP genes during different life stages of Ae. aegypti and in females that received a CHIKV-containing bloodmeal
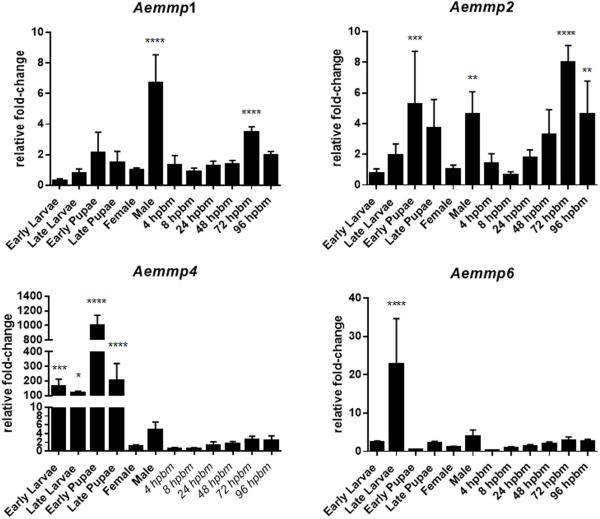
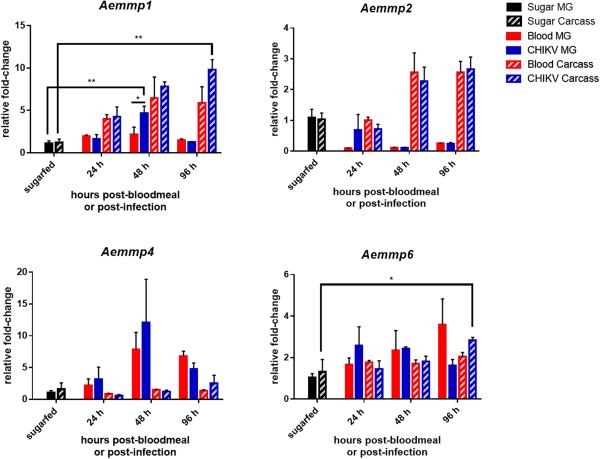
We analysed AeMMP mRNA expression levels during different mosquito life stages via qRT-PCR and compared transcript abundances to those of sugarfed female mosquitoes. We only detected expression of four of the nine AeMMP genes: Aemmp1, Aemmp2, Aemmp4, and Aemmp6 (Fig. 3 and data not shown). Generally, Aemmp1 and Aemmp2 exhibited low expression levels in the mosquito, not changing more than 10-fold irrespective of the developmental stage of the mosquito. Aemmp1 was highly expressed in males and upregulated in females at 72 h post-bloodmeal (hpbm). Aemmp1 expression also responded to the presence of CHIKV (titre in the bloodmeal: 107 pfu/ml) with significantly increased gene expression in midguts at 48 h post-infection and in carcasses at 96 h post-infection (Fig. 4). Aemmp2 was most highly expressed in early pupae and males, and was upregulated in females at 72 and 96 hpbm (Fig. 3). However, Aemmp2 expression in either mosquito midguts or carcasses did not significantly respond to the presence of CHIKV (Fig. 4).
Figure 3.
Expression profiles for Aemmp1, Aemmp2, Aemmp4, and Aemmp6 during different life stages of Ae. aegypti. Total RNA was extracted from HWE mosquitoes at different life cycle stages and from whole-body females, which had received a sugarmeal or a bloodmeal (4-96 hpbm). Relative fold-change transcript abundance was analysed by qRT-PCR for each AeMMP gene by normalizing reads against the abundance of ribosomal protein S7 transcripts. Values were then set in relation to the gene expression level in sugarfed females (baseline value = 1 relative fold-change). Mean values with standard deviations (SD) from three independent experiments are shown. Statistical analysis was performed using a one-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons test (* at p ≤ 0.05).
Figure 4.
Expression profiles for Aemmp1, Aemmp2, Aemmp4, and Aemmp6 in midguts and carcasses of Ae. aegypti females which had received a non-infectious or a CHIKV-containing bloodmeal. Total RNA was extracted at 24, 48, and 96 h post-feeding from midguts and carcasses of HWE mosquitoes, which had received a sugarmeal, a bloodmeal diluted (1:1) with non-infected cell culture medium, or a bloodmeal diluted (1:1) with culture medium from CHIKV-infected Vero cells (titre in the bloodmeal: 107 pfu/ml). Relative fold-change transcript abundance was analysed by qRT-PCR for each AeMMP gene by normalizing reads against the abundance of ribosomal protein S7 transcripts. Values were then set in relation to the gene expression level in sugarfed midguts (baseline value = 1 relative fold-change). Mean values with standard deviations from three independent experiments are shown. Statistical analysis was performed using a one-way analysis of variance followed by Tukey's multiple comparisons test (* at p ≤ 0.05).
Aemmp4 and Aemmp6 were significantly upregulated at earlier stages of mosquito development. Aemmp4 was 100- to 1000-fold upregulated between early larva and late pupa stages, whereas Aemmp6 was upregulated 22-fold during the late larval stage of development.
Expression levels of Aemmp4 and Aemmp6 were not significantly increased in the adult stage irrespectively of whether mosquitoes had received a sugarmeal, a virus-free bloodmeal, or a bloodmeal containing CHIKV (Fig. 3, 4). The only exception was the significant (albeit only ~2-fold) upregulation of Aemmp6 in carcasses of CHIKV-infected mosquitoes at 96 h post-infection. Our observations support the hypothesis that Aemmp4 and Aemmp6 may be predominantly involved in tissue remodelling during metamorphosis, while Aemmp1 and Aemmp2 were responsive to bloodmeal digestion in the female. Aemmp1 expression was also significantly increased in midguts and carcasses when CHIKV was present in the bloodmeal.
Detection of AeMMP1 and AeMMP2 proteins in midguts of Ae. aegypti
Polyclonal antibodies, pAb-mmp1 and pAb-mmp2, were produced against short peptides derived from the hemopexin domains of AeMMP1 and AeMMP2, respectively. In Western blots, pAb-mmp1 and pAb-mmp2 detected their corresponding antigens when expressed as recombinant proteins in Escherichia coli (Fig. S2). Two band signals detected by pAb-mmp1 corresponded to molecular masses of ~75 kDa and ~60 kDa and two band signals detected by pAb-mmp2 were around 75 and 65 kDa. Taking the N-terminal protein tag sequences (Trx-tag, S-tag, and His-tag) originating from the expression vector [pET-32a(+)] into account, the 75 kDa band signals of AeMMP1 and AeMMP2 corresponded to the molecular masses of the complete AeMMP proteins; the predicted molecular masses of the three plasmid-derived tags, AeMMP1, and AeMMP2 were 15kDa, 63 kDA, and 60 kDA, respectively. Detection of AeMMP2 with an anti-His-tag Ab (pAb-HisTag) resulted in a signal pattern similar to that detected with pAb-mmp2, while pAb-HisTag failed to detect AeMMP1 recombinant protein. pAb-mmp1 and pAb-mmp2 did not detect heterologous antigen or antigen in non-IPTG induced bacterial cultures. Furthermore, antigen detected by the preimmune serum (with similar intensities in IPTG- or non-induced cultures) was distinct from that detected by pAb-mmp1 and pAb-mmp2. These observations suggest that both antibodies specifically detected their homologous AeMMP antigens.
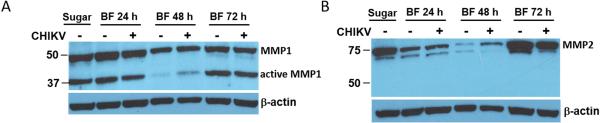
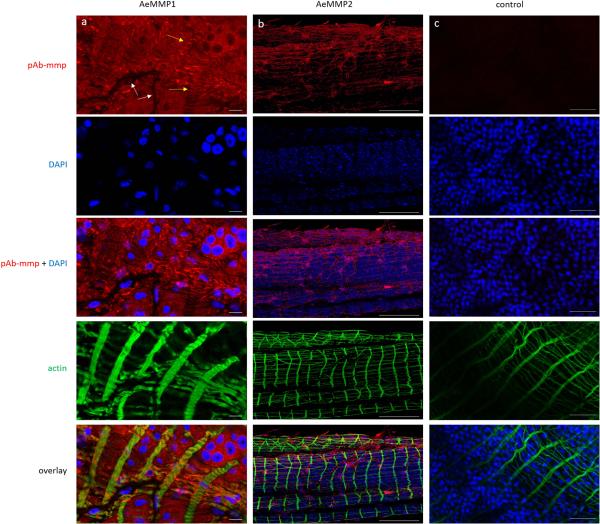
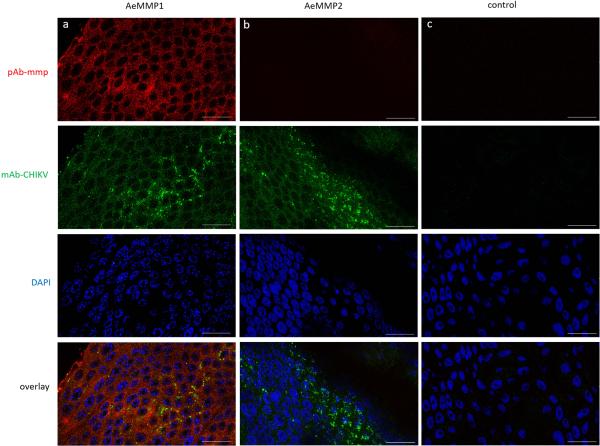
In midguts of Ae. aegypti, AeMMP1 was detected with similar intensities at 24 and 72 hpbm or CHIKV infection (virus titre in the bloodmeal: 107 pfu/ml) (Fig. 5A). At 48 hpbm/CHIKV infection, AeMMP1 antigen seemed to be less abundant though the β-actin loading control was also slightly reduced. Two band signals with molecular masses of 50 and 37 kDa were detected in mosquito midgut extracts by pAb-mmp1. According to molecular mass predications, the 50 kDa band possibly corresponded to the zymogen form of AeMMP1, while the 37 kDa band could represent its active form after cleavage of its ~15 kDa propeptide domain. This indicates that upon its expression in midguts, AeMMP1 may be rapidly proteolytically processed, for example via furin cleavage (see also Fig. 1A) as has been observed for AgMT-MMP1 in mosquito cells (Goulielmaki et al., 2014). pAb-mmp2 detected in midgut tissue two closely migrating band signals of around 75 kDa and 70 kDa, which were larger than the predicted molecular mass of AeMMP2 protein (Fig. 5B). We speculate that post-translational modifications such as protein glycosylation could account for this molecular mass discrepancy. AeMMP2 antigen was less detectable at 24 and 48 hpbm compared to 72 hpbm or in midguts of sugarfed mosquitoes. Similar to AeMMP1, the presence of CHIKV did not obviously affect the abundance of AeMMP2 antigen. In immunofluorescence assays, AeMMP1 was detected in epithelial tissue of the midgut and in midgut-associated muscle fibres but not in tracheal cells, whereas AeMMP2 was only detectable in midgut-associated tracheal cells (Fig. 6). Acquisition of a bloodmeal or the presence of CHIKV in the bloodmeal did not obviously affect the abundance of either AeMMP antigen in midgut-associated tissues or the tissue specificity of either AeMMP (Fig. 7). AeMMP1 antigen, like CHIKV antigen was associated with epithelial tissue and both antigens co-localized in the tissue at 96 h post-infection. AeMMP2 antigen was not detected in epithelial cells where CHIKV infection occurred.
Figure 5.
Detection of AeMMP1 and AeMMP2 antigens by Western blot analysis using pAb generated against peptides derived from each AeMMP. (A) Western blot detection of AeMMP1 antigen by pAb-mmp1 and (B) AeMMP2 antigen by pAb-mmp2 in midguts of mosquitoes that had received a sugarmeal, a bloodmeal diluted 1:1 with non-infected cell culture medium, or a bloodmeal diluted 1:1 with CHIKV-infected cell culture medium (titre in the bloodmeal: 107 pfu/ml) at 24, 48, and 72 h post-feeding (BF). β-actin was used as a loading control. Molecular masses in kDa are indicated. Data in the figure are representative of experiments that have been repeated twice.
Figure 6.
Detection of AeMMP1 and AeMMP2 antigens in midguts of sugarfed Ae. aegypti by indirect immunofluorescence assays using Alexa Fluor 594 labelled (red) anti-rabbit IgG as secondary antibody. Midguts were incubated with either (a) pAb-mmp1, (b) pAb-mmp2, or (c) control with no pAb-mmp. Cell nuclei were stained with DAPI (blue) and actin was stained with Alexa Fluor Phalloidin 488 (green). White and yellow arrows indicate midgut-associated tracheal cells and muscle fibers, respectively. Scale bars: (a, b) = 10 μm, (c) = 50 μm. Confocal microscopy was performed using an inverted spectral confocal microscope (TCP SP8 MP, Leica Microsystems). Selected images are representative of six replicates.
Figure 7.
Detection of AeMMP1 and AeMMP2 antigens in midguts of CHIKV-infected Ae. aegypti by indirect immunofluorescence assays using Alexa Fluor 488 labelled (green) anti-mouse IgG and Alexa Fluor 594 labelled (red) anti-rabbit IgG as secondary antibodies. Midguts were incubated with (a, b) mAb-CHIKV at 96 h post-infection and either (a) pAb-mmp1, (b) pAb-mmp2, or (c) control with no mAb-CHIKV or pAb-mmp. Cell nuclei were stained with DAPI (blue). Scale bars = 25 μm. Confocal microscopy was performed using an inverted spectral confocal microscope (TCP SP8 MP, Leica Microsystems). Selected images are representative of three replicates.
Discussion
In this study, we initially characterized the MMPs of Ae. aegypti to identify those that may play a role in midgut BL remodelling during bloodmeal digestion and arbovirus dissemination.
Arthropods have two to three MMPs in common, whereas mosquitoes of the subfamily Culicinae that have been examined so far possess an additional five (Cx. quinquefasciatus) to seven (Ae. albopictus) MMPs, which all are evolutionally distinct from HuMMPs. In Ae. aegypti, the majority of the nine MMPs contained the canonical propeptide, catalytic, and hemopexin domains. Generally, propeptide and hemopexin domains are highly variable among MMPs and those of Ae. aegypti are no exception (Massova et al., 1998; Overall, 2002). In contrast, the catalytic domains were highly similar among AeMMPs. This may be an indication that according to hemopexin and propeptide domain diversities, each AeMMP is targeting a specific type of extracellular substrate and is activated under a specific circumstance (Knorr et al., 2009; Sternlicht and Werb, 2001).
Overall, MMP activity is tightly regulated at several levels; at the transcriptional level (MMP expression), via propeptide cleavage (MMP activation), via interaction with a tissue inhibitor of MMP protein (TIMP; MMP inhibition), and eventually through protease degradation (Page-McCaw et al., 2007). The Ae. aegypti genome, like that of other arthropods, encodes a single TIMP gene (AAEL013525), which is most similar to human TIMP3 (Knorr et al., 2009; Ra and Parks, 2007; Gomez et al., 1997; Brew et al., 2000). In qRT-PCR experiments, we obtained expression data based on fold-change in transcript abundance for four of the nine AeMMP genes: Aemmp1, Aemmp2, Aemmp4, and Aemmp6. For the remaining five AeMMP genes, transcripts were not detected by RT-PCR in whole body mosquitoes after testing several gene-specific primer combinations (data not shown). Specific detection of Aemmp9 was not possible as its nucleotide sequence, although being truncated, is nearly identical to that of Aemmp1. A previous RNA-seq experiment showed that Aemmp3, Aemmp5, Aemmp7, and Aemmp8 were expressed in whole-body females of the Liverpool strain of Ae. aegypti; however, Aemmp3 and Aemmp8 exhibited only very low expression levels, hardly exceeding 10 fragments per kilobase of transcript per million mapped reads (FPKM) (Bonizzoni et al., 2012). Our qRT-PCR analysis supports the conclusion that Aemmp4 and Aemmp6 expression was strongly induced during metamorphosis, whereas Aemmp1 and Aemmp2 expression was induced in the female during bloodmeal digestion. Thus, AeMMP1 and AeMMP2 could potentially be involved in the dissemination process of CHIKV from the midgut. This prompted us to produce antibodies specific to each of the two AeMMPs, allowing us to further analyse their presence and activity in specific tissues at the protein level.
The Drosophila ortholog of MMP1 is secreted while the An. gambiae ortholog of AeMMP1 is alternatively spliced and either transmembrane-linked (MT-MMP) to be expressed in epithelial tissues when possessing a C-terminal GPI anchor or secreted from hemocytes of the mosquito when lacking the GPI anchor (Llano et al., 2000; Goulielmaki et al., 2014). Our study did not reveal that Ae. aegypti, like An. gambiae, expresses two isoforms of MMP1; however, according to in silico prediction tools, AeMMP1 possesses a GPI anchor. Functional analysis of DmMMP1 and AgMT-MMP1 revealed that both are involved in basement membrane remodelling, including fibronection/collagen IV degradation, tissue repair, and wound healing (Llano et al., 2000; Page-McCaw et al., 2003; Srivastava et al., 2007; Glasheen et al., 2009; Stevens and Page-McCaw, 2012). Upregulation of DmMMP1 during wound healing occurs via the extracellularly regulated jun N-terminal kinase (JNK) signalling pathway.
In An. gambiae, AgMT-MMP1 responded strongly to Plasmodium ookinete invasion of midgut epithelial cells and also to non-parasite-containing bloodmeal acquisition (Goulielmaki et al., 2014). Changes in AgMT-MMP1 activity were manifested at the transcriptional level but also at the level of zymogen cleavage and subcellular activation. As ookinete invasion leads to tissue damage, AgMT-MMP1 may be strongly activated to facilitate wound healing. Furthermore, AgMT-MMP1 activity may be required to enable midgut tissue overstretching during bloodmeal acquisition and/or tissue regeneration following bloodmeal digestion (Goulielmaki et al., 2014). Arboviruses such as CHIKV have not been observed to actively penetrate and damage the ECM during their infection cycle in the mosquito vector. However, midgut infection and dissemination of arboviruses are associated with the ingestion of a bloodmeal. CHIKV dissemination from the midgut has been observed as early as 24 h post-oral acquisition of the virus, a time point at which the epithelial tissue of the blood-filled midgut was still overstretched (Dong et al., 2016). Thus, it appears that the virus was already disseminating from the midgut before transcriptional upregulation of Aemmp1 had reached its peak, which occurred at around 72 hpbm, a time point at which the bloodmeal was already digested. Similarly, Aemmp2 expression levels were significantly increased in bloodfed mosquitoes towards the end of bloodmeal digestion with increased gene expression occurring outside the midgut. In situ studies showed that AeMMP1 was abundant in midgut epithelial cells and muscle fibres whereas AeMMP2 was associated with tracheal cells. Both AeMMP antigens were readily detected in midgut tissues of sugarfed mosquitoes and antigen abundance did not seemingly increase over a 96 h period following ingestion of a bloodmeal containing (or not containing) CHIKV with the exception of AeMMP2 at 72 hpbm. Based on these observations, we speculate that at the end of bloodmeal digestion, increased AeMMP1 and AeMMP2 production may have occurred in tissues outside the midgut epithelium for potential ECM remodelling required after the bloodmeal digestion process to restore overexpanded tissues to their original size. Although we do not see evidence of AeMMP upregulation until late during bloodmeal digestion, we cannot rule out the possibility that AeMMP activity may be essential for CHIKV dissemination from the midgut. Additional, functional studies will be needed to test these hypotheses.
Experimental procedures
Mosquitoes
Ae. aegypti mosquitoes of the RexD (Puerto Rico)-derived, eye-pigment deficient Higgs White Eye (HWE) strain (Wendell et al., 2000; Dong et al., 2016) were reared in a BSL2 insectary under a regime of 28°C with 75-80% relative humidity and a 12 h light/12 h dark cycle. For colony maintenance, mosquitoes received artificial bloodmeals consisting of defibrinated sheep blood (Colorado Serum Company, Denver, CO).
Database searches and phylogenetic analysis
The Ae. aegypti (strain: Liverpool) genome in VectorBase (assembly: AaegL3) was analysed to identify putative MMPs based on the conserved amino acid motif: HEXXGHXXGXXH. We used the NCBI conserved protein domain prediction tool to determine the functional domains for the putative AeMMPs. Phylogenetic analyses were conducted using the LINUX-based T-COFFEE program package (Notredame et al., 2000) for the global MMP alignments or MEGA vs. 5 for the AeMMP/AaMMP alignments. Prior to alignments, the DNA sequences encoding the variable linker motifs between catalytic and hemopexin domains were removed from all MMP sequences. Global MMP alignments were analysed using PhyML (maximum likelihood) under the Hasegawa-Kishino-Yano (HKY) model (Guindon and Gascuel, 2003; Hasegawa et al., 1985) choosing the C. elegans MMP as an outcross group. The AeMMP/AaMMP alignments were further analysed using MEGA vs. 5 using Drosophila MMP1 as an outcross group. Bootstrap values of 1000 were applied in both analyses.
Generation of cDNAs and two-step qRT-PCR
Larvae and pupae of the HWE strain were collected at 2, 4, 6, and 8 days after hatching. Males were collected at 2 days post-emergence; female mosquitoes at 2 days post-emergence and 4, 8, 24, 48, 72, and 96 hpbm. Midguts and carcasses were collected at 24, 48, and 96 h after receiving a CHIKV-containing (titre in the bloodmeal: 107 pfu/ml) or a non-infectious bloodmeal and from sugarfed mosquitoes. Groups of ten larvae, pupae, whole-body mosquitoes, or midguts, and carcasses were collected for total RNA extraction using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesised from 1 μg total RNA using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany) including the random (reverse) primer supplied with the kit. cDNA was diluted 10-fold to be used in quantitative PCR assays. Gene-specific primers were used for qPCR amplification of Aemmps (Table S1). The final reaction volume was 25 μl using the QuantiFast SYBR green PCR kit (Qiagen, Hilden, Germany). qPCR amplification and analysis were carried out using an Applied Biosystems (ABI) 7300 Real-Time PCR System. The PCR program was: hold at 95°C for 10 min and then at 95°C for 15 seconds and 60°C for 1 min, repeating 40 cycles. The relative abundance of Aemmp transcripts was normalised to that of ribosomal protein S7 (AAEL009496) transcripts as an endogenous reference following the 2−ΔΔCT method (Livak and Schmittgen, 2001). Samples were tested as three independent biological replicates.
Expression of recombinant AeMMP1 and AeMMP2 proteins
Full-length Aemmp1 and Aemmp2 cDNAs were amplified by RT-PCR from total RNA of Ae. aegypti. PCR amplicons of Aemmp1 and Aemmp2 cDNAs were inserted into the pET32α expression vector (Novagen, Madison, WI, USA) using XhoI/EcoRI and XhoI/BamHI restriction sites, respectively, to express the MMPs along with the N-terminal Trx-, S-, and His-tags in E. coli BL21 (DE3) cells (New England Biolabs, Ipswich, MA, USA). Protein production was optimal when bacterial cultures were grown at 37°C and shaken at 225 rpm. A 5-ml bacterial "starter" culture from a single colony was grown overnight in LB medium (BD Biosciences, San Jose, CA, USA) containing 100 μg/ml ampicillin. The "starter" culture was then transferred into 500 ml of fresh LB medium and incubated for 2-3 h until the culture reached an OD = 0.6 (at 600 nm wavelength). IPTG was then added at a final concentration of 1mM and the bacterial culture incubated for another 4 h at 37°C and 225 rpm.
Generation of AeMMP1 and AeMMP2-specific antibodies and their use in Western blot analysis
Polyclonal antibodies specific to AeMMP1 and AeMMP2 were produced by GenScript (Piscataway, NJ, USA) based on the peptide amino acid sequences CGTDERPAGDDYNRG and CFEGDSQNRPPVEID (with the N-terminal C being non-specific), respectively. Both amino acid peptides were derived from the hemopexin domain of the MMPs. To prepare samples, five midguts were dissected from mosquitoes at 24, 48, 72 hpbm/post-infection with CHIKV and homogenized in 2x Laemmli sample buffer, boiled for 5 min, and centrifuged at 10,000 rpm for 10 min. For recombinant AeMMP detection, cultures were grown as described above. Two millilitre of the induced culture was centrifuged for 5 min at 13,000 rpm, to pellet the bacteria. Bacteria pellets were then resuspended in 2x Laemmli sample buffer, boiled for 5 min, and centrifuged at 10,000 rpm for 10 min.
Proteins from supernatants were electrophoretically separated by 12% SDS-PAGE and transferred to a nitrocellulose membrane, which was blocked for 1 h in a 5% non-fat dried milk/Tris-buffered saline solution (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.1% Tween 20, pH 7.5; TBST). The membrane was then incubated overnight at 4°C in the blocking solution containing either pAb-mmp1 at 1:3000 dilution or pAb-mmp2 at 1:6000 dilution in TBST. Following overnight incubation with the primary antibody, the membrane was washed three times (10 min/wash) in TBST before being incubated with anti-rabbit IgG-HRP (1:4000, Cell Signaling Technology, Danvers, MA, USA) for 2 h at room temperature. Finally, the membrane was treated for 1 min with SuperSignal West Pico chemiluminescent substrate (Pierce, Waltham, MA, USA). The immunoreactive proteins were visualized by exposing the membrane to an x-ray film. Anti-β-actin-peroxidase antibody (Sigma-Aldrich, St. Louis, MO, USA) was used as a loading control for each lane.
Immunofluorescence assays
Midguts were dissected from sugarfed or CHIKV-infected HWE females at 96 h post-infection. A group of 10 midguts was fixed in 200 μl 4% p-formaldehyde (Sigma-Aldrich) for up to one week at 4°C. Midguts were washed three times with PBS, and permeabilized with 200 μl PBT (1×PBS, 1% BSA, 0.2% Triton X-100) for 1 h on a rocker at room temperature. Samples of sugarfed mosquitoes were incubated overnight at 4°C in 100 μl pAb-mmp1 or pAb-mmp2 at a dilution of 1:100 in PBT. Samples were washed four times with 200 μl PBST (1×PBS, 0.1% Triton X-100) and incubated at 37°C in the dark for 1.5 h with 100 μl anti-rabbit IgG Alexa Fluor 594 (Cell Signaling Technology) diluted 1:200 in PBT. Samples of CHIKV-infected mosquitoes were incubated overnight at 4°C with a mixture of anti-CHIKV monoclonal antibody (mAb) [B1414] (Abcam, Cambridge, UK), diluted 1:100 and pAb-mmp1 or pAb-mmp2 at a dilution of 1:100 in PBT. Samples were washed four times with 200 μl PBST and incubated at 37°C in the dark for 1.5 h with 100 μl anti-mouse IgG Alexa Fluor 488 (Cell Signaling Technology), diluted 1:800 and anti-rabbit IgG Alexa Fluor 594 (Cell Signaling Technology) diluted 1:200 in PBT. To stain nuclei, 5 μl of a 0.5 μg/μl DAPI (Invitrogen) was added to each tube followed by incubation at room temperature for 30 min. Actin filaments of midguts not infected with CHIKV were stained with 200 μl of a 1 unit/ml solution of Alexa Fluor Phalloidin 488 (Invitrogen) for another 30 min at room temperature. After four washes with PBST, midguts were placed individually on glass slides, and mounted with Fluoromount-G (Electron Microscopy Sciences, Hatfield, PA, USA). Samples were viewed and analysed under an inverted spectral confocal microscope (TCP SP8 MP, Leica Microsystems).
5’ RACE
Whole bodies of mosquitoes were collected for total RNA extraction using TRIzol reagent (Invitrogen). Total RNA was treated with DNase I to eliminate DNA contamination. Reverse gene-specific primers (Table S1) were designed to anneal 450 base pairs downstream of the known/anticipated 5' end of the RNA transcript. The 5' RACE System for Rapid Amplification of cDNA Ends, version 2.0 (Invitrogen) was used to perform the analysis based on the protocol provided. Resulting PCR products were cloned into TOPO-TA pCR4.0 for Sanger sequencing.
CHIKV infection of mosquitoes
CHIKV strain 37997 (GenBank accession: AY726732.1; Dong et al., 2016) was propagated in Vero cells in T25 flasks at a multiplicity of infection of 0.01 using Minimum Essential Medium Eagle (MEM) with 7% FBS. For oral challenge of mosquitoes, virus-containing cell culture supernatant was collected at 24-30 h post-infection and mixed with defibrinated sheep blood at a 1:1 ratio including the addition of 10 mM ATP. Seven day-old females that had been sugar-starved overnight were exposed for 1 h to an artificial bloodmeal containing virus infected cell culture/blood mixture or non-infected cell culture/blood mixture (control) using a single glass feeder/carton lined with hog intestine. Fully engorged females were maintained in 1.9 L cartons and offered raisins and water until further analysis. All CHIKV infections were carried out in a Biosafety Level 3 laboratory within the Laboratory for Infectious Disease Research (LIDR) at the University of Missouri.
Supplementary Material
Acknowledgements
The authors thank Dr. Gavin Conant from the Animal Reproductive Group of the Animal Science Research Center of the University of Missouri for his help with the phylogenetic analyses. We also thank Dr. Alexander Jurkevich of the University of Missouri Molecular Cytology Core for his assistance with confocal imaging. The chikungunya virus experiments were conducted in the Laboratory for Infectious Disease Research (LIDR) of the University of Missouri. This study was supported by NIH-NIAID grant R01AI091972 (ALP, RJC, AWEF).
References
- Bode W, Gomis-Ruth FX, Stockler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the 'metzincins'. FEBS Lett. 1993;331(1-2):134–40. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- Bond ND, et al. ßFTZ-F1 and Matrix metalloproteinase 2 are required for fat-body remodeling in Drosophila. Dev Biol. 2011;360(2):286–96. doi: 10.1016/j.ydbio.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Bonizzoni M, et al. Complex modulation of the Aedes aegypti transcriptome in response to dengue virus infection. PLoS One. 2012;7(11):e50512. doi: 10.1371/journal.pone.0050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta. 2000;1477(1-2):267–83. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3(3):207–14. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- Browner MF, Smith WW, Castelhano AL. Matrilysin-inhibitor complexes: common themes among metalloproteases. Biochemistry. 1995;34(20):6602–10. doi: 10.1021/bi00020a004. [DOI] [PubMed] [Google Scholar]
- Cawston TE, Young DA. Proteinases involved in matrix turnover during cartilage and bone breakdown. Cell Tissue Res. 2010;339(1):221–35. doi: 10.1007/s00441-009-0887-6. [DOI] [PubMed] [Google Scholar]
- Dong S, et al. Infection pattern and transmission potential of chikungunya virus in two New World laboratory-adapted Aedes aegypti strains. Sci Rep. 2016;6:24729. doi: 10.1038/srep24729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasciglione GF, et al. The collagenolytic action of MMP-1 is regulated by the interaction between the catalytic domain and the hinge region. J Biol Inorg Chem. 2012;17(4):663–72. doi: 10.1007/s00775-012-0886-z. [DOI] [PubMed] [Google Scholar]
- Franz AW, et al. Tissue Barriers to Arbovirus Infection in Mosquitoes. Viruses. 2015;7(7):3741–67. doi: 10.3390/v7072795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasheen BM, Kabra AT, Page-McCaw A. Distinct functions for the catalytic and hemopexin domains of a Drosophila matrix metalloproteinase. Proc Natl Acad Sci U S A. 2009;106(8):2659–64. doi: 10.1073/pnas.0804171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez DE, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol. 1997;74(2):111–122. A.D. [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Goulielmaki E, Sidén-Kiamos I, Loukeris TG. Functional characterization of Anopheles matrix metalloprotease 1 reveals its agonistic role during sporogonic development of malaria parasites. Infect Immun. 2014;82(11):4865–77. doi: 10.1128/IAI.02080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22(2):160–74. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Houk EJ, Hardy JL, Chiles RE. Permeability of the midgut basal lamina in the mosquito, Culex tarsalis Coquillett (Insecta, Diptera) Acta Trop. 1981;38(2):163–71. [PubMed] [Google Scholar]
- Jackson BC, Nebert DW, Vasiliou V. Update of human and mouse matrix metalloproteinase families. Hum Genomics. 2010;4(3):194–201. doi: 10.1186/1479-7364-4-3-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja P, et al. Assembly of the genome of the disease vector Aedes aegypti onto a genetic linkage map allows mapping of genes affecting disease transmission. PLoS Negl Trop Dis. 2014;8(1):e2652. doi: 10.1371/journal.pntd.0002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr E, et al. MMPs regulate both development and immunity in the tribolium model insect. PLoS One. 2009;4(3):e4751. doi: 10.1371/journal.pone.0004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, et al. Tomato Sl3-MMP, a member of the Matrix metalloproteinase family, is required for disease resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000. BMC Plant Biol. 2015;15:143. doi: 10.1186/s12870-015-0536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llano E, et al. Structural and enzymatic characterization of Drosophila Dm2-MMP, a membrane-bound matrix metalloproteinase with tissue-specific expression. J Biol Chem. 2002;277(26):23321–9. doi: 10.1074/jbc.M200121200. [DOI] [PubMed] [Google Scholar]
- Llano E, et al. Dm1-MMP, a matrix metalloproteinase from Drosophila with a potential role in extracellular matrix remodeling during neural development. J Biol Chem. 2000;275(46):35978–85. doi: 10.1074/jbc.M006045200. [DOI] [PubMed] [Google Scholar]
- Lohi J, et al. Epilysin, a novel human matrix metalloproteinase (MMP-28) expressed in testis and keratinocytes and in response to injury. J Biol Chem. 2001;276(13):10134–44. doi: 10.1074/jbc.M001599200. [DOI] [PubMed] [Google Scholar]
- Lopez-Pelegrin M, et al. A novel mechanism of latency in matrix metalloproteinases. J Biol Chem. 2015;290(8):4728–40. doi: 10.1074/jbc.M114.605956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, et al. Family-wide characterization of matrix metalloproteinases from Arabidopsis thaliana reveals their distinct proteolytic activity and cleavage site specificity. Biochem J. 2014;457(2):335–46. doi: 10.1042/BJ20130196. [DOI] [PubMed] [Google Scholar]
- Massova I, et al. Matrix metalloproteinases: structures, evolution, and diversification. Faseb J. 1998;12(12):1075–95. [PubMed] [Google Scholar]
- Means JC, Passarelli AL. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc Natl Acad Sci U S A. 2010;107(21):9825–30. doi: 10.1073/pnas.0913582107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CM, Page-McCaw A, Broihier HT. Matrix metalloproteinases promote motor axon fasciculation in the Drosophila embryo. Development. 2008;135(1):95–109. doi: 10.1242/dev.011072. [DOI] [PubMed] [Google Scholar]
- Mitten EK, Jing D, Suzuki Y. Matrix metalloproteinases (MMPs) are required for wound closure and healing during larval leg regeneration in the flour beetle, Tribolium castaneum. Insect Biochem Mol Biol. 2012;42(11):854–64. doi: 10.1016/j.ibmb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol. 2004;16(5):558–64. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Woessner JF., Jr. Matrix metalloproteinases. J Biol Chem. 1999;274(31):21491–4. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Overall CM. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22(1):51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A. Remodeling the model organism: matrix metalloproteinase functions in invertebrates. Semin Cell Dev Biol. 2008;19(1):14–23. doi: 10.1016/j.semcdb.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8(3):221–33. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A, et al. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell. 2003;4(1):95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- Passarelli AL. Barriers to success: how baculoviruses establish efficient systemic infections. Virology. 2011;411(2):383–92. doi: 10.1016/j.virol.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra HJ, Parks WC. Control of matrix metalloproteinase catalytic activity. Matrix Biol. 2007;26(8):587–96. doi: 10.1016/j.matbio.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RG, Weiss SJ. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18(11):560–74. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Sato H, et al. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370(6484):61–5. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- Schmidt RL, et al. Cleavage of PGRP-LC receptor in the Drosophila IMD pathway in response to live bacterial infection in S2 cells. Self Nonself. 2011;2(3):125–141. doi: 10.4161/self.17882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Srivastava A, et al. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc Natl Acad Sci U S A. 2007;104(8):2721–6. doi: 10.1073/pnas.0611666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LJ, Page-McCaw A. A secreted MMP is required for reepithelialization during wound healing. Mol Biol Cell. 2012;23(6):1068–79. doi: 10.1091/mbc.E11-09-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoshevsky VA, et al. Genomic composition and evolution of Aedes aegypti chromosomes revealed by the analysis of physically mapped supercontigs. BMC Biology. 2014;12:27. doi: 10.1186/1741-7007-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uria JA, Lopez-Otin C. Matrilysin-2, a new matrix metalloproteinase expressed in human tumors and showing the minimal domain organization required for secretion, latency, and activity. Cancer Res. 2000;60(17):4745–51. [PubMed] [Google Scholar]
- Velasco G, et al. Cloning and characterization of human MMP-23, a new matrix metalloproteinase predominantly expressed in reproductive tissues and lacking conserved domains in other family members. J Biol Chem. 1999;274(8):4570–6. doi: 10.1074/jbc.274.8.4570. [DOI] [PubMed] [Google Scholar]
- Vishnuvardhan S, et al. Identification of a novel metalloproteinase and its role in juvenile development of the tobacco hornworm, Manduca sexta (Linnaeus) J Exp Zool B Mol Dev Evol. 2013;320(2):105–17. doi: 10.1002/jez.b.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14(17):2123–33. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- Wada K, et al. Cloning of three Caenorhabditis elegans genes potentially encoding novel matrix metalloproteinases. Gene. 1998;211(1):57–62. doi: 10.1016/s0378-1119(98)00076-6. [DOI] [PubMed] [Google Scholar]
- Weaver SC. Urbanization and geographic expansion of zoonotic arboviral diseases: mechanisms and potential strategies for prevention. Trends Microbiol. 2013;21(8):360–3. doi: 10.1016/j.tim.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85(2):328–45. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell MD, et al. Chemical and gamma-ray mutagenesis of the white gene in Aedes aegypti. Insect Mol Biol. 2000;9(2):119–25. doi: 10.1046/j.1365-2583.2000.00166.x. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, O'Rear JJ. Basal lamina assembly. Curr Opin Cell Biol. 1994;6(5):674–81. doi: 10.1016/0955-0674(94)90093-0. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, O'Rear JJ. Basement membrane assembly. Methods Enzymol. 1994;245:489–518. doi: 10.1016/0076-6879(94)45025-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.