ABSTRACT
Within the protective outer membrane (OM) fraction of Anaplasma marginale, several vaccine candidates have emerged, including a family of OM proteins (OMPs) 7 to 9, which share sequence identity with each other and with the single protein OMP7 in the vaccine strain A. marginale subsp. centrale. A. marginale OMPs 7 to 9 are logical vaccine candidates because they are surface exposed, present in the OM immunogen and protective cross-linked OM proteins, recognized by immune serum IgG2 and T cells in cattle immunized with OM, and recognized by immune serum IgG2 from cattle immunized with the A. centrale vaccine strain. We report the identification of a globally conserved 9-amino-acid T-cell epitope FLLVDDAI/VV shared between A. centrale vaccine strain OMP7 and the related A. marginale OMPs 7 to 9, where position 8 of the peptide can be isoleucine or valine. The epitope is conserved in American A. marginale strains, in the Australia Gypsy Plains strain, and in multiple field isolates from Ghana. This epitope, together with additional T-cell epitopes that are present within these proteins, should be considered for inclusion in a multivalent vaccine for A. marginale that can provide protection against disease caused by globally distributed bacterial strains.
KEYWORDS: Anaplasma centrale, Anaplasma marginale, outer membrane protein 7, outer membrane protein 8, outer membrane protein 9, T-cell epitopes
INTRODUCTION
Anaplasma marginale is an intraerythrocytic rickettsial pathogen of cattle which, together with other tick-borne pathogens, is responsible for significant economic losses to beef and dairy cattle industries and small-holder farmers worldwide (1). Although it has been repeatedly documented that purified bacterial outer membranes (OMs) can protect cattle against clinical disease and even infection in some individuals (2–4), production of such a vaccine for widespread use is economically unfeasible (5). Therefore, vaccine development ultimately requires the identification of protective antigens and epitopes within the protective OM fraction for inclusion in subunit vaccines (5). Prior studies have identified both IgG2 antibody and CD4 T cells as important for protective immunity against this pathogen, which replicates within erythrocytes devoid of major histocompatibility complex (MHC) molecules (6). Thus, a protective vaccine could consist of multiple proteins and/or their relevant epitopes that stimulate CD4 T cells and B cells.
To identify potentially protective CD4 T-cell antigens, proteins in St. Maries strain A. marginale OMs were separated by two-dimensional electrophoresis, and immunoblots were screened with immune serum IgG2 from either homologous St. Maries OM-immunized cattle or heterologous A. marginale subsp. centrale (here referred to as A. centrale)-immunized cattle (7, 8). A. centrale is a live vaccine currently in use in many countries to protect cattle against heterologous virulent A. marginale strains (5, 9–11). Antigenic proteins identified by mass spectrometry revealed several proteins recognized by both sets of immune sera that were all shown to be surface exposed (4). This provided a candidate list of outer membrane proteins (OMPs) with antibody epitopes shared between A. marginale and A. centrale: Am779, Am854, and OMP7. Recombinant Am854 was recently tested as a vaccine in cattle and did not afford protection (12).
OMP7 is a member of a family of related proteins (OMP7, OMP8, and OMP9) encoded by tandemly arranged genes as part of an operon that share 70 to 75% amino acid sequence identity (13, 14). In A. centrale, this locus is represented by one gene that encodes a protein designated OMP7, which has 64 to 70% identity with OMPs 7 to 9 of A. marginale (15). In addition, OMPs 7 to 9 are highly conserved among North American strains of A. marginale for which sequence information exists (8, 14). Inclusion of conserved T- and B-cell epitopes is an important consideration for a vaccine to provide broad strain cross-protection. Thus, OMPs 8 to 9 were also included in the focused list of five surface-exposed antigens, with antibody epitopes shared by A. centrale and A. marginale, as potential vaccine candidates selected for further study (16).
Incorporation of CD4 T-cell epitopes into a subunit vaccine is critical to provide help to B cells for isotype switching to high-avidity IgG2, so the ability of recombinant OMPs 7 to 9 to stimulate a recall CD4 T-cell response in OM-immunized cattle was investigatrecall CD4 T-cell response in OMed (17). All three proteins were shown to be immunogenic for T cells. In the present study, we test the hypothesis that A. centrale OMP7 and A. marginale OMPs 7 to 9 share at least one T-cell epitope for incorporation in a multivalent subunit vaccine. A vaccine comprised of multiple surface-exposed antigens may recapitulate the protection afforded by vaccination with live A. centrale, A. marginale OM, a complex of OMPs linked by covalent bonds, or the individual proteins within this OM complex (18).
RESULTS
Alignment of A. centrale OMP7 and A. marginale OMPs 7 to 9 predicted amino acid sequences.
In the A. marginale St. Maries strain, OMPs 7 to 9 are encoded by a multigene operon and share 70 to 75% amino acid sequence identity (13, 14). In A. centrale, the locus is represented by one gene, designated omp7, and the encoded OMP7 has 64 to 70% identity with OMPs 7 to 9 of A. marginale (Fig. 1) (15).
FIG 1.
Alignment of the predicted amino acid sequences for A. central OMP7 (OMP7c) and A. marginale St. Maries strain OMPs 7 to 9 (OMP7m, OMP8m, and OMP9m). Underlined sequences contain T-cell epitopes, and the shaded sequences indicate a broadly conserved T-cell epitope. The shaded sequence in A. centrale OMP7 indicates the T-cell epitope not conserved with A. marginale OMPs 7 to 9. Asterisks indicated amino acids that are identical in the four sequences.
Response of cattle with selected DRB3 haplotypes to A. centrale OMP7 and A. marginale OMPs 7 to 9 after immunization.
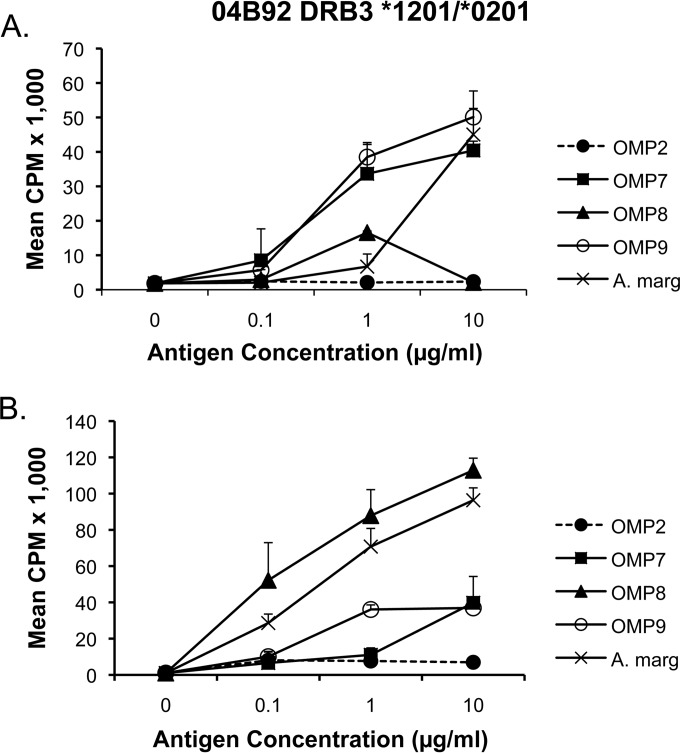
In screening potentially antigenic proteins by immunoblotting A. marginale OM separated by two-dimensional gel electrophoresis with immune bovine IgG2, it was discovered that three OM-immunized cattle (04B90, 04B91, and 04B92) made antibody responses to a protein spot that contained OMP7, identified by mass spectrometry (7). Animal 04B91 expressing DRB3*1201/*2703 had strong T-cell proliferative responses to recombinant OMPs 7 to 9, whereas animal 04B90 expressing DRB3*1101/*1501 had relatively weak responses to OMP7 and OMP9, with no response to OMP8 (17). In previously unpublished data, T-cell lines, enriched for CD4 T cells, from animal 04B92 (DRB3*1201/*0201) were also tested for recall T-cell proliferation to all three recombinant OMPs, and the data from two assays are now presented in Fig. 2. There was a dose-dependent and significant response to A. marginale OM and OMPs 7 to 9 in both assays. Compared to negative control URBC (data not shown), the response was significant for 10 μg/ml OM in the first assay, where P < 0.01 (Fig. 2A) and for all three OM concentrations in the second assay, where P < 0.005 (Fig. 2B). Compared to negative control OMP2, significantly greater responses occurred for OMP8 and OMP9 at 1 μg/ml and for OMP7 and OMP9 at 10 μg/ml in the first assay, where P < 0.05 (Fig. 2A), for OMPs 7 to 9 at 10 μg/ml, and for OMP8 at 0.1 and 1 μg/ml in the second assay, where P < 0.05 (Fig. 2B). The comparable response to OM and individual OMP7, OMP8, and OMP9 is presumably because the response to OM is a polyclonal response to many different OMPs, including OMPs 7 to 9, whereas the response to individual OMP7, OMP8, or OMP9 will be targeted to one or a few epitopes but present at a much higher concentration. Collectively, these studies suggest that cattle expressing the DRB3*1201 haplotype respond strongly to OMPs 7 to 9, although the DRB3*1101, DRB3*1501, and DRB3*2703 haplotypes may also contribute to the T-cell response. For this reason, cattle expressing combinations of these DRB3 haplotypes were selected for identification of T-cell epitopes in the family of related A. marginale OMPs 7 to 9, and A. centrale OMP7 proteins after immunization with A. marginale St. Maries strain OM.
FIG 2.
Proliferative response by animal 04B92 (DRB3*1201/*0101) to A. marginale OMPs 7 to 9. Two-week CD4-T-cell-enriched cell lines from A. marginale OM-immunized animal 04B92 (7) were tested in two separate assays for proliferation against A. marginale OM and recombinant proteins used at 0.1, 1, and 10 μg/ml. The results are presented as the mean cpm of replicate cultures plus one standard deviation of the mean. Responses to OM were compared to URBC, using the Student t test. Responses to OMP7, OMP8, and OMP9 were compared to negative control OMP2 using a one-tailed Student t test with Bonferroni's correction for multiple comparisons, where P < 0.05.
Response of haplotype-selected cattle to A. marginale and peptides spanning A. centrale OMP7 and A. marginale OMPs 7 to 9.
To verify that cattle responded to A. marginale after immunization with OM, peripheral blood mononuclear cells (PBMC) were tested with OM or URBC antigen and TCGF at 1 μg/ml as a positive control for viability. Table 1 shows that all five animals had significant responses to OM postimmunization compared to the response before immunization.
TABLE 1.
PBMC response to A. marginale OM before and after immunization
| Antigena | Proliferative response to antigen reported as the pre- and postimmunization stimulation index (SD) from five test animalsb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 48406 (*2703/*2703) |
48411 (*1201/*1501) |
48422 (*1201/*1201) |
48432 (*1101/*1402) |
48453 (*1501/*1501) |
||||||
| Pre | Post | Pre | Post | Pre | Post | Pre | Post | Pre | Post | |
| URBC | 1.0 (0.4) | 0.3 (0.1) | 2.4 (2.4) | 1.2 (1.6) | 0.6 (0.2) | 2.7 (3.1) | 5.1 (7.2) | 6.9 (6.2) | 1.8 (0.8) | 1.8 (0.5) |
| OM | 1.2 (0.4) | 7.6 (0.4) | 8.3 (2.1) | 182.0 (4.3) | 5.3 (3.2) | 58.6 (6.1) | 27.5 (16.9) | 187.0 (11.9) | 4.9 (0.9) | 8.6 (1.4) |
| TCGF | 13.4 (3.4) | 4.1 (1.5) | 110.9 (2.7) | 92.9 (3.7) | 112.4 (10.4) | 36.7 (14.4) | 91.4 (5.0) | 137.5 (9.8) | 204.9 (9.8) | 24.8 (3.0) |
OMs were purified from the St. Maries strain of A. marginale, and URBC served as a negative control. Both antigens were used at 1 μg/ml. TCGF was used at a final concentration of 10% as a positive control for proliferation.
Animal numbers (DRB3 haplotypes) are indicated in the column subheadings. Boldfaced numbers indicate that the response to antigen or TCGF postimmunization was significantly greater than the response before immunization, i.e., where P < 0.02 (determined by a one-tailed Student t test).
Next, 2-week cell lines were derived from these cattle by stimulating PBMC for 1 week with OM and resting for 1 week. We have shown that this enriches the cell lines for A. marginale-specific CD4 T cells and that the CD4 T cells are the responding population (3, 19–21). These were then used in proliferation assays to screen 30-mer peptides overlapping by 10 amino acids that span A. centrale OMP7 and A. marginale OMPs 7 to 9 (see Table S1 in the supplemental material) to identify those that induce an anamnestic response. Of the five immunized cattle, only three had significant responses to any of the OMP peptides when tested in multiple experiments. Animals 48432 (DRB3*1101/*1402) and 48406 (DRB3*2703/*2703) failed to respond (data not shown), whereas animals 48411 (DRB3*1201/*1501), 48422 (DRB3*1201/*1201), and 48453 (DRB3*1501/*1501) had consistent responses to one or more peptides spanning one protein (48453) or all four proteins (48411 and 48422) (Fig. 3). These data suggest that of our selected cattle, only those expressing MHC class II DRB3*1201 or *1501 haplotypes respond to one or more of the family of related OMPs. Animals 48411 and 48422, but not animal 48453, responded to A. centrale OMP7 peptides 8 and 14 and to A. marginale OMPs 7 to 9 peptide 7 and, to a lesser extent, peptide 8. Comparison of amino acid sequences of the proteins (Fig. 1) and peptides (see Table S1 in the supplemental material) shows that the A. centrale OMP7 peptide 8 and A. marginale OMPs 7 to 9 overlapping peptides 7 and 8 all share a similar 10-amino-acid sequence (FLLVDDAI/VVR), suggesting that this may contain a conserved T-cell epitope shared among these related OMP proteins. This is likely presented by DRB3*1201 or a DQ molecule expressed by this haplotype. In addition, these two cattle, but not 48453, recognized A. marginale OMP8 peptides 3 and 9 and A. marginale OMP9 peptides 11 and 12, again likely presented by DRB3*1201 or a DQ protein expressed by this haplotype. Animal 48411 recognized A. marginale OMP9 peptide 10, as did animal 48453, which responded only to this peptide among all of the tested peptides (Fig. 3 and data not shown). These cattle share the DRB3*1501 haplotype, suggesting DRB3*1501 or a haplotype-expressed DQ molecule presents this peptide to T cells.
FIG 3.
Proliferative responses of T-cell lines to peptides spanning A. centrale (Ac) OMP7 and A. marginale (Am) OMPs 7 to 9. Synthetic 30-amino-acid peptides overlapping by 10 amino acids were tested in triplicate for the induction of proliferation of 2-week T-cell lines from each indicated animal. The stimulation indices (SI) are indicated on the y axis, and the peptide number is indicated on the x axis. Results are presented only where there was a positive response to a peptide spanning one of the proteins. Asterisks indicate an SI of >4.0, which was considered to be significant. The results are representative of two or three separate assays for each animal.
Mapping the T-cell epitopes shared by A. centrale OMP7 and A. marginale OMPs 7 to 9.
Since A. centrale is used in many countries as a vaccine strain to protect against disease from A. marginale infection (5, 11), it is of interest to identify antigens and epitopes shared among A. centrale OMP7 and A. marginale OMPs 7 to 9 that may constitute part of a protective vaccine formulation. The sequence shared by the four most immunogenic peptides, A. centrale OMP7 peptide 8 and A. marginale OMPs 7 to 9 peptide 7, is GDSL/M/IFLLVDDAI/VR. A. centrale OMP7 peptide 7 was not stimulatory (Fig. 3) and overlaps A. centrale OMP7 peptide 8 by 10 amino acids (GDSLFLLVDD), suggesting that the 9-mer core epitope lacks at least the N-terminal amino acids GD. To refine the T-cell epitope, truncated synthetic peptides from A. centrale OMP7 peptide 8 lacking GD and corresponding peptides in A. marginale OMPs 7 to 9 peptides 7 and 8 were synthesized and tested for induction of proliferation by T-cell lines from DRB3*1201 homozygous animal 48422 (Table 2). The results show that the core 9-mer epitope is FLLVDDAI/VV, as predicted. Position 8 of this core epitope can contain either isoleucine or valine. Although programs that predict MHC class II binding peptides are not available for cattle, analysis of the predicted class II-restricted T-cell epitopes within A. centrale peptide 8 and A. marginale peptide 7 using the Immune Epitope Database (IEDB) Analysis and Resource (22, 23) revealed the sequences FLLVDDAIV and FLLVDDAVV are predicted to be strong binders to the randomly selected DRB1*01:01 molecule (data not shown).
TABLE 2.
Mapping T-cell epitopes on A. centrale OMP7 and A. marginale OMPs 7 to 9
| Peptide | Sequencea | T-cell proliferation (SI)b |
|---|---|---|
| A. marginale OM | 26.0 (1.3) | |
| A. centrale OMP7 peptide 8, A. marginale OMP 7 to 9 peptide 7 | ||
| 7C_7 | LEAVRERFPIWKVGGRTWTKGDSLFLLVDD | 1.4 (0.8) |
| 7C_8 | GDSLFLLVDDAIVRLATGQSDHDDPAAKAL | 15.4 (4.5) |
| 7C_8.1 | SLFLLVDDAIVR | 20.7 (1.0) |
| 7C_8.2 | LFLLVDDAIVR | 15.7 (8.4) |
| 7C/7M_8.3 | FLLVDDAIVR | 13.7 (8.0) |
| 7C_8.4 | LLVDDAIVR | 0.7 (0.1) |
| 7C_8.5 | FLLVDDAIV | 33.6 (0.2) |
| 8/9M_7.1 | FLLVDDAVVR | 26.2 (6.9) |
| A. centrale OMP7 peptide 14 | ||
| 7C_14 | KLSFIEAALSRIGGHKIEIPAVVANTFGAN | 14.9 (2.1) |
| 7C_14.1 | KLSFIEAALSRIG | 11.8 (4.0) |
| 7C_14.2 | LSFIEAALSRIG | 28.4 (5.4) |
| 7C_14.3 | SFIEAALSRIG | 9.3 (5.0) |
| 7C_14.4 | GHKIEIPAVVANT | 0.5 (0.1) |
| 7M_14.4 | GYKIEIPAVAANT | 1.2 (1.1) |
| 8M_14.4 | GHKIEIPAVAANT | 0.6 (0.1) |
| 9M_14.4 | GYRIKIPAVVANT | 0.3 (0.1) |
| Similar sequences | ||
| AM1137 | VLSYIEAAGLRIV | 22.3 (1.9) |
| AM171 | SLAFILAVLSV | 2.1 (2.7) |
| AM1313 | KSFIELAVLSK | 0.8 (0.9) |
Overlapping 10-amino-acid peptide sequences are underlined.
A 2-week-old T-cell line from animal 48422 (DRB3*1201/*1201) was used. Standard deviations are indicated in parentheses. Responses were considered significant if the stimulation index (SI) was >4.0 (indicated by boldfaced type).
Conservation of the A. centrale OMP7/A. marginale OMPs 7 to 9 T-cell epitope FLLVDDAI/VV among geographically distant strains.
BLASTP analysis using A. centrale OMP7 as the query showed that the epitope FLLVDDAI/VV is conserved among multiple North American strains of A. marginale in addition to the St. Maries strain, including OMPs 7 to 9 from the Florida strain (14), OMP8 and OMP9 from the Virginia strain, two membrane proteins from the Oklahoma strain, one membrane protein from the Mississippi strain, three membrane proteins from the South Idaho strain, and three membrane proteins from the Washington Okanogan (Washington 'O) strain (Table S2). The peptide is also conserved in OMP7 and OMP8 of two geographically distant isolates, one from northeastern Brazil and another from Australia (Gypsy Plains). Complete sequence data for OMP9 is not available for these two isolates (Table S2).
With some exceptions (noted above), most sequence data for A. marginale OMPs is derived from North American isolates. Overall, little is known about the genetic variability of primary vaccine candidates from strains circulating in tropical regions where infection rates of A. marginale are high and genetic variation is also expected to be high (24, 25). To determine whether the T-cell epitope is highly conserved in more distant geographic regions, omp7, omp8, and omp9 were amplified, cloned, and sequenced, and the predicted amino acids were aligned from A. marginale-infected cattle in two regions of Ghana, Ashiaman and Juapong, in the greater Accra and Volta regions, respectively (26). The 9-mer epitope FLLVDDAI/VV was completely conserved in all sequences (Fig. S1 to S3). Thus, this core T-cell epitope is broadly conserved in globally disbursed A. marginale strains.
Mapping the A. centrale OMP7 peptide 14 epitope and potential epitopes of AM1137, AM171, and AM1313.
Interestingly, animals 48411 and 48422 responded to A. centrale OMP7 peptide 14, which is not conserved in A. marginale OMPs 7 to 9 (Fig. 1 and Table S1). Testing truncated peptides of OMP7 peptide 14 verified that A. centrale OMP7 peptide 14-derived peptide SFIEAALSRIG (peptide 7C_14.3) contains a T-cell epitope (Table 2). This result is intriguing as the cattle were immunized with A. marginale St. Maries strain OM, suggesting that the specific response to A. centrale OMP7 peptide 14 is mediated by T cells that were primed by a cross-reactive epitope found in the OM preparation. BLASTP analysis of the A. marginale St. Maries strain genome identified sequences from three proteins with significant identity to A. centrale OMP7 peptide 14: AM1137, a nucleoside diphosphate kinase (NDK); AM171, a major facilitator superfamily (MFS) transporter; and AM131, the VirB11 protein member of the type IV secretion system. VirB11 was previously shown to be a CD4 T-cell antigen for OM-immunized cattle (27). Peptides from these proteins corresponding to the A. centrale OMP7 peptide 14 sequence were synthesized and tested in a proliferation assay (Table 2) with DRB3*1201 homozygous animal 48422. There was no response to the AM171 (MFS transporter) or AM1313 (VirB11) derived peptides. However, peptide VLSYIEAAGLRIV derived from AM1137 (NDK) stimulated a response similar to that stimulated by A. centrale OMP7 peptides 14 and 7C_14.2 (LSFIEAALSRIG). Interestingly, the NDK protein sequence is completely conserved in Florida, Australia Dawn, and Australia Gypsy Plains strains (NCBI protein identifiers B9KGY6.1, AGZ79964.1, and AGZ79164.1, respectively).
DISCUSSION
The specific antigens that induce protective immunity against A. marginale are not known. Immunization with individual OMPs has, to date, failed to provide protection against homologous strain challenge in the natural bovine host equivalent to that achieved by immunizing with OM (6, 12, 28–31). In contrast, complexes of cross-linked OM proteins, and a mixture of individual proteins from the complexes were able to provide protection against disease in challenged cattle (18). In addition, immunization with the live A. centrale vaccine strain can provide protection against disease caused by some strains of A. marginale but does not prevent infection (5, 11). Thus, the key to inducing protective immunity with a subunit vaccine may be to provide a constellation of immunogenic surface proteins, including those with epitopes shared by A. marginale and A. centrale. This does not rule out the possibility that secreted or cytoplasmic proteins may also be useful vaccine candidates. In this study, we have focused on identifying T-cell epitopes shared by A. centrale OMP7 and A. marginale OMPs 7 to 9 that may be useful as part of such a multivalent vaccine.
A. marginale OMPs 7 to 9 are promising vaccine candidates because they are recognized by IgG2 and T cells from cattle immunized with the protective A. marginale OM fraction (7, 17) and by IgG2 from cattle immunized with live A. centrale (8). The production of IgG2 generally requires the induction of CD4 T-cell help to promote isotype switching. Although we did not phenotype the 2-week lymphocyte cell lines in the present study, prior studies have consistently identified the responding cell population from A. marginale-immune cattle to stimulation with A. marginale OM and surface proteins to be CD4 T cells (3, 19–21). Although γδ T cells might contribute to a proliferative response, these cells are not MHC restricted (32). Because A. marginale OMPs 7 to 9 are immunogenic for T cells from cattle that express DRB3*1201 (17) (Fig. 2), we hypothesized that these antigenic proteins share a T-cell epitope with A. centrale OMP7, and our hypothesis is accepted. Importantly, the shared T-cell epitope FLLVDDAI/VV is conserved among annotated OMPs 7 to 9 and predicted membrane proteins of seven North American strains, as well as strains from geographically distant Brazil, Australia, and Ghana.
It has been recently reported that in cattle populations experiencing high A. marginale infection prevalence, such as those in Ghana, there is increased diversity among the isolates compared to populations with low infection prevalence (25). In the United States, the prevalence of A. marginale infections is relatively low compared to that of more tropical regions (11). Thus, in spite of overall higher levels of diversity in Ghanaian A. marginale field isolates, this T-cell epitope is highly conserved and may be a useful component of a multivalent vaccine if linked to B-cell epitopes on OMPs 7 to 9.
In addition to the globally conserved T-cell epitope FLLVDDAI/VV, a minimum of four other epitopes is present within the peptides spanning the A. marginale OMP 8 to 9 proteins. These are found in peptides 3 and 9 in OMP8, peptides 11 and 12 that could contain one or more epitopes in OMP9 (both restricted by DRB3*1201 or a DQ molecule expressed by this haplotype), and peptide 10 in OMP9 that is apparently restricted by DRB3*1501 or a haplotype-linked DQ molecule. These sequences are also highly conserved within multiple A. marginale strains from the Americas (see Table S3 in the supplemental material), so some may also be useful components of a vaccine. However, compared to the Ghanaian sequences, these are less conserved (Fig. S2 and S3). For example, OMP8 peptides 3 and 9 are highly conserved in Ghana variants 1, 2, 4, 5, and 7 but not as conserved in the other variants, and OMP9 peptide 10 to 12 sequences are only highly conserved in peptide 12 due to substitutions and deletions in OMP9 of the three Ghana variants.
In the present study, we identified cattle with only two haplotypes, DRB3*1201 and DRB3*1501, that responded to the OMP 7 to 9 peptides, although this does not preclude recognition of these or other T-cell epitopes on A. centrale OMP7/A. marginale OMPs 7 to 9 by cattle with different MHC class II molecules. Furthermore, these are common haplotypes among cattle in Washington (unpublished observations), with frequencies similar to those reported for the Holstein breed in Canada, Japan, and Iran, where the frequencies of DRB3*1201 ranged from 13.0 to 20.0% and the frequencies of DRB3*1501 ranged from 9.0 to 13.4% (33–37). A multivalent vaccine that affords protection in a population of outbred cattle should include epitopes recognized by these common DRB3*1201 or DRB3*1501 molecules or associated DQ molecules, as well as those recognized by other commonly expressed BoLA class II molecules. This strengthens the rationale for the inclusion of these OMP 7 to 9 T-cell epitopes in a vaccine that would be broadly protective for cattle against anaplasmosis.
An interesting finding from this study was the strong response to an epitope within A. centrale OMP7 peptide 14 that was not present in A. marginale. Because the T cells responding to this epitope were derived from cattle immunized with A. marginale St. Maries strain OM, we reasoned that there must be an A. marginale protein that was responsible for priming the T cells. The A. centrale epitope was mapped to LSFIEAALSRIG, and a similar epitope (VLSYIEAAGLRIV) identified in AM1137, an NDK protein, induced a comparable T-cell response. The epitope is restricted by DRB3*1201 or a linked DQ molecule, and the 9-mer core epitope may be F/YIEAAL/GS/LRI, where anchor residues that bind to the MHC class II molecule are found at position 1 (P1), P4, P6, and P9 (38). If this is the case, an aromatic, nonpolar phenylalanine or tyrosine occupies P1, a conserved nonpolar alanine occupies P4, a nonpolar leucine or glycine occupies P6, and a conserved nonpolar isoleucine occupies P9. These amino acids are either identical or structurally similar. The other mapped T-cell epitope restricted by this haplotype and described above is FLLVDDAI/VV, where an aromatic nonpolar phenylalanine also occupies P1, a conserved nonpolar valine occupies P4, an acidic aspartic acid occupies P6, and a conserved nonpolar valine occupies P9. T-cell receptor (TCR) contact residues are found at P2, P5, and P8 (38). These TCR contact residues within A. centrale OMP 7 peptide LSFIEAALSRIG and NDK peptide VLSYIEAAGLRIV would be identical for the two peptides if a phenylalanine occupies P1, an isoleucine occupies P2, an alanine occupies P5, and an arginine occupies P8 (indicated in boldface). Thus, it is feasible that the TCR primed by the AM1137 NDK protein binds both peptides that are presented by the same MHC class II molecule. However, without cloning T cells specific for the epitope and verifying that the clones respond to both peptides, this can only be speculative.
Another question regarding the potential T-cell priming by the AM1137 NDK protein is that AM1137 is predicted by psort (http://psort.hgc.jp) to be a cytoplasmic membrane protein, and cattle were immunized with purified OM that are relatively free of cytoplasmic proteins (39). However, the cytoplasmic proteins PepA cytosol amino peptidase and elongation factor Tu were both recognized by sera from cattle immunized with purified A. marginale OM (7), suggesting that certain cytoplasmic proteins are associated with the bacterial outer membrane. If NDK is associated with the inner bacterial membrane, it may have localized with the OM fraction.
In summary, we have identified a broadly conserved 9-mer CD4 T-cell epitope shared between A. centrale vaccine strain OMP7 and the related A. marginale family of proteins OMPs 7 to 9. These proteins are considered to be logical vaccine candidate antigens because they are surface exposed, present in protective OM and cross-linked OM proteins, recognized by immune serum IgG2 and T cells in cattle immunized with the protective OM antigen preparation, and recognized by immune serum from cattle immunized with the A. centrale vaccine strain (8, 16–18). This epitope, together with additional T-cell epitopes that we have shown are present within these proteins, may be important for inclusion in a multivalent vaccine for A. marginale that can provide protection against disease caused by geographically distant bacterial strains in cattle that express common BoLA class II haplotypes DRB3*1201 and *1501.
MATERIALS AND METHODS
Selection and immunization of cattle.
Four age-matched Holstein calves were selected based on MHC class II haplotypes previously identified in two cattle (04B91 and 04B92) that had strong T-cell proliferative responses to OMPs 7 to 9 and one animal (04B90) with weaker responses to recombinant A. marginale St. Maries OMP7 and OMP9 (17; the present study). The DRB3 haplotypes of animals used in the present study were determined by PCR-RFLP analysis (40) and verified by sequencing (12). The animals selected and their haplotypes (in parentheses) are as follows: 48406 (DRB3*2703/*2703), 48411 (DRB3*1201/*1501), 48422 (DRB3*1201/*1201), 48432 (DRB3*1101/*1401), and 48453 (DRB3*1501/1501). Calves were immunized four times subcutaneously at 3-week intervals with 60 μg of purified A. marginale St. Maries strain OM (7) emulsified in 6 mg of saponin diluted in 1.3 ml of phosphate-buffered saline (PBS). Animal studies were conducted using an approved Institutional Animal Care and Use Committee (Washington State University, Pullman, WA) protocol.
Expression and purification of recombinant proteins.
Recombinant A. marginale OMPs 7 to 9 and negative control OMP2, which is not expressed in the bovine erythrocyte stage (14), were expressed as T-7 and His-tagged fusion proteins from the pET28 vector (Novagen, Inc., EMD Biosciences, Darmstadt, Germany) and purified on nickel NTA agarose columns (Qiagen, Inc., Valencia, CA) and by anti-T7 tag affinity chromatography (Novagen) as described previously (17). The recombinant proteins were stored at −20°C.
Design and synthesis of peptides.
Starting at the N-terminal methionine, 30-mer peptides with a 10-residue C-terminal overlap between the next adjacent peptide were designed for the full length of OMP7 from A. centrale and OMPs 7 to 9 from A. marginale St. Maries strain. A total of 19 30-mer peptides plus a 20th peptide constituting the remaining C-terminal amino acids, all overlapping by 10 amino acids and spanning each OMP, were synthesized and purified. Using a CLUSTALW alignment of the full-length proteins, A. centrale OMP7 peptides inducing T-cell proliferation were compared to A. marginale OMP 7 to 9 peptides noting similar/dissimilar amino acids at analogous positions. Position-specific amino acid information was used to design new peptides with smaller overlaps to identify the core T-cell epitopes. All peptides were synthesized and purified to >80% purity by NeoBiolab. Peptides were solubilized in 10% dimethyl sulfoxide (DMSO) in PBS to a concentration of 1 mg/ml and stored at −20°C. They were evaluated for the ability to induce T-cell proliferation from A. marginale OM-immunized cattle.
Isolation of PBMC and generation of 2-week T-cell lines.
PBMC were purified from whole blood samples by Histopaque gradient centrifugation, repeatedly washed with Hanks buffered salt solution (Gibco), and suspended in complete RPMI (cRPMI; RPMI 1640 medium [Sigma-Aldrich] supplemented with 10% fetal bovine serum [FBS], 100 mM l-glutamine, 50 μM β2-mercaptoethanol, 24 mM HEPES buffer, and 50 μg of gentamicin sulfate/ml) as described previously (19, 20). Lymphocytes were either used immediately in proliferation assays or cryopreserved in heat-inactivated FBS containing 10% DMSO. To establish 2-week T-cell lines, 4 × 106 PBMC were cultured in 1.5 ml of cRPMI with A. marginale OM at 1 μg/ml in 24-well plates, followed by incubation in 5% CO2 at 37°C for 1 week, allowing an enrichment (expansion) of OM-specific T cells. For experiments using PBMC from animal 04B92, 2-week cell lines were similarly established from PBMC depleted of CD8+ T cells and γδ T cells to enrich for CD4+ T cells (39). T cells were collected, washed, resuspended in cRPMI, and cultured in 24-well plates at 7 × 105 cells/well with 2 × 106 cells/well of irradiated (3,000 rads) autologous PBMC as a source of antigen-presenting cells (APC) without antigen (rested) for 1 week in 5% CO2 at 37°C. The cells were then centrifuged, washed, and resuspended in cRPMI and either used in proliferation assays or cryopreserved.
Lymphocyte proliferation assays.
PBMC were resuspended in cRPMI and cultured in triplicate 100-μl volumes in 96-well round-bottom plates at a final concentration of 2 × 106 cells/ml. The plates were incubated at 5% CO2 in air at 37°C for 6 days, pulsed with 0.25 μCi of [3H]thymidine per well, and allowed to incubate for an additional 18 h before they were harvested with a Tomtec Harvester96 and quantified with a Perkin-Elmer 1450 TriLux MicroBeta liquid scintillation counter. The antigens included purified A. marginale OM and control uninfected bovine red blood cell (URBC) membranes, both used at a final concentration of 1 μg/ml.
For experiments using cell lines, 2 × 104 lymphocytes and 2 × 105 APC were cultured per well in total volume of 100 μl. For animal 04B92, recombinant OMPs 7 to 9 and negative control OMP2 and A. marginale OM were used at 0.1, 1, and 10 μg/ml. For animals 48406, 48411, 48422, 48432, and 48453, the antigens were A. marginale OM used at 1 μg/ml and peptides used at 10 μg/ml.
For all assays, the cells with medium only served as a negative control, and cells with 10% T-cell growth factor (TCGF) (19) served as a positive control. The cells were incubated in 5% CO2 in air at 37°C for 3 days, radiolabeled with 0.25 μCi of [3H]thymidine per well and allowed to incubate for an additional 18 h before they were harvested and counted. The results are presented as the mean counts per minute (cpm) of triplicate culture wells or as the stimulation index (SI), calculated as the mean cpm of cells cultured with antigen/mean cpm of cells cultured with medium. Independent assays were performed two or more times.
Statistical analyses.
A one-tailed Student t test was used to determine the significance of the proliferative response to URBC, OM, or TCGF in individual animals before and after immunization with OM, where P < 0.05. The same test was used with a Bonferroni correction for multiple comparisons when determining the significance of the proliferative response to OM compared to the response to URBC and to OMP7, OMP8, or OMP9 compared to the response to OMP2. In peptide screening assays, a response was considered significant if the SI was >4.0.
Protein similarity searches to identify epitopes conserved in related A. marginale proteins.
The Basic Local Alignment Search Tool for Proteins (BLASTP) version 2.3.1 (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify A. marginale protein sequences in the NCBI protein database with similarity to sequences we identified as T-cell epitopes from A. centrale and A. marginale St. Maries strain (41).
Accession number(s).
The NCBI accession numbers for the A. marginale omp7, omp8, and omp9 sequences for the reported Ghanaian strains are KX591891 to KX591921 and protein identifiers are APB02966 to APB02996.
Supplementary Material
ACKNOWLEDGMENTS
We thank Deb Alperin for excellent technical assistance and Emma Karel for animal care.
This research was supported by NIH NIAID grants AI053692 (to W.C.B.) and AI44005 (to G.H.P.), USDA ARS CRIS project 5348-32000-003-00D (to M.W.U. and S.M.N.), the Wellcome Trust for the Fellowship in Public Health and Tropical Medicine grant 097171/Z/11/Z (to J.E.F.), and a Colombian scholarship for a doctorate abroad, Colciencias, Convocatoria 568/2012 (to. E.G.F.-B.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CVI.00406-16.
REFERENCES
- 1.Marcelino I, de Almeida AM, Ventosa M, Pruneau L, Meyer DF, Martinez D, Lefrançois T, Vachiéry N, Coelho AV. 2012. Tick-borne diseases in cattle: applications of proteomics to develop new generation vaccines. J Proteomics 75:4232–4250. doi: 10.1016/j.jprot.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Tebele N, McGuire TC, Palmer GH. 1991. Induction of protective immunity using Anaplasma marginale initial body membranes. Infect Immun 59:3199–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown WC, Shkap V, Zhu D, McGuire TC, Tuo W, McElwain TF, Palmer GH. 1998. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect Immun 66:5406–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noh SM, Brayton KA, Brown WC, Norimine J, Munske GR, Davitt CM, Palmer GH. 2008. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect Immun 76:2219–2226. doi: 10.1128/IAI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noh SM, Brown WC. 2012. Adaptive immune responses to infection and opportunities for vaccine development, p 328–363. In Palmer GH, Azad AF (ed), Intracellular pathogens II: Rickettsiales. ASM Press, Washington, DC. [Google Scholar]
- 6.Palmer GH, Rurangirwa FR, Kocan KM, Brown WC. 1999. Molecular basis for vaccine development against the ehrlichial pathogen Anaplasma marginale. Parasitol Today 15:281–286. doi: 10.1016/S0169-4758(99)01469-6. [DOI] [PubMed] [Google Scholar]
- 7.Lopez JE, Siems WF, Brayton KA, Palmer GH, McGuire TC, Brown WC. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect Immun 73:8109–8118. doi: 10.1128/IAI.73.12.8109-8118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agnes JT, Brayton KA, LaFollett M, Norimine J, Brown WC, Palmer GH. 2010. Identification of Anaplasma marginale outer membrane protein antigens conserved between A. marginale sensu stricto strains and the live A. marginale subsp. centrale vaccine. Infect Immun 79:1311–1318. doi: 10.1128/IAI.01174-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pipano E. 1995. Live vaccines against hemoparasitic diseases in livestock. Vet Parasitol 57:213–231. doi: 10.1016/0304-4017(94)03122-D. [DOI] [PubMed] [Google Scholar]
- 10.Bock RE, de Vos AJ. 2001. Immunity following use of Australian tick fever vaccine: a review of the evidence. Aust Vet J 79:832–839. doi: 10.1111/j.1751-0813.2001.tb10931.x. [DOI] [PubMed] [Google Scholar]
- 11.Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. 2010. The natural history of Anaplasma marginale. Vet Parasitol 167:95–107. doi: 10.1016/j.vetpar.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Ducken DR, Brown WC, Alperin DC, Brayton KA, Reif KE, Turse JE, Palmer GH, Noh SM. 2015. Subdominant outer membrane antigens in Anaplasma marginale: conservation, antigenicity, and protective capacity using recombinant protein. PLoS One 10:e0129309. doi: 10.1371/journal.pone.0129309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, McGuire TC, Knowles DP. 2005. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci U S A 102:844–849. doi: 10.1073/pnas.0406656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noh S, Brayton KA, Knowles DP, Agnes JT, Dark MJ, Brown WC, Baszler TV, Palmer GH. 2006. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect Immun 74:3471–3479. doi: 10.1128/IAI.01843-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herndon DR, Palmer GH, Shkap V, Knowles DP Jr, Brayton KA. 2010. Complete genome sequence of Anaplasma marginale subsp. centrale. J Bacteriol 192:379–380. doi: 10.1128/JB.01330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer GH, Brown WC, Noh S, Brayton KA. 2012. Genome-wide screening and identification of antigens for rickettsial vaccine development. FEMS Microbiol Immunol 64:115–119. doi: 10.1111/j.1574-695X.2011.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez JE, Beare PA, Heinzen RA, Norimine J, Lahmers KK, Palmer GH, Brown WC. 2008. High-throughput identification of T-lymphocyte antigens from Anaplasma marginale expressed using in vitro transcription and translation. J Immunol Methods 332:129–144. doi: 10.1016/j.jim.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Noh SM, Turse JE, Brown WC, Norimine J, Palmer GH. 2013. Linkage between Anaplasma marginale outer membrane proteins enhances immunogenicity but is not required for protection from challenge. Clin Vaccine Immunol 20:651–656. doi: 10.1128/CVI.00600-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han S, Norimine J, Palmer GH, Mwangi W, Lahmers KK, Brown WC. 2008. Rapid deletion of antigen-specific CD4+ T cells following infection represents a strategy of immune evasion and persistence for Anaplasma marginale. J Immunol 181:7759–7769. doi: 10.4049/jimmunol.181.11.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Norimine J, Brayton KA, Palmer GH, Scoles GA, Brown WC. 2010. Anaplasma marginale infection with persistent high-load bacteremia induces a dysfunctional memory CD4+ T lymphocyte response but sustained high IgG titers. Clin Vaccine Immunol 17:1881–1890. doi: 10.1128/CVI.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbott JR, Palmer GH, Howard CJ, Hope JC, Brown WC. 2004. Anaplasma marginale major surface protein 2 CD4+-T-cell epitopes are evenly distributed in conserved and hypervariable regions (HVR), whereas linear B-cell epitopes are predominantly located in the HVR. Infect Immun 72:7360–7366. doi: 10.1128/IAI.72.12.7360-7366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. 2008. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. 2010. Peptide binding predictions for HLA DR, DP, and DQ molecules. BMC Bioinformatics 11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueti MW, Tan Y, Broschat SL, Castaneda Ortiz EJ, Camacho-Nuez M, Mosqueda JJ, Scoles GA, Grimes M, Brayton KA, Palmer GH. 2012. Expansion of variant diversity associated with a high prevalence of pathogen strain superinfection under conditions of natural transmission. Infect Immun 80:2354–2360. doi: 10.1128/IAI.00341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castañeda-Ortiz EJ, Ueti MW, Camacho-Nuez M, Mosqueda JJ, Mousel MR, Johnson WC, Palmer GH. 2015. Association of Anaplasma marginale strain superinfection with infection prevalence within tropical regions. PLoS One 10:e0120748. doi: 10.1371/journal.pone.0120748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beckley CS, Shaban S, Palmer GH, Hudak AT, Noh SM, Futse JE. 2016. Disaggregating tropical disease prevalence by climatic and vegetative zones within tropical West Africa. PLoS One 11:e0152560. doi: 10.1371/journal.pone.0152560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutten EL, Norimine J, Beare PA, Heinzen RA, Lopez JE, Morse K, Brayton KA, Gillespie JJ, Brown WC. 2010. Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine. Infect Immun 78:1314–1325. doi: 10.1128/IAI.01207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer GH, Barbet AF, Davis WC, McGuire TC. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 231:1299–1302. doi: 10.1126/science.3945825. [DOI] [PubMed] [Google Scholar]
- 29.Palmer GH, Oberle SM, Barbet AF, Goff WL, Davis WC, McGuire TC. 1988. Immunization of cattle with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect Immun 56:1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer GH, McElwain TF. 1995. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet Parasitol 57:233–253. doi: 10.1016/0304-4017(94)03123-E. [DOI] [PubMed] [Google Scholar]
- 31.Abbott JR, Palmer GH, Kegerreis KA, Hetrick PF, Howard CJ, Hope JC, Brown WC. 2005. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J Immunol 174:6702–6715. doi: 10.4049/jimmunol.174.11.6702. [DOI] [PubMed] [Google Scholar]
- 32.Lahmers KK, Norimine J, Abrahamsen MS, Palmer GH, Brown WC. 2005. The CD4+ T cell immunodominant Anaplasma marginale major surface protein 2 stimulates γδ T cell clones that express unique T cell receptors. J Leuk Biol 77:199–208. [DOI] [PubMed] [Google Scholar]
- 33.Sharif S, Mallard BA, Wilkie BN, Sargeant JM, Scott HM, Dekkers JC, Leslie KE. 1998. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with occurrence of disease and milk somatic cell score in Canadian dairy cattle. Anim Genet 29:185–193. doi: 10.1111/j.1365-2052.1998.00318.x. [DOI] [PubMed] [Google Scholar]
- 34.Schwab AE, Geary TG, Baillargeon P, Schwab AJ, Fecteau G. 2009. Association of BoLA DRB3 and DQA1 alleles with susceptibility to Neospora caninum and reproductive outcome in Quebec Holstein cattle. Vet Parasitol 165:136–140. doi: 10.1016/j.vetpar.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Takeshima S, Saitou N, Morita M, Inoko H, Aida Y. 2003. The diversity of bovine MHC class II DRB3 genes in Japanese Black, Japanese Shorthorn, Jersey, and Holstein cattle in Japan. Gene 316:111–118. doi: 10.1016/S0378-1119(03)00744-3. [DOI] [PubMed] [Google Scholar]
- 36.Nassiry MR, Sadeghi B, Tohidi R, Afshari JT, Khosravi M. 2008. Comparison of bovine lymphocyte antigen DRB3.2 allele frequencies between two subpopulations of Iranian Holstein cattle. Afr J Biotechnol 7:2671–2675. [Google Scholar]
- 37.Pashmi M, Qanbari S, Ghorashi SA, Sharifi AR, Simianer H. 2009. Analysis of relationship between bovine lymphocyte antigen DRB3.2 alleles, somatic cell count and milk traits in Iranian Holstein population. J Anim Breed Genet 126:296–303. doi: 10.1111/j.1439-0388.2008.00783.x. [DOI] [PubMed] [Google Scholar]
- 38.Mohan JF, Unanue ER. 2012. Unconventional recognition of peptides by T cells and the implications for autoimmunity. Nat Rev Immunol 12:721–727. doi: 10.1038/nri3294. [DOI] [PubMed] [Google Scholar]
- 39.Lopez JE, Palmer GH, Brayton KA, Dark MJ, Leach SE, Brown WC. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect Immun 75:2333–2342. doi: 10.1128/IAI.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Eijk M, Stewart-Haynes J, Lewin H. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim Genet 23:483–496. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schäffer AA, Zhang J, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.