Abstract
Under no-strand bias conditions, each genomic DNA strand should present equimolarities of A and T and of G and C. Deviations from these rules are attributed to asymmetric properties intrinsic to DNA mutation–repair processes. In bacteria, strand biases are associated with replication or transcription. In eukaryotes, recent studies demonstrate that human genes present transcription-coupled biases that might reflect transcription-coupled repair processes. Here, we study strand asymmetries in intron sequences of evolutionarily distant eukaryotes, and show that two superimposed intron biases can be distinguished. (i) Biases that are maximum at intron extremities and decrease over large distances to zero values in internal regions, possibly reflecting interactions between pre-mRNA and splicing machinery; these extend over ∼0.5 kb in mammals and Arabidopsis thaliana, and over 1 kb in Caenorhabditis elegans and Drosophila melanogaster. (ii) Biases that are constant along introns, possibly associated with transcription. Strikingly, in C.elegans, these latter biases extend over intergenic regions that separate co-oriented genes. When appropriately examined, all genomes present transcription-coupled excess of T over A in the coding strand. On the opposite, GC skews are either positive (mammals, plants) or negative (invertebrates). These results suggest that transcription-coupled asymmetries result from mutation–repair mechanisms that differ between vertebrates and invertebrates.
INTRODUCTION
During genome evolution, mutations do not occur at random as illustrated by the diversity of the nucleotide substitution rate values (1–4). This non-randomness is considered as a by-product of the various DNA mutation and repair processes acting on genomic DNA, and the study of mutation effects on genome composition can shed light on these processes. This is exemplified by the nucleotide compositional asymmetries observed in genome sequences. Under no-strand bias conditions, i.e. when mutation rates are identical for complementary nucleotide substitutions (e.g. A→G and T→C), one would expect equimolarities of adenine and thymine and of guanine and cytosine (5,6), a property often referred to as the Chargaff's second parity rule (7,8). On the opposite, strand compositional biases, i.e. A ≠ T and G ≠ C, have been observed in prokaryotes and extensively studied in bacterial, organelle and viral genomes. These compositional skews have been attributed to asymmetries intrinsic to the replication or to the transcription processes. In models based on replication, the leading and lagging replicating strands are subject to different mutational and repair pressures resulting in asymmetric nucleotide compositions (9,10). The leading strand is relatively enriched in G over C and to a lesser extent in T over A in positions under weak selective pressure (intergenic regions and third codon position). These properties are now routinely used to identify the bacterial replication origins (10–15). In other models, transcription would increase single-strand deamination of cytosines (C→T) (16,17) and favor the transcription-coupled repair mechanisms (18) leading to pronounced strand asymmetries (19). In eukaryotes, the existence of compositional biases has been debated. Unlike eubacterial genomes, the yeast genome did not present clear replication-coupled strand asymmetry except in subtelomeric regions of chromosomes (20). A comparative study of the β-globin replication origin in primates concluded that the replication-coupled mutational bias was absent (21). In contrast, other works suggested that transcription or replication might contribute to compositional asymmetry (22–24). Recently, several studies helped clarifying this contrasted situation. First, in a comparative study of mammalian orthologous regions, asymmetries of nucleotide substitution rates were evidenced in transcribed regions (25). These asymmetries were characterized by an excess of (A→G) transitions relatively to (T→C) transitions (differing from the excess of (C→T) transitions observed in bacteria), which may rather be a by-product of the transcription-coupled repair mechanism acting on uncorrected substitution errors during replication. Further genome-scale analyses of human genes established the existence of transcription-coupled nucleotide biases (26,27). The TA and GC skews are both specific to transcribed regions and their values suggest that they result from mutational processes including not only transitions but also transversions (26).
Do transcription-coupled biases also exist in other eukaryotic genomes? To address this issue, we examined the strand compositional asymmetries in evolutionarily distant eukaryotes. In each genome, we compared the skews in transcribed and non-transcribed region. In transcribed regions, analyses were performed with intron sequences, which are considered as weakly selected regions. However, intron extremities contain selected motifs involved in the splicing process. We observed that strand asymmetries differed strongly in border regions and in central regions of introns, indicating that the splicing machinery interacts with intron sequences at larger distances from splice sites than previously expected. After appropriate removal of intron borders, we observed transcription-coupled TA and GC strand asymmetries in all eukaryotic genomes examined. We discuss the possibility that these asymmetries result from mutation–repair mechanisms that differ between vertebrates and invertebrates.
MATERIALS AND METHODS
Sequences
The sequence and annotation data were retrieved from the Genome Browser of University of California for the human (April 2003), the mouse (October 2003), the Drosophila melanogaster (January 2003) and the Caenorhabditis elegans (May 2003) genomes. The sequence and annotation data for Arabidopsis thaliana, Schizosaccharomyces pombe and Plasmodium falciparum were downloaded from NCBI (November 2003). All analyses were carried out using the RefSeq gene annotation. When several genes presented identical exonic sequences, only the longest one was retained. All intron sequences were considered on the coding strand and 30 bp were removed at both intron extremities; to avoid irrelevant stochastic fluctuations, the sequences that were <100 bp were not considered; consequently, introns shorter than 160 bp (100 bp + 30 bp at both ends) were not considered. Repeated elements were removed with RepeatMasker for human, mouse, D.melanogaster and A.thaliana sequences. Two subsets of human introns were retrieved from the UCSC Genome Browser (July 2003). (i) Introns for which there is no splice variant known (17 319 introns): (ii) introns for which splice variants are known: the introns of this class were chosen as situated on both sides of cassette exons, which led to a set of 7940 introns.
Intron skew profiles
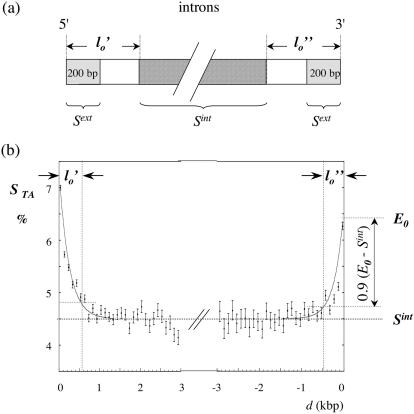
The TA and GC skews were calculated as STA = (T − A)/(T + A) and SGC = (G − C)/(G + C). Introns were divided into two halves; for each half, the mean values of the skews were calculated in adjacent 100 bp windows and plotted as functions of the distance to the corresponding intron extremity. For a given skew profile, each border region was fitted by a curve E decreasing from E0 at the corresponding intron extremity to the plateau value measured in internal regions Sint (Figure 1). For all genomes except for C.elegans and D.melanogaster, E was an exponential decreasing asymptotically to Sint; for C.elegans and D.melanogaster, the curve was a straight line for the 5′ and 3′ borders of the STA profile as well as for the 5′ border of the SGC profile (data not shown). The skew potentially associated with splicing decreased from the extremum value E0 − Sint (e.g. at the 3′ end of the human STA profile in Figure 1b) to close to zero values in internal regions. In order to separate the border regions from internal regions efficiently, the length of the border was chosen such that it corresponded to 90% of E0 − Sint (see Figure 1). These values were determined for the 5′ and 3′ ends of the STA and SGC profiles and the maximum of these four values was retained as the final value l0 (Table 1). An estimation of the skew potentially associated with splicing was given by the mean value of the difference Δspl = Sext − Sint calculated for introns larger than 2l0 + 160 (Sext was the mean value of the skew calculated in the first and last 200 bp of introns, excluding 30 bp at both extremities).
Figure 1.
Intron skew profiles. (a) Scheme of the intron analysis. The 5′ and 3′ intron halves are represented, with the regions in which the mean skews Sint and Sext were calculated; the 30 bp situated at intron extremities, as well as the repeated elements, were excluded from the analyses (see Materials and Methods). (b) Distribution of the TA skews in human introns. The introns were separated in two halves and the skews were calculated in 100 bp windows; the mean skew calculated for all introns (in %) was plotted as a function of the distance d to the intron extremities (this distance was the same as in the unmasked sequences); vertical bars, SEMs. Each curve (grey) represents a fit of the 5′ (3′) border profile by an exponential. The length of the 5′ (l0′) and of the 3′ (l0″) border regions were measured as described in Materials and Methods.
Table 1. Strand asymmetries in intronic sequences of the indicated eukaryotic genomes.
| n | l | (G+C)% | STAint | SGCint | STAext | SGCext | STAspl | SGCspl | l0 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Human (all introns) | 126 217 | 6.2 | 43.5 | 4.49 ± 0.01 | 3.29 ± 0.01 | 6.24 ± 0.03 | 5.08 ± 0.03 | 2.49 ± 0.01 | 1.52 ± 0.01 | 0.53 |
| human (alternative introns) | 7490 | 6.7 | 42.3 | 5.22 ± 0.02 | 3.05 ± 0.02 | 8.53 ± 0.02 | 4.40 ± 0.04a | 3.30 ± 0.03 | 1.27 ± 0.04a | 0.55a |
| mouse | 114 340 | 5.3 | 43.7 | 4.25 ± 0.01 | 2.56 ± 0.01 | 6.00 ± 0.03 | 4.68 ± 0.04 | 2.44 ± 0.01 | 1.6 ± 0.01 | 0.49 |
| D.melanogaster | 14 677 | 2.2 | 37.8 | 0.98 ± 0.02 | −0.99 ± 0.01 | 0.95 ± 0.10 | −1.77 ± 0.11 | 0.23 ± 0.03 | −1.78 ± 0.03 | 0.96 |
| C.elegans | 36 741 | 0.65 | 32.3 | 3.08 ± 0.03 | −2.97 ± 0.03 | −0.85 ± 0.06 | 3.0 ± 0.1a | −2.95 ± 0.04 | 5.9 ± 0.1a | 1.0 |
| A.thaliana | 27 055 | 0.30 | 33.2 | 1.95 ± 0.06 | 0.64 ± 0.07 | 11.46 ± 0.09 | 5.51 ± 0.11 | 4.58 ± 0.06 | 0.40 ± 0.09 | 0.56 |
Intron sequences were analyzed as described in Materials and Methods (when indicated, the repeated elements were removed from the analysis): n, number of introns examined in each genome (>160 bp); l, mean value of the length of introns in kb; (G+C)%, mean value of the G+C content of introns; STAint and SGCint, mean values of the TA and GC skews measured after removal of border regions of length l0 (at both intron extremities); STAext and SGCext, mean values of the TA and GC skews at intron extremities; Δspl, mean value of the difference of the skews in extremities and in internal regions of introns; the skews are given in % (± standard error of the means); l0, length of intron border regions in kb.
aIndicates that the mean was calculated only for the 5′ end.
Gene skew profiles
In transcribed regions, the mean values of the skews were calculated in adjacent 100 bp windows for internal regions of introns: only the introns with internal regions totally overlapping the window (not overlapping with border regions of length l0) were considered (when indicated, repeated elements were removed; the distances of these windows to the gene extremities were not modified); the mean value of 10 consecutive windows was plotted as a function of the distance d to the 5′ or 3′ end of the gene (Figure 4); in intergenic regions, the repeated elements were removed and when the remaining sequence was <160 bp, the gene and the intergene were not considered.
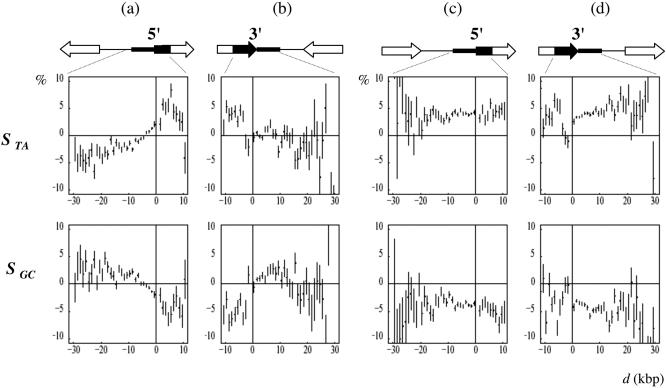
Figure 4.
Strand asymmetries in the regions surrounding the 5′ and 3′ gene extremities. The values of the TA and GC skews STA and SGC were calculated (in %) in adjacent windows starting from each gene extremity in both directions; in transcribed regions, only internal regions of introns were analyzed after removal of border regions of length l0 (see Materials and Methods). In abscissa is figured the distance d (kb) of each 1 kb window to the indicated gene extremity, zero values of abscissa corresponding to 5′ (left panels) or to 3′ (right panels) gene extremities (the distance d was the same as in the unmasked sequences). In ordinate, the mean value, for all genes, of the skews calculated at the corresponding abscissa; arrows indicate the genes; red profiles were performed without removal of intron border regions for C.elegans and A.thaliana; vertical bars, SEM.
RESULTS AND DISCUSSION
Studies of human genes have shown the existence of strand biases that might result from asymmetries intrinsic to transcription-coupled DNA repair (25–27). In order to determine if such biases also exist in other eukaryotes, we performed genome-wide analyses of the TA and GC strand asymmetries in transcribed and non-transcribed regions of distantly related organisms. Strand biases associated with transcription can only be observed in those transcribed regions that are free from selective constraints. Analyses were thus performed for intron sequences, which are usually considered as weakly selected regions except for the splice sites covering at most few tens of nucleotides at extremities. However, due to their interactions with the splicing machinery, intron extremities also contain oligonucleotide motifs recognized on the pre-mRNA by constitutive and/or regulatory components of the splicing machinery [for reviews see (28,29)]. These could result in specific skews decreasing (in modulus) with the distance to splice sites. In contrast, the strand asymmetries associated with transcription should result in constant skews in the remaining internal intron sequences. To differentiate between these two types of biases, we generated the profiles of the TA and GC skews, STA and SGC, along intron sequences and compared intron borders to internal regions. In a second step, to examine the skews in transcribed and non-transcribed regions, we generated the profiles of STA and SGC calculated in internal regions of introns along gene sequences, and along the neighbor 5′ and 3′ intergenic sequences.
Intron borders and splicing-coupled skews
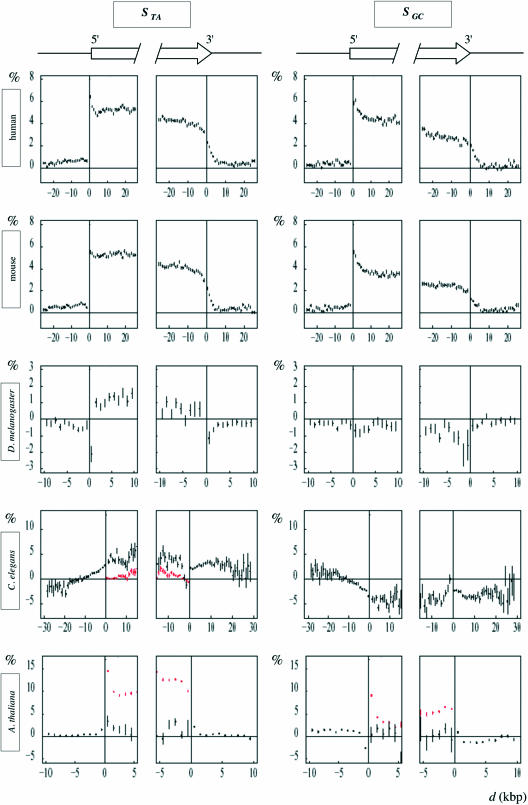
The mean values of STA and SGC calculated in 0.1 kb adjacent windows (see Materials and Methods) were plotted as a function of the window distance to the 5′ or to the 3′ intron ends (Figure 2). In all the genomes analyzed, we observed direct or inverse U-shape profiles that allowed us to distinguish between two types of regions: (i) intron borders where the skews strongly varied with the distance to the splice sites, and (ii) internal regions where the skews remained constant. In human, mouse and A.thaliana, the skew profiles decreased from intron ends to central regions. An opposite situation was observed with the SGC profile in D.melanogaster and with the STA profile in C.elegans, which increased toward central regions. In a few cases, no border region could be identified, as in D.melanogaster where rather constant values were observed for STA along both 5′ and 3′ intron halves, and in C.elegans where SGC remained constant along the 3′ intron halves. The skew potentially associated with transcription was measured as the plateau value in internal regions, Sint. In border regions, the skew could be considered as the sum of Sint and of an additional skew, Δspl, potentially associated with splicing (either positive as in human or negative as in C.elegans for STA). Δspl varied from extreme values at the 5′ and 3′ intron ends to close to zero values in internal regions. Mean values of these skews (measured in the 200 bp intron extremities) were calculated for each organism (Materials and Methods and Table 1). The border length l0 was estimated for the 5′ and 3′ intron extremities and was of the order of 0.5 kb in human, mouse and A.thaliana, and of 1 kb in C.elegans and D.melanogaster (Materials and Methods and Table 1). An interesting observation was that the U-shape (as well as the plateau value Sint) of the intron profiles did not depend significantly on the intron size, when introns presented internal regions i.e. when their length l was larger than 2l0 (data not shown).
Figure 2.
Intron skew profiles. For each indicated genome, the skews were calculated for all introns in 100 bp windows, and plotted as a function of the distance d to the intron extremities as in Figure 1; vertical bars, SEM.
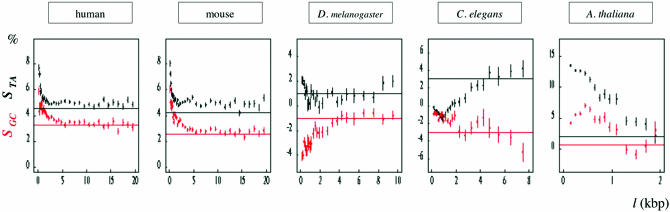
It results from the previous observations that if one measures the skew of intron sequences without removing the border regions, these values will strongly depend on the intron size. This is exemplified in Figure 3 where we observed that the skews of large introns (l > 2l0) mainly reflected the value of the skews measured in internal regions, Sint (potentially associated with transcription) but that the skews of small introns (l < 2l0) presented values that, in most cases, strongly differed from Sint and reflected the skews measured in border regions (potentially associated with splicing). For example, in human and mouse STA and SGC strongly decreased with the intron length l, for introns shorter than 1 kb, and then remained rather constant for larger introns: the TA skew of mouse introns shorter than 0.2 kb was STA = 8.1 ± 0.2%, and it decreased to STA = 4.9 ± 0.03% for l > 3 kb, consistently with STAint = 4.25 ± 0.01%. Strong differences between short and large introns were also observed in D.melanogaster (except for the TA skew) and C.elegans. The most extreme case was observed for A.thaliana. In this genome, 86% of introns were shorter than 0.5 kb and the mean TA skew for these introns was STA = 13.1 ± 0.1%, consistently with STAext = 11.46 ± 0.09%; it decreased by an order of magnitude for large introns to STA = 1.75 ± 0.78% for l > 2 kb, consistently with STAint = 1.95 ± 0.06%. Consequently, the intron skew must be measured either in internal regions or in border regions depending on the mechanism under study.
Figure 3.
Histograms of the mean values of the skews calculated in intron sequences (of length l > 160 bp) as a function of the intron length l (the skews were calculated in masked intron sequences but the length l corresponded to unmasked introns); black, TA skew; red, GC skew; vertical bars, SEM; horizontal lines represent the mean skews in internal regions of all introns, STAint (black) and SGCint (red) (see Figure 1).
A surprising result was that the size of intron borders and the corresponding values of the skews strongly differed among organisms (Table 1). Are these results compatible with our knowledge of intronic splicing motifs? In human and mouse, intron borders presented positive TA and GC skews. A number of sequence motifs identified in constitutive splicing are enriched in T and G residues. In mammals, G-rich motifs (G triplets and quartets) frequently found in introns (30,31) were proposed to increase the efficiency of constitutive splice site selection (32,33). In addition, systematic analyses of the over-represented motifs in human intron borders showed that they mainly consist in U-rich motifs and GGG motifs (and to a lesser extent in CCC motifs) that are found in the first and last 200 bp of introns (excluding the 3′ polypyrimidine tract) (27,34). A number of intronic motifs have also been identified in alternative splicing regulation, and these could participate in the observed skews of intron borders. Did alternatively spliced introns present intron skew profiles similar to those of constitutively spliced introns? To address this question, we analyzed two subsets of human introns (i) introns for which there is no splice variant known and (ii) introns for which splice variants are known (see Materials and Methods). Constitutive introns which represent the majority of introns (35) presented U-shape profiles similar to those observed for all human introns (data not shown). The skew profiles of alternative introns did not strongly differ from those of constitutive introns (see Table 1) except for the 3′ extremity of the GC skew profile for which rather constant values were observed along the 3′ intron halves (ΔGCspl(3′) = −0.39 ± 0.04%). Various intronic motifs have been identified as regulators of alternative splicing of human introns [note that intronic sequences flanking alternatively spliced introns are conserved between human and mouse (36)]. A number of these motifs are G-rich and/or U-rich such as the GGGA motif binding to the hnRNP F/H family (37), U/G-rich motifs binding to ETR3 and CUG-BP [(38) and references therein], (U)n repeats binding to TDP43 (39), UGCAUG over-represented in proximal (100 bp) intron sequences downstream of alternative exons (33) possibly binding to the Fox-1 factor (40), (A/U)GGG modulating the splicing of an alternative exon (41), U-rich motifs situated near the 5′ splice site binding TIA-1 and TIAR (42). The absence of border region observed in the 3′ end of the GC profile might result from the presence of similar amounts of C-rich and G-rich motifs as those identified by computer analysis in alternatively spliced introns (43). These lists of intronic motifs involved in constitutive and/or alternative splicing are not exhaustive, but they strongly sustain the hypothesis that the positive peaks of the TA and GC skews observed in human and mouse intron borders result from splicing-related motifs.
The other genomes examined presented intron border profiles that differed from the human and mouse profiles. There is no fundamental difference for the splicing mechanism in vertebrates and in invertebrates; however, differences have been shown including sequence motifs recognized on the pre-mRNA by the splicing machinery (44). Such differences could clearly result in mean skews differing among organisms. For D.melanogaster, the difference between the skews in intron borders and in internal regions was negative for SGC with an inversed U-shape profile (ΔGCspl = −1.78 ± 0.03%). This suggested that the peaks in the border regions resulted from C-enriched sequence motifs involved in splicing. For C.elegans, the skew profiles presented unique features: STA was negative at both ends progressively increasing to positive internal values (ΔTAspl = −2.95 ± 0.04%) and SGC was positive at the 5′ end and negative in internal regions (ΔGCspl = 5.9 ± 0.1%). This would result from splicing motifs enriched in A over T at both ends, and/or enriched in G over C at the 5′ end. However, only few intronic splicing motifs have been identified in D.melanogaster and these are regulators of alternative splicing like the U-rich motif regulating the female-specific Msl2 intron retention (45), the U-rich motif regulating the neuron-specific nrg transcript splicing (46) and the G/A-rich motif involved in the splicing regulation of the prospero twintron (47). To our knowledge, no such splicing motif is known to date in C.elegans. The small number of splicing motifs known in invertebrates, and the fact that these motifs are involved in specific mechanisms, did not allow us to further investigate if their sequences are consistent with the observed skew profiles. For A.thaliana, a large excess of T over A was observed in intron borders. Although the mechanism of splicing is basically similar in plants and in other eukaryotes, distinguishing features have been identified (48). Essentially, plant introns contain AU- and U-rich sequences required for efficient intron recognition and splicing (49–52) like the U-rich fragments promoting splicing at 5′ intron borders (53) and binding to the hnRNP-like UBP1 (54) or controlling the chlAPX gene alternative splicing (55). This sustained our hypothesis that the peaks of the TA skew observed in A.thaliana 5′ and 3′ introns ends were associated with splicing factors. Similarly, SGC decreased (although with lower values than STA) from intron ends to internal regions suggesting that, in this organism, the sequence elements that contribute to the splicing efficiency contained more G than C residues.
Overall, the skews observed in border regions of introns were fully consistent with the sequence of the already known splicing motifs. This analysis provided a new insight in the nucleotide composition and in the distribution of the motifs involved in constitutive and/or regulatory splicing processes: when considered in all introns, the abundance of these motifs decreased regularly from splice junctions to internal regions. Despite the similarities of the splicing mechanism among eukaryotes, our results showed that the distribution of splicing motifs along introns differ between organisms, extending at distances that vary from 0.5 kb in vertebrates to 1 kb in invertebrates.
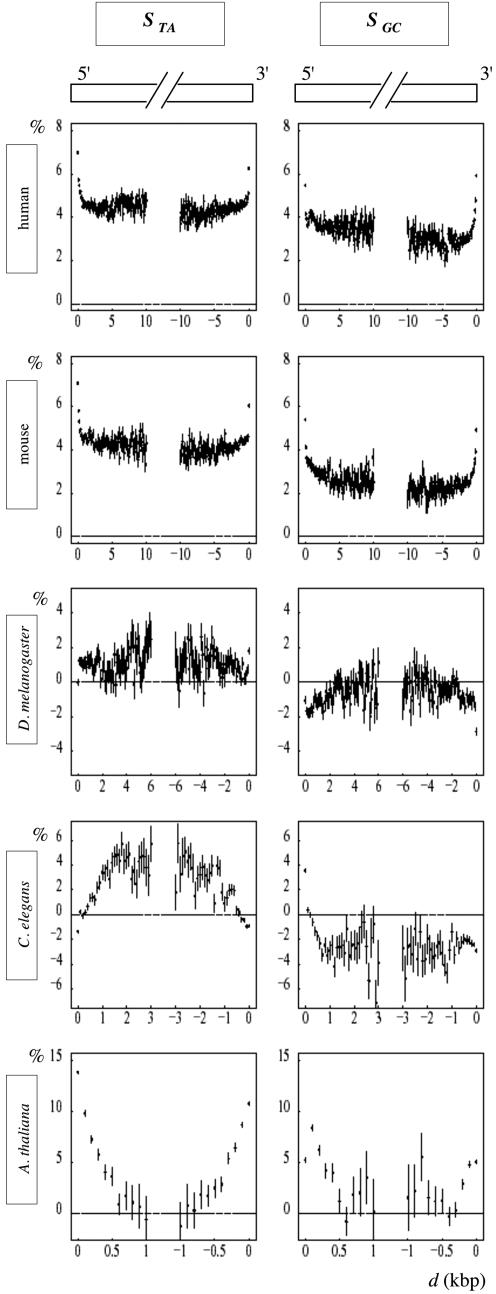
Gene skew profiles and transcription-coupled skews
Analysis of intron border regions allowed us to delineate the internal regions of introns (presenting constant TA and GC skews). We then tested the hypothesis that the skews observed in these regions, Sint, resulted from mutations associated with transcription (in the germline cells). These skews calculated for all genes were expected to be specific of transcribed regions. To generate the gene profiles of the skews, STA and SGC were calculated in adjacent windows along the gene sequences, only in intronic regions (after removal of introns' borders and repeated elements), and along the upstream and downstream intergenic regions (Figure 4). For all genomes except for C.elegans, the profiles showed sharp transitions at gene extremities and then reached plateau values in transcribed regions. This step-like pattern was fully consistent with strand biases associated with transcription. In both human and mouse 5′ gene extremities, the skews raised abruptly from intergenic to transcribed regions but in 3′ gene extremities, they decreased less rapidly over 2–3 kb after the polyadenylation site. This pattern possibly resulted from the fact that transcription does not stop precisely at the polyA site, but downstream of this site at distances that can extend over several kb [for review see (56)]. The transcription of these regions would then lead to the observed broadening of the 3′ end profiles. For C.elegans, the skew patterns presented unique features. At the 5′ extremities, the TA skew in intergenic regions increased progressively on ∼10 kb from close to zero values to reach the plateau value (∼3%) at the transcription start site. At the 3′ extremities, STA remained at the plateau values in the downstream intergenic regions over at least 20 kb. Similar properties were observed at the 5′ and 3′ gene extremities for the GC skew, which presented profiles with signs opposed to those of the STA profiles. To illustrate the importance of the removal of intron extremities, skew profiles were also generated with complete intron sequences for C.elegans and for A.thaliana (red profiles).
The analysis of strand asymmetries was also performed with the genomes of P.falciparum and S.pombe. For P.falciparum, the mean intron size was 202 bp (3403 introns analyzed) and these introns presented positive TA and GC skews (calculated for complete intron sequences) STA = 4.82 ± 0.31% and SGC = 2.11 ± 0.58%. For S.pombe, the mean size of introns was 190 bp (519 introns analyzed) with STA = 15.00 ± 0.52% and SGC = 3.15 ± 0.59% (complete intron sequences). For both genomes, the intron skew profiles were determined but, due to the small intron sizes, the border regions could not be determined. Consequently, the splicing and transcription components of these profiles could not be separated from each other, precluding further analysis of these genomes in the present study. However, we observed that both genomes clearly exhibited positive TA and GC skews specifically associated with transcribed regions (data not shown).
Could the step-like pattern observed in most genomes be explained not by transcription, but by replication in the germline cells? Although very little is known about germline replication origins, a possibility would be that they coincide with gene promoters as already observed for replication origins associated with CpG islands (57). Such origins would produce strand asymmetries in the leading strand resulting in sharp transitions at the 5′ gene extremities similar to those observed here. However, they would also produce negative biases in the opposite direction, except if we suppose that all these origins promote unidirectional replication (in the same direction as transcription). In addition, the replication should terminate precisely at the 3′ gene extremities (to reproduce the pattern observed here). Altogether, this makes the hypothesis very unlikely. Another possibility would be multiple replication origins located more or less uniformly (randomly) in the intergenic regions, as observed in Xenopus embryos (58). This would produce the constant skews observed in transcribed regions. However, the skew profiles at the gene extremities would not present sharp transitions but would rather increase progressively to reach the plateau values at both gene ends. The fact that replication could not produce the observed skew patterns did not exclude the possibility that strand asymmetries associated with replication exist in the genomes examined. If the genes were positioned randomly, relative to the replication origins, replication biases would cancel each other in the skew profiles. The mean skews would be zero in intergenic regions and they would have no effect in transcribed regions. In such a model, the small positive skews observed in the 5′ and 3′ non-transcribed regions of human and mouse genomes (Figure 4) could result from some co-orientation of transcription and of replication. If the TA and GC skews associated with replication were positive (24), preferential orientation of (a small proportion of) genes with the leading strand would then produce small positive skews in intergenic regions. This situation would preclude the measurement of the transcription-coupled skews of individual genes but it would only have little effect on our estimation of the mean values of the transcription-coupled biases.
A particular organization of C.elegans genes
The unique patterns of the skew profiles observed for C.elegans might be a by-product of a particular gene organization. In this organism, a large number of genes are organized in operons, i.e. multigenic units transcribed into polycistronic pre-mRNAs that are further processed into monocistronic RNAs (59). DNA regions that separate the genes of an operon should possibly be transcribed with similar rates than the genes themselves and the TA and GC skews should be similar in intergenes and in introns (internal regions) of the adjacent genes. However, STA and SGC did not decrease over several kilobases downstream of the gene 3′ ends, a property which was not compatible with the small size (at most few hundreds of nucleotides) of the intergenic regions that separate the successive genes in operons (60). Another possibility was that the skews observed here would result from replication. As discussed previously, multiple replication origins distributed in the regions upstream of the 5′ gene extremities would produce patterns similar to those observed here, e.g. TA skew increasing progressively (over several kilobases) upstream of the gene 5′ end, and then remaining constant along the transcribed (intronic) regions. However, this should lead to profiles decreasing progressively to zero values downstream of the gene 3′ end (at a rate comparable to that observed upstream of the gene 5′ end), a property that was not observed here.
An alternative hypothesis was that in C.elegans, transcription would continue over large distances after the polyadenylation site, at least in a subclass of genes, leading to the observed 3′ intergenic profiles. In this case, the profile of 3′ intergenic regions would depend on the orientation of the neighbor genes: in the region situated between two converging genes, the skews would cancel each other. To further investigate this question, we examined the gene skew profiles around the extremities of diverging, converging or co-oriented, adjacent genes (Figure 5). For the first two groups of genes (converging and diverging, Figure 5a–b), the TA and GC skew presented step-like patterns reminiscent of those observed, e.g. for human genes in Figure 4. In contrast, the skew profiles of the co-oriented genes presented an unprecedented, rather flat and constant patterns: these genes were preceded and followed by intergenes with skews similar to those observed in transcribed regions (Figure 5c–d). These results suggested that the intergenic regions separating co-oriented genes were transcribed in the same direction as those of the surrounding genes and with similar transcription rates. Among co-oriented genes, 2579 (respectively 2532) were preceded (respectively followed) by intergenic regions >1 kb indicating that these genes differ from those situated in the already identified C.elegans operons, in which the successive genes are separated by at most few hundreds of nucleotides (60) (in our analysis the genes separated by intergenes smaller than 100 bp were not considered). As discussed earlier, a possibility that would explain the profiles observed for co-oriented genes was that transcription would continue over large distances after the polyadenylation site. However, this hypothesis was not in agreement with the 3′ end profiles of converging genes which were close to zero immediately after the 3′ gene extremities (Figure 5b). To explain these profiles, the transcription of each gene should, for instance, terminate precisely at the polyadenylation site of the associated converging gene. Another transcription-based hypothesis was that intergenic regions contain a large proportion of still undiscovered transcribed regions that would be oriented in the same direction as their neighbor upstream and downstream co-oriented genes. In intergenes between convergent or divergent genes, such transcripts would not exist, or they would be randomly oriented, leading to zero mean skews. This hypothesis could explain all the profiles observed here, but it relied on the fact that all intergenic regions between co-oriented genes, or a large proportion of them, would be covered by unknown transcripts. A last transcription-based hypothesis was that co-oriented genes would be co-transcribed, raising the possibility of existence of a still unknown class of C.elegans transcription units characterized by large (>1 kb) intergenes, in addition to the already described operons.
Figure 5.
Strand asymmetries in the regions surrounding the 5′ and 3′ C.elegans gene extremities associated with particular gene orientations. Each of the genes studied is (a) preceded (converging genes) or (b) followed by a gene transcribed in the opposed orientation (diverging genes); each of the gene studied is (c) preceded or (d) followed by a gene transcribed in the same orientation (co-oriented genes); the values of the TA and GC skews STA and SGC were calculated and plotted as in Figure 4.
Alternatively, we supposed that the skews observed in C.elegans would be associated with replication, not transcription. In this model, replication origins would be flanked on either side by genes transcribed in the same direction as the leading strand: each replication origin would separate two diverging blocks of co-oriented genes. Consequently, these genes and their intergenes would present constant skews as observed in Figure 5c–d. Preferential gene orientation relative to replication has been observed in bacteria (61) but not in eukaryotes, and the present results would show the first example of such organization of eukaryotic genome. In this model, the origins (respectively the terminators) of replication would be situated between diverging (respectively converging) genes. However, the step-like pattern of the corresponding skew profiles observed in Figure 5a–b suggested that replication origins and terminators would coincide precisely with 5′ and 3′ gene extremities, respectively, making this model rather unlikely. In conclusion, the analysis of the skew profiles revealed that in C.elegans, distant co-oriented genes are separated by intergenic regions presenting unique transcription properties or alternatively, would follow each other in replicon units. Besides the well-known particular genomic architecture of C.elegans chromosomes [duplicate genes, operons (60,62)], our results suggest an additional level of gene organization either related to transcription or to replication. Although the latter possibility cannot be excluded, we favor the hypothesis that the biases observed in intergenic regions between co-oriented genes are associated with transcription.
CONCLUSION
The study of compositional asymmetries of intronic sequences in mammalian, invertebrate and plant genomes has revealed the presence of two different types of biases. We suggest that one of these two types is the result of the presence of sequence motifs involved in splicing in intron extremities. The sequence composition and the distribution of these motifs would result in the particular skews observed in the 5′ and 3′ intron ends. These skews can be related to other compositional biases, like those resulting from amino acid composition or codon bias (63) in the sense that they result from selection-driven processes. In contrast, the second type of biases results from mutation-driven processes and is consequently only detected in sequences that are free from selection, i.e. in internal regions of introns. The step-like profiles of the skews (measured in internal regions of introns), coinciding with the gene positions, clearly established that all genomes presented strand asymmetries specifically associated with transcription. The presence of transcription-coupled TA and GC biases in evolutionarily distant eukaryotes raised the possibility that they result from common mutation–repair processes. The TA and GC skews observed in human and mouse were consistent with a previous comparative study of mammalian genomes showing transcription-coupled asymmetry of nucleotide transition rates (25). According to these authors, the asymmetry would not result from cytosine deamination as proposed for bacterial genomes (9,14,16), but from transcription-coupled DNA repair (18) acting on mismatched base pairs due to uncorrected replication errors. Asymmetries intrinsic to these mechanisms would be responsible for the compositional skews specifically observed in mammalian transcribed regions. Basically, the transcription-coupled repair process is conserved in eukaryotes and prokaryotes (64) and this mechanism acting in germline cells during evolution could finally produce the transcription-coupled strand asymmetries observed here. However, our results showed a wide spectrum of the mean transcription-coupled TA and GC skews among eukaryotic genomes. Human and mouse genomes presented positive mean TA and GC skews (excess of T over A and excess of G over C on the coding strand). This could result from an excess of A→G over T→C transitions that might be common to vertebrates. In contrast, invertebrates presented positive mean TA skews but negative mean GC skews (excess of C over G). Such negative GC biases could result from mutational patterns differing between invertebrates and vertebrates like, for instance, a strong asymmetry of transversion rates that would be specific to invertebrates. Differences of mutation patterns between Drosophila species and mammals have been reported previously, showing a larger transversion/transition ratio in Drosophila than in mammals (3). In this context, biased transversion rates could produce the various compositional skews observed here. As a result, the differences between compositional bias profiles observed in internal regions of introns, in vertebrate and in invertebrate genomes would reflect different mutation/repair processes at work in the germline cells of these organisms.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Action Bioinformatique Inter EPST 2002, by the Centre National de la Recherche Scientifique (CNRS) and by the French Ministère de l'Education et de la Recherche. We thank Joëlle Marie, Olivier Hyrien and Maria Gallego for very helpful and constructive discussions. We also thank one of the Referees for his (her) comments on the interpretation of the different skews observed in vertebrates and invertebrates.
REFERENCES
- 1.Gojobori T., Li,W.H. and Graur,D. (1982) Patterns of nucleotide substitution in pseudogenes and functional genes. J. Mol. Evol., 18, 360–369. [DOI] [PubMed] [Google Scholar]
- 2.Li W.H., Wu,C.I. and Luo,C.C. (1984) Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J. Mol. Evol., 21, 58–71. [DOI] [PubMed] [Google Scholar]
- 3.Petrov D.A. and Hartl,D.L. (1999) Patterns of nucleotide substitution in Drosophila and mammalian genomes. Proc. Natl Acad. Sci. USA, 96, 1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z. and Gerstein,M. (2003) Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res., 31, 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobry J.R. (1995) Properties of a general model of DNA evolution under no-strand-bias conditions. J. Mol. Evol., 40, 326–330. [DOI] [PubMed] [Google Scholar]
- 6.Baisnee P.F., Hampson,S. and Baldi,P. (2002) Why are complementary DNA strands symmetric? Bioinformatics, 18, 1021–1033. [DOI] [PubMed] [Google Scholar]
- 7.Chargaff E. (1951) Structure and function of nucleic acids as cell constituents. Fed. Proc., 10, 654–659. [PubMed] [Google Scholar]
- 8.Rudner R., Karkas,J.D. and Chargaff,E. (1968) Separation of B. subtilis DNA into complementary strands. 3. Direct analysis. Proc. Natl Acad. Sci. USA, 60, 921–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank A.C. and Lobry,J.R. (1999) Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene, 238, 65–77. [DOI] [PubMed] [Google Scholar]
- 10.Tillier E.R. and Collins,R.A. (2000) The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol., 50, 249–257. [DOI] [PubMed] [Google Scholar]
- 11.Grigoriev A. (1998) Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res., 26, 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mrazek J. and Karlin,S. (1998) Strand compositional asymmetry in bacterial and large viral genomes. Proc. Natl Acad. Sci. USA, 95, 3720–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salzberg S.L., Salzberg,A.J., Kerlavage,A.R. and Tomb,J.F. (1998) Skewed oligomers and origins of replication. Gene, 217, 57–67. [DOI] [PubMed] [Google Scholar]
- 14.Francino M.P. and Ochman,H. (1997) Strand asymmetries in DNA evolution. Trends Genet., 13, 240–245. [DOI] [PubMed] [Google Scholar]
- 15.Lobry J.R. and Sueoka,N. (2002) Asymmetric directional mutation pressures in bacteria. Genome Biol., 3, RESEARCH0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beletskii A. and Bhagwat,A.S. (1998) Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol. Chem., 379, 549–551. [PubMed] [Google Scholar]
- 17.Francino M.P. and Ochman,H. (2001) Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol. Biol. Evol., 18, 1147–1150. [DOI] [PubMed] [Google Scholar]
- 18.Svejstrup J.Q. (2002) Mechanisms of transcription-coupled DNA repair. Nature Rev. Mol. Cell Biol., 3, 21–29. [DOI] [PubMed] [Google Scholar]
- 19.Francino M.P., Chao,L., Riley,M.A. and Ochman,H. (1996) Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science, 272, 107–109. [DOI] [PubMed] [Google Scholar]
- 20.Gierlik A., Kowalczuk,M., Mackiewicz,P., Dudek,M.R. and Cebrat,S. (2000) Is there replication-associated mutational pressure in the Saccharomyces cerevisiae genome? J. Theor. Biol., 202, 305–314. [DOI] [PubMed] [Google Scholar]
- 21.Francino M.P. and Ochman,H. (2000) Strand symmetry around the beta-globin origin of replication in primates. Mol. Biol. Evol., 17, 416–422. [DOI] [PubMed] [Google Scholar]
- 22.Shioiri C. and Takahata,N. (2001) Skew of mononucleotide frequencies, relative abundance of dinucleotides, and DNA strand asymmetry. J. Mol. Evol., 53, 364–376. [DOI] [PubMed] [Google Scholar]
- 23.Duret L. (2002) Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev., 12, 640–649. [DOI] [PubMed] [Google Scholar]
- 24.Niu D.K., Lin,K. and Zhang,D.Y. (2003) Strand compositional asymmetries of nuclear DNA in eukaryotes. J. Mol. Evol., 57, 325–334. [DOI] [PubMed] [Google Scholar]
- 25.Green P., Ewing,B., Miller,W., Thomas,P.J. and Green,E.D. (2003) Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet., 33, 514–517. [DOI] [PubMed] [Google Scholar]
- 26.Touchon M., Nicolay,S., Arneodo,A., d'Aubenton-Carafa,Y. and Thermes,C. (2003) Transcription-coupled TA and GC strand asymmetries in the human genome. FEBS Lett., 555, 579–582. [DOI] [PubMed] [Google Scholar]
- 27.Louie E., Ott,J. and Majewski,J. (2003) Nucleotide frequency variation across human genes. Genome Res., 13, 2594–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black D.L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem., 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 29.Dreyfuss G., Kim,V.N. and Kataoka,N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol., 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 30.Nussinov R. (1988) Conserved quartets near 5′ intron junctions in primate nuclear pre-mRNA. J. Theor. Biol., 133, 73–84. [DOI] [PubMed] [Google Scholar]
- 31.Engelbrecht J., Knudsen,S. and Brunak,S. (1992) G+C-rich tract in 5′ end of human introns. J. Mol. Biol., 227, 108–113. [DOI] [PubMed] [Google Scholar]
- 32.McCullough A.J. and Berget,S.M. (1997) G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell Biol., 17, 4562–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brudno M., Gelfand,M.S., Spengler,S., Zorn,M., Dubchak,I. and Conboy,J.G. (2001) Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res., 29, 2338–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majewski J. and Ott,J. (2002) Distribution and characterization of regulatory elements in the human genome. Genome Res., 12, 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 36.Sorek R. and Ast,G. (2003) Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res., 13, 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caputi M. and Zahler,A.M. (2001) Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem., 276, 43850–43859. [DOI] [PubMed] [Google Scholar]
- 38.Singh G., Charlet,B.N., Han,J. and Cooper,T.A. (2004) ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res., 32, 1232–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagani F., Buratti,E., Stuani,C., Romano,M., Zuccato,E., Niksic,M., Giglio,L., Faraguna,D. and Baralle,F.E. (2000) Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem., 275, 21041–21047. [DOI] [PubMed] [Google Scholar]
- 40.Jin Y., Suzuki,H., Maegawa,S., Endo,H., Sugano,S., Hashimoto,K., Yasuda,K. and Inoue,K. (2003) A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J., 22, 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirand-Pugnet P., Durosay,P., Brody,E. and Marie,J. (1995) An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res., 23, 3501–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dember L.M., Kim,N.D., Liu,K.Q. and Anderson,P. (1996) Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem., 271, 2783–2788. [DOI] [PubMed] [Google Scholar]
- 43.Miriami E., Margalit,H. and Sperling,R. (2003) Conserved sequence elements associated with exon skipping. Nucleic Acids Res., 31, 1974–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCullough A.J. and Schuler,M.A. (1993) AU-rich intronic elements affect pre-mRNA 5′ splice site selection in Drosophila melanogaster. Mol. Cell. Biol., 13, 7689–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forch P., Puig,O., Kedersha,N., Martinez,C., Granneman,S., Seraphin,B., Anderson,P. and Valcarcel,J. (2000) The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell, 6, 1089–1098. [DOI] [PubMed] [Google Scholar]
- 46.Lisbin M.J., Qiu,J. and White,K. (2001) The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev., 15, 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scamborova P., Wong,A. and Steitz,J.A. (2004) An intronic enhancer regulates splicing of the twintron of Drosophila melanogaster prospero pre-mRNA by two different spliceosomes. Mol. Cell. Biol., 24, 1855–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson G.G. and Filipowicz,W. (1996) Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol., 32, 1–41. [DOI] [PubMed] [Google Scholar]
- 49.Goodall G.J. and Filipowicz,W. (1989) The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell, 58, 473–483. [DOI] [PubMed] [Google Scholar]
- 50.Gniadkowski M., Hemmings-Mieszczak,M., Klahre,U., Liu,H.X. and Filipowicz,W. (1996) Characterization of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in Nicotiana plumbaginifolia. Nucleic Acids Res., 24, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baynton C.E., Potthoff,S.J., McCullough,A.J. and Schuler,M.A. (1996) U-rich tracts enhance 3′ splice site recognition in plant nuclei. Plant J., 10, 703–711. [DOI] [PubMed] [Google Scholar]
- 52.Ko C.H., Brendel,V., Taylor,R.D. and Walbot,V. (1998) U-richness is a defining feature of plant introns and may function as an intron recognition signal in maize. Plant Mol. Biol., 36, 573–583. [DOI] [PubMed] [Google Scholar]
- 53.McCullough A.J. and Schuler,M.A. (1997) Intronic and exonic sequences modulate 5′ splice site selection in plant nuclei. Nucleic Acids Res., 25, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambermon M.H., Simpson,G.G., Wieczorek Kirk,D.A., Hemmings-Mieszczak,M., Klahre,U. and Filipowicz,W. (2000) UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. Embo J., 19, 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshimura K., Yabuta,Y., Ishikawa,T. and Shigeoka,S. (2002) Identification of a cis element for tissue-specific alternative splicing of chloroplast ascorbate peroxidase pre-mRNA in higher plants. J. Biol. Chem., 277, 40623–40632. [DOI] [PubMed] [Google Scholar]
- 56.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antequera F. and Bird,A. (1999) CpG islands as genomic footprints of promoters that are associated with replication origins. Curr. Biol., 9, R661–667. [DOI] [PubMed] [Google Scholar]
- 58.Blow J.J., Gillespie,P.J., Francis,D. and Jackson,D.A. (2001) Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J. Cell Biol., 152, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blumenthal T. (1998) Gene clusters and polycistronic transcription in eukaryotes. Bioessays, 20, 480–487. [DOI] [PubMed] [Google Scholar]
- 60.Blumenthal T., Evans,D., Link,C.D., Guffanti,A., Lawson,D., Thierry-Mieg,J., Thierry-Mieg,D., Chiu,W.L., Duke,K., Kiraly,M. et al. (2002) A global analysis of Caenorhabditis elegans operons. Nature, 417, 851–854. [DOI] [PubMed] [Google Scholar]
- 61.McLean M.J., Wolfe,K.H. and Devine,K.M. (1998) Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol., 47, 691–696. [DOI] [PubMed] [Google Scholar]
- 62.Lercher M.J., Blumenthal,T. and Hurst,L.D. (2003) Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res., 13, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karlin S., Campbell,A.M. and Mrazek,J. (1998) Comparative DNA analysis across diverse genomes. Annu. Rev. Genet., 32, 185–225. [DOI] [PubMed] [Google Scholar]
- 64.Hanawalt P.C. (2001) Controlling the efficiency of excision repair. Mutat. Res., 485, 3–13. [DOI] [PubMed] [Google Scholar]