Abstract
The specificity of DNA-mediated protein assembly was studied in two in vitro systems, based on (i) the DNA-binding domain of bacteriophage 434 repressor cI (amino acid residues 1–69), or (ii) the DNA-binding domain of the yeast transcription factor GCN4, (amino acids 1–34) and their respective oligonucleotide cognates. In vivo, both of these peptides are part of larger protein molecules that also contain dimerization domains, and the resulting dimers recognize cognate palindromic DNA sequences that contain two half-sites of 4 bp each. The dimerization domains were not included in the peptides tested, so in solution—in the presence or absence of non-cognate DNA oligonucleotides—these molecules did not show appreciable dimerization, as determined by pyrene excimer fluorescence spectroscopy and oxidative cross-linking monitored by mass spectrometry. Oligonucleotides with only one 4 bp cognate half-site were able to initiate measurable dimerization, and two half-sites were able to select specific dimers even from a heterogeneous pool of molecules of closely related specificity (such as DNA-binding domains of the 434 repressor and their engineered mutants that mimic the binding helix of the related P22 phage repressor). The fluorescent technique allowed us to separately monitor the unspecific, ionic interaction of the peptides with DNA which produced a roughly similar signal in the case of both cognate and non-cognate oligonucleotides. But in the former case, a concomitant excimer fluorescence signal showed the formation of correctly positioned dimers. The results suggest that DNA acts as a highly specific template for the recruitment of weakly interacting protein molecules that can thus build up highly specific complexes.
INTRODUCTION
Transcriptional regulation relies on the formation of large multicomponent protein–DNA complexes (1–3). The protein complexes recognize short stretches of DNA with remarkable specificity, in spite of the large number of competing non-specific protein–DNA and protein–protein interactions. In general, the components of these complexes are transcription factors with no tendency to associate in the absence of DNA. Moreover, the DNA-binding affinities of some of the individual factors may be very weak (3). Complex formation is believed to follow a recruitment model, which predicts DNA acting as a template where the components sequentially bind to each other to form the assembly. The specificity of this process ultimately depends both on DNA recognition and on the interactions of the protein components (2,3).
The ternary complexes formed by dimeric repressors and DNA are the simplest models of multicomponent DNA-recognition. Formation of dimeric repressor/DNA complexes is thought to follow predominantly either the so-called monomeric pathway, in which a repressor monomer binds first to DNA, or the so-called dimeric pathway, in which the repressor subunits dimerize before binding to DNA. Members of the bZIP and bHLH protein families like activating transcription factor 2 (ATF-2) and Max (4) as well as the Arc repressor of the P22 phage (5) and some type II endonucleases (6–8) bind as monomers to their target sites. In this case, the sequential binding of monomers seems to be faster and more specific than the alternative pathway wherein the monomers dimerize before binding to DNA (4,5,9–16). On the other hand, typical type II restriction endonucleases exist in solution as homodimers and are known to bind to and cleave DNA as homodimers (6,17), and the same mechanism is followed by some DNA helicases (18).
Since an initial dimer formation step is not always a prerequisite for the formation of these ternary complexes, our goal was to follow the recruitment and assembly of weakly interacting protein components on the nucleic acid. We used two different model systems, the N-terminal region of the bacteriophage 434 cI repressor and the basic motif of the yeast transcription factor GCN4, both devoid of their dimerization domains.
A 434 cI repressor monomer is composed of an N-terminal DNA-binding domain (residues 1–69, denoted here as R69), which contains a helix–turn–helix (HTH) motif, and a C-terminal domain, which is involved in dimerization (19–21). The 434 repressor binds to its tightest natural operator site OR1 with a Kd of ∼10−9 M (22–24). When the C-terminal dimerization domain is removed, the DNA-binding affinity is reduced by about two orders of magnitude, but tight DNA-binding is restored when two R69 domains are covalently connected (24–26). A set of contacts between monomers has been observed in the crystal structure of R69 bound to a 20 bp DNA fragment containing the OR1 binding site: a patch of hydrophobic residues from both monomers is involved plus two salt bridges and a hydrogen bond (27). The inter-domain contacts are strong enough to define the structure of the bound dimer but, when not associated with DNA, R69 is a monomer in solution even at millimolar concentrations necessary for NMR measurement (28) and folding cooperativity (a manifestation of subunit interactions) cannot be observed even in the covalently connected dimer (26).
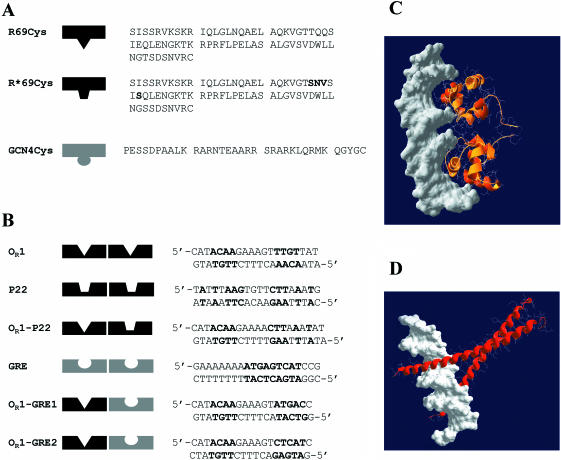
Another advantage of the 434 repressor system is the wealth of information available on specificity changes. By replacing the DNA-contacting residues at the −1,1,2 and 5 positions in the recognition helix with the corresponding residues of the Salmonella phage P22 repressor c2, the binding specificity of the 434 repressor can be altered so as to bind to the P22 operator (29). Later studies on covalent dimers (24,30) showed that the mutants are highly selective, even though the operator sites of the 434 and the P22 repressor hold some sequence similarity. We engineered wild-type R69 with a single cysteine appended at the C-terminus (R69Cys) and a mutant specific for the operator of the P22 repressor c2 (R*69Cys) (Figure 1).
Figure 1.
(A) Amino acid sequences of R69Cys and R*69Cys. R69Cys and R*69Cys differ in the sequence of the recognition helix (the mutated residues in R*69Cys are shown in bold) (B) Sequences of various synthetic oligonucleotides used in the study. The nucleotides shown in bold represent the consensus sequence of the specific binding sites. OR1 corresponds to the OR1 site of the operator sequence of 434 repressor. P22 is the sequence of the operator OR1 site of phage P22 repressor c2. GRE contains the specific operator sequence of GCN4 repressor. OR1–P22 represents a hybrid DNA with one 434-specific and one P22-specific half-sites. OR1–GRE1 and OR1–GRE2 represent hybrid DNAs with one 434-specific and one GCN4-specific half-sites: they differ in the orientation of the GRE half-site. (C) Computer model of two R69Cys monomers in complex with DNA. The coordinates of residues 1–69 were taken from the NMR structure of R69 in solution (Brookhaven PDB 1PRA) and superimposed (1–63 residues) on the peptide backbone of the resolved X-ray R69/OR2 complex (1RPE). The model was constructed using the program Swiss-PdbViewer v3.6b3. (D) Computer model of two GCN4 monomers in complex with DNA (Brookhaven PDB 1YSA).
Our second experimental model is the basic domain of yeast transcription factor GCN4. GCN4 binds as a dimer to a specific DNA recognition sequence (GRE, Figure 1) using a leucine zipper as a dimerization motif. We synthesized by solid phase peptide synthesis (SPPS) the DNA-binding domain (residues 1–34) lacking the dimerization sequence and with an extra cysteine at the C-terminus (Figure 1). In contrast to R69, this domain has no ordered structure in solution, but the helices contacting the DNA major groove, form readily upon DNA binding and there is no interaction between the two DNA-binding motifs in the three-dimensional (3D) structure of the bound dimer (31–33).
The experimental methodologies chosen to detect the assembly event consist of site-specific fluorescence spectroscopy and oxidative cross-linking. From the crystal structures data, we engineered a Cys residue into a suitable sequence position (the C-terminus in both our models) that is supposed to be sufficiently near when two monomers form a dimer (Figure 1). Then a pyrene label is attached to the Cys residues, followed by monitoring the emergence of excimer fluorescence resulting from the two eventually interacting labels. Pyrene fluorescence was often used as a proximity label both for protein–protein interaction and nucleic acid hybridization studies (17,34,35). Our second experimental strategy is based on monitoring by liquid chromatography coupled to mass spectrometry (LC-MS), the rate of formation of disulfide-linked dimers and their homo- or hetero-composition, to check whether the nucleic acid acts as a template for the ligation of two monomers. Preformed disulfide cross-linked DNA-binding domain dimers with strong DNA-binding properties have been constructed both from the 434 repressor (26) and from the GCN4 (36), and we have used the same strategies for constructing our Cys-modified monomers.
Here, we show that the cognate site specificity plays a central role in recruiting the right monomers from a pool of polypeptides of related specificity and slight changes in the cognate sequence lead to drastic changes in complex formation. A half-cognate site is enough to recruit a detectable amount of complexes in both our systems. The experimental techniques employed in this work allow a quick and sensitive detection of the complex formation (pyrene excimer fluorescence) and a precise determination of the composition of the dimer (LC-MS).
MATERIALS AND METHODS
Materials
N-(1-pyrenyl) maleimide, Tris(2-carboxyethyl) phosphine hydrochloride (TCEP), 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB), heparin (from porcine intestinal mucosa, average MW 5000) and polyacrylic acid (average MW 1200) were purchased from Sigma-Aldrich Corp. (St Louis, MO, USA). The synthetic oligodeoxyribonucleotides were purified by high-performance liquid chromatography (HPLC). Escherichia coli tRNA was purchased from Roche (Germany) and an average MW 28 000 was used in the calculations. Single-stranded DNA (ssDNA) (A)25 was purchased from Primm (Milan, Italy). 9-fluorenylmethyloxycarbonyl (Fmoc)-protected amino acids and 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium tetrafluoroborate (TBTU) were purchased from Calbiochem-Novabiochem AG (Switzerland). Diisopropylethylamine (DIPEA), trifluoroacetic acid (TFA), ethanedithiol (EDT), triisopropylsilane (TIPS) from Fluka Chemie AG (Switzerland). All other reagents and solvents were reagent grade and were purchased from Fluka, Sigma-Aldrich (USA), Biosolve Ltd (Netherlands), Alexis (USA) and Advanced ChemTech (USA).
Peptide synthesis of GCN4Cys
Automatic peptide synthesis of GCN4Cys was performed on a Protein Technologies PS3 peptide synthesizer (Rainin Instrument Co. Inc., USA).
The peptide was synthesized according to standard solid-phase Fmoc chemistry with TBTU as peptide coupling reagent using four equivalents of TBTU/Fmoc-Xaa-OH/DIPEA (0.95/1.00/1.90). The peptide-resin was cleaved/deprotected with TFA/EDT/H2O/TIPS (90/5/2.5/2.5, 20 ml/g resin) for 3 h at RT. The peptide was precipitated and washed with diethyl ether, dissolved in 5% acetic acid (25 ml/g peptide-resin), extracted with diethyl ether (10 ml six times) and freeze-dried. The peptide was purified by reverse phase HPLC (RP-HPLC) and freeze-dried. The calculated molecular weight was confirmed by mass spectrometry [GCN4Cys: 3959.54 (calc.), 3959.12 (electrospray ionization mass spectroscopy, ESI-MS)].
Preparation of R69Cys and R*69Cys
R69Cys was prepared and expressed as described (24,26). A similar approach was taken to construct the engineered molecule R*69Cys (24). The proteins (sequences shown in Figure 1) were purified as disulfide dimers by fast performance liquid chromatography (FPLC) on a SP-Sepharose ion-exchange column followed by a Mono-S HR10/10 column from Amersham Pharmacia Biotech (Uppsala, Sweden). The samples were found homogenous by SDS-PAGE and RP-HPLC and the calculated molecular weights were confirmed by ESI-MS [R69Cys: 7654.72 (calc.), 7654.4 (ESI-MS); R*69Cys: 7541.65 (calc.), 7542.0 (ESI-MS)]. The reduced subunits were prepared via incubation with 5 mM TCEP for at least 12 h and the complete reduction checked by LC-MS. The proteins were finally desalted by gel filtration on an FPLC desalting column (HiPrep 26/10 from Pharmacia). The collected fractions were quantified by UV absorbance (ɛ280 = 5690) and stored frozen.
Site-directed fluorescence spectroscopy
The R69Cys and GCN4Cys molecules were covalently labeled with the fluorescent probe pyrene at their cysteine residue (R69Cys-py and GCN4Cys-py). Ten milligrams of N-(1-pyrenyl) maleimide, dissolved in 0.5 ml of dimethylsulfoxide were added to 1 ml of protein solution [2 mg/ml, in 25 mM sodium potassium phosphate buffer (pH 7.4) containing 5 mM TCEP]. The mixture was incubated at room temperature (25°C) for 24 h. Insoluble material was removed by centrifugation. The labeled protein was separated from the reagent by gel filtration on an FPLC desalting column (HiPrep 26/10 from Pharmacia). The labeled protein fractions were pooled and concentrated using Centricon-3 filters from Millipore (Bedford, MA, USA) or by RP-HPLC and freeze-drying (GCN4Cys). The concentration of the protein stock solution was determined by amino acid analysis. The labeling was found quantitative as determined by LC-MS and by measuring the optical density of the probe at 344 nm, using an extinction coefficient of 44 mM−1 cm−1 (37).
Fluorescence spectra were recorded using a Hitachi F-4000 fluorescence spectrophotometer. The excitation and emission bandpasses were set at 5 and 3 nm, respectively. Fluorescence spectra were recorded at room temperature in corrected spectrum mode with the excitation wavelength set at 335 nm.
The proteins in 0.5 ml of 20 mM sodium phosphate buffer, pH 7.2, were titrated with increasing concentrations of the different oligonucleotides or polyanions. To study the effect of NaCl, a mixture of 1 μM of R69Cys-py and 5.5 μM of the oligonucleotide in 20 mM sodium phosphate buffer, pH 7.2, was adjusted to different salt concentrations.
Disulfide cross-linking and ESI-MS analysis
R69Cys, R*69Cys or GCN4Cys (100 μM) in 20 mM ammonium acetate buffer (pH 7.0) were incubated with 5 mM TCEP for at least 12 h to ensure complete reduction of the subunits, then purified and desalted by RP-HPLC, and stored at 4°C in acidic conditions to minimize thiol oxidation. An aliquot of the reduced subunit (900 pmol) was incubated at room temperature in the absence or in the presence of cognate and non-cognate oligonucleotides, in 900 μl (or 12 ml for the experiment at 75 nM concentration) of 100 mM ammonium acetate buffer (pH 7.0). The reaction was initiated by adding concentrated DTNB to a final amount of 450 pmol. Small aliquots (≈90 pmol as monomer) of the samples were withdrawn at different time intervals and quenched by adding 2 μl TFA. The aliquots were immediately analyzed on a Waters HPLC system equipped with a microbore column Zorbax SB300 C18 1 × 150 mm from Agilent Technologies, Inc. (DE, USA) thermostated at 70°C and detected by ESI-MS on an API-150EX from Applied Biosystems (CA, USA). Six multiply charged molecular ions in the total ion chromatogram (TIC) were monitored: for R69 disulfide dimers, we monitored (M+11H)11+ and (M+13H)13+ ions; for monomers, we followed (M+6H)6+ and (M+7H)7+ which partially overlap with (M+12H)12+ and (M+14H)14+ of the dimers, respectively, but can be distinguished on the basis of the retention time. For GCN4Cys monomers, we monitored (M+4H)4+, (M+3H)3+, (M+2H)2+; for GCN4Cys dimers, we used the (M+7H)7+, (M+5H)5+ signals. The areas of the dimer peaks in TIC were integrated using the software Biomultiview from Applied Biosystems and fitted on linear calibration curves constructed on seven standards ranging from 5 to 340 pmol of preformed disulfide dimers (correlation coeff. R = 0.9946 for R69Cys dimer; R = 0.9931 for R*69Cys dimer; R = 0.9919 for R69Cys–R*69Cys; R = 0.9847 for GCN4Cys dimer). The rate constant k was calculated by fitting the dimerization kinetics to the hyperbolic equation
![]()
where [M] is the monomer concentration, M0 is the initial concentration of monomer and t is the time expressed in minutes.
RESULTS
Fluorescence spectroscopy: the fluorescent molecules
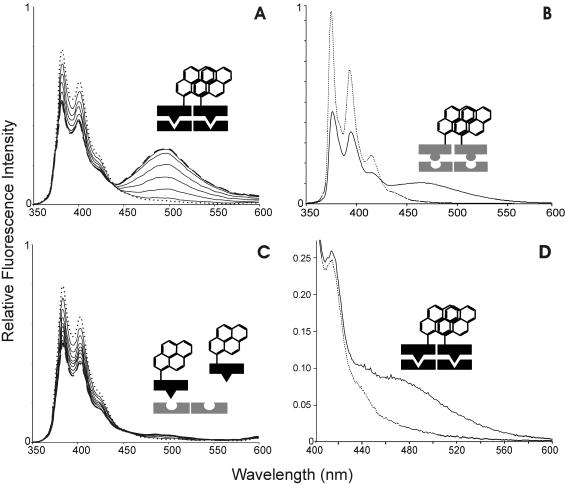
Monomeric pyrene molecules attached to proteins exhibit a fluorescence spectrum consisting of two peaks around 380 and 400 nm, respectively (38). If two pyrene labels stack, excimer fluorescence emerges between 450 and 600 nm. The pyrene absorption spectrum for R69Cys-py presents the typical pyrene absorption peaks at around 325 and 345 nm (17,38). In the absence of DNA, the fluorescence spectrum of a 1 μM solution of R69Cys-py shows two prominent sharp peaks centered at 383 and 405 nm, respectively, and exhibits almost no fluorescence between 450 and 600 nm (Figure 2A, dotted line), the same spectrum is shown by a solution of the pyrene labeled GCN4Cys (Figure 2B, dotted line). This result indicates that, in the absence of DNA, the pyrene labels of R69Cys-py and GCN4Cys-py are not in a stacking excimer configuration. Furthermore, we can also rule out the presence of ground-state stacking interactions because the absorbance spectra of the modified proteins are not broadened with respect to the spectrum of pyrene labeled N-acetyl cysteine (data not shown).
Figure 2.
(A) Fluorescence spectra of R69Cys-py (1 μM) in the absence (dotted line) and in the presence (continuous lines and dashed line) of the specific OR1. Spectra correspond to increasing DNA concentrations: 0.00, 0.48, 0.94, 1.40, 1.85, 2.29, 2.73 and 3.15 μM (dashed line), respectively. (B) Spectrum of 2 μM GCN4Cys-py in the absence (dotted line) and in the presence (continuous line) of the specific oligonucleotide GRE. (C) Fluorescence spectra of R69Cys-py (1 μM) in the absence (dotted line) and in the presence (continuous lines and dashed line) of the non-specific (GRE) DNAs obtained at DNA concentrations as in (A) and also at 4.90 and 6.66 μM (dashed line). (D) Fluorescence spectra of the excimer region of 80 nM R69Cys-py in the absence (dotted line) and in the presence (continuous line) of OR1.
Fluorescence in the presence of the specific operator
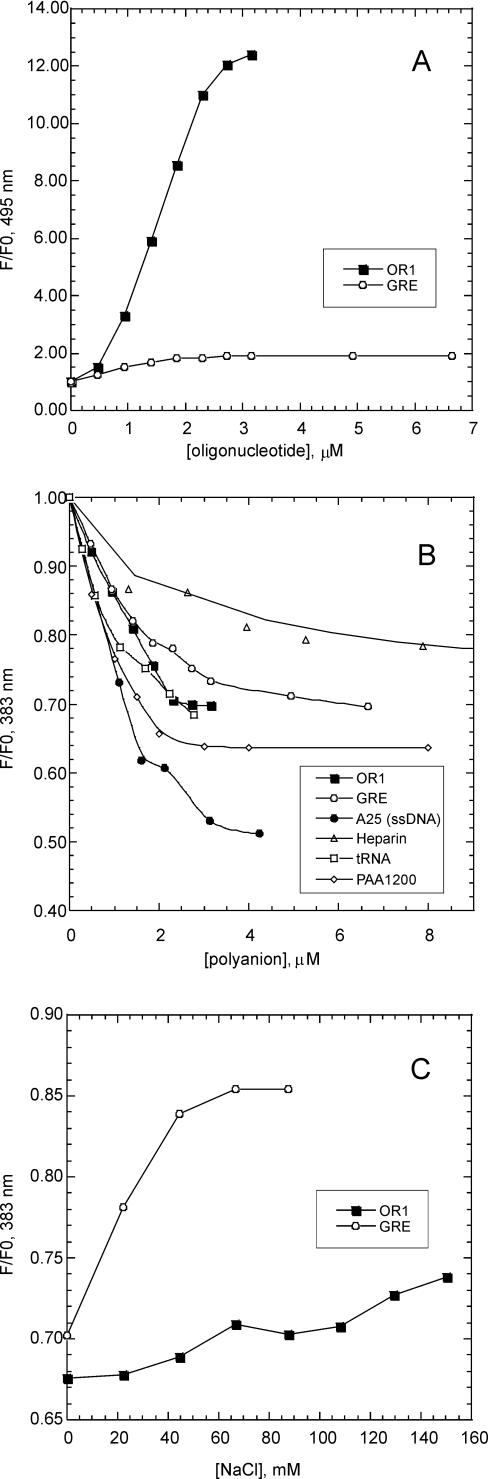
Addition of the specific oligonucleotide OR1, (see Figure 1A for the sequences) to a 1 μM solution of R69Cys-py results in a concentration-dependent increase of the excimer fluorescence measured at 495 nm (Figure 2A and 3A) and a concomitant decrease in the monomeric pyrene signal measured at 383 nm (Figure 2A and 3B); both modifications in the spectrum reach a steady state when DNA concentration is around 2 μM with ratio F/F0 ≈ 12 at 495 nm. The appearance of excimer bands is consistent with the formation of an intermolecular dimer indicating a close proximity of the two C-termini.
Figure 3.
(A) Variation of relative fluorescence intensity (F/F0) of R69Cys-py (1 μM) at 495 nm as a function of the cognate oligonucleotide, OR1 (filled square) and the non-cognate oligonucleotide, GRE (empty circle) DNA concentrations. F and F0 are the fluorescence intensities in the presence and in the absence of DNA. (B) Variation of relative fluorescence intensity of R69Cys-py (1 μM) at 383 nm as a function of OR1 and GRE oligonucleotides (spectra shown in Figure 2A and 2C, respectively), as non-cognate ssDNA (A25), heparin and polyacrylate. (C) Effect of NaCl on the interaction of R69Cys-py with the specific oligonucleotide, OR1 (filled square) and the non-cognate oligonucleotide, GRE (empty circle). Concentrations of the peptide and DNA are 1 and 5.5 μM, respectively. F and F0 are the fluorescence intensities at 383 nm in the presence and in the absence of salt.
We also performed fluorescence experiments to test the monomer recruitment at lower concentration values. Figure 2D shows the excimer fluorescence region of 80 nM R69Cys-py in the absence and in the presence of 125 nM OR1. The excimer band suggests that recruitment and complex formation is still consistent at this concentration. A series of additional fluorescence measurements at 1, 0.5, 0.25, 0.125 μM R69Cys-py (data not shown) allowed us to estimate the Kd of the R69Cys-py/OR1 complex at around 0.25 μM, a value in agreement with data previously measured with R69 monomers by gel-retardation assays (24).
The appearance of the excimer signal is also evident when the interacting partners are GCN4Cys-py (3.5 μM) and its specific binding site GRE (3.5 μM); as in the case of R69Cys, we also observe a decrease in the 383 nm band and the total lack of excimer in the absence of specific DNA (Figure 2B). It must be noted that the concentration range at which GCN4Cys-py showed excimer fluorescence in our experiments (>1 μM) is considerably higher than the R69Cys-py range (≥80 nM); this probably could be not only due to a different binding strength of the monomeric GCN4Cys-py for its DNA half-site, but also due to the lack of the stabilizing effect exerted by the intermolecular contacts present in the assembled R69 dimer.
Non-specific operators
When R69Cys is in the presence of a non-specific oligonucleotide (GRE), this does not result in appreciable excimer fluorescence even when increasing the nucleic acid concentration to 6.66 μM (Figure 2C and 3A). It is apparent, on the other hand, that addition of non-operator DNA leads anyway to a concentration-dependent decrease of the 383 nm signal which levels off at around 2.5 μM DNA (Figure 2C and 3B). As the intensity of pyrene monomer fluorescence is known to decrease in the vicinity of nucleic acids (34,35,39,40), we tested the effect of a number of other polyanions on the spectrum of R69Cys-py (Figure 3B). It appears that tRNA, single-stranded DNA (ssDNA), polyacrylate, and to a lesser extent, heparin all produce a concentration-dependent decrease in the monomeric pyrene spectrum quite similar to the one produced by non-cognate double-stranded DNA (dsDNA); this phenomenon suggests that the decrease of monomeric pyrene fluorescence could be partly due to electrostatic interactions, i.e. the presence of negative charges in the vicinity of the fluorescent group might be responsible for quenching the pyrene fluorescence. Monitoring the signal at 383 nm in the presence of increasing ionic strength shows that the decrease is indeed salt sensitive, the intensity of monomeric pyrene fluorescence can be partially restored by increasing the NaCl concentration, and this effect is much more pronounced in the interaction with non-specific oligonucleotides than with the specific operator (Figure 3C). We also measured the excitation spectrum of R69Cys-py in the absence and in the presence of GRE. The spectrum in the presence of GRE is not broadened in comparison to the R69Cys-py alone (data not shown). This is a further indication that the lack of excimer fluorescence upon non-specific DNA interaction is probably due to a real lack of pyrene proximity and not to the impossibility of two pyrene rings in a stacked ground-state configuration to reorient in an excimer configuration.
The oligonucleotide P22 contains two properly spaced P22-operator half-sites (Figure 1). Examining the P22 half-site sequence, we see two A • T base-pairs in position 3 and 4 of the half-site, the same sequence being present in the specific OR1 half-site. In the crystal structure of R69, the two A • T base pairs are involved in H-bonds and van der Waals contacts with the R69 amino acid residues (27); in this respect, it could be expected that a correctly positioned R69Cys-py would, to some extent, bind to the P22 subsite but in a previously published work, single-chain R69 dimers (up to 200 nM concentration) did not give detectable bandshift in the presence of a P22 operator (24).
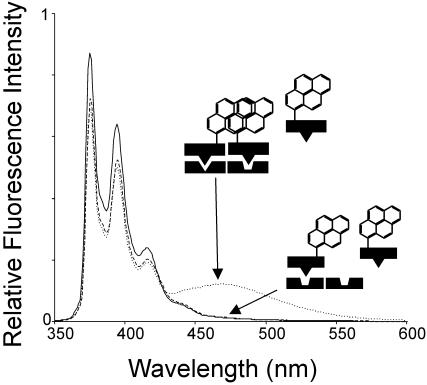
As expected, when we tested a 1 μM solution of R69Cys-py in the presence of the P22 oligonucleotide, no excimer fluorescence was detectable (Figure 4); in this case the situation was similar to that observed with GRE, the symmetric P22 operator behaving as a non-specific DNA and again causing a significant decrease in the monomeric pyrene fluorescence signal at 383 nm.
Figure 4.
Fluorescence spectra of R69Cys-py in the absence (continuous line) and in the presence of 2 μM hybrid oligonucleotides OR1–P22 (dotted line) and 2 μM P22 (dashed line).
Hybrid operator sites
We investigated the assembly of R69Cys-py on a hybrid operator DNA (OR1–P22) composed of one 434 half-site and one consensus P22 half-site (sequence in Figure 1B). Figure 4 shows that R69Cys-py exhibits significant excimer fluorescence in the presence of the hybrid operator. The calculated F/F0 value at 495 nm increases to a maximum value of 5.2 (at 2 μM oligonucleotide), which is significantly smaller than the value of ≈12 (Figure 3A) obtained in the presence of the OR1 oligonucleotide. This result suggests that the OR1 operator half-site is sufficient to promote the formation of a productive dimer since the characteristic excimer fluorescence is readily detectable when a properly spaced, partly similar half-site is present. Similar to the previous experiments, addition of the hybrid OR1–P22 operator induced a decrease in the monomeric pyrene fluorescence signal at 383 nm.
Disulfide cross-linking studies
As demonstrated by the fluorescence studies, the cysteine residues of two DNA-bound monomers are close enough to enable pyrene stacking. As a consequence, disulfide dimer formation also could be greatly facilitated upon DNA-binding. We carried out R69Cys and GCN4Cys dimerization experiments in the absence and in the presence of various specific and non-specific oligonucleotides. The formation of the disulfide bond is a limiting step in this process because spontaneous oxidation proceeds slowly. It must be noted that in the fluorescence studies, the excimer band was readily visible as soon as the sample was available for the monitoring of the spectrum (30–60 s after the DNA addition and mixing).
We promoted the disulfide formation by adding 5,5′-dithio-bis-(2-nitrobenzoic acid) in a molar ratio 1:2 with respect to the proteins in order to form in situ an activated mixed disulfide intermediate (Protein–TNB), which would readily react with a proximal monomer carrying a free thiol. We found this approach preferable to the use of a conventional chemical cross-linker (for instance bis-maleimidoethane) because the DTNB ensures the shortest distance of reactivity.
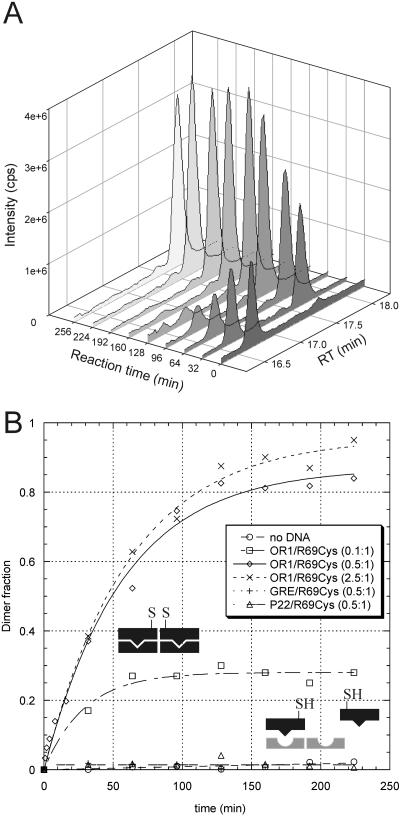
We followed the reaction by LC-MS, which allowed a fine monitoring of the different molecular species present in the reaction (Figure 5A). The quantity of disulfide dimer was calculated by area integration of the extracted ion chromatogram given by its characteristic ions. The areas were fitted on linear calibration curves composed of standards of preformed disulfide dimers.
Figure 5.
(A) Disulfide cross-linking experiment with 1 μM R69Cys in the presence of 0.5 μM OR1. The figure shows the TIC from RP-HPLC of aliquots taken at increasing reaction times. The compound with retention time RT = 16.7 min is monomeric R69Cys, the compound with RT = 17.7 min is R69Cys disulfide dimer. (B) Disulfide cross-linking experiment with 1 μM R69Cys in the absence or in the presence of specific and non-specific oligonucleotides at different protein–DNA ratios. The points are the average of three independent experiments and represent the fraction of dimer formed (determined dimer concentration divided by half the initial monomer concentration) at subsequent intervals.
Specific operators promote disulfide cross-linking
In our experimental conditions (1 μM R69Cys), the formation of disulfide dimers proceeds very slowly in the absence of DNA; even after 300 min from the addition of DTNB (Figure 5B), no substantial increase can be observed. The addition of non-specific oligonucleotides GRE or P22 to the reaction mixture in various stoichiometric ratios (0.5- to 3-fold of the R69Cys concentration) does not substantially influence the rate of dimer formation (kR69Cys+GRE = 0.0003 μM−1 min−1) (Figure 5).
On the other hand, addition of the specific symmetric operator OR1, in stoichiometric amounts, has a dramatic effect on the rate of dimerization (kR69Cys+OR1 = 0.03 μM−1 min−1, kR69Cys+OR1/kR69Cys+GRE = 100): after 30 min, the fraction of R69Cys dimer is around 40% and at 150 min, this raises to >80% (Figure 5). Addition of operator DNA in higher ratios (2.5-fold) does not modify substantially the rate of dimerization (kR69Cys+OR1/kR69Cys+OR1(×5) = 0.87) while a sub-stoichiometric amount (0.1-fold) reduces the reaction rate almost proportionally (kR69Cys+OR1/kR69Cys+OR1(×0.2) = 4.30) (Figure 5). Furthermore, we observed that oligonucleotides corresponding to various natural operator sites (OR1, OR2 and OR3), promoted subunit assembly to a very similar extent (data not shown). As we did in the fluorescence studies, we also tested the disulfide formation at lower concentrations using 75 nM R*69Cys (shown in Figure 6); the experiments confirmed a strong enhancement of the disulfide formation rate in the presence of a stoichiometric amount of P22. GCN4Cys shows a slightly higher rate, with respect to R69Cys, of spontaneous disulfide formation in the absence of DNA (% of GCN4Cys dimer formed = ∼10% after 105 min). This can be due to the higher concentration at which we run most of the experiments (2 μM). However, in this case also, the addition of the cognate oligonucleotide GRE strongly accelerates the disulfide dimer formation rate reaching >90% of dimer after 105 minutes (data not shown).
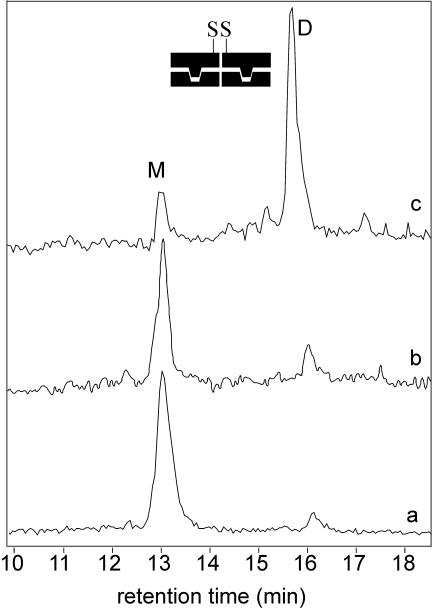
Figure 6.
Disulfide cross-linking experiment with 75 nM R*69Cys in the presence of 37.5 nM P22. The figure shows the TIC from RP-HPLC of aliquots taken at increasing reaction times. The peak M belongs to monomeric R*69Cys, the peak D belongs to R*69Cys disulfide dimer. Trace ‘a’ is the reaction mixture in the absence of DNA at time 0. Trace ‘b’ is the reaction mixture in the absence of DNA at 180 min. Trace ‘c’ is the reaction mixture in the presence of P22 at 150 min.
Hybrid operators promote heterodimer formation
While the oligonucleotide P22 does not promote the dimerization of R69Cys (Figure 5), evident dimer formation could be detected in the presence of the hybrid operator OR1–P22 (kR69Cys+OR1/kR69Cys+OR1–P22 = 8.75) (Figure 7). This result confirms what has been suggested by the fluorescence studies: an R69Cys molecule bound at one 434 operator half-site might be sufficient to promote a measurable assembly of a second subunit on an adjacent (and partly similar) P22 half-site. Other hybrid operators OR1–GRE1 and OR1–GRE2 (sequences in Figure 1) also promoted evident R69Cys dimer formation supporting the hypothesis that one bound subunit can stabilize the assembly of a second subunit more loosely bound (% of R69Cys dimer formed = 10 and 15%, respectively with OR1–GRE1 and OR1–GRE2 after 60 min). The two latter oligos also efficiently increase the disulfide formation rate of GCN4Cys (% of GCN4Cys dimer formed = 42 and 46%, respectively with OR1–GRE1 and OR1–GRE2 after 60 min); this effect, even if not comparable to the one exerted by the symmetric GRE site is anyway consistent and more pronounced than in the R69 system.
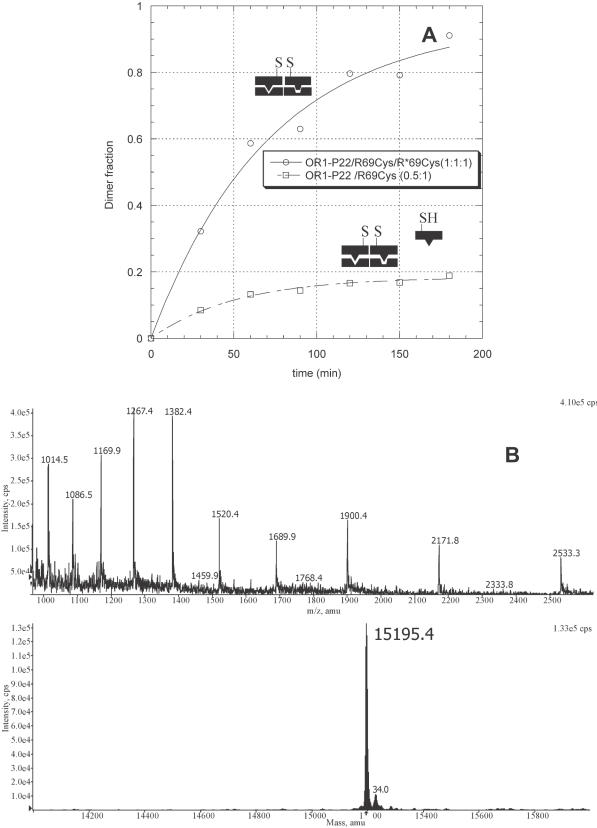
Figure 7.
(A) Disulfide cross-linking experiment with R69Cys alone (1 μM) and both R69Cys and R*69Cys (1 μM) in the presence of the hybrid oligonucleotide OR1–P22 (1 μM). The points represent the fraction of the dimer formed at increasing reaction times. (B) ESI-MS spectrum and deconvoluted mass of the main product (the heterodimer) of the reaction mixture containing R69Cys and R*69Cys (1 μM) in the presence of the hybrid oligonucleotide OR1–P22 (1 μM).
In solution and in the absence of DNA, both R69Cys and its engineered mutant R*69Cys can be oxidized to the respective homodimers ([R69Cys]2 and [R*69Cys]2). When a 1:1 mixture of R69Cys and R*69Cys is oxidized in the absence of DNA, the result is a statistical mixture (1:1:2) of the two homodimers and a heterodimer that can be all distinguished by LC-MS.
If specific DNA really acts as a template for disulfide-mediated cross-linking, then the presence of specific DNA in the reaction mixture would influence the distribution of the dimeric products.
When hybrid OR1–P22 operator was added to a solution containing a 1:1 mixture of R69Cys and R*69Cys, it promoted the formation of a mixture of dimers composed of >96% (Figure 7) of the heterodimer [R69Cys]–[R*69Cys] as evidenced by mass spectrometric analysis [ESI-MS: 15195.4; 15194.3 (calc.)]. This result thus shows that sequences as short as the operator half-sites are sufficient to specifically recruit protein molecules. According to the results previously described (Figure 4 and Figure 7), we mention that R69Cys itself is capable of binding to the P22 half-site of the hybrid operator to a certain extent as a result of such recruitment. The fact that almost no R69Cys homodimers could be observed by mass spectroscopy shows that R*69Cys successfully competes for the binding when applied in a 1:1 ratio.
As suggested from the decrease of the 383 nm signal in the fluorescence experiments, R69Cys seems to interact to some extent with non-specific DNA, but this interaction is non-productive either for pyrene excimer formation or for disulfide dimerization. To verify this hypothesis, we tested the interaction of R69Cys with OR1 in the presence and in the absence of a high excess of non-specific GRE (60-fold with respect to OR1 concentration). The disulfide formation rate decreases slightly upon GRE competition (kR69Cys+OR1/kR69Cys+OR1+GRE = 1.46) suggesting that the R69Cys molecules could get partially trapped in non-specific interactions (data not shown).
DISCUSSION
The 3D structures of the 434 repressor DNA-binding domain (R69)/operator DNA complex (27) and of GCN4 bound to the ATF/CREB site (41) show that in both systems the C-termini of the two bound monomers are in close proximity. This is in principle sufficient both for C-terminally attached pyrene residues to produce a stacking interaction and excimer fluorescence, and for the C-terminally attached cysteines to form an intermolecular disulfide bridge that leads to covalent dimerization. We hypothesized that such a conformation—which we term productive dimerization–would be promoted by operator DNA.
The appearance of pyrene excimer fluorescence and disulfide dimer formation in the presence of the cognate sites OR1 (for R69Cys-py) and GRE (for GCN4Cys-py) support evidence that operator DNA induces a productive assembly, while non-operator DNA does not. In agreement with the published data about the monomeric state of R69 (26,28) and of the GCN4 basic motif in solution (16,31), the absence of excimer fluorescence and the slow rate of spontaneous disulfide formation indicate that these molecules in the absence of DNA do not show appreciable levels of productive dimerization. In principle, the dimerization states can be categorized as productive or non-productive. Non-productive dimers would be those in which the two C-termini cannot get in the spatial arrangement required for either disulfide formation or pyrene stacking. Fluorescence spectroscopy allows the detection of non-productive, imperfectly stacked pyrene labels via a broadening and a red-shift of the absorbance spectrum. We did not observe such a peak broadening both in the absence of DNA and when non-cognate DNA was added. We believe that non-productive dimers, if existing, are probably reaction intermediates with a very short half-life. Consequently, we discuss our results on productive dimerization only.
There is no data available in the literature about the preferred assembly pathway of R69 dimers or similarly structured DNA-binding domains devoid of dimerization domains. According to our experimental data, we envision a model where R69 monomers interact with the nucleic acid and associate in a dimeric complex directly on the DNA platform, hence preferring a monomer pathway. The same assembly mechanism is employed by GCN4Cys as already demonstrated for this and other bZIP proteins even if the coexistence of the dimeric pathway is still under debate (11,16,42).
In this respect, we need to mention the decrease of monomeric pyrene fluorescence that was detected with R69Cys (and GCN4Cys) in the presence of cognate DNA, non-cognate DNA, as well as a number of linear polyanions. It is well documented that the fluorescence of monomeric pyrene is quenched as the label enters a polar environment (34,35,39,40). Polyanions as diverse as tRNA, ssDNA, polyacrylate and to a lesser extent, heparin, produced a concentration-dependent decrease very similar to that produced by cognate DNA; in addition, the effect is salt-sensitive. The interaction and structural modifications of GCN4 in the presence of non-specific DNA are described (32,43) and our experiments, an enhancement in the rate of disulfide cross-linking upon interaction with non-specific oligos, support this model. A certain degree of interaction of R69Cys with non-specific DNA is also suggested by the diminished rate of disulfide formation when R69Cys interacts with OR1 in the presence of an excess of the non-specific GRE oligonucleotides. These observations lead us to suggest that the decrease of monomer fluorescence may be primarily due to the pyrene label entering a polar environment like that of the DNA backbone. As non-specific interactions are primarily electrostatic in nature their disruption by an increase in ionic strength may partially release the R69Cys-py from the neighborhood of the DNA restoring the signal at 383 nm. On the other hand, the specific interaction with the cognate operator would be only slightly affected by the NaCl concentration keeping the pyrene label in the vicinity of the nucleic acid. Such non-specific interactions may be expected to guide the sequential assembly of monomers via 1D diffusion along the DNA chain (16). Transient interactions and conformational changes induced by non-specific DNA have been described both for R69 covalent dimers (25) and for the entire 434 repressor (44). One-dimensional diffusion, specific DNA recognition, dimer assembly and subsequent stabilization of the dimerization interface might thus form hierarchical steps in this process.
Further strong support for a DNA-mediated assembly model comes from the experiments with the hybrid operators. The operator seems to promote and select the assembly of homo- or heterodimers according to the availability and specificity of the protein species present in solution. Two adjacent heterologous half-sites can guide the assembly of specific dimers as was demonstrated by the almost exclusive formation of heterodimer when R69Cys and R*69Cys are in the presence of P22–OR1 in a disulfide cross-linking experiment. The selectivity of dimer recruitment is noteworthy considering that the P22 oligo contains a palindromic P22 recognition site, whose sequence is partly similar to the cognate OR1 sites. This similarity is shown by a modest formation of homodimers when only R69Cys is in the presence of the hybrid operator, in support of a model where very weak interactions (protein–protein and protein–DNA) can also be stabilized on the nucleic acid platform. This weak affinity is anyway competed out when a better binder, R*69Cys, is present in solution and shows how forming a dimeric complex by a monomer pathway can greatly increase the specificity.
The results presented here therefore indicate for the first time that the sequential binding of monomers, previously suggested for other transcription factors such as ATF-2 and Max (4) as well as for the P22 Arc repressor (5), is a plausible pathway also for the N-terminal domain of the 434 repressor, R69.
The 3D structure of the protein–DNA complexes available so far indicate that a recognition complex contains an intricate network of atomic interactions that connects the protein partners with each other as well as with the DNA. The contribution of the individual interactions to recognition specificity is difficult to decipher since in many complexes the number of H-bonds between the protein and the DNA base pairs is not dramatically higher than those directed to the sugar-phosphate backbone. To summarize, we can point out that specificity of nucleic acid binding might critically depend on such apparently insignificant differences, i.e. on weak links that add up to a robust network of atomic interactions.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks are due to Professor A. Hermetter (Technische Universitaet, Graz) and Dr Daniele Arosio regarding the interpretation of the fluorescence experiments and M. Rusnati (University of Brescia) for his advise on heparin chemistry.
REFERENCES
- 1.Tijan R. and Maniatis,T. (1994) Transcriptional activation: a complex puzzle with few easy pieces. Cell, 77, 5–8. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M. and Gann,A. (1998) Imposing specificity by localization: mechanism and evolvability. Curr. Biol., 8, R812–R822. [DOI] [PubMed] [Google Scholar]
- 3.Beckett D. (2001) Regulated assembly of transcription factors and control of transcription initiation. J. Mol. Biol., 314, 335–352. [DOI] [PubMed] [Google Scholar]
- 4.Kohler J.J., Metallo,S.J., Schneider,T.L. and Schepartz,A. (1999) DNA specificity enhanced by sequential binding of protein monomers. Proc. Natl Acad. Sci. USA, 96, 11735–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rentzeperis D., Jonsson,T. and Sauer,R.T. (1999) Acceleration of the refolding of Arc repressor by nucleic acids and other polyanions. Nat. Struct. Biol., 6, 569–573. [DOI] [PubMed] [Google Scholar]
- 6.Bitinaite J., Wah,D.A., Aggarwal,A.K. and Schildkraut,I. (1998) FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10570–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedhoff P., Lurz,R., Luder,G. and Pingoud,A. (2001) Sau3AI, a monomeric type II restriction endonuclease that dimerizes on the DNA and thereby induces DNA loops. J. Biol. Chem., 276, 23581–23588. [DOI] [PubMed] [Google Scholar]
- 8.Soundararajan M., Chang,Z., Morgan,R.D., Heslop,P. and Connolly,B.A. (2002) DNA binding and recognition by the IIs restriction endonuclease MboII. J. Biol. Chem., 277, 887–895. [DOI] [PubMed] [Google Scholar]
- 9.Rastinejad F., Perlmann,T., Evans,R.M. and Sigler,P.B. (1995) Structural determinants of nuclear receptor assembly on DNA direct repeats [see comments]. Nature, 375, 203–211. [DOI] [PubMed] [Google Scholar]
- 10.Wu X., Spiro,C., Owen,W.G. and McMurray,C.T. (1998) cAMP response element-binding protein monomers cooperatively assemble to form dimers on DNA. J. Biol. Chem., 273, 20820–20827. [DOI] [PubMed] [Google Scholar]
- 11.Metallo S.J. and Schepartz,A. (1997) Certain bZIP peptides bind DNA sequentially as monomers and dimerize on the DNA [letter]. Nat. Struct. Biol., 4, 115–117. [DOI] [PubMed] [Google Scholar]
- 12.Holmbeck S.M.A., Dyson,H.J. and Wright,P.E. (1998) DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J. Mol. Biol., 284, 533–539. [DOI] [PubMed] [Google Scholar]
- 13.Wendt H., Thomas,R.M. and Ellenberger,T. (1998) DNA-mediated folding and assembly of MyoD-E47 heterodimers. J. Biol. Chem., 273, 5735–5743. [DOI] [PubMed] [Google Scholar]
- 14.Kim B. and Little,J.W. (1992) Dimerization of a specific DNA-binding protein on the DNA. Science, 255, 203–206. [DOI] [PubMed] [Google Scholar]
- 15.Bray D. and Lay,S. (1997) Computer-based analysis of the binding steps in protein complex formation. Proc. Natl Acad. Sci. USA, 94, 13493–13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger C., Piubelli,L., Haditsch,U. and Bosshard,H.R. (1998) Diffusion-controlled DNA recognition by an unfolded, monomeric bZIP transcription factor [Erratum (1998) FEBS Lett., 429, 221.]. FEBS Lett., 425, 14–18. [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Chen,Y., Watrob,H., Bartlett,S.G., Jen-Jacobson,L. and Barkley,M.D. (1998) N-termini of EcoRI restriction endonuclease dimer are in close proximity on the protein surface. Biochemistry, 37, 15457–15465. [DOI] [PubMed] [Google Scholar]
- 18.Maluf N.K., Ali,J.A. and Lohman,T.M. (2003) Kinetic mechanism for formation of the active, dimeric UvrD helicase–DNA complex. J. Biol. Chem., 278, 31930–31940. [DOI] [PubMed] [Google Scholar]
- 19.Pabo C.O. and Sauer,R.T. (1992) Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem., 61, 1053–1095. [DOI] [PubMed] [Google Scholar]
- 20.Harrison S.C. and Aggarwal,A.K. (1990) DNA recognition by proteins with the helix–turn–helix motif. Annu. Rev. Biochem., 59, 933–969. [DOI] [PubMed] [Google Scholar]
- 21.Sauer R.T., Ross,M.J. and Ptashne,M. (1982) Cleavage of the lambda and P22 repressors by recA protein. J. Biol. Chem., 257, 4458–4462. [PubMed] [Google Scholar]
- 22.Bell A.C. and Koudelka,G.B. (1993) Operator sequence context influences amino acid–base-pair interactions in 434 repressor–operator complexes. J. Mol. Biol., 234, 542–553. [DOI] [PubMed] [Google Scholar]
- 23.Donner A.L., Paa,K. and Koudelka,G.B. (1998) Carboxyl-terminal domain dimer interface mutant 434 repressors have altered dimerization and DNA binding specificities. J. Mol. Biol., 283, 931–946. [DOI] [PubMed] [Google Scholar]
- 24.Simoncsits A., Chen,J., Percipalle,P., Wang,S., Toro,I. and Pongor,S. (1997) Single-chain repressors containing engineered DNA-binding domains of the phage 434 repressor recognize symmetric or asymmetric DNA operators. J. Mol. Biol., 267, 118–131. [DOI] [PubMed] [Google Scholar]
- 25.Percipalle P., Simoncsits,A., Zakhariev,S., Guarnaccia,C., Sanchez,R. and Pongor,S. (1995) Rationally designed helix–turn–helix proteins and their conformational changes upon DNA binding. EMBO J., 14, 3200–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-Sanz J., Simoncsits,A., Toro,I., Pongor,S., Mateo,P.L. and Filimonov,V.V. (1999) A thermodynamic study of the 434-repressor N-terminal domain and of its covalently linked dimers. Eur. J. Biochem., 263, 246–253. [DOI] [PubMed] [Google Scholar]
- 27.Aggarwal A.K., Rodger,D.W., Drottar,M., Ptashne,M. and Harrison,S.C. (1988) Recognition of DNA operator by the repressor of phage 434: a view at high resolution. Science, 242, 899–907. [DOI] [PubMed] [Google Scholar]
- 28.Neri D., Billeter,M. and Wuthrich,K. (1992) Determination of the nuclear magnetic resonance solution structure of the DNA-binding domain (residues 1 to 69) of the 434 repressor and comparison with the X-ray crystal structure. J. Mol. Biol., 223, 743–767. [DOI] [PubMed] [Google Scholar]
- 29.Wharton R.P. and Ptashne,M. (1985) Changing the binding specificity of a repressor by redesigning an alpha-helix. Nature, 316, 601–605. [DOI] [PubMed] [Google Scholar]
- 30.Chen J., Pongor,S. and Simoncsits,A. (1997) Recognition of DNA by single-chain derivatives of the phage 434 repressor: high affinity binding depends on both the contacted and non-contacted base pairs. Nucleic Acids Res., 25, 2047–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saudek V., Pasley,H.S., Gibson,T., Gausepohl,H., Frank,R. and Pastore,A. (1991) Solution structure of the basic region from the transcriptional activator GCN4. Biochemistry, 30, 1310–1317. [DOI] [PubMed] [Google Scholar]
- 32.O'Neil K.T., Shuman,J.D., Ampe,C. and DeGrado,W.F. (1991) DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry, 30, 9030–9034. [DOI] [PubMed] [Google Scholar]
- 33.Konig P. and Richmond,T.J. (1993) The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J. Mol. Biol., 233, 139–154. [DOI] [PubMed] [Google Scholar]
- 34.Talavera E.M., Afkir,M., Salto,R., Vargas,A.M. and Alvarez-Pez,J.M. (2000) Fluorescence-labelled DNA probes to detect complementary sequences in homogeneous media. J. Photochem. Photobiol. B, 59, 9–14. [DOI] [PubMed] [Google Scholar]
- 35.Yguerabide J., Talavera,E., Alvarez,J.M. and Afkir,M. (1996) Pyrene-labeled DNA probes for homogeneous detection of complementary DNA sequences: poly(C) model system. Anal. Biochem., 241, 238–247. [DOI] [PubMed] [Google Scholar]
- 36.Talanian R.V., McKnight,C.J. and Kim,P.S. (1990) Sequence-specific DNA binding by a short peptide dimer. Science, 249, 769–771. [DOI] [PubMed] [Google Scholar]
- 37.Hangland R.P. (1992) Handbook of Fluorescent Probes and Research Chemicals. Molecular Probes, Inc., Eugene, OR. [Google Scholar]
- 38.Lehrer S.S. (1997) Intramolecular pyrene excimer fluorescence: a probe of proximity and protein conformational change. Meth. Enzymol., 278, 286–295. [DOI] [PubMed] [Google Scholar]
- 39.Preuss R., Dapprich,J. and Walter,N.G. (1997) Probing RNA-protein interactions using pyrene-labeled oligodeoxynucleotides: Qbeta replicase efficiently binds small RNAs by recognizing pyrimidine residues. J. Mol. Biol., 273, 600–613. [DOI] [PubMed] [Google Scholar]
- 40.Ohtani H., Masuko,M., Wada,Y. and Kodama,T. (2000) Dynamics of the fluorescence properties of pyrene residues appended to oligonucleotide hybridization probes. Nucleic Acids Symp. Ser., 51–52. [DOI] [PubMed] [Google Scholar]
- 41.Keller W., Konig,P. and Richmond,T.J. (1995) Crystal structure of a bZIP/DNA complex at 2.2 A: determinants of DNA specific recognition. J. Mol. Biol., 254, 657–667. [DOI] [PubMed] [Google Scholar]
- 42.Cranz S., Berger,C., Baici,A., Jelesarov,I. and Bosshard,H.R. (2004) Monomeric and dimeric bZIP transcription factor GCN4 bind at the same rate to their target DNA site. Biochemistry, 43, 718–727. [DOI] [PubMed] [Google Scholar]
- 43.Kohler J.J. and Schepartz,A. (2001) Effects of nucleic acids and polyanions on dimer formation and DNA binding by bZIP and bHLHZip transcription factors. Bioorg. Med. Chem., 9, 2435–2443. [DOI] [PubMed] [Google Scholar]
- 44.Ciubotaru M., Bright,F.V., Ingersoll,C.M. and Koudelka,G.B. (1999) DNA-induced conformational changes in bacteriophage 434 repressor. J. Mol. Biol., 294, 859–873. [DOI] [PubMed] [Google Scholar]