Abstract
Approximately half of all cancers harbor chromosomal translocations that can either contribute to their origin or govern their subsequent behavior. Chromosomal translocations by definition can only occur when there are two DNA double-strand breaks (DSBs) on distinct chromosomes that are repaired heterologously. Thus, chromosomal translocations are by their very nature problems of DNA DSB repair. Such DNA DSBs can be from internal or external sources. Internal sources of DNA DSBs that can lead to translocations can occur are inappropriate immune receptor gene maturation during V(D)J recombination or heavy-chain switching. Other internal DNA DSBs can come from aberrant DNA structures, or are generated at collapsed and reversed replication forks. External sources of DNA DSBs that can generate chromosomal translocations are ionizing radiation and cancer chemotherapy. There are several known nuclear and chromatin properties that enhance translocations over homologous chromosome DSB repair. The proximity of the region of the heterologous chromosomes to each other increases translocation rates. Histone methylation events at the DSB also influence translocation frequencies. There are four DNA DSB repair pathways, but it appears that only one, alternative non-homologous end-joining (a-NHEJ) can mediate chromosomal translocations. The rate-limiting, initial step of a-NHEJ is the binding of poly−adenosine diphosphate ribose polymerase 1 (PARP1) to the DSB. In our investigation of methods for preventing oncogenic translocations, we discovered that PARP1 was required for translocations. Significantly, the clinically approved PARP1 inhibitors can block the formation of chromosomal translocations, raising the possibility for the first time that secondary oncogenic translocations can be reduced in high risk patients.
INTRODUCTION
Karl Sax first described chromosomal translocations from ionizing radiation (IR) in a seminal publication in 1938 entitled Chromosome Aberrations Induced by X-rays (1). Sax studied the plant Tradescantia reflexa, which had large and easily visualized chromosomes. He found a high correlation between IR and translocations, and was the first to postulate that IR caused chromosome damage. He observed multiple “fusions between different chromosomes” after IR, which was the first description of chromosomal translocations (1).
Three decades later, Nowell, Rowley, and Baltimore showed that translocations not only defined cancer types, but they could contribute to oncogenesis themselves (2). These investigators, working in distinct laboratories, found that nearly half of all cancers have distinctive chromosomal translocations which define their clinical behavior (3−6). Nowell performed his studies with a simple light microscope on his desk in a tiny office. Rowley would spread her cytogenetic photomicrographs across her dining room table, and her family would eat dinner in the kitchen. Both are testaments to the power of questioning the origins of a strange observation, not letting it slip by, and then testing possible answers. In most hematologic malignancies and in many solid tumors, these distinctive chromosomal translocations have prognostic significance, and they define treatment plans that specifically target the products of those translocations (5−12). However, despite their seminal significance for malignancy, the molecular mechanisms of chromosomal translocations has been poorly defined until just recently (3−6). Given the enormous success of therapy that can be targeted to products of these translocations, an increased understanding of the mechanisms of these translocations could not only reveal novel insight into oncogenesis, but could also identify targets for future directed therapies.
ABERRANT FUNCTION OF THE IMMUNE RECEPTOR NUCLEASES CAN GENERATE TRANSLOCATIONS
It is intuitively obvious and experimentally demonstrated that all translocations are generated from two simultaneous DNA double-stranded breaks (DSBs) on distinct chromosomes that are heterologously repaired (3−6,13−14). Therefore, the spatiotemporal location of translocations is mediated by when, where, and how DSBs occur. In addition, these dangerous DSBs must also occur in cells at risk for malignant transformation; for example, a post-mitotic neutrophil is not at risk for an oncogenic translocation. An antigen-encountering lymphocyte, poised to clonally expand, on the other hand, is a cell at risk for neoplastic translocations.
Worse, lymphocytes are not only able to rapidly proliferate, but they generate endogenous DSBs normally during the maturation of their response to antigen (14,15). Endogenous DSBs normally occur in lymphocytes during immune receptor V(D)J recombination or heavy-chain class switch recombination, as B cells mature. The DNA DSBs required for these two normal immune receptor maturation steps are introduced by three nucleases, the recombination activating genes (RAG1 and 2), and activation-induced cytosine deaminase (AID) (14−17, Table 1). Given the organismal pressure to respond quickly to infection, these maturation steps are rapid and occur in large cell numbers. However, there is an error rate for the repair of these immune nuclease-generated DSBs required for lymphocyte maturation that can produce translocations, which if they occur within a cell at risk and within a gene that can stimulate proliferation, can lead to neoplastic transformation of the lymphocyte (18). Aberrant immune receptor DSB repair leading to translocations are much more common than the incidence of lymphocyte malignancy, but most of these translocations do not involve genes that can stimulate cell growth. In addition, aberrant immune receptor rearrangements often stimulate apoptosis in maturing lymphocytes, so the cell obliterates itself before it can harm the organism (18).
The immune receptor nucleases RAG1 and 2 recognize recombination signal sequences (RSSs) that lead to V(D)J recombination during immunoglobulin (Ig) and T-cell receptor gene rearrangement. These nucleases generate a single-strand (ss) nick at the RSS that is converted to a DSB by the DNA-PKcs/Artemis nuclease complex (14,16). Chromosomal translocations can arise when RAG1 or 2 nicks RSS-like sequences in other genes located on distinct chromosomes during V(D)J recombination (18−20). These nicks in RSS-like sequences on other chromosomes can also be turned into DSBs by DNA PKcs/Artemis, just like the RSS sequences in the V(D)J region. These nicks can also be converted to a DSB by an oxidative nick on the adjacent DNA strand, or by the passage of a replication fork (18,19).
In this case, the lymphocyte will have two simultaneous DSBs, one in the immune receptor gene, and one in the chromosome with the RSS-like sequence. This DSB can be re-ligated to the normal DSB in the immune receptor gene. Since the lymphocyte will heavily transcribe the immune receptor locus as an appropriate consequence of antigen stimulation, then whatever sequence is adjacent to the RSS-like sequence will produce large quantities of mRNA and protein. If the RSS-like sequence is in a gene whose role is to stimulate proliferation or prevent apoptosis, then neoplasia can result (19−23). This is the most important molecular mechanism responsible for multiple types of lymphocytic cancers, depending on the maturation stage of the lymphocyte in which it occurs. For example, acute lymphoblastic leukemia of both B- and T-cell origins has a translocation at the early V(D)J recombination, whereas myeloma has a translocation at the class switch locus, consistent with its more mature origin (14,16,20,21).
Although RAG1 and 2 usually recognize RSS sequences, they can also be abnormally attracted to non-canonical DNA structures, where they can also nick single strands. If there are two of these RAG1/2 nicks that are converted to DSBs simultaneously, then these nicks can also lead to oncogenic translocations (19−23). Interestingly, the 5’ region of both the c-myc and bcl2 genes contain sequences that can form DNA G-quadruplex structures, which are quite distinct and much less stable than canonical double helical B-DNA. These structures are located at the sites of known translocation junctions, and for at least bcl2, they can be cleaved by RAG1/2 (23−27). RAG1/2 can also nick other aberrant DNA structures such as bubbles and loops which can form during transcription and/or replication (20,22,23,27−29). The fact that the RAGs cut non-canonical DNA structures, regardless of whether that sequence resembles an RSS, can explain why occasional translocation junctions in acute lymphoblastic leukemia and non-Hodgkin lymphoma do not have consensus RSS sequences (3−5,20).
After Ig V(D)J gene rearrangements for initial lymphocyte maturation, diversity of immunoglobulins can be further enhanced by somatic mutations in the hyper-variable region, and also class switch from IgG to IgM (6,20). The nuclease AID is essential for both somatic mutation and class switch recombination, both of which can also generate oncogenic chromosomal translocations. AID deaminates cytosines to uracil, which is recognized by base excision DNA repair components. The uracil is deglycosylated by uracil glycosylase, and the abasic site is then cleaved by apurinic/apyrimidine endonuclease (APE1), leaving an ss nick (15,17). If ss nicks occur on opposite strands, this will result in a DSB, which if repaired aberrantly to another chromosome could lead to a translocation (30). Aberrant repair of DSBs generated by AID during class switch recombination is the most important mechanism for the origins of the more mature lymphoid cancers, such as non-Hodgkin lymphoma and myeloma, consistent with the developmental stage of lymphocytes in which heavy-chain class switch takes place (20,30−32).
Recently, AID has been found to play a role in generating the DSBs that lead to oncogenic chromosomal translocations in several types of non-lymphoid cancers. AID is normally expressed only in lymphocytes, to generate immune receptor diversity in response to the continued presence of antigen, but its expression can also be induced by radiation or steroid hormones (33). Interestingly, androgen steroid hormones can induce AID expression in prostate cells. When aberrantly expressed in these cells, AID can lead to DSBs in the TMPRSS2 androgen receptor and the ERG gene, which can result in a translocation that causes prostate cancer (33−36). ERG is a member of a large transcription factor family termed the Ets gene family that is known for its oncogenic capability after translocations in many cancers, such as Ewing’s sarcoma (the EWS-Fli1 translocation).
TMPRSS2 can also translocate with other members of the Ets family besides ERG, although ERG is the most common partner. The mechanism of this oncogenic translocation event is complex; androgen engagement of its steroid hormone receptor not only induces AID expression, but recruits AID to an intronic region in both TMPRSS2 and Erg, where it generates DSBs and subsequent translocation (34−36). The mechanistic theme here is the same as the immune receptor translocations, where the normal transcription of the immune receptor drives constitutive expression of the translocated gene to generate neoplastic transformation (30−32). Here, the normal transcription of TMPRSS2 in prostate cells drives the constitutive expression of ERG, which leads to prostate cell proliferation without any regulation. This recently described translocation event may be more common in prostate cancer than previously thought, perhaps even up to one half of some types. This leads to the hypothesis that solid-tumor oncogenic translocations may be just as common as in hematologic malignancies. Rather, in solid tumors, the driver translocations just have not been defined yet, given the massively jumbled genome of many of these cancers (3,5−7,37).
UNSTABLE NON-CANONICAL DNA STRUCTURES CAN LEAD TO TRANSLOCATIONS
Besides G-quadruplex DNA, there are other non-canonical DNA structures which are unstable, and can lead to the DSBs that mediate translocations (Table 1). When the junctions from MLL translocations in acute myeloid leukemia and BCR-ABL translocations in chronic myeloid leukemia were sequenced, it was discovered that alu elements were often present (38−41). Alu elements are made up of repeating sequences that can be found in many other places in the genome. Thus, they can anneal and homologously recombine with similar alu sequences on heterologous chromosomes, leading to translocations if the crossed-over DNA structure during the recombination is not properly resolved. Significantly, alu elements within introns of the MLL gene itself can recombine with other alu elements within the same MLL allele, but in other introns. This can produce the leukemogenic MLL duplications seen in some types of acute myeloid leukemia (38−40). Indeed, the internal tandem duplications of Flt3, which result in unregulated Flt3 tyrosine kinase activity, and subsequent leukemia cell proliferation, can also be the result of homologous recombination between alu sequences within the same gene (42).
Also, inherited translocations such as the t(11;22)(q23;q11), which are not oncogenic, are generated from palindromic AT-rich repeat sequences (43,44). The hypothesis here is that these palindromic sequences form cruciform structures that are the target of nucleases that resolve cruciform structures (termed Holliday junctions) that occur during repair and restart of stressed replication fork (45). These nucleases include the Holliday junction resolvases, such as MUS81 and Gen1, which nick cruciform structures to permit their resolution into productive replication forks (45,46). However, these nicks in cruciform structures can degenerate into DSBs that result in translocations (46,47).
The DSBs that lead to chromosomal translocations can also occur when there are internal or external DNA insults. Internal DNA insults resulting in DSBs can occur from reactive oxygen species that split phosphodiester binds on adjacent opposing strands. They can also be generated from replication forks that stall and then collapse to form free double-strand ends (3−6,13,48−50). This is seen in BRCA1 or 2 mutant breast and ovarian cancers, where collapsed replication forks cannot be repaired. These collapsed replication forks can reverse to form chicken-foot structures, which have one free double-strand end that can ligate to a free double-strand end on a heterologous chromosome (49,51). Also, chromosomal translocations can be one result of failed decatenation, or untangling of sister chromatids before anaphase. Anaphase of non-decatenated sister chromatids can cause DNA DSBs, which if on distinct chromosomes, can be re-ligated to heterologous chromosomes (3−6,52).
External DNA damage such as IR, ultraviolet light, or cytotoxic cancer chemotherapy, can also generate DNA DSBs in heterologous chromosomes that can be re-ligated to form translocations (3−6,48−50,52,53). One translocated chromosome may share two centromeres, while the other partner may have none. The translocated chromosome structure with two centromeres would fail to segregate at mitosis, being pulled equally to distinct mitotic poles, which may result in mitotic catastrophe and apoptosis, or in micronuclei formation (52,53). The only way to rescue this mitosis is for the shear stress to generate a DSB in that chromosome, allowing the centromeres to pull each chromosome fragment to the different mitotic poles. If that happens on two chromosomes, then the resulting DSBs can be re-ligated improperly, and a translocation can result. Thus, the cell with these DSBs can survive its first mitosis. If so, the cell is at risk for transformation to malignancy, if the products of translocations can confer unregulated proliferative status (3−6,13,49−53).
THE ROLE OF CHROMATIN IN TRANSLOCATIONS
Chromosomes are not naked DNA within the nucleus, rather they are packaged in nucleosomes made up of octamers of histones, with two copies each of histone 2A, 2B, 3, and 4 (6,49,54,55). The packaging of DNA within the nucleus is termed chromatin. Access to chromosomal DNA by the replication, transcription, or repair machinery is regulated by modifications of these histones, such as methylation, acetylation, SUMOylation, or ubiquitination (6,54,55). We and others have shown that histone methylation promotes non-homologous end-joining (NHEJ) DNA DSB repair (55). We found that histone 3 lysine 36 dimethylation recruits and stabilizes NHEJ components such as the Ku or MRN complexes to a newly formed DSB (55).
Given the role that histone methylation plays in promoting proper NHEJ, it is not surprising that histone methylation regulates chromosomal translocations (56,57) (Table 1). The monomethylation of histone 3 lysine 4 (H3K4me1) appears to promote translocations in multiple models (56,57). However, the trimethylation of histone 3 lysine 9 (H3K9me3) represses chromosomal translocations. Interestingly, methylation of H3K4 has been reported to open chromatin to increase access to local DNA, whereas the methylation of H3K9 is thought to close chromatin around DNA (56,57). This leads to the hypothesis that for chromosomal translocations to occur, there must not only be two simultaneous DSBs that are spatially adjacent (58), but the DNA surrounding these DSBs must also be accessible to the other DSB DNA. In addition, it is possible that given the open and accessible DNA adjacent to the two independent DSBs, the distinct DSBs share one repair apparatus, and it is this shared DSB repair apparatus that results in the chromosomal translocation (58). This brings us to the question of which of the four major DSB repair pathways mediates chromosomal translocations.
TABLE 1.
DNA Repair Pathways and Sequence Elements That Regulate Chromosomal Translocations*
| Inhibits Translocations | Enhances Translocations |
| c-NHEJ (DNA Ligase 4, Ku70/80, Metnase), homologous recombination | Single-strand annealing, a-NHEJ (Sirt6, CtIP, DNA Ligase 3, PARP1) |
| GC-rich B-DNA | AT-rich repeat sequences, G quadroplexes, alu sequences, RSS-like sequences |
| PARP1 repression (olaparib, rucaparib, velaparib), Sirt6 repression | DNA-PKcs inhibition (e.g., Nu7441) |
| Productive decatenation, progressive replication forks | Anaphase before completed decatenation, collapsed replication forks, IR, cancer chemotherapy |
| Appropriate RAG1/2 sequence recognition | Over-expression of AID |
| Histone 3 lysine 9 tri-methylation | Histone 3 lysine 4 mono-methylation |
* In sequences without repeat elements, a-NHEJ is required for oncogenic translocations. When similar repeat elements are present in the sequence adjacent to two DSBs on heterologous chromosomes, SSA can mediate chromosomal translocations. Unstable non-canonical DNA structures and improper activity of the immune receptor maturation nucleases can lead to DNA DSBs that result in chromosomal translocations.
Abbreviations: c-NHEJ, classical non-homologous end-joining; a-NHEJ, alternative non-homologous end-joining; RSS, recombinant signal sequence; PARP1, polyadenosine diphosphate ribose polymerase 1; DNA PKcs, DNA Protein Kinase CS; IR, ionizing radiation; RAG, recombinant activating gene; AID, activation-induced cytosine deaminase; DSB, double-strand break.
TYPES OF DSB REPAIR PATHWAYS
Because translocations result from the aberrant repair of two simultaneous DSBs, a DSB repair pathway must play a key role in the generation of chromosomal translocations. After a DSB occurs, its repair can be accomplished by one of four major pathways: Homologous recombination (HR), single-strand annealing (SSA), or NHEJ (53,54,59−61), which is comprised of two distinct pathways. The two pathways of NHEJ are classical NHEJ (c-NHEJ) and alternative NHEJ (a-NHEJ).
All DSB repair pathways initiate when Mre11/RAD50/NBS1 (termed the MRN complex) recognize a free DNA DSB end, and activate the ATM kinase, which auto-phosphorylates to increase its kinase activity. ATM then phosphorylates histone H2Ax, forming H2Ax-γ (54,60). H2Ax-γ is then responsible for the recruitment of further DNA repair components to the site of the DSB. The HR pathway and the two NHEJ pathways, classical and alternative, diverge from one another subsequent to H2Ax-γ formation (61,62).
Of these four DNA repair pathways, HR is the only conservative pathway in which the original sequence is maintained without error because the HR repair process uses the sister chromatid as a template (54,60−62). Both the SSA repair pathway and the two NHEJ pathways are non-conservative, in that there are always sequence alterations at the re-ligated DSB junctions (3-6,20,39,54,60−62). While these sequence alterations are most commonly deletions, they could also be insertions.
Several lines of evidence, including the sequencing of translocation junctions in leukemic patients, indicated that in the absence of alu repeat sequences, translocations are most likely mediated by an NHEJ pathway, given that there are frequently deletions or insertions at the translocation site (3-6,13,20,63−66). The presence of deletions or insertions in the sequence at the translocation junction is evidence against HR being the DNA DSB repair pathway that mediates translocations. If repeat sequences are present, however, SSA could mediate translocations, given that such repeats can be found in the sequences of some translocation junctions (38,39). Normally, c-NHEJ is the most common pathway for normal physiologic mammalian DSB repair (67). This holds true even in cells having already undergone replication, where there is a sister chromatid that can serve as a homologous template for HR repair (67). c-NHEJ has fewer components, requires less DNA synthesis, and thus would be faster than HR in the repair of a DSB. Its drawback is that it is not conservative, and leaves either insertions or deletions at the site of the DSB. If these are in coding or regulatory sequences then they can alter the behavior of the cell (3−6).
CLASSICAL VERSUS ALTERNATIVE NHEJ IN CHROMOSOMAL TRANSLOCATIONS
c-NHEJ begins when the Ku complex recognizes the free ends at a DSB, and recruits DNA-PKcs to that region to initiate processing of the free ends (68,69). DNA-PKcs helps recruit and activate nucleases such as Artemis, Mre11, and Metnase to process the free ends, which are often damaged by the breaking of the DNA, to clean, ligatable blunt ends (61,62,70−74). Cleaving off an overhang with one of the nucleases above or filling it in with a DNA polymerase to obtain a ligatable blunt end create the deletions or insertions that occur in the repaired DSB. DNA-PKcs then recruits the Ligase IV/XRCC4/XLF complex to the DSBs, and this complex rejoins the processed blunt ends (60,62,70−75).
There is increasing data that suggests that c-NHEJ protects against chromosomal translocations (76−78). Genetically deleting c-NHEJ repair pathway components such as Ku70 or Ligase 4 resulted in increased translocations, indicating that the c-NHEJ pathway normally inhibits translocations (76,78). Additionally, the over-expression of the c-NHEJ component Metnase decreased translocation rates in an embryonic stem cell reporter assay (77). In like manner, over-expression of dominant-negative Metnase mutants increased the frequency of translocations in this same reporter assay (77).
Historically, these results were surprising to those of us working in this field. Many questioned how there could there be insertions or deletions at translocation junctions, which would indicate an NHEJ pathway repair process, yet c-NHEJ pathway components protect against translocations? Thus, those reports seemed counter to the previous junctional sequence data that an NHEJ pathway mediates translocations (63−66). At that time, the a-NHEJ pathway was poorly characterized, and was not considered as a candidate to mediate translocations. Those reports implied that the c-NHEJ pathway is required to maintain genomic stability, not disrupt it via chromosomal translocations, which could result in oncogenesis (20,76−78).
The components and mechanisms of the a-NHEJ pathway still remain much less characterized compared to the c-NHEJ pathway. The a-NHEJ pathway is also known by many other names, including microhomology-mediated EJ, Ku-independent NHEJ, error-prone NHEJ, and back-up NHEJ (68,69,75,79). The first and rate-limiting step in a-NHEJ is when PARP1 successfully out-competes the Ku complex for the ends of a DSB (68,69,80). If this is a competitive process, then Ku wins much more frequently, since c-NHEJ repairs the majority of DSBs within the cell (60,70−75). Most of the time, the a-NHEJ pathway functions as a back-up DSB repair pathway (68,74,75,76,79). So why does the cell need more than one NHEJ DSB repair pathway? One hypothesis is that a-NHEJ is used to rapidly restart replication forks when HR fails (48,81). Another is that it rescues DSB repair when HR stalls after 5’ end resection, since a-NHEJ can also use 5’ end resection (48,81).
The historical paradox of how NHEJ mediated translocations, when many c-NHEJ components were found to repress translocations, were resolved when Simsek et al discovered that DNA Ligase III (Lig III), the final component of the a-NHEJ pathway, was required for translocations (76). a-NHEJ became recognized as the crucial pathway through which chromosome translocations occur. Zhang et al and Lee-Theilen et al demonstrated that other a-NHEJ components were also essential for chromosomal translocations (83,84). They found that the a-NHEJ component CtlP was required for chromosomal translocations (83,84). Together, these reports provided the evidence that c-NHEJ repressed translocations and promoted genomic stability, whereas the a-NHEJ pathway is the major DSB repair pathway through which chromosomal translocations occur.
PARP1 AS A PREVENTABLE STEP IN TRANSLOCATIONS
PARP1 is the initial, rate-limiting step of a-NHEJ, by out-competing the Ku complex for free DNA ends of a DSB (68,69). Upon DNA DSB occurrence, PARP1 is activated by mono-ribosylation via Sirt6 (85) and later turned off by autologous poly-ribosylation (86). The role of PARP1 in a-NHEJ was first characterized in biochemical assays in vitro (87), and then later studies demonstrated that depleting PARP1 caused severe defects in NHEJ repair of DSBs in Ku-deficient cells. Just finding NHEJ in Ku-deficient cells was a surprise, and was an important finding demonstrating the existence of a-NHEJ in vivo. This report also provided evidence that PARP1 was essential for a-NHEJ in vivo as well as in vitro (88).
PARP1 has several roles in the initiation of a-NHEJ. It binds to the free DNA ends and stabilizes the MRN complex at the DSB, which itself enhances the activation of ATM (86,87). PARP1 then activates 5’ end resection at the DSB via the nuclease Mre11 from the MRN complex (70,89). PARP1 also promotes BRCA1 recruitment of CtIP, which although not a nuclease itself also assists in 5’ end resection (74,83,84,87,90). a-NHEJ uses 5’ end resection to create free single-stranded DNA overhang ends at the DSB junction, and these single-strand overhangs search for short homologies in the opposing strand. These short homologies, or microhomologies, anneal to each other, and after trimming of excess sequence, mediate re-ligation (70,74,75,79). End resection commits the DSB to either HR or a-NHEJ repair, as the 3’ single-strand ends inhibit c-NHEJ blunt end re-ligation. The overlapping single-strand flaps that occur from microhomology annealing are ultimately trimmed by an unknown nuclease, the resulting single-strand gaps between the microhomology and the undamaged double-strand DNA surrounding the DSB site are filled in, and then Lig III re-ligates the breaks (82,90). a-NHEJ is defined by deletions at the repaired DSB, which are also common in c-NHEJ, and microhomologies at the repaired DSB junctions, which are rare in c-NHEJ. The presence of both deletions and microhomologies at the repaired DSB junction are often unique to a-NHEJ, and differentiate a-NHEJ from other forms of DNA DSB repair when translocation junctions are sequenced (74,75,79,83,84).
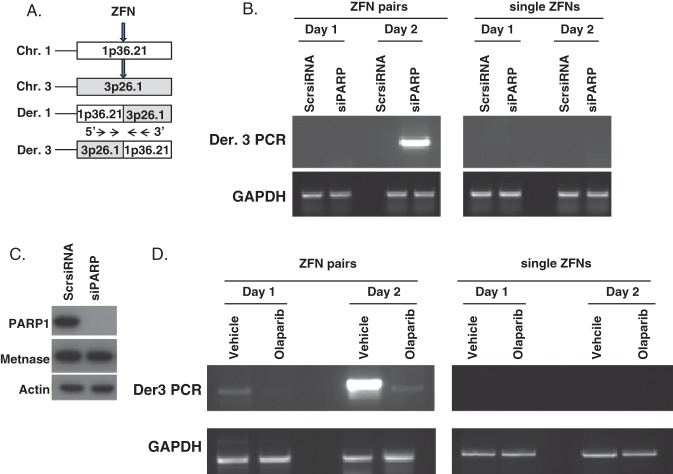
Recently, several PARP1 small molecule inhibitors have been tested in clinical trials for cancer therapy of BRCA1 or 2 mutant breast and ovarian cancers (91). One, olaparib, has been US Food and Drug Administration approved for relapsed BRCA1 or 2 mutated ovarian cancers, with several others thought to be approved soon (91). In an exciting new clinical trial report, olaparib was found to be a highly effective treatment for metastatic BRCA1/2 mutant prostate cancer as well (37). Given the role of PARP1 in the a-NEHJ pathway, and the importance of a-NHEJ in mediating chromosomal translocations, we sought to investigate whether the PARP1 inhibitors olaparib and rucaparib could inhibit chromosomal translocations. These small molecule PARP1 inhibitors have been extensively tested in clinical trials, and were found to be well-tolerated in patients (37,91). Using two distinct translocation reporter systems first synthesized by Simsek and Jasin (76) and Weinstock et al (78), we discovered that PARP1 inhibition with olaparib or rucaparib markedly decreased chromosomal translocation rates (Figure 1) (92). Translocations were also abrogated when PARP1 itself was depleted using small interfering RNA, which implied that the decrease in chromosomal translocation rates observed with olaparib and rucaparib were due to PARP1 inhibition, rather than off-target effects of these drugs (Figure 1) (92). However, these translocation reporter assays measured chromosomal translocations after induced DSB using a restriction enzyme, and not physiologic or clinically relevant oncogenic DNA DSB generation. Thus, we also tested the extent of chromosomal translocations after VP16 or IR in the untransformed murine hematopoietic cell line 32D and in normal human WI38 fibroblasts. Chromosomal translocation events as assessed by G-banded cytogenetics were also reduced by olaparib after exposure to either VP16 or IR (92). These data provided further evidence that chromosomal translocations induced by physiological DNA stressors, and they result from the a-NHEJ pathway rather than c-NHEJ, and that PARP1 itself is crucial in this process.
Fig. 1.
Olaparib inhibition of polyadenosine diphosphate ribose polymerase 1 (PARP1) or PARP1 small interfering RNA (siRNA) decrease zinc finger nuclease-induced translocations. (A) Olaparib-treated HEK-293T cells transfected with siRNA and zinc finger nucleases (ZFN) to induce double-strand breaks (DSBs) for a 1;3 translocation at the times indicated. (B) PARP1 siRNA repressed ZFN-induced translocations, as assayed by polymerase chain reaction (PCR) of the der[3] product. Translocations are only generated by two ZFN pairs, required to create two simultaneous DSBs in target chromosomes, but not by the negative control single ZFNs of each pair. That siRNA against PARP1 represses translocations indicates that the translocation effect is specific to PARP1 inhibition, and not an off-target effect of olaparib. (C) siRNA against PARP1 represses the expression of PARP1 protein, but has no effect on Metnase protein, indicating its specificity. (D) Quantification of ZFN–induced translocations with or without olaparib using PCR to detect the der[3] translocation product. These data raise the possibility that oncogenic translocations can be prevented in high risk situations by treatment with the clinical PARP1 inhibitors. From Wray et al (92); used by permission. Abbreviations: Chr, Chromosome; Der, Derivative; Scr, Scrambled; GAPDH, Glyceraldehyde phosphate dehydrogenase.
One reason that a-NHEJ mediates chromosomal translocations is that PARP1 may not effectively tether free DNA ends together as compared to the Ku complex (93). Or, PARP1 may simply be far less efficient at promoting end-joining of a DNA DSB compared to Ku70/80 (68,69,80). Either of these mechanisms could result in increased drifting apart of free ends, and would allow a greater chance for them to be re-joined to free ends on a heterologous chromosome. Another reason why PARP1 promotes chromosomal translocations could be that PARP1 enhances the activities of other a-NHEJ components, such as Mre11, CtlP, or DNA Ligase III, and these mediate the translocation activity of a-NHEJ (82−84,90).
CONCLUSION
The etiology of the DSBs that produce chromosome translocations are widely variable, but the end results are the same, and the consequences are severe. Ironically, some of the most effective cancer therapies that oncologists use can induce DNA DSBs which generate chromosomal translocations that cause secondary cancers. Mounting evidence indicates that the a-NHEJ DNA DSB repair pathway, although less common in repairing normal DSBs, is the major DSB repair pathway through which chromosomal translocations occur. The related NHEJ DSB repair pathway, c-NHEJ, suppresses chromosomal translocations.
Many cancers that occur after radiation or chemotherapy have defined chromosomal translocations (11,12). Such secondary cancers are especially troubling as they occur in patients who are likely cured of their original cancer, and have long assumed a normal life. These secondary cancers are also extraordinarily difficult to treat, rapidly acquiring resistance to all conventional therapy (11,12,93,94). To be caught once again by malignancy, and a malignancy that is most often incurable, when the patient thought they had escaped and survived, is a horrific problem. If one could predict who is likely to develop oncogenic chromosomal translocations, then one could at least increase monitoring, although it is not clear that early diagnosis would be helpful because most of these secondary malignancies are not responsive to conventional therapy (11,12,93,94). Perhaps PARP1 levels in the marrow CD34+ cells of a patient undergoing high-dose DNA damaging therapy could predict an oncogenic translocation (92). Preventing such translocations from occurring in the first place would be a significant advance in the clinical practice of oncology, and decrease the suffering associated with these secondary cancers.
Inhibition of PARP1, a rate-limiting step in a-NHEJ confirms the role of a-NHEJ in the development of chromosome translocations, and demonstrates that such deleterious genomic events can be prevented. This raises the intriguing possibility that secondary oncogenic translocations that could occur during otherwise curative cancer therapy can be decreased by pre-treatment with a PARP1 inhibitor. For example, in a short, high-intensity course of cancer therapy, such during a stem cell transplant conditioning regimen, which has a defined risk of generating oncogenic chromosomal translocations, could a PARP1 inhibitor be given before conditioning, and thereby abrogate the risk of secondary malignancies (92)? This raises for the first time the possibility that oncogenic chromosomal translocations can be prevented.
Footnotes
Potential Conflicts of Interest: None to disclose.
DISCUSSION
Billings, Baton Rouge: That’s very interesting, in that all of us who take care of cancer patients may wonder that if earlier, they may have sown seeds of their destruction. And the patient that you described, it looked to me like he had had a 50-year hiatus between his first hit and his second hit. How long would you treat these patients to prevent that cancer? When would you start? Who would you start on? And how do you know the effect of what you’re doing when you have a 50-year hiatus?
Hromas, Gainesville: So, as you know we have about 20,000 to 50,000 stem cells sitting quiescent in our marrow. Our hypothesis is that you develop the insult from the radiation at the time of exposure. However, it wasn’t until that stem cell was pushed into developmental proliferation that this patient developed leukemia. Our hypothesis was that the translocation that caused his leukemia was an early event that did not become manifest until that quiescent stem cell started to proliferate and differentiate. That’s a great question and difficult to address without a clinical trial, but the hypothesis is that you could immediately, after their DNA damaging insult, reduce the incidence of oncogenic or leukemogenic translocations. I don’t think you have to treat forever. I think just treating after the initial event would be sufficient. Interestingly, when we give mice olaparib before radiation, they actually have further chromosome abnormalities. However, giving it after the event prevents the translocations that could cause leukemia. We feel that these translocations occur when a DNA double-strand break occurs on two separate chromosomes, and each is resistant to repair by non-homologous end-joining repair. Microhomology-mediated end-joining is a rescue pathway for those double-strand breaks that fail to repair, and this pathway can lead to translocations. This rescue pathway would only have to be blocked for a short period of time in order for the DNA breaks to be repaired by the proper pathway. So we don’t think you need to treat forever, just immediately after the event for a short period of time.
Billings, Baton Rouge: Would it be better to treat right before the radiation insult?
Hromas, Gainesville: Probably not, Because PARP1 is required for base excision repair, and plays a role in replication restart as well. So you can actually induce toxicity in that situation. We want to stop those double-strand breaks that are persistent from repairing through the rescue pathway. Microhomology-mediated end-joining is initiated by PARP1, so blocking PARP1 in that situation, after the insult, would prevent chromosomal translocations without harming other DNA repair pathways.
Garber, Boston: That was a very provocative talk, and thank you. You know there has been concern about myelodysplastic syndromes with the PARP inhibitors and I wondered how that might affect your thinking about this? Are there other mechanisms that could be considered to prevent chromosomal translocations with the use of these drugs? Of course everybody on these trials has had chemotherapy before because they only get to the trials when they have recurred after their treatment. They still all had chemotherapy so you can’t separate the effect of the PARP inhibitor, but there is concern about it.
Hromas, Gainesville: In some of the trials there has been a worry about that. In our situation we are talking about only a short course of treatment. These mice were treated for a day. They got just a few injections, and then we are done. We are talking about just immediately after high-dose chemotherapy or radiation prior to an autologous peripheral blood stem cell transplant. I would not treat before chemotherapy; I would do it immediately after. The thought is that you would not need to treat long, just to prevent the aberrant repair of those DNA double-strand breaks over a few days. I don’t think you need to treat long enough to induce the risk of myelodysplastic syndromes.
Berger, Cleveland: When you inhibit PARP1, you are inhibiting the initiating of a DNA repair pathway. So what happens to those cells with the damage done chromosomes? Have you looked to see whether those cells with damaged chromosomes in the treated animals survived?
Hromas, Gainesville: That is an excellent question. We have not done this. We are now examining several mouse models of radiation-induced leukemia to test whether PARP1 inhibition prevents leukemia formation after radiation. There is a certain species that’s derived from C57B6 mice that forms a radiation-induced translocation that develops into acute leukemia. We are testing right now whether we can prevent that radiation-induced acute leukemia by treating after the radiation dose for a short period of time. The other interesting question is, can you induce cell survival with olaparib? My thought would be that chromosomal translocation selects for cell survival, and selects for proliferation, so actually the olaparib may prevent that cell from surviving the aberrant translocation. By preventing the translocation with olaparib, you might prevent the cell proliferation required for survival, secondarily. We haven’t looked at that—it’s a wonderful question, and single cell tracking is now possible in some systems.
Michael Gershon, New York City: I was going to ask a similar question. I wondered if instead of inhibiting the translocation, if you could try to kill the cells that have the chromosome break? That is, I wonder what happens after you inhibit PARP1, what happens to the damaged cell?
Hromas, Gainesville: That’s a good question: after the chromosomal translocation occurs, can you selectively target that cell, for example, as a synthetically lethal event? A lot of people have tried that, and right now we don’t have any mechanism to do that. It’s an excellent thought and I think with the advent of the first synthetically lethal cancer drug, olaparib, there is going to be a large number of other PARP1 inhibitors, even more potent, becoming available. I wonder if there would be a way to target another area of DNA repair that would promote cell death in the oncogenic cell. Let me just mention Simon Powell and Thomas Skorski have data showing that Rad 52 can be an alternative double-strand break repair pathway in BRCA1 and 2 mutant breast or ovarian cancers. So they have thought that perhaps targeting Rad 52 might induce cell death in that cancer cell. There are people working on targeting Rad 52 for that reason. Thank you very much.
REFERENCES
- 1.Sax K. Chromosome aberrations induced by X-rays. Genetics. 1938;23:494–516. doi: 10.1093/genetics/23.5.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandra HS, et al. Philadelphia Chromosome Symposium: commemoration of the 50th anniversary of the discovery of the Ph chromosome. Cancer Genet. 2011;204:171–9. doi: 10.1016/j.cancergen.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aplan PD. Causes of oncogenic chromosomal translocation. Trends Genet. 2006;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nickoloff JA, et al. Mechanisms of leukemia translocations. Curr Opin Hematol. 2008;15:338–45. doi: 10.1097/MOH.0b013e328302f711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das K, Tan P. Molecular cytogenetics: recent developments and applications in cancer. Clin Genet. 2013;84:315–25. doi: 10.1111/cge.12229. [DOI] [PubMed] [Google Scholar]
- 6.Wang JH. Mechanisms and impacts of chromosomal translocations in cancers. Front Med. 2012;6:263–74. doi: 10.1007/s11684-012-0215-5. [DOI] [PubMed] [Google Scholar]
- 7.Pillai RN, Ramalingam SS. The biology and clinical features of non–small cell lung cancers with EML4-ALK translocation. Curr Oncol Rep. 2012;14:105–10. doi: 10.1007/s11912-012-0213-4. [DOI] [PubMed] [Google Scholar]
- 8.Chesi MP, Bergsagel L. Molecular pathogenesis of multiple myeloma: basic and clinical updates. Int J Hematol. 2013;97:313–23. doi: 10.1007/s12185-013-1291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salas C, Pérez-Vera P, Frías S. Genetic abnormalities in leukemia secondary to treatment in patients with Hodgkin’s disease. Rev Invest Clin. 2011;63:53–63. [PubMed] [Google Scholar]
- 10.Harrison CJ. Targeting signaling pathways in acute lymphoblastic leukemia: new insights. Hematology Am Soc Hematol Educ Program. 2013:118–25. doi: 10.1182/asheducation-2013.1.118. [DOI] [PubMed] [Google Scholar]
- 11.Leone G, et al. Therapy related leukemias: susceptibility, prevention and treatment. Leuk Lymphoma. 2001;41:255–76. doi: 10.3109/10428190109057981. [DOI] [PubMed] [Google Scholar]
- 12.Hromas R, et al. A novel syndrome of radiation-associated acute myeloid leukemia involving AML1 gene translocations. Blood. 2000;95:4011–3. [PubMed] [Google Scholar]
- 13.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–80. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fugmann SD, et al. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 15.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 16.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 17.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 18.Marculescu R, et al. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J Exp Med. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnal SM, et al. Non-consensus heptamer sequences destabilize the RAG post-cleavage complex, making ends available to alternative DNA repair pathways. Nucleic Acids Res. 2010;38:2944–54. doi: 10.1093/nar/gkp1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alt FW, et al. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–29. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–21. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Posey JE, et al. Target DNA structure plays a critical role in RAG transposition. PLoS Biol. 2006;4:e350. doi: 10.1371/journal.pbio.0040350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghavan SC, et al. The structure-specific nicking of small heteroduplexes by the RAG complex: implications for lymphoid chromosomal translocations. DNA Repair (Amst) 2007;6:751–9. doi: 10.1016/j.dnarep.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghavan SC, et al. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J Biol Chem. 2005;280:22749–60. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 25.Raghavan SC, et al. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 26.Nambiar M, et al. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39:936–48. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, et al. DNA structure-induced genomic instability in vivo. J Natl Cancer Inst. 2008;100:1815–7. doi: 10.1093/jnci/djn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howarth KD, et al. Large duplications at reciprocal translocation breakpoints that might be the counterpart of large deletions and could arise from stalled replication bubbles. Genome Res. 2011;21:525–34. doi: 10.1101/gr.114116.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit Rev Biochem Mol Biol. 2012;47:222–35. doi: 10.3109/10409238.2012.655374. [DOI] [PubMed] [Google Scholar]
- 30.Robbiani DF, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–41. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–38. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koduru S, et al. Dendritic cell-mediated activation-induced cytidine deaminase (AID)-dependent induction of genomic instability in human myeloma. Blood. 2012;119:2302–9. doi: 10.1182/blood-2011-08-376236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okazaki IM, Kotani A, Honjo T. Role of AID in tumorigenesis. Adv Immunol. 2007;94:245–73. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- 34.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casey OM, et al. TMPRSS2- driven ERG expression in vivo increases self-renewal and maintains expression in a castration resistant subpopulation. PLoS One. 2012;7:e41668. doi: 10.1371/journal.pone.0041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mateo J, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolomietz E, et al. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- 39.Elliott BC, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17:885–94. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 40.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–7. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Jeffs AR, Wells E, Morris CM. Nonrandom distribution of interspersed repeat elements in the BCR and ABL1 genes and its relation to breakpoint cluster regions. Genes Chromosomes Cancer. 2001;32:144–54. doi: 10.1002/gcc.1176. [DOI] [PubMed] [Google Scholar]
- 42.Christiansen DH, Pedersen-Bjergaard J. Internal tandem duplications of the FLT3 and MLL genes are mainly observed in atypical cases of therapy-related acute myeloid leukemia with a normal karyotype and are unrelated to type of previous therapy. Leukemia. 2001;15:1848–51. doi: 10.1038/sj.leu.2402246. [DOI] [PubMed] [Google Scholar]
- 43.Kurahashi H, et al. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–45. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurahashi H, et al. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–70. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–27. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coté AG, Lewis SM. Mus81-dependent double-strand DNA breaks at in vivo–generated cruciform structures in S. cerevisiae. Mol Cell. 2008;31:800–12. doi: 10.1016/j.molcel.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 47.Kurahashi H, et al. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J Biol Chem. 2004;279:35377–83. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen C, et al. More forks on the road to replication stress recovery. J Mol Cell Biol. 2001;3:4–12. doi: 10.1093/jmcb/mjq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fenech M, et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–32. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 50.Bower JJ, et al. Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene. 2010;29:4787–99. doi: 10.1038/onc.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaufmann WK. Dangerous entanglements. Trends Mol Med. 2006;12:235–7. doi: 10.1016/j.molmed.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Povirk LF. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair (Amst) 2006;5:1199–212. doi: 10.1016/j.dnarep.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 54.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Fnu S, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108(2):540–5. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burman B, et al. Histone modifications predispose genome regions to breakage and translocation. Genes Dev. 2015;29:1393–402. doi: 10.1101/gad.262170.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makova KD, Hardison RC. The effects of chromatin organization on variation in mutation rates in the genome. Nat Rev Genet. 2015;16:213–23. doi: 10.1038/nrg3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–54. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaheen M, et al. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood. 2011;117:6074–82. doi: 10.1182/blood-2011-01-313734. [DOI] [PubMed] [Google Scholar]
- 60.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Brandsma I, Gent DC. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr. 2012;3:9. doi: 10.1186/2041-9414-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 63.Rowley JD. The role of chromosome translocations in leukemogenesis. Semin Hematol. 1999;36:59–72. [PubMed] [Google Scholar]
- 64.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5:1282–97. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 65.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–98. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 66.Mattarucchi E, et al. Microhomologies and interspersed repeat elements at genomic breakpoints in chronic myeloid leukemia. Genes Chromosomes Cancer. 2008;47:625–32. doi: 10.1002/gcc.20568. [DOI] [PubMed] [Google Scholar]
- 67.Shahar OD, et al. Live imaging of induced and controlled DNA double-strand break formation reveals extremely low repair by homologous recombination in human cells. Oncogene. 2012;31:3495–504. doi: 10.1038/onc.2011.516. [DOI] [PubMed] [Google Scholar]
- 68.Cheng Q, et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res. 2011;39:9605–19. doi: 10.1093/nar/gkr656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–82. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2006;8:983–95. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–51. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 72.Beck BD, et al. Biochemical characterization of metnase’s endonuclease activity and its role in NHEJ repair. Biochemistry. 2011;50:4360–70. doi: 10.1021/bi200333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–71. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 74.Bothmer A, et al. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J Exp Med. 2013;210:115–23. doi: 10.1084/jem.20121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 76.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–6. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wray J, et al. The transposase domain protein Metnase/SETMAR suppresses chromosomal translocations. Cancer Genet Cytogenet. 2010;200:184–90. doi: 10.1016/j.cancergencyto.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–81. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–38. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guirouilh-Barbat J, Huck S, Lopez BS. S-phase progression stimulates both the mutagenic KU-independent pathway and mutagenic processing of KU-dependent intermediates, for nonhomologous end joining. Oncogene. 2008;27:1726–36. doi: 10.1038/sj.onc.1210807. [DOI] [PubMed] [Google Scholar]
- 81.Truong LN, et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:7720–5. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–4. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee-Theilen M, et al. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18:75–9. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mao Z, et al. SIRT6 Promotes DNA Repair Under Stress by Activating PARP1. Science. 2011;332:1443–6. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–24. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 87.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 88.Mansour WY, Rhein T, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010;38:6065–77. doi: 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rahal EA, et al. ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining. Cell Cycle. 2010;9:2866–77. doi: 10.4161/cc.9.14.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Della-Maria J, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem. 2011;286:33845–53. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee JM, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol. 2014;25:32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wray J, et al. PARP1 is required for chromosomal translocations. Blood. 2013;121:4359–65. doi: 10.1182/blood-2012-10-460527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brown JR, et al. Increasing incidence of late second malignancies after conditioning with cyclophosphamide and total-body irradiation and autologous bone marrow transplantation for non-Hodgkin’s lymphoma. J Clin Oncol. 2005;23:2208–14. doi: 10.1200/JCO.2005.05.158. [DOI] [PubMed] [Google Scholar]
- 94.Leiper AD. Late effects of total body irradiation. Arch Dis Child. 1995;72:382–5. doi: 10.1136/adc.72.5.382. [DOI] [PMC free article] [PubMed] [Google Scholar]