Abstract
Background
While recent advances in next generation sequencing technologies have enabled researchers to readily identify countless microbial species in soil, rhizosphere, and phyllosphere microbiomes, the biological functions of the majority of these species are unknown. Functional studies are therefore urgently needed in order to characterize the plethora of microorganisms that are being identified and to point out species that may be used for biotechnology or plant protection. Here, we used a dual culture assay and growth analyses to characterise yeasts (40 different isolates) and their antagonistic effect on 16 filamentous fungi; comprising plant pathogens, antagonists, and saprophytes.
Results
Overall, this competition screen of 640 pairwise combinations revealed a broad range of outcomes, ranging from small stimulatory effects of some yeasts up to a growth inhibition of more than 80% by individual species. On average, yeasts isolated from soil suppressed filamentous fungi more strongly than phyllosphere yeasts and the antagonistic activity was a species-/isolate-specific property and not dependent on the filamentous fungus a yeast was interacting with. The isolates with the strongest antagonistic activity were Metschnikowia pulcherrima, Hanseniaspora sp., Cyberlindnera sargentensis, Aureobasidium pullulans, Candida subhashii, and Pichia kluyveri. Among these, the soil yeasts (C. sargentensis, A. pullulans, C. subhashii) assimilated and/or oxidized more di-, tri- and tetrasaccharides and organic acids than yeasts from the phyllosphere. Only the two yeasts C. subhashii and M. pulcherrima were able to grow with N-acetyl-glucosamine as carbon source.
Conclusions
The competition assays and physiological experiments described here identified known antagonists that have been implicated in the biological control of plant pathogenic fungi in the past, but also little characterised species such as C. subhashii. Overall, soil yeasts were more antagonistic and metabolically versatile than yeasts from the phyllosphere. Noteworthy was the strong antagonistic activity of the soil yeast C. subhashii, which had so far only been described from a clinical sample and not been studied with respect to biocontrol. Based on binary competition assays and growth analyses (e.g., on different carbon sources, growth in root exudates), C. subhashii was identified as a competitive and antagonistic soil yeast with potential as a novel biocontrol agent against plant pathogenic fungi.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-016-0908-z) contains supplementary material, which is available to authorized users.
Keywords: Antagonism, Biocontrol, Candida, Fungal plant pathogen, Mitochondrial genome, Plant protection, Yeast
Background
The fungal kingdom includes important plant pathogens that cause a plethora of diseases in all crops worldwide. Of particular concern are rot and wilt diseases caused by soilborne fungi, fungal spots, blights and blotches, rusts, mildews, cankers and anthracnoses, as well as postharvest decay of fruits and vegetables [1, 2]. Infestations by aggressive, fungal pathogens can severely constrain agricultural production and often the only resort is crop rotation, fallow, or even an abandonment of the cropland [3, 4].
Soil, roots, and the phyllosphere harbour complex microbiomes consisting of thousands of bacterial and fungal species that may suppress diseases, act as pathogens, or affect plant health and growth by various other mechanisms [5–8]. Yet, microbiomes are still a largely untapped resource for protecting crop plants against pathogens and for increasing agricultural productivity [9, 10]. Considerable efforts are therefore undertaken to harness and use microbiota for novel applications in agriculture [11–14]. Microbiomes of plants, rhizosphere, or soil have been elucidated by large-scale, DNA sequencing-based metagenomics approaches [7, 15, 16], but the contributions and functions of the large majority of the species are still mostly unknown. Microbiota thus consist predominantly of yet uncharacterized bacteria and fungi, tritagonists, that regulate microbial interactions [17].
Yeast-like fungi inhabit all aerobic environments; from the arctic and glaciers to the tropics or even the desert and from dry to saline and high-sugar habitats [18–24]. Many yeast species are particularly well known for their biotechnological applications or medical relevance. In agriculture, yeasts have been identified as powerful antagonists of fungal pathogens causing postharvest and storage diseases and of microorganisms attacking flowers and leaves [25–31]. Few yeast species have reached the market as commercial products for the postharvest control of pathogens (e.g., Aureobasidium pullulans as BoniProtect, Candida oleophila strain 1-182 as AspireTM, Candida sake as Candifruit, Metschnikowia fructicola as Shemer, or Cryptococcus albidus as YieldPlus) or against fireblight (e.g., A. pullulans as BlossomProtect); some of which are not marketed anymore or only registered locally [32–37]. Yeasts suppressing soilborne pathogens have been described rarely and a commercial application has not been considered yet. Candida valida, Rhodotorula glutinis and Trichosporon asahii protected sugar beet against the soil pathogen Rhizoctonia solani [38]. In another study, Saccharomyces unispora and Candida steatolytica antagonised Fusarium oxysporum causing wilt disease in kidney beans [39] and Saccharomyces cerevisiae controlled a Fusarium infection of sugar beet [40]. In a successful example of postharvest biocontrol, M. fructicola has been employed as part of a combined strategy to control the soilborne pathogen Thielaviopsis basicola on carrots [41]. These examples clearly document the potential of yeasts to suppress and antagonise soilborne pathogens, but also highlight the limited knowledge on their biological functions in soil.
The genus Candida comprises several species that have been studied extensively with respect to biotechnological applications, biocontrol, but also as human pathogens. Candida guilliermondii is a ubiquitously present, saprophytic yeast that has received particularly broad attention because of its presence in clinical samples, the biotechnological production of metabolites and enzymes, applications in bioremediation, or the control of plant pathogenic fungi [42]. The antagonistic potential of C. guilliermondii against diverse fungal pathogens (e.g., Botrytis cinerea, Colletotrichum capsici, Penicillium expansum, Penicillium digitatum, Rhizopus stolonifer) has been demonstrated in various cultures such as apple, citrus, nectarine, peach, or tomato ([42], and references therein). Other Candida species have also been studied for their biocontrol potential and as commercial plant protection agents against postharvest decay of fruits, based on Candida species, have been developed (see above) [29, 43, 44].
In the course of the work described here, we used binary competition assays to determine the antagonistic activity of soil and phyllosphere yeasts from Switzerland against a range of pathogenic and saprophytic filamentous fungi. Among the six most antagonistic yeasts out of a collection of 40 different isolates (A. pullulans, Candida subhashii, Cyberlindnera sargentensis, Hanseniaspora sp., Metschnikowia pulcherrima, and Pichia kluyveri), C. subhashii was the only one that has so far not been studied with respect to biocontrol. This species has only been reported from a patient suffering from peritonitis during a long-term peritoneal dialysis treatment and an isolate highly similar to this type strain (99.8% identity in the 26S rDNA D1/D2 domain, 1.3% sequence difference for the 5.8S-ITS region) was obtained from a soil sample from East Japan [45, 46]. Except for these two reports, only one additional publication reporting the mitochondrial genome of C. subhashii has appeared [47]. Here, we describe C. subhashii as a common and frequent soil fungus that has broad metabolic capabilities, grows in root exudates, and that strongly antagonizes a wide range of filamentous fungi (all species tested in this study, including notorious plant pathogens, saprophytes, but also other antagonists of the genus Trichoderma). Since it has not been experimentally confirmed that C. subhashii is indeed a pathogen, and based on its broad distribution in different soils and the apparent adaptations to the soil environment, it is concluded that C. subhashii is a competitive soil fungus and potential candidate for the biological control of soilborne fungal pathogens.
Methods
Isolation and cultivation of fungi
Soil or plant material (e.g., apple leaves, flowers, bark, skin) was diluted 10-fold (w/v) with peptone water (1 g/L Bacto Peptone) [48], vigorously mixed, and shaken (20 min, 25 °C, 250 rpm, on an orbital shaker). The resulting suspensions were diluted and different dilutions (usually 1:50 and 1:100) were plated on DifcoTM potato dextrose agar (PDA; Becton, Dickinson and Company, Le Pont de Claix, France) supplemented with 5 ml chloramphenicol and tetracycline HCl (5 mg/ml in ethanol or water, respectively), and incubated at 22 °C for 2–4 days. Single fungal colonies were transferred to PDA agar plates without antibiotics and repeatedly streaked out until pure cultures were obtained. Isolates were maintained on PDA agar plates and stored in 15% (v/v) glycerol at −80 °C.
Identification of fungal isolates
Species identification was first attempted by MALDI-TOF as previously described [49]. In cases where MALDI-TOF did not allow species identification, the fungal ITS region was amplified with primers ITS1f [50] and ITS4 [51], PCR products were directly used for sequencing, and all isolates were assigned a species hypothesis according to the UNITE database [52, 53] (see also Table 1). Crude protein extracts of isolates that were identified based on their ITS sequence were used to generate reference MALDI-TOF spectra for future identifications of the same species [49]. All isolates generated in the course of this study have been deposited and are available at the Culture Collection of Switzerland (CCoS; https://www.ccos.ch; Table 1).
Table 1.
Yeasts and filamentous fungi used in this study. All strains that were isolated in the course of this study are deposited and available at the Culture Collection of Switzerland (CCoS; https://www.ccos.ch)
| Culture Collection | Isolate | SH-number | Name | Source | Origin/Reference | |
|---|---|---|---|---|---|---|
| 40 yeast isolates tested in this study | CCOS995 | BC 1.01 | SH216366.07FU | Rhodosporidium sphaerocarpum | Agricultural soil | This study, Switzerland |
| CCOS996 | BC 1.03 | SH218818.07FU | Candida subhashii | Agricultural soil | This study, Switzerland | |
| CCOS997 | BC 1.06 | SH196641.07FU | Trichosporon dehoogii | Agricultural soil | This study, Switzerland | |
| CCOS998 | BW 2.02 | SH195538.07FU | Trichosporon ovoides | Agricultural soil | This study, Switzerland | |
| CCOS999 | BW 5.01 | SH196643.07FU | Trichosporon moniliiforme | Agricultural soil | This study, Switzerland | |
| CCOS1000 | BW 7.01 A | SH190095.07FU | Schwanniomyces yamadae | Agricultural soil | This study, Switzerland | |
| CCOS1001 | BW 7.02 | SH182010.07FU | Trichosporon gracile | Agricultural soil | This study, Switzerland | |
| CCOS1009 | SHA 10.3 | SH175136.07FU | Candida sp | Agricultural soil | This study, Switzerland | |
| CCOS1010 | SHA 15.4 | SH205045.07FU | Cryptococcus laurentii | Agricultural soil | This study, Switzerland | |
| CCOS1011 | SHA 17.2 | SH195578.07FU | Cyberlindnera saturnus | Agricultural soil | This study, Switzerland | |
| CCOS1012 | SHA 25.3 | SH031361.07FU | Barnettozyma vustinii | Agricultural soil | This study, Switzerland | |
| CCOS1013 | SHA 43.1 | SH216362.07FU | Rhodotorula graminis | Agricultural soil | This study, Switzerland | |
| CCOS1014 | SHA 51.1 | SH212824.07FU | Guehomyces pullulans | Agricultural soil | This study, Switzerland | |
| CCOS1015 | SHA 7.1 | SH175136.07FU | Candida sp | Agricultural soil | This study, Switzerland | |
| CCOS1008 | NBB 7.2.1 | SH195774.07FU | Aureobasidium pullulans | Orchard soil | This study, Switzerland | |
| CCOS1004 | FGA 2.2 | SH218818.07FU | Candida subhashii | Potting soil | This study, Switzerland | |
| CCOS1005 | FGA 3.3 | SH196641.07FU | Trichosporon dehoogii | Potting soil | This study, Switzerland | |
| CCOS1006 | KS 1/d7.18 | SH192275.07FU | Candida boidinii | Old compost | This study, Germany | |
| CCOS1003 | F 2.6 | SH198057.06FU | Cryptococcus heimaeyensis | Irrigation water | This study, Switzerland | |
| CCOS1002 | Dip141103.2 | SH199823.07FU | Pichia membranifaciens | Insect (Drosophila) | This study, Switzerland | |
| CCOS976 | APC 1.1 | SH194776.07FU | Rhodotorula slooffiae | Apple flowers | This study, Switzerland | |
| CCOS977 | APC 1.10 | SH005240.07FU | Dioszegia sp | Apple flowers | This study, Switzerland | |
| CCOS978 | APC 1.2 | SH180747.07FU | Metschnikowia pulcherrima | Apple flowers | This study, Switzerland | |
| CCOS979 | APC 1.5 | SH221435.07FU | Cryptococcus wieringae | Apple flowers | This study, Switzerland | |
| CCOS980 | APC 1.7 | SH192046.07FU | Basidiomycota sp | Apple flowers | This study, Switzerland | |
| CCOS981 | APC 10.2 | SH207120.07FU | Basidiomycota sp | Apple leaves | This study, Switzerland | |
| CCOS982 | APC 11.10 B | SH204094.07FU | Pichia kluyveri | Apple bark | This study, Switzerland | |
| CCOS983 | APC 11.3 | SH019470.07FU | Tremella moriformis | Apple bark | This study, Switzerland | |
| CCOS984 | APC 12.1 | SH177122.07FU | Hanseniaspora sp. | Apple bark | This study, Switzerland | |
| CCOS985 | APC 13.2 | SH194503.07FU | Sporidiobolales sp | Apple bark | This study, Switzerland | |
| CCOS986 | APC 18.3 | SH194739.07FU | Erythrobasidium hasegawianum | Apple leaves | This study, Switzerland | |
| CCOS987 | APC 19.2 | SH194775.07FU | Rhodotorula pinicola | Apple leaves | This study, Switzerland | |
| CCOS988 | APC 2.3 | SH204123.07FU | Starmerella bombicola | Apple flowers | This study, Switzerland | |
| CCOS989 | APC 27.4 | SH206552.07FU | Cryptococcus cerealis | Apple bark | This study, Switzerland | |
| CCOS990 | APC 3.4 | SH205935.07FU | Sporobolomyces oryzicola | Apple flowers | This study, Switzerland | |
| CCOS991 | APC 6.7 | SH193763.07FU | Leucosporidiella creatinivora | Apple leaves | This study, Switzerland | |
| CCOS992 | APC 9.2 | SH181628.07FU | Cryptococcus victoriae | Apple leaves | This study, Switzerland | |
| CCOS993 | AS 1.02 | SH181630.07FU | Cryptococcus sp | Apple fruit | This study, Germany | |
| CCOS994 | AS 1.06 | SH190089.07FU | Debaryomyces prosopidis | Apple fruit | This study, Germany | |
| EUROSCARF | BY4741 | - | Saccharomyces cerevisiae | - | [59] | |
| 16 test strains (filamentous fungi) | CCOS1018 | BC 4.14 | SH216250.07FU | Mycosphaerella tassiana | Agricultural soil | This study, Switzerland |
| CCOS1019 | BC 8.11 | SH207825.07FU | Trichoderma ghanense | Agricultural soil | This study, Switzerland | |
| CCOS1020 | BC 8.14 | SH213620.07FU | Gibberella fujikuroi | Agricultural soil | This study, Switzerland | |
| CCOS1022 | SHA 18.1 | SH188374.07FU | Mucor moelleri | Agricultural soil | This study, Switzerland | |
| CCOS1023 | SHA 9.1 | SH185778.07FU | Mucor circinelloides | Agricultural soil | This study, Switzerland | |
| CCOS1007 | NBB 2.4.2 | SH190868.07FU | Trichoderma spirale | Orchard soil | This study, Switzerland | |
| CCOS1021 | F 2.1 | SH181342.07FU | Trichoderma viride | Irrigation water | This study, Switzerland | |
| CCOS1017 | Asp 1.1 | SH219673.07FU | Fusarium proliferatum | Infected asparagus | This study, Switzerland | |
| A 06.5 | SH215493.07FU | Alternaria eichhorniae | Diseased, stored apple | Laimburg, Italy | ||
| - | FP13013 | Fusarium poae | Oat | S. Vogelgsang, Agroscope | ||
| - | FL13014 | Fusarium langsethiae | Oat | S. Vogelgsang, Agroscope | ||
| CBS 121292 | FG0410 | Fusarium graminearum | Wheat | S. Vogelgsang, Agroscope | ||
| - | FCr11115 | Fusarium crookwellense | Wheat | S. Vogelgsang, Agroscope | ||
| - | 11SD14 | - | Monilinia fructicola | Infected apricot | [49] | |
| - | 106 | - | Rhizoctonia solani | [107] | ||
| ARSEF 1095 | F52/Met52 | - | Metarhizium brunneum | Cydia pomonella | Austria |
Quantification of yeast antagonism against filamentous fungi in vitro
Yeasts were collected from a PDA plate (less than 2 weeks old), diluted in water, and adjusted to an OD600 of 0.1. Fifteen microlitre of this suspension was plated on PDA plates (5.5 cm in diameter) in quadruples. Conidia of filamentous fungi were collected in water, diluted (OD600 = 0.1), and 5 μl were inoculated in the centre of the plates (previously overlaid with yeasts or fresh PDA plates as a control). Plates were incubated at 22 °C for 3 to 15 days depending on the fungal species. Growth of the filamentous fungus was quantified before it reached the edge of the control plate (plate without yeasts) with the help of a planimeter (Planix 5, Tamaya Technics Inc., Tokyo, Japan). The average of the relative growth (growth in presence of yeast/growth on control plate) of four replicates for each of the 640 combinations was calculated, log2-transformed, and all data were clustered using EPCLUST (http://www.bioinf.ebc.ee/EP/EP/EPCLUST/) for visualisation (correlation measure based distance (uncentered), complete linkage).
Growth analysis of yeasts at different temperatures
Yeasts were collected from a PDA plate, resuspended in sterile water, adjusted to an OD600 of 1, and 10-fold dilutions were prepared in a microtiter plate. The dilutions were spotted onto PDA plates with a multi-blot replicator (delivered volume approx. 3 μl) (V & P Scientific, Inc., San Diego, USA). The plates were incubated at temperatures ranging from 15 to 37 °C and the maximal dilution to which the yeast grew was recorded. Each experiment was performed at least twice for each isolate and the average fold-dilution is indicated as reflective of the growth.
Microarray phenotype analysis
Overnight liquid cultures were grown in DifcoTM potato dextrose broth (PDB; Becton, Dickinson and Company, Le Pont de Claix, France). Cells were pelleted by centrifugation (4 °C, 10 min, 650 g), the supernatant was discarded, and the cells were washed twice with sterile water. For each yeast isolate, a suspension with an OD600 of 1 was prepared and 100 μl of this solution were inoculated in each well of a Biolog YT MicroPlateTM (Endotell AG, Allschwil, Switzerland) [54]. The absorption at 590 nm was determined in a plate reader (Infinite® 200 Pro; Tecan Group Ltd., Switzerland) daily for 3 days. All data were normalized with the corresponding water control and growth was expressed relative to the initial measurement at day 0. The maximal relative growth at any of the three time-points was recorded (rounded to the first integer). For each yeast, the experiment was performed twice and the average of the two measurements is shown. Substrates that did not lead to detectable growth for any of the yeasts are not shown. For four carbon sources (glucose, maltose, N-acetyl-glucosamine, melezitose), the microarray phenotype results were confirmed by performing growth analyses in defined medium. Yeast nitrogen base (with amino acids and ammonium sulphate) was supplemented with glucose, maltose, N-acetylglucosamine or melezitose (stock solutions were filter sterilized, final concentration 10 g/L) and growth was followed by measuring the OD600 in a plate reader (Infinite® 200 Pro; Tecan Group Ltd., Switzerland). The final measurement (mean of five replicates and standard error) after 42 h is shown.
Growth in root exudates
Mung bean (Vigna radiata) root exudates were collected according to Barbour et al. [55] and used at a final concentration of 0.1 mg/ml. Yeasts were inoculated to an initial OD600 of 0.1 and growth was measured in a plate reader (Infinite® 200 Pro; Tecan Group Ltd., Switzerland) for 3 days. The mean of six replicates and the standard error are shown.
Sequencing and analysis of the C. subhashii FGA 2.2 mitochondrial genome
Candida subhashii strain FGA 2.2 genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit and sequenced on the PacBio RS II platform (performed at the Functional Genomics Center Zurich). Subsequent de novo genome assembly and resequencing were performed using PacBio SMRT Portal 2.3.0 [56]. Assembly was generated using protocol RS_HGAP_Assembly.3. The contig corresponding to the mitochondrial genome revealed a linear DNA molecule. Manual curation was performed to extend both telomeres to their full length of 729 bp, resulting in a mitochondrial DNA (mtDNA) assembly of 29,930 bp. One additional resequencing step was performed using SMRT portal protocol RS_Resequencing.1, which resulted in a mean coverage depth of 567-fold. The C. subhashii strain FGA 2.2 mitochondrial genome was annotated by reference to the C. subhashii type strain CBS10753 [47].
To construct a phylogenetic tree, the mtDNA sequences of 22 diverse yeast species, selected based on previous studies and the availability of complete and annotated mitochondrial genomes [47], were obtained from NCBI (Table 2). The amino acid sequences of the conserved proteins Atp6, Atp8, Atp9, Cob, Cox1, Cox2 and Cox3 were extracted from the downloaded sequences as well as from the C. subhashii mtDNA assembly. Multiple sequence alignment (MSA) using MUSCLE 3.8 [57] and trimming of overhanging sequences ensured that the amino acid sequences of all genes and all 22 strains were of similar length. The amino acid sequences of all proteins were concatenated for every strain and a final MSA with MUSCLE was performed. The resulting alignment of 1743 amino acids was used to create a phylogenetic tree by RAxML 8.1 applying the JTT + Γ model [58]. The phylogeny was tested by performing 100 bootstrap replicates.
Table 2.
Strain designations and accession numbers of the mitochondrial genomes used for calculating the maximum likelihood phylogeny
| Species | Strain | Accession | Comment |
|---|---|---|---|
| Candida albicans | L757 | JQ864233 | CTG clade |
| Candida metapsilosis | MCO448 | AY962591 | CTG clade |
| Candida neerlandica | NRRL Y-27057 | EU334437 | CTG clade |
| Candida parapsilosis | CBS 7157 (SR 23) | X74411 | CTG clade |
| Candida sake | CBS 159 | KC993194 | CTG clade |
| Candida subhashii | FR 392/CBS10753 | GU126492 | CTG clade |
| Candida subhashii | FGA 2.2/CCOS1004 | KX781248 | CTG clade |
| Candida tropicalis | CBS 94 | KC993185 | CTG clade |
| Debaryomyces hansenii | CBS767 | DQ508940 | CTG clade |
| Meyerozyma guilliermondii | CBS 2030 | KC993176 | CTG clade |
| Pichia farinosa | CBS7064 | FN356025 | CTG clade |
| Candida glabrata | ATCC 2001 | AJ511533 | WGD clade |
| Saccharomyces castellii | NRRL Y-12630 | AF437291 | WGD clade |
| Saccharomyces cerevisiae | S288c | KP263414 | WGD clade |
| Saccharomyces pastorianus | Weihenstephan 34/70 | EU852811 | WGD clade |
| Saccharomyces servazzii | NRRL Y-12661 | AJ430679 | WGD clade |
| Barnettozyma californica | CBS 252 | KC993183 | |
| Cyberlindnera jadinii | CBS 1600 | KC993189 | |
| Kluyveromyces lactis | CBS2359 | AY654900 | |
| Kluyveromyces thermotolerans | CBS 6340 | AJ634268 | |
| Wickerhamomyces pijperi | CBS 2887 | KC993192 | |
| Yarrowia lipolytica | W29 | AJ307410 | outgroup |
Results
The antagonistic activity of naturally occurring yeasts against filamentous fungi in vitro
From a collection of yeasts naturally occurring in agricultural environments, a subset of 40 species was selected (Table 1). These isolates represented the taxonomic diversity in our collection and mostly originated from soil samples (agricultural soil, orchard soil, potting soil, compost; 18 isolates), and the apple phyllosphere (flowers, leaves, fruits, bark; 19 isolates) (Table 1). In addition, one isolate each from irrigation water or a Drosophila species (collected in Wädenswil, Switzerland) was included. Finally, for comparison, a reference strain of Saccharomyces cerevisiae (BY4741) [59] was included. The antagonistic activity of these 40 yeasts against 16 fungal test strains (a broad selection of commonly isolated, pathogenic, antagonistic, or saprophytic filamentous fungi) (Table 1) was quantified by determining the relative growth of each filamentous fungus in the presence of each yeast (relative to the growth in the absence of yeasts) (Fig. 1; Additional file 1).
Fig. 1.
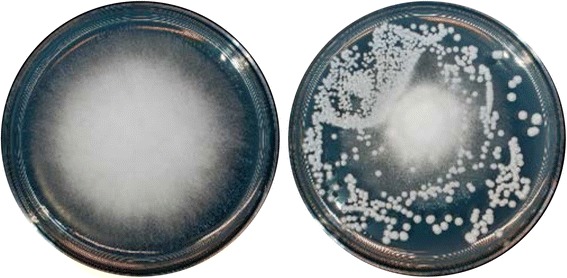
Example of the binary competition assay that was used to quantify the interactions of 40 yeast isolates with 16 filamentous test fungi. Competition assays were performed by quantifying the growth area of a filamentous fungus (e.g., the plant pathogen Gibberella fujikuroi BC 8.14) on control plates (left) and in the presence of a yeast isolate (e.g., C. subhashii, right). Overall, 640 competition assays were carried out and quantified (in quadruples)
All data were clustered based on the outcome of the pairwise interactions of all filamentous fungi with each yeast isolate (Fig. 2a). Overall, the majority of yeast isolates reduced the growth of filamentous fungi, but in a few interactions a small stimulatory effect of a yeast isolate was detected (Fig. 2a; Additional file 1). Based on their growth profiles in the presence of all 40 yeast isolates, the three Trichoderma isolates were clustered together with the two Mucor isolates, while nine plant pathogenic species (six Fusarium isolates, Alternaria eichhorniae, Mycosphaerella tassiana, Monilinia fructicola) formed a second, broad cluster (Fig. 2a). The growth profiles of R. solani and Metarhizium brunneum in the presence of yeasts strongly differed from each other and from all other filamentous fungi. Clustering of the different yeasts based on their effect on the growth of all 16 filamentous fungi lead to a clear separation of isolates obtained from the apple phyllosphere and those isolated from soil (Fig. 2a).
Fig. 2.
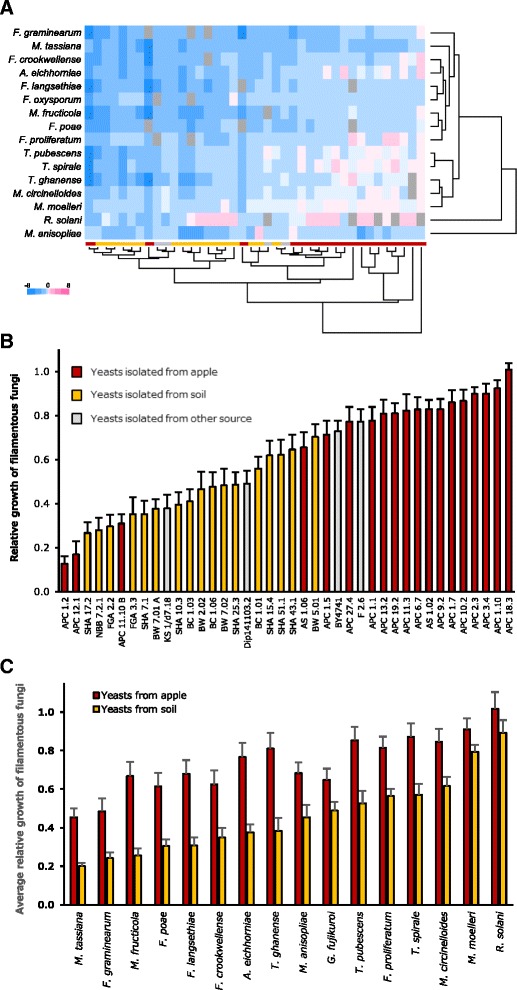
Binary competition assays identify strongly antagonistic yeasts with potential for biocontrol applications. a The average relative growth (four replicates) of 16 filamentous fungi in the presence of 40 different yeasts was log2-transformed and all data were clustered (correlation measure based distance (uncentered), complete linkage). Colours (see legend) range from strong inhibition (−8; dark blue), via no effect (white) to strong growth promotion (8; dark pink). Missing data are indicated by grey squares. b The overall average relative growth of filamentous fungi (over all 16 test strains used in this study) in the presence of each yeast isolate. The strain S. cerevisiae BY4741 is included as a reference. c The average relative growth of each filamentous fungus (average of relative growth in presence all apple or soil yeast isolates). Data obtained with yeasts that were isolated from the apple phyllosphere or from soil are marked in red and yellow, respectively
The overall average relative growth of filamentous fungi (over all 16 isolates used in this study) in the presence of each yeast isolate revealed a broad spectrum of responses (Fig. 2b). While, on average, some yeast isolates (e.g., APC 18.3) exhibited no detectable effect on filamentous fungi, others (e.g., APC 1.2) reduced their growth by more than 80%. The variance of this measure, for each yeast, was small and similar over the entire range of overall relative growth, suggesting that the average antagonistic activity, against a broad range of filamentous fungi, is an inherent property of a particular yeast isolate. The same effect was documented by ranking all 40 yeasts according to their effect on the relative growth of all 16 filamentous fungi (most antagonistic yeast ranked as “1”; least antagonistic isolated as “40”) (not shown). Based on both measures, the overall average relative growth and the average rank for all filamentous fungi, the same six yeast isolates were identified as having the highest antagonistic activity (APC 1.2: Metschnikowia pulcherrima, APC 12.1: Hanseniaspora sp., SHA 17.2: Cyberlindnera sargentensis, NBB 7.2.1: A. pullulans, FGA 2.2: C. subhashii, APC 10.11 B: Pichia kluyveri). In 10 interactions with filamentous fungi, M. pulcherrima (APC 1.2) was the most antagonistic yeast isolate among those tested here (average relative growth of 0.1, average rank of 1.9). Although the two most antagonistic yeast isolates were obtained from apple (APC 1.2 and APC 12.1), overall the results indicated weaker antagonism of yeasts isolated from apple as compared to the isolates obtained from soil samples (Fig. 2b). Comparing the average relative growth of each filamentous fungus in the presence of yeasts from soil (17 isolates) or from apple (19 isolates) documented this finding: as compared to the apple yeasts, soil yeasts more strongly reduced the growth of all tested filamentous fungi (Fig. 2c). The overall relative growth of the 16 filamentous fungi ranged from 0.3 to 0.9 (average 0.6) and above-ground plant pathogens (e.g., M. tassiana, F. gramineaurm, F. poae, M. fructicola, F. langsethiae, F. crookwellense, A. eichhorniae) were generally more sensitive to inhibition by yeasts than soil fungi (Fig. 2c). Fast-growing, saprophytic soil fungi such as Mucor circinelloides, Mucor moelleri, and the soil pathogen R. solani were least inhibited in their growth by yeasts.
Physiological characteristics of strongly antagonistic yeasts from soil or apple
The six overall strongest antagonists comprised three yeasts from apple (APC 1.2: M. pulcherrima; APC 12.1: Hanseniaspora sp.; APC 10.11 B: P. kluyveri) and soil each (SHA 17.2: C. sargentensis; NBB 7.2.1: A. pullulans; FGA 2.2: C. subhashii). In order to identify common and distinguishing characteristics that may affect the potential as biocontrol agents, these six most antagonistic yeasts were further characterized with respect to their growth requirements.
All six yeast isolates grew well at temperatures up to 30 °C and two isolates, one isolate each from apple and soil (APC 11.10 B: P. kluyveri; FGA 2.2: C. subhashii, respectively), were able to multiply at 37 °C (Fig. 3a). Microarray phenotype analysis, using the Biolog YT MicroPlateTM, revealed a broader metabolic versatility of the three soil yeasts as compared to the three isolates obtained from the apple phyllosphere (Fig. 3b). Most noteworthy were a number of di-, tri- and tetrasaccharides (e.g., maltose, melebiose, palatinose, sucrose, maltotriose, melezitose, raffinose, stachyose) that were assimilated and/or oxidized by at least one soil yeast, while none of these carbon sources were utilized by any of the three yeast isolates obtained from the apple phyllosphere. In particular the two yeasts A. pullulans (NBB 7.2.1) and C. subhashii (FGA 2.2) assimilated and/or oxidized a large number of compounds (34 and 20, respectively), including different acids (e.g., acetic, formic, aspartic, fumaric, malic acids) (Fig. 3b). In contrast, P. kluyveri (APC 11.10 B) only grew with glucose and M. pulcherrima (APC 1.2) and Hanseniaspora sp. (APC 12.1) only showed detectable growth with 9 and 11 carbon sources, respectively. Interestingly, however, the phyllosphere yeast M. pulcherrima, as well as C. subhashii and A. pullulans, were able to utilize N-acetyl-glucosamine (GlcNac), a component of bacterial and fungal cell walls and insect exoskeletons. The broad metabolic versatility observed, for example for A. pullulans, did not go along with the ability to grow with root exudates as the sole source of nutrients (Fig. 3c). Aureobasidium pullulans (NBB 7.2.1) and Hanseniaspora sp. (APC 12.1) did not grow solely in root exudates (0.1%). In contrast, P. kluyveri (APC 11.10 B), which grew only in the presence of glucose in the phenotype microarray analysis, was able to multiply in 0.1% (w/w) mung bean root exudate. Of the six yeast isolates tested here, the soil isolate SHA 17.2 of C. sargentensis grew best in root exudates.
Fig. 3.
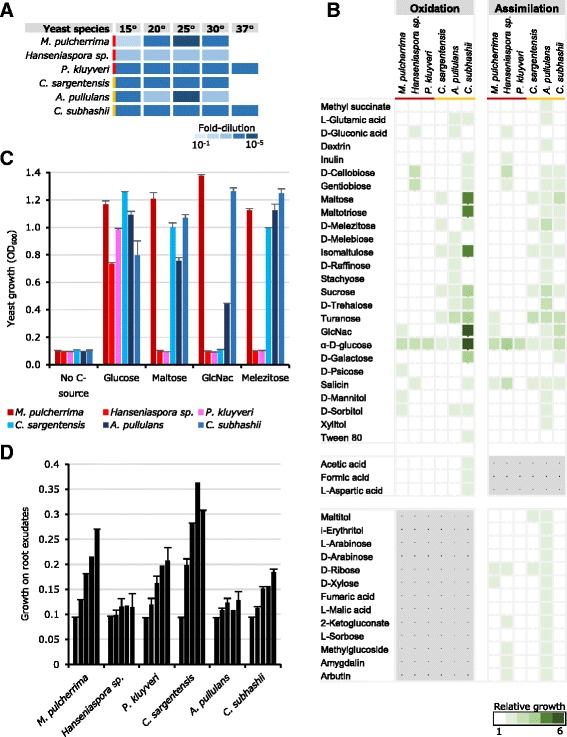
Physiological characteristics of strongly antagonistic yeasts. The six most strongly antagonistic yeasts were characterized by determining their growth at different temperatures (a), the assimilation and oxidation of different carbon sources (b) and the growth with selected sugars (c) or with root exudates (growth at days 0, 1, 2, 3, and 6 is depicted) (d). All experiments were repeated at least twice and the mean and standard errors are shown
Candida subhashii is an abundant soil fungus
One of the overall strongest antagonists was C. subhashii, a species that has previously only been described in a patient sample in Canada and was considered a human pathogen [45]. During our collection of fungal isolates from Swiss agricultural samples, C. subhashii was repeatedly isolated from agricultural soil and from commercially available potting substrates. In one of the latter, C. subhashii constituted approx. 50,000 CFU per gram of soil and was the fourth most frequent taxon based on ITS barcode sequencing (data not shown). To further confirm that the C. subhashii soil isolate was indeed the same species as the clinical isolate, the mitochondrial genome of the Swiss C. subhashii isolate was sequenced (available at NCBI under the accession number KX781248) and phylogenetic analyses were performed.
The mitochondrial genome sequence of the C. subhashii soil isolate FGA 2.2 was identical to the C. subhashii type strain (FR 392/CBS 10753), except that the former had an insertion of 135 bp in a non-coding region between two genes (bases 15,872 to 16,006 in the assembly of FGA 2.2). Consequently, the FGA 2.2 mitochondrial genome exhibited the same peculiarities as the corresponding genome of the type strain: exceptionally high GC content (52.7%), a lack of introns in coding sequences, and telomere-like termini of the linear molecules. A maximum likelihood phylogenetic tree of seven mitochondrial proteins (Atp6, Atp8, Atp9, Cob, Cox1, Cox2, Cox3) revealed the C. subhashii sequences as a group basal to the C. parapsilosis/C. albicans/C. tropicalis cluster, within the CTG clade. The CTG clade comprises the majority of Candida species and forms a monophyletic group of yeasts that exhibit a genetic code transition, causing the codon CTG to be translated as serine instead of leucine [60–62] (Fig. 4). Based on these results it was concluded that the two C. subhashii isolates indeed belong to the same species, are virtually identical despite the vastly different sources of origin, and that soil is a natural habitat of C. subhashii.
Fig. 4.
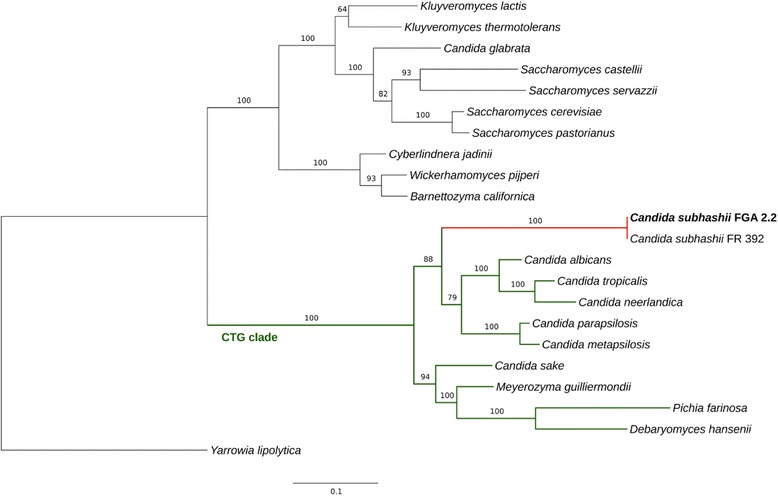
Maximum likelihood phylogeny constructed using a concatenated alignment of conserved mitochondrial protein sequences. The concatenated amino acid alignment of the conserved protein coding genes Atp6, Atp8, Atp9, Cob, Cox1, Cox2 and Cox3 was used to construct a phylogenetic tree. The isolate C. subhashii FGA 2.2 is shown in bold and the CTG clade is marked in green. Yarrowia lipolytica was used as the outgroup. As the concatenated protein sequences encoded by the seven genes of the previously published and the Swiss C. subhashii strains were identical, the branch length on the respective node was zero (in red). Bootstrap scores for all nodes are shown (percentage of 100 bootstrap runs). The bar represents the number of amino acid substitutions per site. Further information about the selected sequences is reported in Table 2
Discussion
Soil yeasts are generally more antagonistic and metabolically versatile than apple phyllosphere yeasts
Our competition experiments indicated that, on average and under the in vitro conditions tested here, yeasts isolated from soil suppress filamentous fungi more strongly than phyllosphere yeasts. This was the case irrespective of whether the filamentous fungus was isolated from soil or the phyllosphere, or if it was a pathogen or saprophyte. Furthermore, the comparison of three strongly antagonistic yeasts from soil and from the apple phyllosphere suggested a higher metabolic diversity of soil yeasts.
Due to rapidly fluctuating temperatures, low humidity, scarce nutrient availability, and UV irradiation, the phyllosphere is considered a harsh environment [63], but likely features a lower niche complexity as compared to soil. Consequently, interspecific competition between phyllosphere microorganisms is strong and favours the evolution of antagonistic activities to ward off competing microbes. Soil, in contrast, is a highly heterogeneous and rich habitat with a plethora of niches and thus hosts a complex microbiome [64]. In addition to environmental factors and interspecific competition, plants release root exudates and thereby also shape the microbial community in the rhizosphere [6, 65, 66]. The ability to metabolize root exudates may thus indicate adaptation of the corresponding yeast to soil. Indeed, soil yeasts were able to grow in the presence of various sugars and organic acids (e.g., maltose, sucrose, raffinose, acetic, formic, aspartic, fumaric, malic acids) that have been detected in root exudates of higher plants [67], while the tested, strongly antagonistic phyllosphere yeasts were unable to utilize these substrates. Nevertheless, the two phyllosphere yeasts M. pulcherrima and P. kluyveri were both able to multiply in root exudates, suggesting that on one hand plants likely release factors that allow these species to grow and that on the other hand M. pulcherrima and P. kluyveri may have the potential to colonize the rhizosphere, even though they were usually isolated from the phyllosphere. This finding is particularly relevant with respect to potential biocontrol applications against soilborne fungal pathogens, where rhizosphere competence is a factor that can contribute to a successful control [68–70].
Binary competition assays identify strongly antagonistic yeasts with potential for biocontrol
The dual culture assays employed here revealed the antagonistic activity of 40 yeast isolates against 16 filamentous fungi. The level of inhibition ranged from no effect at all (even slight stimulatory activities were detected in some interactions) to a growth reduction of more than 80% as compared to growth on the control plates (in the absence of yeasts). The most strongly inhibitory yeasts were M. pulcherrima, Hanseniaspora sp., C. sargentensis, A. pullulans, C. subhashii, and P. kluyveri. Except for C. subhashii, these species, or close relatives thereof, are known antagonists and have been implicated in the biological control of plant pathogenic fungi in the past. The general nature of the antagonistic activity observed under the experimental conditions used here suggests that yeasts inhibited filamentous fungi based on their strong competitiveness for micro- and/or macro-nutrients or due to indirect effects, which is an advantageous property for a potential biocontrol agent. Further studies will have to reveal the mode of antagonism and to decipher the contribution of competition, indirect effects of metabolites, or specific antagonistic factors, in each interaction, in more detail.
In the experiments described here, M. pulcherrima was the overall most strongly antagonistic yeast. Pulcherrimin, an iron-binding pigment produced by M. pulcherrima, is believed to mediate this antagonistic activity against other fungi [71–74]. In the past, M. pulcherrima has been studied as an antagonist of fruit rot diseases (for example caused by Alternaria alternata, B. cinerea, P. expansum) [31, 75, 76] and a related species, M. fructicola, is being used for postharvest biocontrol applications against storage diseases of sweet potatoes and carrots [77]. A strong antagonistic activity against soilborne fungal pathogens and species of Fusarium has not been reported. Hanseniaspora species are widespread and frequent in the environment, mostly studied with respect to their occurrence on grapes and winemaking, and their antagonistic activity against green mould of citrus or B. cinerea was shown [78–82]. Cyberlindnera sargentensis (synonym Williopsis sargentensis) belongs to a genus of yeasts that have been shown to promote plant growth, produce volatile sulphur compounds, and kill other fungi or bacteria via killer proteins [83–88]. The basidiomycetous yeast A. pullulans is a cosmopolitan species that is used in biotechnology and acts as an antagonist against fungal and bacterial plant pathogens such as postharvest diseases or fire blight [25, 89–95]. Pichia kluyveri and related species (e.g., Wickerhamomyces anomalus, P. fermentans, etc.) are widely studied with respect to wine fermentation as well as biological control, mostly of fungal postharvest diseases of fruits [30, 96–101].
Besides the identification of known antagonists (as well as at least one new antagonist; C. subhashii), this study also identified soilborne pathogens and several Fusarium species as new, potential targets of antagonistic yeasts. The results presented also suggest that yeast antagonism is an isolate-/species-specific property and little dependent on the target organism: a strongly antagonistic yeast exhibits this activity against a broad range of fungi. This finding has important implications for using and studying such yeasts with respect to their application in biocontrol. For example, it may be more promising to optimize the activity of demonstrably strong antagonists than isolating “new” antagonists for each pathogenic fungus to be controlled. An intriguing possibility for optimizing the activity of biocontrol organisms are communities of compatible strains that may achieve better control of plant pathogens than single strains. Initial experiments with mixtures of weakly antagonistic yeasts show that such synergistic effects can indeed be observed (data not shown). With respect to research, these results emphasize the need to study and reveal modes of antagonism that will enable translating strong antagonistic activity in the laboratory to an effective and reliable control in the field. Reliable, biological assays, but also 3rd generation DNA sequencing technologies and bioinformatics tools that have become available as of late, are the foundations for characterizing potential biocontrol strains and for identifying modes of antagonism goal-oriented and rapidly.
Candida subhashii is an antagonistic soil fungus
Among the strongly antagonistic yeasts, C. subhashii was the least studied species and not described as an antagonist of saprophytic and pathogenic, filamentous fungi. In fact, C. subhashii was considered a human pathogen because it has been isolated from a patient sample [45]. Nevertheless, it must be noted that only one case report of a C. subhashii infection exists: a patient on a long-term peritoneal dialysis treatment developed a peritonitis that was ascribed to a C. subhashii infection and successfully treated with fluconazole, ampicillin, and amoxicillin [45]. Whether or not C. subhashii can indeed colonise and cause symptoms in a healthy mammalian host has not been tested. The thermotolerance (growth at 37 °C) of C. subhashii, also observed for the isolates described here and for an isolate similar to C. subhashii described from Japan [46], is a requirement for human pathogenicity, but many isolates exhibiting this property, more frequently found within the Ascomycota than the Basidiomycota, have not yet been described as mammalian, let alone human pathogens [102].
Here, C. subhashii was repeatedly isolated from soil samples and from commercially available potting substrates, where it occurred in large concentrations (among the most frequent fungi in potting substrate: approx. 50,000 CFU per gram of substrate, the fourth most frequent taxon based on ITS barcode sequencing (data not shown)). In addition, C. subhashii was highly competitive against different soil fungi, metabolized carbohydrates commonly found in the rhizosphere, and grew in root exudates as well as on roots and in soil. The metabolic profile of the Swiss C. subhashii isolate FGA 2.2 was comparable to the one of Candida sp. NY7122, a pentose-fermenting soil yeast that is similar to C. subhashii and that was isolated from a Japanese soil. However, the latter isolate was able to assimilate L-arabinose and D-xylose [46], which was not the case for C. subhashii FGA 2.2 under the conditions tested here. Based on these results it was concluded that soil is the natural habitat for C. subhashii, where this species is a common and competitive organism. Specifically, the particular large number of C. subhashii cells in potting substrate, comprised of white and black peat (of European origin), suggests that either or both of these components are a natural reservoir of this antagonistic soil yeast.
The extremely high similarity of the mitochondrial genomes of the Swiss and clinical (Canadian) C. subhashii isolates is surprising and unexpected, particularly when considering the vastly different origins of the two isolates. However, identical or almost identical mitochondrial genomes have also been reported, for example, in Penicillium isolates from Spain and China, respectively, and may indicate a rapid, global spread of one particular isolate of C. subhashii [103]. On the other hand, the identical mitochondrial genomes are in contrast to studies reporting considerable intra-species variation in size, intron content, and recombination in fungal mitochondrial genomes [104–106]. At present, it is not clear why the C. subhashii mitochondrial genome is so conserved and future studies will have to address the conservation and evolution of the mitochondrial genome in more detail as well as reveal the entire genome sequence of C subhashii as a basis for identifying genes mediating antagonistic functions.
Conclusions
The work presented here combines a broad screening of the antagonistic activity of naturally occurring yeasts against saprophytic and pathogenic filamentous fungi with growth analyses to compare the metabolic potential of the most antagonistic yeasts. Among the most strongly antagonistic yeasts were M. pulcherrima, A. pullulans, Hanseniaspora sp., C. sargentensis, P. kluyveri and C. subhashii. Competition assays indicated that the antagonistic activity of yeasts is an inherent property of particular yeast isolates and species and little dependent on the interacting filamentous fungus. Among the strongly antagonistic yeasts, soil yeasts were generally more antagonistic and metabolically versatile as compared to yeasts isolated from the phyllosphere. The identification of C. subhashii as a strongly antagonistic soil yeast is particularly noteworthy, because previously the natural habitat of this species was unknown and it was described, in one publication, as a human pathogen. The results presented here thus define C. subhashii as a common and competitive soil yeast.
Acknowledgements
Kenneth H. Wolfe and Geraldine Butler are greatly acknowledged for the annotation of the C. subhashii mitochondrial genome and for helpful comments during the planning and preparation of this manuscript. Ina Schlathölter and Daniel Prata provided experimental support. Interesting discussions and conceptual input by Jürg E. Frey, Eduard Holliger and Mitja Remus-Emsermann contributed to and improved this work. Susanne Vogelgsang, Klaus Marschall, and Matthias Lutz provided fungal strains that were used in this study.
Funding
This research was supported by Agroscope.
Availability of data and material
All strains obtained in the course of this study are deposited at the Culture Collection of Switzerland (CCOS; https://www.ccos.ch) and the corresponding accession numbers are listed in Table 1 of this manuscript. The sequence of the Swiss C. subhashii isolate FGA 2.2 is deposited at NCBI under the accession number KX781248.
Authors’ contributions
MHB and FMF performed the experimental work. FMF planned the study and wrote the manuscript. MS and CHA sequenced and assembled the C. subhashii mitochondrial genome and wrote the corresponding sections of the manuscript. All authors have read the final manuscript.
Competing interests
The authors declare that they have no competing interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional file
The average relative growth (four replicates) of all binary competition experiments (16 filamentous fungi in the presence of 40 different yeasts) performed in the course of this study. (XLSX 21 kb)
Contributor Information
Maja Hilber-Bodmer, Email: maja.hilber-bodmer@agroscope.admin.ch.
Michael Schmid, Email: michael.schmid@agroscope.admin.ch.
Christian H. Ahrens, Email: christian.ahrens@agroscope.admin.ch
Florian M. Freimoser, Email: florian.freimoser@agroscope.admin.ch
References
- 1.Koike ST, Subbarao KV, Davis RM, Turini TA. Vegetable diseases caused by soilborne pathogens. University of California ANR Publications; 2003. 8099
- 2.Baumgartner K, Coetzee MP, Hoffmeister D. Secrets of the subterranean pathosystem of Armillaria. Mol Plant Pathol. 2011;12(6):515–534. doi: 10.1111/j.1364-3703.2010.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison WR, Tuell JK, Hausbeck MK, Szendrei Z. Constraints on Asparagus production: the association of Ophiomyia simplex (Diptera: Agromyzidae) and Fusarium spp. Crop Sci. 2011;51:1414–1423. doi: 10.2135/cropsci2011.01.0032. [DOI] [Google Scholar]
- 4.Wu H-S, Gao Z-G, Zhou X-D, Shi XB, Wang M-Y, Shang X-X, Liu Y-D, Gu D-L, Wang W-Z. Microbial dynamics and natural remediation patterns of Fusarium-infested watermelon soil under 3-yr of continuous fallow condition. Soil Use Manag. 2013;29(2):220–229. doi: 10.1111/sum.12029. [DOI] [Google Scholar]
- 5.Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fert Soils. 2012;48(5):489–499. doi: 10.1007/s00374-012-0691-4. [DOI] [Google Scholar]
- 6.Lakshmanan V, Selvaraj G, Bais HP. Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol. 2014;166(2):689–700. doi: 10.1104/pp.114.245811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel R. The metagenomics of soil. Nat Rev Microbiol. 2005;3(6):470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- 9.Schnitzer SA, Klironomos JN, Hillerislambers J, Kinkel LL, Reich PB, Xiao K, Rillig MC, Sikes BA, Callaway RM, Mangan SA, et al. Soil microbes drive the classic plant diversity-productivity pattern. Ecology. 2011;92(2):296–303. doi: 10.1890/10-0773.1. [DOI] [PubMed] [Google Scholar]
- 10.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37(5):634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 11.Broadfoot M. Microbes added to seeds could boost crop production. Scientific American 2016 (January). https://www.scientificamerican.com/article/microbes-added-to-seeds-could-boost-crop-production/.
- 12.Reid A, Greene SE. How microbes can help feed the world. Report from the American Academy of Microbiology 2013 [PubMed]
- 13.Quiza L, St-Arnaud M, Yergeau E. Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front Plant Sci. 2015;6:507. doi: 10.3389/fpls.2015.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vrieze M, Pandey P, Bucheli TD, Varadarajan AR, Ahrens CH, Weisskopf L, Bailly A. Volatile organic compounds from native potato-associated Pseudomonas as potential anti-oomycete Agentsa. Front Microbiol. 2015;6:1295. doi: 10.3389/fmicb.2015.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner TR, James EK, Poole PS. The plant microbiome. Genome Biol. 2013;14(6):209. [DOI] [PMC free article] [PubMed]
- 16.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17(8):478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Freimoser FM, Pelludat C, Remus-Emsermann MN. Tritagonist as a new term for uncharacterised microorganisms in environmental systems. ISME J. 2016;10(1):1–3. doi: 10.1038/ismej.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzzini P, Branda E, Goretti M, Turchetti B. Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential. FEMS Microbiol Ecol. 2012;82(2):217–241. doi: 10.1111/j.1574-6941.2012.01348.x. [DOI] [PubMed] [Google Scholar]
- 19.Lai X, Cao L, Tan H, Fang S, Huang Y, Zhou S. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007;1(8):756–762. doi: 10.1038/ismej.2007.51. [DOI] [PubMed] [Google Scholar]
- 20.Bass D, Howe A, Brown N, Barton H, Demidova M, Michelle H, Li L, Sanders H, Watkinson SC, Willcock S, et al. Yeast forms dominate fungal diversity in the deep oceans. Proc Biol Sci. 2007;274(1629):3069–3077. doi: 10.1098/rspb.2007.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rikhvanov EG, Varakina NN, Sozinov DY, Voinikov VK. Association of bacteria and yeasts in hot springs. Appl Environ Microbiol. 1999;65(9):4292–4293. doi: 10.1128/aem.65.9.4292-4293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams KM, Liu P, Fay JC. Evolution of ecological dominance of yeast species in high-sugar environments. Evolution. 2015;69(8):2079–2093. doi: 10.1111/evo.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oro L, Ciani M, Comitini F. Yeasts from xerophilic environments reveal antimicrobial action against fruit pathogenic molds. J Food Safety. 2015;36(1):100–108. doi: 10.1111/jfs.12217. [DOI] [Google Scholar]
- 24.Cantrell SA, Dianese JC, Fell J, Gunde-Cimerman N, Zalar P. Unusual fungal niches. Mycologia. 2011;103(6):1161–1174. doi: 10.3852/11-108. [DOI] [PubMed] [Google Scholar]
- 25.Seibold A, Viehrig M, Jelkmann W. Yeasts as antagonists against Erwinia amylovora. Acta Hortic. 2006;704:367–369. doi: 10.17660/ActaHortic.2006.704.56. [DOI] [Google Scholar]
- 26.Punja ZK, Utkhede RS. Using fungi and yeasts to manage vegetable crop diseases. Trends Biotechnol. 2003;21(9):400–407. doi: 10.1016/S0167-7799(03)00193-8. [DOI] [PubMed] [Google Scholar]
- 27.Pusey PL, Stockwell VO, Mazzola M. Epiphytic bacteria and yeasts on apple blossoms and their potential as antagonists of Erwinia amylovora. Phytopathology. 2009;99(5):571–581. doi: 10.1094/PHYTO-99-5-0571. [DOI] [PubMed] [Google Scholar]
- 28.Sharma RR, Singh D, Singh R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: A review. Biol Control. 2009;50(3):205–221. doi: 10.1016/j.biocontrol.2009.05.001. [DOI] [Google Scholar]
- 29.Liu J, Sui Y, Wisniewski M, Droby S, Liu Y. Review: Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int J Food Microbiol. 2013;167(2):153–160. doi: 10.1016/j.ijfoodmicro.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Schnurer J, Jonsson A. Pichia anomala J121: a 30-year overnight near success biopreservation story. Antonie Van Leeuwenhoek. 2011;99(1):5–12. doi: 10.1007/s10482-010-9509-2. [DOI] [PubMed] [Google Scholar]
- 31.Parafati L, Vitale A, Restuccia C, Cirvilleri G. Biocontrol ability and action mechanism of food-isolated yeast strains against Botrytis cinerea causing post-harvest bunch rot of table grape. Food Microbiol. 2015;47:85–92. doi: 10.1016/j.fm.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Sundh I, Melin P. Safety and regulation of yeasts used for biocontrol or biopreservation in the food or feed chain. Antonie Van Leeuwenhoek. 2011;99(1):113–119. doi: 10.1007/s10482-010-9528-z. [DOI] [PubMed] [Google Scholar]
- 33.Bar-Shimon M, Yehuda H, Cohen L, Weiss B, Kobeshnikov A, Daus A, Goldway M, Wisniewski M, Droby S. Characterization of extracellular lytic enzymes produced by the yeast biocontrol agent Candida oleophila. Curr Genet. 2004;45(3):140–148. doi: 10.1007/s00294-003-0471-7. [DOI] [PubMed] [Google Scholar]
- 34.Droby S, Vinokur V, Weiss B, Cohen L, Daus A, Goldschmidt EE, Porat R. Induction of resistance to Penicillium digitatum in grapefruit by the yeast biocontrol agent Candida oleophila. Phytopathology. 2002;92(4):393–399. doi: 10.1094/PHYTO.2002.92.4.393. [DOI] [PubMed] [Google Scholar]
- 35.Lahlali R, Serrhini MN, Jijakli MH. Efficacy assessment of Candida oleophila (strain O) and Pichia anomala (strain K) against major postharvest diseases of citrus fruits in Morocco. Commun Agric Appl Biol Sci. 2004;69(4):601–609. [PubMed] [Google Scholar]
- 36.Lima G, Ippolito A, Nigro F, Salerno M. Effectiveness of Aureobasidium pullulans and Candida oleophila against postharvest strawberry rots. Postharvest Biol Tec. 1997;10(2):169–178. doi: 10.1016/S0925-5214(96)01302-6. [DOI] [Google Scholar]
- 37.Calvo-Garrido C, Vinas I, Elmer P, Usall J, Teixido N. Candida sake CPA-1 and other biologically based products as potential control strategies to reduce sour rot of grapes. Lett Appl Microbiol. 2013;57(4):356–361. doi: 10.1111/lam.12121. [DOI] [PubMed] [Google Scholar]
- 38.El-Tarabily KA. Suppression of Rhizoctonia solani diseases of sugar beet by antagonistic and plant growth-promoting yeasts. J Appl Microbiol. 2004;96(1):69–75. doi: 10.1046/j.1365-2672.2003.02043.x. [DOI] [PubMed] [Google Scholar]
- 39.El-Mehlawy AA. The rhizosphere yeast fungi as biocontrol agents for wild disease of kidney bena caused by Fusarium oxysporum. Int J Agricult Biol. 2004;6(2):310–316. [Google Scholar]
- 40.Shalaby ME-S, El-Nady MF. Application of Saccharomyces cerevisiae as a biocontrol agent against Fusarium infection of sugar beet plants. Acta Biologica Szegediensis. 2008;52(2):271–275. [Google Scholar]
- 41.Eshel D, Regev R, Orenstein J, Droby S, Gan-Mor S. Combining physical, chemical and biological methods for synergistic control of postharvest diseases: a case study of black root rot of carrot. Postharvest Biol Tec. 2009;54(1):48–52. doi: 10.1016/j.postharvbio.2009.04.011. [DOI] [Google Scholar]
- 42.Papon N, Savini V, Lanoue A, Simkin AJ, Creche J, Giglioli-Guivarc’h N, Clastre M, Courdavault V, Sibirny AA. Candida guilliermondii: biotechnological applications, perspectives for biological control, emerging clinical importance and recent advances in genetics. Curr Genet. 2013;59(3):73–90. doi: 10.1007/s00294-013-0391-0. [DOI] [PubMed] [Google Scholar]
- 43.Sharma N. Biological control for preventing food deterioration: strategies for pre- and postharvest management. John Wiley & Sons; 2014
- 44.Sundh I, editor. Beneficial microorganisms in agriculture, food and the environment: safety assessment and regulation. CABI; 2012.
- 45.Adam H, Groenewald M, Mohan S, Richardson S, Bunn U, Gibas CF, Poutanen S, Sigler L. Identification of a new species, Candida subhashii, as a cause of peritonitis. Med Mycol. 2009;47(3):305–311. doi: 10.1080/13693780802380545. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe I, Ando A, Nakamura T. Characterization of Candida sp. NY7122, a novel pentose-fermenting soil yeast. J Ind Microbiol Biotechnol. 2012;39(2):307–315. doi: 10.1007/s10295-011-1033-5. [DOI] [PubMed] [Google Scholar]
- 47.Fricova D, Valach M, Farkas Z, Pfeiffer I, Kucsera J, Tomaska L, Nosek J. The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology. 2010;156(Pt 7):2153–2163. doi: 10.1099/mic.0.038646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mian MA, Fleet GH, Hocking AD. Effect of diluent type on viability of yeasts enumerated from foods or pure culture. Int J Food Microbiol. 1997;35(2):103–107. doi: 10.1016/S0168-1605(96)01208-1. [DOI] [PubMed] [Google Scholar]
- 49.Freimoser FM, Hilber-Bodmer M, Brunisholz R, Drissner D. Direct identification of Monilinia brown rot fungi on infected fruits by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. Chem Biol Technol Agricult. 2016;3:7. doi: 10.1186/s40538-016-0058-4. [DOI] [Google Scholar]
- 50.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 51.White TJ, Bruns TD, Lees S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 52.Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22(21):5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 53.Abarenkov K, Nilsson RH, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, et al. The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytol. 2010;186(2):281–285. doi: 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- 54.DeNittis M, Querol A, Zanoni B, Minati JL, Ambrosoli R. Possible use of Biolog methodology for monitoring yeast presence in alcoholic fermentation for wine-making. J Appl Microbiol. 2010;108(4):1199–1206. doi: 10.1111/j.1365-2672.2009.04547.x. [DOI] [PubMed] [Google Scholar]
- 55.Barbour WM, Hattermann DR, Stacey G. Chemotaxis of Bradyrhizobium japonicum to soybean exudates. Appl Environ Microbiol. 1991;57(9):2635–2639. doi: 10.1128/aem.57.9.2635-2639.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 57.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 60.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gabaldon T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, Lespinet O, Arnaise S, Boisnard S, Aguileta G, Atanasova R, et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14:623. [DOI] [PMC free article] [PubMed]
- 62.Massey SE, Moura G, Beltrao P, Almeida R, Garey JR, Tuite MF, Santos MA. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 2003;13(4):544–557. doi: 10.1101/gr.811003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindow SE, Leveau JH. Phyllosphere microbiology. Curr Opin Biotech. 2002;13(3):238–243. doi: 10.1016/S0958-1669(02)00313-0. [DOI] [PubMed] [Google Scholar]
- 64.Garbeva P, van Veen JA, van Elsas JD. Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- 65.Zheng Y, Chen L, Luo CY, Zhang ZH, Wang SP, Guo LD. Plant identity exerts stronger effect than fertilization on soil arbuscular mycorrhizal fungi in a sown pasture. Microb Ecol 2016 [DOI] [PubMed]
- 66.Chapelle E, Mendes R, Bakker PA, Raaijmakers JM. Fungal invasion of the rhizosphere microbiome. ISME J. 2016;10(1):265–268. doi: 10.1038/ismej.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neumann G, Römheld V. The release of root exudates as affected by the plant physiological status. In: Pinton R, Varanini Z, Nannipieri Z, editors. The rhizosphere: Biochemistry and organic substances at the soil-plant interface. Marcel Dekker; 2000.
- 68.Schreiter S, Sandmann M, Smalla K, Grosch R. Soil type dependent rhizosphere competence and biocontrol of two bacterial inoculant strains and their effects on the rhizosphere microbial community of field-grown lettuce. PLoS One. 2014;9(8):e103726. doi: 10.1371/journal.pone.0103726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whipps JM. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot. 2001;52(Spec Issue):487–511. doi: 10.1093/jexbot/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 70.Ghirardi S, Dessaint F, Mazurier S, Corberand T, Raaijmakers JM, Meyer JM, Dessaux Y, Lemanceau P. Identification of traits shared by rhizosphere-competent strains of fluorescent pseudomonads. Microb Ecol. 2012;64(3):725–737. doi: 10.1007/s00248-012-0065-3. [DOI] [PubMed] [Google Scholar]
- 71.Saravanakumar D, Clavorella A, Spadaro D, Garibaldi A, Gullino ML. Metschnikowia pulcherrima strain MACH1 outcompetes Botrytis cinerea, Alternaria alternata and Penicillium expansum in apples through iron depletion. Postharvest Biol Tec. 2008;49(1):121–128. doi: 10.1016/j.postharvbio.2007.11.006. [DOI] [Google Scholar]
- 72.Sipiczki M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl Environ Microb. 2006;72(10):6716–6724. doi: 10.1128/AEM.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spadaro D, Vola R, Piano S, Gullino ML. Mechanisms of action and efficacy of four isolates of the yeast Metschnikowia pulcherrima active against postharvest pathogens on apples. Postharvest Biol Tec. 2002;24(2):123–134. doi: 10.1016/S0925-5214(01)00172-7. [DOI] [Google Scholar]
- 74.Turkel S, Ener B. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z Naturforsch C. 2009;64(5-6):405–410. doi: 10.1515/znc-2009-5-618. [DOI] [PubMed] [Google Scholar]
- 75.Piano S, Neyrotti V, Migheli Q, Gullino ML. Biocontrol capability of Metschnikowia pulcherrima against Botrytis postharvest rot of apple. Postharvest Biol Tec. 1997;11(3):131–140. doi: 10.1016/S0925-5214(97)00022-7. [DOI] [Google Scholar]
- 76.Kinay P, Yildiz M. The shelf life and effectiveness of granular formulations of Metschnikowia pulcherrima and Pichia guilliermondii yeast isolates that control postharvest decay of citrus fruit. Biol Control. 2008;45(3):433–440. doi: 10.1016/j.biocontrol.2008.03.001. [DOI] [Google Scholar]
- 77.Droby S, Wisniewski M, Macarisin D, Wilson C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol Tec. 2009;52(2):137–145. doi: 10.1016/j.postharvbio.2008.11.009. [DOI] [Google Scholar]
- 78.Taqarort N, Echairi A, Chaussod R, Nouaim R, Boubaker H, Benaoumar AA, Boudyach E. Screening and identification of epiphytic yeasts with potential for biological control of green mold of citrus fruits. World J Microbiol Biotechnol. 2008;24:3031. doi: 10.1007/s11274-008-9849-5. [DOI] [Google Scholar]
- 79.Liu HM, Guo JH, Cheng YJ, Liu P, Long CA, Deng BX. Inhibitory activity of tea polyphenol and Hanseniaspora uvarum against Botrytis cinerea infections. Lett Appl Microbiol. 2010;51(3):258–263. doi: 10.1111/j.1472-765X.2010.02888.x. [DOI] [PubMed] [Google Scholar]
- 80.Albertin W, Setati ME, Miot-Sertier C, Mostert TT, Colonna-Ceccaldi B, Coulon J, Girard P, Moine V, Pillet M, Salin F, et al. Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering. Front Microbiol. 2015;6:1569. doi: 10.3389/fmicb.2015.01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruiz-Moyano S, Martin A, Villalobos MC, Calle A, Serradilla MJ, Cordoba MG, Hernandez A. Yeasts isolated from figs (Ficus carica L.) as biocontrol agents of postharvest fruit diseases. Food Microbiol. 2016;57:45–53. doi: 10.1016/j.fm.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 82.Cadez N, Raspor P, de Cock AW, Boekhout T, Smith MT. Molecular identification and genetic diversity within species of the genera Hanseniaspora and Kloeckera. FEMS Yeast Res. 2002;1(4):279–289. doi: 10.1111/j.1567-1364.2002.tb00046.x. [DOI] [PubMed] [Google Scholar]
- 83.Goretti M, Turchetti B, Buratta M, Branda E, Corazzi L, Vaughan-Martini A, Buzzini P. In vitro antimycotic activity of a Williopsis saturnus killer protein against food spoilage yeasts. Int J Food Microbiol. 2009;131(2-3):178–182. doi: 10.1016/j.ijfoodmicro.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 84.Nassar AH, El-Tarabily KA, Sivasithamparam K. Promotion of plant growth by an auxin-producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biol Fertil Soils. 2005;42:97–108. doi: 10.1007/s00374-005-0008-y. [DOI] [Google Scholar]
- 85.Tan AW, Lee PR, Seow YX, Ong PK, Liu SQ. Volatile sulphur compounds and pathways of L-methionine catabolism in Williopsis yeasts. Appl Microbiol Biotechnol. 2012;95(4):1011–1020. doi: 10.1007/s00253-012-3963-x. [DOI] [PubMed] [Google Scholar]
- 86.Hodgson VJ, Button D, Walker GM. Anti-Candida activity of a novel killer toxin from the yeast Williopsis mrakii. Microbiology. 1995;141(8):2003–2012. doi: 10.1099/13500872-141-8-2003. [DOI] [PubMed] [Google Scholar]
- 87.Minter DW. Cyberlindnera, a replaement name for Lindnera Kurtzman et al., nom. illegit. Mycotaxon. 2009;110:473–476. doi: 10.5248/110.473. [DOI] [Google Scholar]
- 88.Walker GM, McLeod AH, Hodgson VJ. Interactions between killer yeasts and pathogenic fungi. FEMS Microbiol Lett. 1995;127(3):213–222. doi: 10.1111/j.1574-6968.1995.tb07476.x. [DOI] [PubMed] [Google Scholar]
- 89.Mari M, Martini C, Guidarelli M, Neri F. Postharvest biocontrol of Monilinia laxa, Monilinia fructicola and Monilinia fructigena on stone fruit by two Aureobasidium pullulans strains. Biol Control. 2012;60(2):132–140. doi: 10.1016/j.biocontrol.2011.10.013. [DOI] [Google Scholar]
- 90.Mari M, Martini C, Spadoni A, Rouissi W, Bertolini P. Biocontrol of apple postharvest decay by Aureobasidium pullulans. Postharvest Biol Tec. 2012;73:56–62. doi: 10.1016/j.postharvbio.2012.05.014. [DOI] [Google Scholar]
- 91.Chi Z, Wang F, Chi Z, Yue L, Liu G, Zhang T. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl Microbiol Biotechnol. 2009;82(5):793–804. doi: 10.1007/s00253-009-1882-2. [DOI] [PubMed] [Google Scholar]
- 92.Mounir R, Durieux A, Bodo E, Allard C, Simon JP, Achbani EH, El-Jaafari S, Douira A, Jijakli MH. Production, formulation and antagonistic activity of the biocontrol like-yeast Aureobasidium pullulans against Penicillium expansum. Biotechnol Lett. 2007;29(4):553–559. doi: 10.1007/s10529-006-9269-2. [DOI] [PubMed] [Google Scholar]
- 93.Castoria R, de Curtis F, Lima G, Caputo E, Pacifico S, de Cicco V. Aureobasidium pullulans (LS30) an antagonist of postharvest pathogens of fruits: study on its modes of action. Postharvest Biol Technol. 2001;22:7–17. doi: 10.1016/S0925-5214(00)00186-1. [DOI] [Google Scholar]
- 94.Leibinger W, Breuker B, Hahn M, Mendgen K. Control of postharvest pathogens and colonization of the apple surface by antagonistic microogranisms in the field. Phytophathology. 1997;87:1103–1110. doi: 10.1094/PHYTO.1997.87.11.1103. [DOI] [PubMed] [Google Scholar]
- 95.Kunz S, Haug P. 12th International Conference on Cultivation Technique and Phytopathological Probelm in Organic Fruit-Growing. Weinsberg: Fördergemeinschaft ökologischer Obstbau; 2006. Development of a strategy for fire blight control in organic fruit growing; pp. 113–117. [Google Scholar]
- 96.Buzzini P, Martini A. Large-scale screening of selected Candida maltosa, Debaryomyces hansenii and Pichia anomala killer toxin activity against pathogenic yeasts. Med Mycol. 2001;39(6):479–482. doi: 10.1080/mmy.39.6.479.482. [DOI] [PubMed] [Google Scholar]
- 97.Giobbe S, Marceddu S, Scherm B, Zara G, Mazzarello VL, Budroni M, Migheli Q. The strange case of a biofilm-forming strain of Pichia fermentans, which controls Monilinia brown rot on apple but is pathogenic on peach fruit. FEMS Yeast Res. 2007;7(8):1389–1398. doi: 10.1111/j.1567-1364.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- 98.Haissam JM. Pichia anomala in biocontrol for apples: 20 years of fundamental research and practical applications. Antonie Van Leeuwenhoek. 2011;99(1):93–105. doi: 10.1007/s10482-010-9541-2. [DOI] [PubMed] [Google Scholar]
- 99.Masneuf-Pomarede I, Bely M, Marullo P, Albertin W. The genetics of non-conventional wine yeasts: current knowledge and future challenges. Front Microbiol. 2015;6:1563. doi: 10.3389/fmicb.2015.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jolly NP, Varela C, Pretorius IS. Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014;14(2):215–237. doi: 10.1111/1567-1364.12111. [DOI] [PubMed] [Google Scholar]
- 101.Restuccia C, Giusino F, Licciardello F, Randazzo C, Caggia C, Muratore G. Biological control of peach fungal pathogens by commercial products and indigenous yeasts. J Food Prot. 2006;69(10):2465–2470. doi: 10.4315/0362-028X-69.10.2465. [DOI] [PubMed] [Google Scholar]
- 102.Robert V, Cardinali G, Casadevall A. Distribution and impact of yeast thermal tolerance permissive for mammalian infection. BMC Biol. 2015;13:18. doi: 10.1186/s12915-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marcet-Houben M, Ballester AR, de la Fuente B, Harries E, Marcos JF, Gonzalez-Candelas L, Gabaldon T. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics. 2012;13:646. doi: 10.1186/1471-2164-13-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jung PP, Friedrich A, Reisser C, Hou J, Schacherer J. Mitochondrial genome evolution in a single protoploid yeast species. G3 (Bethesda) 2012;2(9):1103–1111. doi: 10.1534/g3.112.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fritsch ES, Chabbert CD, Klaus B, Steinmetz LM. A genome-wide map of mitochondrial DNA recombination in yeast. Genetics. 2014;198(2):755–771. doi: 10.1534/genetics.114.166637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Torriani SF, Penselin D, Knogge W, Felder M, Taudien S, Platzer M, McDonald BA, Brunner PC. Comparative analysis of mitochondrial genomes from closely related Rhynchosporium species reveals extensive intron invasion. Fungal Genet Biol. 2014;62:34–42. doi: 10.1016/j.fgb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 107.Tamm L, Thürig B, Bruns C, Fuchs JG, Köpke U, Laustela M, Leifert C, Mahlberg N, Nietlispach B, Schmidt C, et al. Soil type, management history, and soil amendments influence the development of soil-borne (Rhizoctonia solani, Pythium ultimum) and air-borne (Phytophthora infestans, Hyaloperonospora parasitica) diseases. Eur J Plant Pathol. 2010;127:465–481. doi: 10.1007/s10658-010-9612-2. [DOI] [Google Scholar]


