Abstract
Background
Our previous study reported that MEG3 is an important tumor suppressor gene that is inactivated in cervical cancer. However, the diagnostic and prognostic values of MEG3, as well as the molecular mechanism of MEG3 inactivation in cervical cancer, remain unclear. In this study, we aimed to further elucidate the role and potential inactivation mechanism of MEG3 in cervical cancer.
Methods
ROC curve and Cox regression analyses were used to assess the diagnostic and prognostic value of MEG3 in patients with cervical cancer. The methylation status of the MEG3 promoter in cervical cancer tissue samples was tested using methylation-specific PCR. Furthermore, we altered the methylation status of the MEG3 promoter in two cervical cancer cell lines (HeLa and CaSki) using a DNA methylation transfer enzyme inhibitor (5-Aza-CdR), to investigate whether promoter hypermethylation is a potential cause of MEG3 inactivation. Finally, we used CCK-8 and colony formation assays to evaluate the cell proliferation ability of HeLa and CaSki cells that had been treated with 5-aza-CdR, to investigate whether downregulation of MEG3 caused by promoter hypermethylation had biological effects.
Results
ROC curve analysis indicated that MEG3 status showed sufficient sensitivity and specificity for prediction of tumor size and lymph node metastasis in patients with cervical cancer. In addition, our follow-up data showed that low MEG3 expression was correlated with recurrence and short overall survival. Moreover, hypermethylation of the MEG3 promoter was observed in most cervical cancer tissue samples, and demethylation of the MEG3 promoter led to re-expression of MEG3 and inhibited proliferation of HeLa and CaSki cells.
Conclusions
MEG3 is a powerful tool for diagnosis and prognosis of patients with cervical cancer, and low expression of MEG3 is likely to be related to promoter hypermethylation in cervical cancer.
Electronic supplementary material
The online version of this article (doi:10.1186/s13046-016-0472-2) contains supplementary material, which is available to authorized users.
Keywords: Cervical cancer, Diagnosis, Long noncoding RNA, MEG3, Methylation, Prognosis
Background
Cervical cancer is one of the most commonly diagnosed cancers among women worldwide, accounting for over 500,000 new cases and 260,000 cases of death annually [1]. Although the incidence and mortality rates of cervical cancer have declined over the past 30 years, the 5-year survival rate of advanced-stage patients has remained below 40% [2, 3]. Therefore, it is important to explore the molecular mechanisms of cervical cancer in order to identify effective prognostic markers and design improved therapeutic strategies.
Long noncoding RNA (lncRNA) is usually defined as an RNA molecule that is longer than 200 nucleotides and lacks significant protein-coding capacity [4]. Studies have reported that lncRNAs regulate various biological processes such as gene expression, transcription, and cellular proliferation, among others [5, 6]. More importantly, abnormal expression of lncRNAs was also found to be involved in metastasis, recurrence, and prognosis of various human cancers [7]. Maternally expressed gene 3 (MEG3), a lncRNA that has attracted much research interest, is aberrantly expressed in several human cancers including gastric cancer [8, 9], colorectal cancer [10], retinoblastoma [11], and ovarian cancer [12] according to recent studies. Moreover, our previous study showed that MEG3 is associated with the progression of cervical cancer via regulation of cell proliferation and apoptosis [13]. However, the diagnostic and prognostic value of MEG3 for cervical cancer remains unknown.
In addition to inducing abnormal expression of tumor suppressor genes and oncogenes, epigenetic alterations play an important role in the development of cancers. Many studies have confirmed that hypermethylation of gene promoter regions can directly cause a decrease in gene expression levels [14]. Hypermethylation of the promoter region of tumor suppressor genes, including genes that encode lncRNAs, is a common cause of cancer [15]. However, no reports have been published to date on the relationship between promoter methylation and MEG3 expression in cervical cancer.
In this study, we investigated the relationship between MEG3 and cervical cancer based on the results of our previous study. We first performed ROC curve and Cox regression analyses to determine the clinical value of MEG3 in patients with cervical cancer. Then we identified the epigenetic regulation of MEG3 expression in cervical cancer tissues and cells. These results deepen our understanding of the role of MEG3 in cervical cancer.
Methods
Patient samples
Seventy-two cervical cancer tissue and its corresponding normal tissues were obtained from patients admitted to our hospital for surgery from April 2012 to March 2013. The specimens were immediately placed in liquid nitrogen after surgical resection. All the patients were newly diagnosed as cervical cancer without receiving any treatment. The samples which considered as cervical cancer or normal tissues were determined by pathologic examination. Written consent was obtained from each patient before tissue collection. The protocol was approved by the Institutional Research Ethics Committee of our hospital. The patients’ clinico-pathological characteristics are summarized in Additional file 1: Table S1.
Cell lines and culture
Cervical cancer cell lines HeLa and CaSki were purchased from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). HeLa cells were maintained in DMEM (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and CaSki cells were cultured in RPMI 1640 medium (Gibco) with 10% FBS at 37 °C in a humidified incubator of 5% CO2.
Transfection of siRNA
SiRNA for this study was the same as our previous study. Briefly, the siRNA for knockdown MEG3 (si-MEG3) and its negative control (si-NC) were designed and synthesized by GenePharma (Shanghai, China). SiRNA was transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocol. 48 h after transfection, cells were harvested and subjected to qRT-PCR analyses and functional assays.
RNA isolation and qRT-PCR
Total RNA was isolated from tissues and cells using Trizol reagent (TaKaRa, Otsu, Japan) according to the manufacturer’s instructions. Concentration and purification of RNA were determined by measuring its optical density using NanoDrop 2000 Spectrophotometer (1.8 < A260/280 < 2.0, Thermo Scientific Wilmington, DE, USA). Reverse transcription was carried out using PrimeScriptTM RT reagent Kit following the manufacturer’s protocol (TaKaRa). QRT-PCR was performed using a SYBR Premix Ex Taq II kit (TaKaRa). Briefly, reactions were loaded into a 96-well plate in duplicate and firstly incubated at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 30 s, 1 min of annealing at 60 °C and extension at 60 °C for 1 min on CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, California, US). The β-Actin was chosen as the endogenous normalizer and the 2−ΔΔCt method was used to determine relative expression of MEG3. The primers used for qRT-PCR in this study are the same as our previous study.
Methylation specific PCR for promoter of MEG3
First, DNA from cells and tissues was extracted using the QIAmp DNA Mini kit (Qiagen, German), according to the manufacturer’s recommendations. Next, we used the EpiTect Bisulfite kit (Qiagen) for bisulfate conversion following the product manual. At last, Methylation specific PCR (MSP) was performed on Bisulfite-treated DNA using EpiTect MSP kit (Qiagen). The primers of MEG3 promoter which specific for methylated and unmethylated were as follows: the methylated pair (M) was 5′-GTT AGT AAT CGG GTT TGT CGG C (forward) and 5′-AAT CAT AAC TCC GAA CAC CCG CG (reverse); the unmethylated pair (U) was 5′-GAG GAT GGT TAG TTA TTG GGG T (forward) and 5′-CCA CCA TAA CCA ACA CCC TAT AAT CAC A (reverse). The PCR reaction was conducted in DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) under the following conditions: 95 °C 15 min; 94 °C 30s, 70 °C 30s, 72 °C 30s, 5 cycles; 94 °C 30s, 65 °C 30s, 72 °C 30s, 5 cycles; 94 °C 30s, 60 °C 30s, 72 °C 30s, 30 cycles; 72 °C 7 min. The PCR products were identified by electrophoresis through 2.5% agarose gel, stained with ethidium bromide, visualized under UV and compared by densitometry.
Treatment with 5-Aza-CdR and DZNep
The DNA demethylating agent 5-aza-2-deoxy-cytidine (5-Aza-CdR) was obtained from Sigma-Aldrich (MO, USA) and diluted in dimethyl sulfoxide (DMSO, MP Biomedicals, California, USA). Total 1 × 105 cells were seeded in six-well culture plate and treated with 0, 5, 10 μmol/L 5-aza-CdR for next 5 days. The medium was replaced with the same concentration of 5-Aza-CdR every day. Then cells were collected and prepared for qRT-PCR and MSP to measure the expression of MEG3 and the methylation status of the MEG3 promoter respectively. For CCK-8 assay, we treated cells with 10 μmol/L 5-aza-CdR for 5 days, following transfection of si-MEG3 and compared proliferation ability with cells which were treated with 5-Aza-CdR only.
3-Deazaneplanocin A (DZNep), an inhibitor of the histone methyltransferase EZH2, was purchased from Sigma-Aldrich (MO, USA). The cells were seeded at 1 × 105 cells per well and treated with DZNep at 0, 1 and 5 μmol/L for 5 days. After that, cells were harvested for qRT-PCR and Western Blot.
CCK-8 assay
Cell proliferation ability was evaluated using Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) following the manufacturer’s instructions. Briefly, Hela and Caski cells were harvested 48 h post-transfection before seeding into a 96-well plate at a density of 3x103 cells per well. After 6 h of incubation, 10 μl CCK-8 was added to each well at 0, 24, 48 or 72 h time point. Cells were incubated for 1.5 h at 37 °C and the absorbance was read at 450 nm using SoftMax pro5 Microplate Reader (Molecular Devices, California, USA).
Colony formation assay
For Colony formation assay, 1x103 cells per well were seeded into the 6-well plates and cultured for 7 days in complete growth media. Afterwards, the adherent cells were washed with PBS, fixed with 10% paraformaldehyde for 10 min, stained with 1% Crystal violet for 5 min, photographed and counted.
Western blot
The cells were lysed with RIPA (Thermo Scientific, USA) supplemented with protease inhibitors (Roche, Switzerland) according to the manufactures protocol. Fifty micrograms of proteins were separated by SDS-PAGE and transferred to PVDF membranes (Roche, Switzerland). Afterwards, the membranes were blocked and incubated with rabbit anti-human EZH2 antibody (1:1,000; CST, USA) at 4 °C overnight. Then membranes were washed with TBST and probed with HRP Goat-anti-Rabbit (1:2000; Santa Cruz Biotechnology) at room temperature for 2 h. At last, the proteins were measured semiquantitatively with ECL (Thermo Scientific) and normalized to the expression of β-Actin (1:1,000; CST, USA).
Statistical analysis
Each experiment was performed in triplicate and the results were represented as mean ± SEM. The SPSS software (version 11.0, SPSS Inc, USA) was used for the statistical analyses. Differences between groups were compared using the Student’s t test or LSD test for continuous variables and the Pearson’s chi-square test for qualitative variables. Statistical significance was considered at P ≤ 0.05.
Results
Diagnostic value of MEG3 for patients with cervical cancer
The results of qRT-PCR analysis showed that MEG3 expression was significantly lower in cervical cancer tissues compared to corresponding normal tissues, consistent with our previous study (Fig. 1a). Because MEG3 was shown to be significantly associated with tumor size and lymph node metastasis in our previous study, we conducted receiver operating characteristic (ROC) curve analysis to test its diagnostic value. The results showed that MEG3 could be a candidate to discriminate between tumors < 4 cm and tumors ≥ 4 cm, with an AUC (area under the ROC curve) of 0.745, a sensitivity of 56.1%, and a specificity of 80.6% at a cut-off value of 0.705 (P < 0.001, Fig. 1b). ROC curve analysis also revealed that MEG3 could serve as a biomarker for lymph node metastasis, with an AUC of 0.716. At a cut-off value of 0.475, the sensitivity and the specificity were 70.5% and 67.9%, respectively (P = 0.002, Fig. 1c).
Fig. 1.
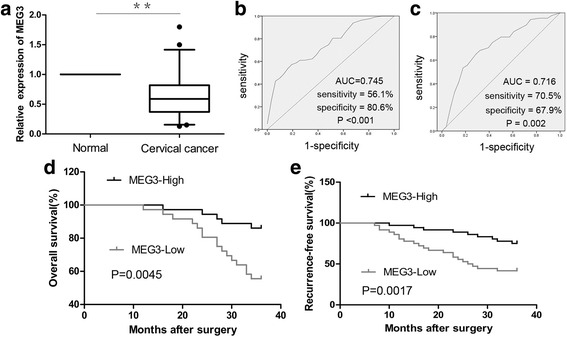
The diagnostic and prognostic value of MEG3 in cervical cancer. a Relative expression of MEG3 in cervical cancer tissues (n = 72) and corresponding normal tissues (n = 72). ROC curve of MEG3 expression predicts the tumor size (b) and presence of lymph nodes metastasis (c) in terms of sensitivity and specificity. d, e Patients in MEG3-Low group (n = 36) had significantly shorter recurrence-free and overall survival than those in MEG3-High group (n = 36). P value was calculated by Log-rank test.** P < 0.01
In addition, we tested the ROC model above using data collected from 108 patients with cervical cancer in our previous study. The AUCs for tumor size and lymph node metastasis were 0.753 and 0.862, respectively (P < 0.001, Fig. 2). Furthermore, at a cut-off value of 0.705, the sensitivity and specificity for diagnosis of tumor size (<4 cm or ≥ 4 cm) were 54.8% and 84.8%, respectively (Fig. 2a). When the cut-off value was 0.475, the sensitivity and specificity for prediction of lymph node metastasis were 76.1% and 85.4%, respectively (Fig. 2b).
Fig. 2.
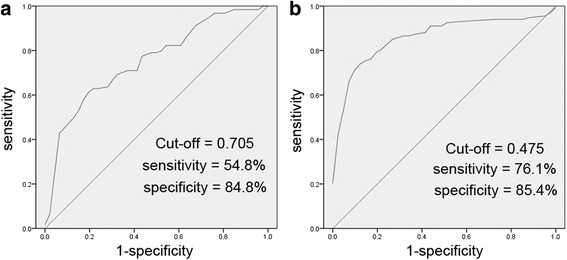
MEG3 cut-off values for tumor size (a) and presence of lymph nodes metastasis (b) setting before were tested by other 108 patients with cervical cancer from our previous study. The AUC was 0.753 (P < 0.001, a) and 0.862 (P < 0.001, b), respectively
Relationship between MEG3 expression and prognosis
To further evaluate whether MEG3 expression is linked to survival, patients with cervical cancer (n = 72) were classified into the MEG3-Low (n = 36) and MEG3-High groups (n = 36) according to the median value of MEG3 expression. Kaplan–Meier analysis showed that patients in the MEG3-Low group had significantly shorter recurrence-free and overall survival than those in the MEG3-High group (P < 0.01, Fig. 1d, e).
Furthermore, we tested the predictive value of MEG3 for relevant clinical and pathological parameters, including tumor size, FIGO stage, lymph node metastasis, HR-HPV infection, age, menopause, histology, depth of invasion, differentiation, and lymphatic vascular space invasion (LVSI). Univariate analysis showed that MEG3 expression, differentiation, FIGO stage, and lymph node metastasis were all prognostic factors for recurrence-free survival in patients with cervical cancer (Table 1). In a multivariate analysis based on the Cox proportional hazards regression model, only MEG3 expression, FIGO stage, and lymph node metastasis were found to be independent prognostic markers for recurrence-free survival (Table 1).
Table 1.
Univariate and Multivariate analyses for recurrence-free survival
| Risk factors | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | P value | 95% CI | HR | P value | 95% CI | |
| MEG3 expression | 0.111 | 0.002 | 0.027 ∼ 0.451 | 0.159 | 0.040 | 0.028 ∼ 0.919 |
| FIGO stage, (I, II) | 3.687 | <0.001 | 1.780 ∼ 7.634 | 3.033 | 0.005 | 1.400 ∼ 6.571 |
| Lymph nodes metastasis (Negative, Positive) | 2.095 | 0.043 | 1.023 ∼ 4.291 | 2.259 | 0.048 | 1.007 ∼ 5.070 |
| Differentiation (Well/Moderately, Poorly) | 2.134 | 0.040 | 1.036 ∼ 4.396 | 0.670 | 0.406 | 0.201 ∼ 1.722 |
| Depth of invasion (≤2/3, >2/3) | 1.358 | 0.403 | 0.663 ∼ 2.783 | |||
| HR HPV infection (Negative, Positive) | 1.501 | 0.347 | 0.643 ∼ 3.502 | |||
| Tumor size | 1.243 | 0.132 | 0.936 ∼ 1.652 | |||
| LVSI (Negative, Positive) | 0.797 | 0.538 | 0.387 ∼ 1.643 | |||
| Age | 0.998 | 0.894 | 0.967 ∼ 1.030 | |||
| Histology (Squamous, Adenocarcinoma) | 0.927 | 0.961 | 0.412 ∼ 2.240 | |||
| Menopause (Yes, No) | 1.426 | 0.333 | 0.696 ∼ 2.921 | |||
HR hazard ratio
Methylation status of the MEG3 promoter in cervical cancer tissues
The methylation status of the MEG3 promoter was assessed by MSP. The unmethylated pattern (U), partially methylated pattern (M and U), and methylated pattern (M) were observed in 12.5%, 22.2%, and 65.3% of cervical cancer tissues, respectively. In comparison, the unmethylated pattern (70.8%) was more common than the methylated (13.9%) or partially methylated patterns (15.3%) in corresponding normal tissues (Fig. 3a, Table 2). The difference in promoter methylation status between cervical cancer tissues and corresponding normal tissues was statistically significant (P < 0.001, Table 2). More importantly, there was a positive correlation between cervical cancer and promoter hypermethylation (R = 0.592, Table 2). MEG3 methylation was a risk factor for cervical cancer (OR = 17, Table 2).
Fig. 3.
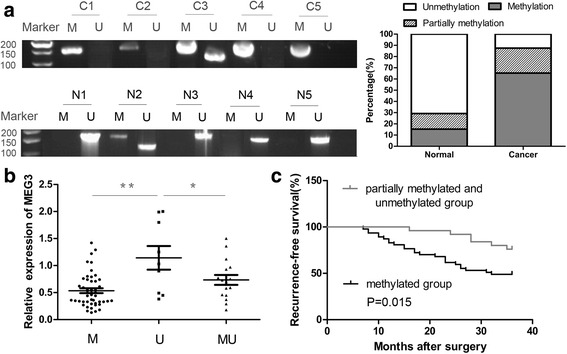
Methylation status of the MEG3 promoter in cervival cancer. a Methylation status of MEG3 promoter in cervival cancer tissues (C1-C5 as example) and its corresponding normal tissues (N1-N5 as example) was tested by MSP. The methylated pattern (M) of MEG3 was 160 bp and the unmethylated pattern (U) was 120 bp. b Patients were classified into methylation group (M), partially methylated group (MU) and unmethylated group (U) based on the methylation levels of MEG3 promoter. MEG3 expression level was associated with the methylation level of MEG3 promoter. c Hypermethylation of MEG3 promoter indicated poor recurrence-free survival. * P < 0.05, ** P < 0.01
Table 2.
Correlation between cervical cancer and methylation status of MEG3 promoter
| Tissues | Methylation status | χ2 | P | R | OR (95% CI) | |
|---|---|---|---|---|---|---|
| M, MU | U | |||||
| Cervical cancer | 63 | 9 | 50.4 | <0.001 | 0.592 | 17.000 (7.167–40.324) |
| Normal | 21 | 51 | ||||
OR odds ratio
To better understand the role of methylation, we also analyzed the correlation between MEG3 expression and promoter methylation level in cervical cancer tissues. As shown in Fig. 3b, expression of MEG3 was significantly higher in the unmethylated group than in the methylated or partially methylated groups.
Furthermore, HR-HPV infection and lymph node metastasis were significantly correlated with MEG3 methylation (Table 3). The Chi-square test also indicated that MEG3 methylation was a risk factor for HR-HPV infection and lymph node metastasis (Table 3). However, MEG3 methylation was not significantly correlated with age, menopause, histology, differentiation, FIGO stage, tumor size, depth of invasion, or LVSI in cervical cancer (Table 3). Moreover, Kaplan–Meier analysis showed that the promoter methylation level of MEG3 was significantly correlated with recurrence-free survival time: patients in the methylated group had significantly shorter recurrence-free survival times than those in the partially methylated or unmethylated groups (P = 0.015, Fig. 3c).
Table 3.
Relationship between MEG3 methylation and clinical pathological characteristics in patients with cervical cancer
| Clinical pathological characteristics | P | R | OR (95% CI) |
|---|---|---|---|
| HR HPV infection (Positive, Negative) | 0.013 | 0.291 | 3.830 (1.279–11.467) |
| Lymph nodes metastasis (Positive, Negative) | 0.025 | 0.264 | 3.520 (1.131–10.953) |
| FIGO stage (II, I) | 0.056 | ||
| Depth of invasion (>2/3, ≤2/3) | 0.106 | ||
| Differentiation (Well/Moderately, Poorly) | 0.741 | ||
| Tumor size (>4 cm, ≤4 cm) | 0.703 | ||
| LVSI (Positive, Negative) | 0.675 | ||
| Age (>50, ≤50) | 0.371 | ||
| Histology (Squamous, Adenocarcinoma) | 0.899 | ||
| Menopause (Yes, No) | 0.658 |
OR odds ratio
Epigenetic regulation of MEG3 in cervical cancer cells
As the MSP assay showed, the promoter of MEG3 was hypermethylated in HeLa and CaSki cells (0 μmol/L, Fig. 4a). To investigate the role of promoter methylation in regulation of MEG3 expression in cervical cancer cells, we examined the effect of 5-aza-CdR on promoter methylation levels and MEG3 expression. Compared with the control treatment, 5-aza-CdR treatment significantly decreased the promoter methylation level in HeLa and CaSki cells (Fig. 4a). Furthermore, qRT-PCR analysis indicated that expression of MEG3 was significantly higher in the 5-aza-CdR treatment group than in the control group (Fig. 4b, P < 0.05).
Fig. 4.
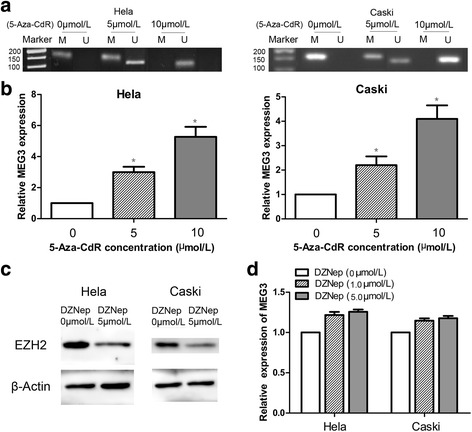
The effects of 5-aza-CdR and DZNeP on MEG3 expression. a MEG3 promoter was hypermethylation in HeLa and CaSki cells (0 μmol/L). MEG3 promoter could be demethylation by different concentrations of 5-aza-CdR (5 and 10 μmol/L) in HeLa and CaSki cells. b MEG3 was re-expressed in the HeLa and CaSki cells treated with different concentrations of 5-aza-CdR (5 and 10 μmol/L), compared to cells of the control group (0 μmol/L 5-aza-CdR). c EZH2 protein levels in cells treated with DZNep (0, 5 μmol/L) were examined by western blot d MEG3 expression was analyzed by RT-qPCR in cells treated with DZNep (0, 1, 5 μmol/L) for 5 days and there was no significant statistical difference among the groups
In addition, we also examined the effects of DZNep treatment based on HeLa and CaSki cells. We first confirmed the significant reduction of EZH2 protein levels (Fig. 4c). However, no significant change in MEG3 expression levels was observed after treatment with DZNep (Fig. 4d).
Methylation status of the MEG3 promoter influenced proliferation of cervical cancer cells
As shown by the CCK-8 assay, 5-aza-CdR significantly decreased the proliferation ability of HeLa and CaSki cells, compared with control cells (Fig. 5a, P < 0.05). According to the colony formation assay, 5-Aza-CdR (10 μmol/L 5-Aza-CdR + si-NC group) also resulted in a notable decrease in the number of colonies, as compared with the control groups (0 μmol/L 5-Aza-CdR + si-NC group) (Fig. 5d P < 0.05). To prevent excessive use of si-MEG3, qRT-PCR confirmed that there was no significant difference in MEG3 expression between the 5-aza-CdR (10 μmol/L) + si-MEG3 and the si-NC groups (Fig. 5b). The efficiency of si-MEG3 was tested in our previous study. Furthermore, the CCK-8 and colony formation assays indicated that the proliferation ability of HeLa and CaSki cells in the 5-aza-CdR (10 μmol/L) + si-MEG3 group was significantly higher than that in the 5-aza-CdR (10 μmol/L) + si-NC group (Fig. 5c, d, P < 0.05).
Fig. 5.
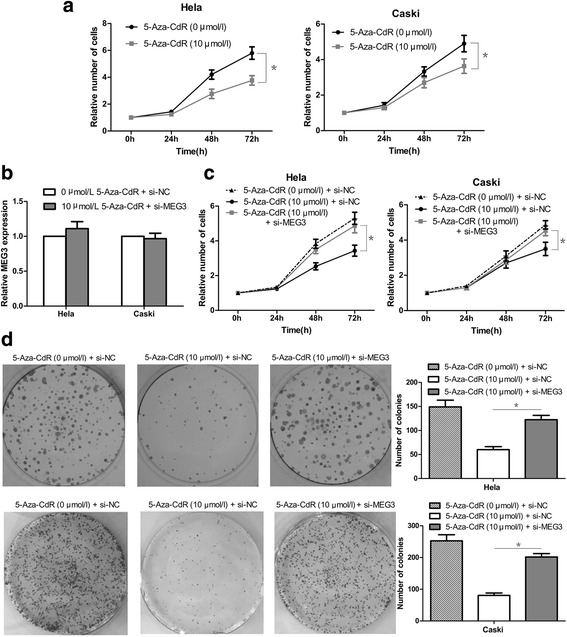
The effects of 5-aza-CdR on cervical cancer cells proliferation in vitro. a 5-aza-CdR dramatically decreased the proliferation ability of HeLa and CaSki cells by CCK-8 assay. b si-MEG3 was just canceled out the promoting effect of 5-aza-CdR on MEG3 expression. c and d si-MEG3 could partly reverse the inhibitory effect of 5-aza-CdR on proliferation of HeLa and CaSki cells, as shown by CCK-8 assays and Colony formation assays. * P < 0.05
Discussion
Our previous study showed that MEG3 expression levels are related to HR-HPV infection, tumor size, FIGO stage, and lymph node metastasis in patients with cervical cancer [13]. However, the value of MEG3 in clinical practice is generally unknown. In the present study, we identified MEG3 as a potential marker for lymph node metastasis. Then we tested the ROC model using data from 108 other patients with cervical cancer from our previous study. The results confirmed that the MEG3 expression cut-off value that we set previously for prediction of lymph node metastasis was still effective, supporting our previous results. The presence or absence of lymph node metastasis is a crucial factor in therapy decision-making for patients with cervical cancer and our study found that MEG3 is a good candidate to predict it. That indicates that MEG3 shows excellent potential value for clinical work, especially for cervical cancer, in which biopsy is routine before therapy.
The lack of valid prognostic prediction models makes it difficult to apply individualized therapy to patients with cervical cancer. Developing a method for identifying patients at high risk of treatment failure is important. For these high-risk patients, modified therapies such as neoadjuvant radiation or chemotherapy could potentially be applied to improve patient survival. Although extensive effort has been devoted to understanding the prognostic value of MEG3 in human tumors [8, 10, 11], little was known about its value in cervical cancer until now. In the present study, we evaluated the prognostic value of MEG3 using Kaplan–Meier and Cox regression analyses. Kaplan–Meier analysis of patients with cervical cancer indicated that MEG3 expression level was associated with recurrence-free and overall survival. Furthermore, MEG3 was confirmed to be an independent prognostic marker for recurrence-free survival by multivariate analysis. Therefore, MEG3 may represent a novel indicator of prognosis in cervical cancer, helping to identify high-risk patients before treatment.
Emerging results indicate that epigenetic aberrations, especially DNA methylation, regulate tumor growth by silencing tumor suppressor genes [16, 17]. The promoter region as well as the intergenic germ line-derived differentially methylated region (IG-DMR) of MEG3 is rich in CpG dinucleotides [18]. Thus, many studies have reported that promoter methylation plays an important role in loss of MEG3 expression in tumors [8, 11, 19]. In this study, we tested the hypothesis that promoter hypermethylation may be able to reduce MEG3 levels in cervical cancer, by evaluating the methylation level of the MEG3 promoter and its effects on MEG3 expression. Similar to other findings, low MEG3 expression was shown to be related to hypermethylation of the MEG3 promoter in cervical cancer tissues.
Further, we endeavored to reveal the role of MEG3 methylation in the progress of cervical cancer. We found that MEG3 methylation is not only a risk factor for cervical cancer, but also for HR-HPV infection and lymph node metastasis. More importantly, patients with hypermethylation of the MEG3 promoter showed poor recurrence-free survival. Collectively, these results indicate the importance of MEG3 methylation in cervical cancer progression. Interestingly, we noted that the role of MEG3 methylation was consistent with that of MEG3 expression in cervical cancer, and the clinical characteristics of patients in the methylated group were similar to those of patients in the MEG3-Low group, according to our previous study. These results provide strong evidence for the relevance of MEG3 inactivation and promoter hypermethylation.
In addition, we showed, using cell-based experiments, that promoter methylation and MEG3 expression are correlated. We revealed that the MEG3 promoter is hypermethylated in cervical cancer cells, and that demethylation of the promoter resulted in re-expression of MEG3. More importantly, as the promoter methylation level decreased, the expression of MEG3 increased in cervical cancer cells, which indicated that loss of MEG3 expression in cervical cancer cells was the result of promoter hypermethylation, at least in part. Moreover, our results indicated that a decrease in the methylation level resulted in not only re-expression of MEG3, but also reduction of the proliferation potential in cervical cancer cells. Furthermore, repression of MEG3 re-expression could partly reverse the inhibitory effect of 5-aza-CdR on proliferation of HeLa and CaSki cells, revealing that MEG3 re-expression may play a crucial role in 5-aza-CdR functions. Based on the above results, we have a clear indication that MEG3 re-expression owing to DNA demethylation could inhibit proliferation of cervical cancer cells. The results also indicate that inactivation of MEG3 via promoter hypermethylation plays an important role in regulation of cervical cancer cell proliferation. In addition, it confirms the results of our previous study, that down-regulation of MEG3 could suppress the proliferation of cervical cancer cells in another way. In summary, we propose that the following process may occur in cervical cancer cells: promoter hypermethylation → inactivation of MEG3 → malignant cell proliferation.
In addition to DNA methylation, we are also interested in the effect of histone methylation on MEG3 expression. DZNep has been shown to reduce expression of EZH2, inhibit methylation of H3K27, and affect histone methylation [20]. However, DZNep did not significantly affect MEG3 expression in cervical cancer cells, implying that histone methylation may not be a major mechanism of MEG3 inactivation. However, this is just the first step in study of histone methylation of MEG3 and much remains to be done.
Conclusions
MEG3 may be a useful diagnostic tool and prognostic marker for cervical cancer, and its inactivation in cervical cancer may be due to promoter hypermethylation. MEG3 shows potential for use in diagnostic applications and therapeutic interventions in cervical cancer.
Acknowledgements
Not applicable.
Funding
The Project Supported by National Natural Science Foundation of China (Grant No. 81572575) and Guangdong Natural Science Foundation (Grant No. 2014A030313109).
Availability of data and materials
All data generated during this study are included in this article and its supplementary information files.
Authors’ contributions
ZJ, GY, YT and LZ conceived and designed the project. ZJ, GY, YT and LZ performed patient collection and clinical data interpretation. ZJ and GY participated performed the statistical analysis. ZJ and GY performed the experiments and wrote the manuscript. ZJ, GY, YT and LZ contributed to the writing and to the critical reading of the paper. All authors read and approved the final manuscript. All authors read and gave their approval for the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Informed consent was obtained from all individual participants included in the study.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Abbreviations
- 5-Aza-CdR
5-aza-2-deoxy-cytidine
- AUC
The areas under the ROC curve
- CCK-8
Cell Counting Kit-8
- lncRNA
Long noncoding RNA
- LVSI
Lymphatic vascular space invasion
- MEG3
Maternally expressed gene 3
- MSP
Methylation specific PCR
- ROC
Receiver operating characteristic curve
Additional file
Patients’ clinico-pathological characteristics. (DOC 40 kb)
Contributor Information
Jun Zhang, Email: zhangjunsysu@126.com.
Zhongqiu Lin, Email: linzhongqiulzq@126.com.
Yali Gao, Phone: +86-0755-2294-2840, Email: Gaoylsz@126.com.
Tingting Yao, Phone: +86-20-3407-1153, Email: tingtingyaotty@126.com.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.SEER Cancer Statistics Review, 1975-2000. National Cancer Institute. 2003. https://seer.cancer.gov/csr/1975_2000/.
- 3.Cancer Facts and Figures 2012. American Cancer Society, 2012. http://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2012/.
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calore F1, Lovat F, Garofalo M. Non-coding RNAs and cancer. Int J Mol Sci. 2013;14(8):17085-110. [DOI] [PMC free article] [PubMed]
- 8.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumor Biol. 2014;35(2):1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 9.Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36(6):4851–9. doi: 10.1007/s13277-015-3139-2. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Lu X. Decreased expression of MEG3 contributes to retinoblastoma progression and affects retinoblastoma cell growth by regulating the activity of Wnt/β-catenin pathway. Tumour Biol. 2016;37(2):1461–9. doi: 10.1007/s13277-015-4564-y. [DOI] [PubMed] [Google Scholar]
- 12.Sheng X, Li J, Yang L, Chen Z, Zhao Q, Tan L, Zhou Y, Li J. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol Rep. 2011;32(1):277–85. doi: 10.3892/or.2014.3208. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17(1):104–13. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 15.Laird PW. The power and the promise of DNA methylation makers. Nat Rev Cancer. 2003;3(4):253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg962. [DOI] [PubMed] [Google Scholar]
- 17.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;30(11):5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab. 2005;90:2179–2186. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- 19.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during this study are included in this article and its supplementary information files.


